Abstract
Diet, host gene composition, and alterations in the intestinal microbiota can contribute to obesity. In microbe-induced obesity, metabolic changes stem from primary perturbation of the microbiota, consequent to modern changes in human biology. Microbiota disruption during early development can result in syndromes of metabolic dysfunction. We focus on the pathways involved in these interactions, particularly related to energy extraction and the role of inflammation in the metabolic phenotypes. Model physiologic systems and perturbations including gastric bypass surgery, pregnancy, and hibernation provide insight into the respective roles of the critical participants.
INTRODUCTION
Obesity has markedly increased in the US in the late 20th century (Flegal et al., 2012), as well as worldwide (Haslam and James, 2005). Much focus on the origins of obesity has centered on dietary excesses [diet-induced obesity (DIO)] or on host genes [gene-induced obesity (GIO)] (Hollopeter et al., 1998) (Figure 1). However, in addition to increased calories or alterations in host metabolism, the composition of the intestinal microbiota can contribute to the progression of obesity (Bäckhed et al., 2004; Turnbaugh et al., 2008; Turnbaugh et al., 2006), thus DIO and GIO have both microbe-independent and microbe-dependent mechanisms. Alternately, metabolic changes can stem from a primary perturbation of the microbiota (Cho et al., 2012), consequent to modern changes in human biology (Blaser and Falkow, 2009; Dominguez-Bello et al., 2011) in which the initiating factors lack microbe-independent effects [microbe-induced obesity (MIO)]. Epidemiological studies in humans have shown that antibiotic treatment during the first six months of life (Trasande et al., 2013), or disrupted colonization from Caesarian section delivery (Blustein et al., 2013; Huh et al., 2012) can increase the risk of being overweight later in life. These two interventions have no direct contribution to host caloric intake or metabolism (Coates et al., 1963) but have large effects on the microbiome (Dethlefsen et al., 2008; Dominguez-Bello et al., 2010).
Figure 1. Pathways in microbe-induced obesity.
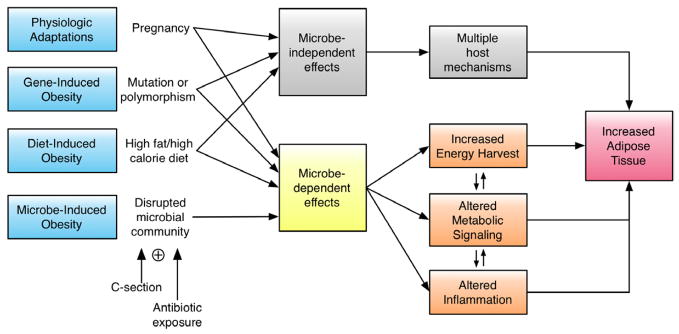
Normal physiological adaptations (i.e. pregnancy), genetic mutations or polymorphisms (gene-induced obesity, GIO), or a diet with excessive fat or calories (diet-induced obesity, DIO) can promote weight gain and increase adiposity through microbe-independent and microbe-dependent mechanisms. Disrupting the founding microbial community through C-section and/or early-life antibiotic exposure also can lead to increased weight gain and adiposity (microbe-induced obesity, MIO). All effects are microbe-dependent since the initiating factors do not contribute to obesity independent of microbes (e.g. antibiotics have no caloric value). For each pathway that leads to obesity, the altered microbiota can contribute to adiposity through increased energy harvest or by altering metabolic signals, inflammation, or immunity.
The microbes that colonize humans substantially out-number human cells (Savage, 1977) and the collective unique microbial genes outnumber human genes by more than a factor of 100 (Arumugam et al., 2011; HMPConsortium, 2012; Qin et al., 2010). These co-evolved microbes have a complex role in maintaining health (HMPConsortium, 2012), but can also contribute to disease (Cho and Blaser, 2012; Holmes et al., 2012). Model systems have been used to examine the effects of microbiome change in the context of different diets or on early life development. In this Perspective, we examine mechanistic pathways in which the disrupted microbiome can contribute to obesity through altering energy extraction from food or altering inflammation and immunity (Figure 2). The microbiome is dynamic and resilient, but subject to perturbations that can impact metabolism. We explore major recovery strategies in the microbiome in response to host metabolic perturbations (Figure 3), and discuss metabolic adaptations that aid particular classes of microbiota during recovery in the competitive environments of well-established ecosystems (Figure 4).
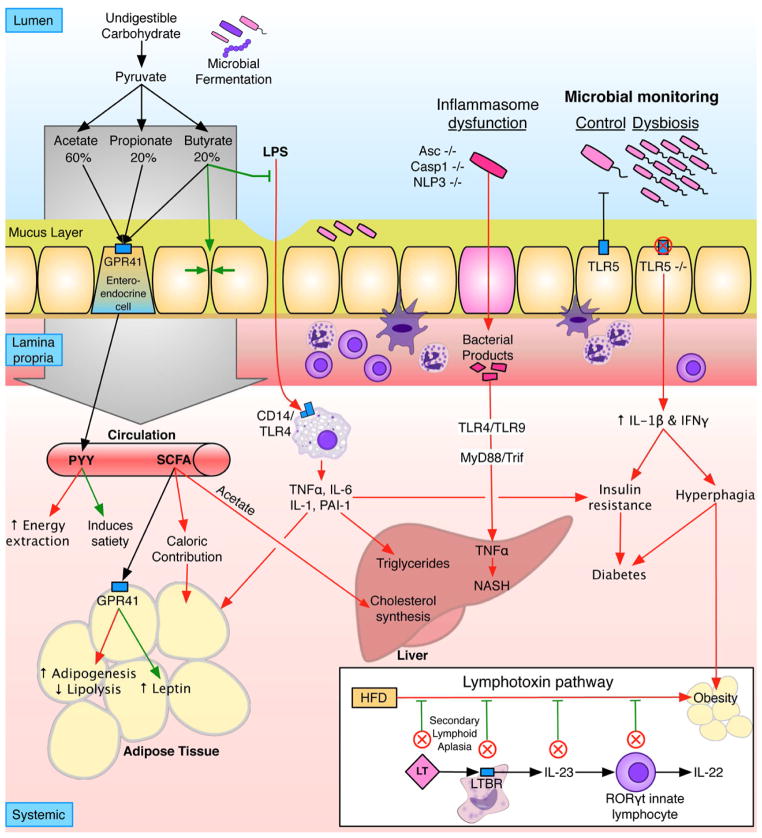
Figure 2. Pathways involving microbiome and immunity that contribute to obesity.
Red lines indicate pathways that promote obesity; green lines indicate pathways that protect. Undigested carbohydrates are fermented by the intestinal microbiota to short-chain fatty acids (SCFA), primarily acetate (60%), propionate (20–25%), and butyrate (15–20%). SCFA can signal through G protein-coupled receptor 41 (GPR41) on enteroendocrine cells, inducing the secretion of peptide YY, which can contribute to obesity through slowing transit time thus increasing energy extraction from food, or can protect against obesity by increasing satiety. SCFA are readily absorbed and contribute up to 10% of caloric intake, can increase adipogenesis by binding GPR41 on adipocytes. In the liver, acetate is a substrate for cholesterol synthesis and lipogenesis. Locally, butyrate increases the epithelial tight junctions, blocking the translocation of LPS, a potent inflammatory mediator that increases weight gain, total body and liver adiposity, and insulin resistance. Similarly, the mucus layer can also block LPS translocation. Inflammasome dysfunction allows translocation of bacterial products, which induce hepatic TNFα expression and drive non-alcoholic steatohepatitis (NASH) in a TLR4/TLR9-dependent manner, in mice with other risk factors (e.g. methionine choline deficient diet). Loss of TLR5, which senses bacterial flagellin, leads to a dysbiosis in the microbiome, increased inflammatory cytokines, hyperphagia, insulin resistance, and diabetes in the context of a high fat diet. The NASH and metabolic syndrome phenotypes in inflammasome- and TLR5- deficient mice are transferrable via the microbiome to germ-free wild-type (wt) recipients, demonstrating the roles of the altered microbiome in adiposity. Inset: Deleting any component of the lymphotoxin pathway results in loss of control of the microbiome and blocks the ability of mice to gain weight on high-fat diet. Co-housing lymphotoxin receptor-deficient and wt mice restores diet-induced obesity and partially restores a wt/HFD microbiota.
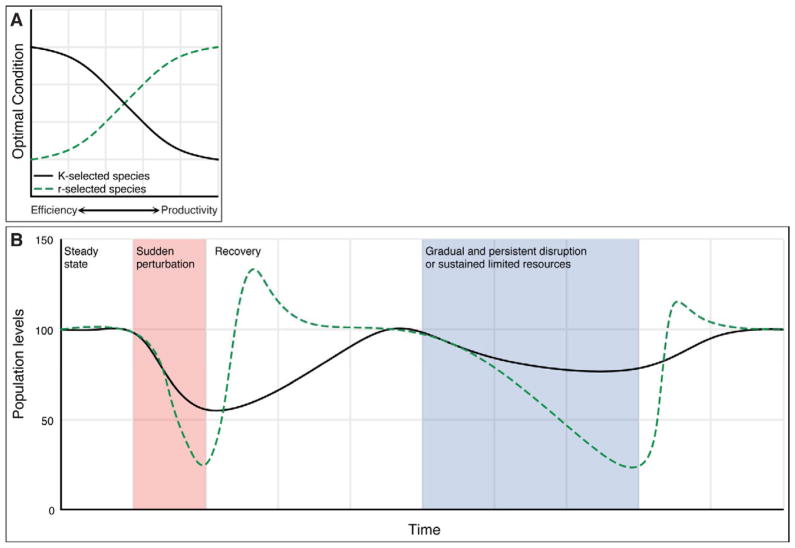
Figure 3. Response of r- and K-selected species to environmental stress.
Panel A: r-species recover from environmental stress optimizing growth rate and productivity, while K-selected species resist environmental change by optimizing their efficiency when resources are scarce. Panel B: Following a large and sudden perturbation in which both r- and K-selected species suffer diminished in population levels, the r-selected species can rapidly bloom and dominate the environment due to utilization of unused resources, whereas the recovery from K-selected species is slower. During a longer, sustained environmental stress, K-selected species can resist large population losses because of their high-efficiency adaptation strategies, whereas r-selected species are more constrained to maintain populations, and suffer losses.
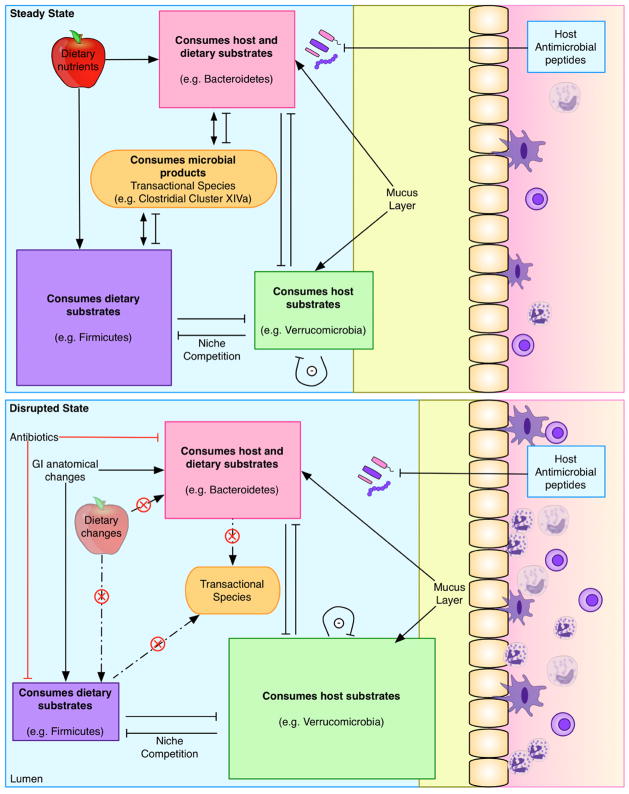
Figure 4. Microbial equilibrium and host effects in relation to energy substrates.
Microbes in the intestine can derive energy from the diet, host, or compounds secreted by other members of the consortium. Four classes of microbes may be considered, based on their predominant energy source: diet (purple), host (green), either (pink), or neither (orange). Competition for niche colonization is affected by differential growth rates (r- and K-selected species), utilization of exclusive substrates, susceptibility to host defenses (e.g. antimicrobial peptides), production of inhibitory products, and community organization (e.g. auto-inducers). Perturbation may occur, for example, via dietary change (e.g. changes in a micro or macronutrient or long-term fasting), by antibiotics that differentially select the microbiota, or changes in gastrointestinal tract structure (e.g. gastric bypass). Importantly, in this model, competition between the major classes affects host mucus layer, epithelial permeability, and inflammation, which has downstream metabolic consequences.
1. ENERGY EXTRACTION FROM FOOD
The diet provides nutrients to both the host and microbial consortium, which have co-evolved to derive and impart benefits to each other across a wide array of vertebrate lineages (Ley et al., 2008a; Ley et al., 2008b). The microbiota improve the fitness of the host by increasing resistance to pathogens, providing essential vitamins, and increasing caloric extraction from food. The very presence of microbiota influences weight gain and fat storage as germ-free mice have significantly lower weight and body fat percent, despite eating more calories than conventionalized mice (Bäckhed et al., 2004).
Current technologies have allowed more efficient production of food; consequently, the caloric intake has increased in both men and women in the last 30 years (Wright et al., 2004). Increased caloric intake for the host translates to increased energy availability for the microbiota, altering the environment they inhabit (Kau et al., 2011). In addition, readily available processed foods commonly substitute complex carbohydrates for simple refined sugars and contain food preservatives, which affect the composition and diversity of the microbiota (Bernbom et al., 2006; Kau et al., 2011; Payne et al., 2012). In obesity, both reduced microbial diversity and interconnectivity have been observed (Greenblum et al., 2012; Turnbaugh et al., 2009), consistent with our a priori notions of the importance of microbiota diversity to health.
An increase in caloric intake, whether from a high fat diet (DIO) (Turnbaugh et al., 2008), or from overeating normal chow driven by the absence of the satiety hormone leptin in ob/ob mice (GIO) (Turnbaugh et al., 2006), selects for obesogenic microbiota. This was demonstrated by transferring the obesity phenotype to germ-free recipient mice via microbiome transplantation, providing evidence that the microbiota can contribute to obesity in addition to increased caloric consumption (Figure 1). Both DIO and ob/ob GIO caused phylum-level shifts in the intestinal microbiota, increasing Firmicutes and decreasing Bacteroidetes (Ley et al., 2005; Ley et al., 2006; Turnbaugh et al., 2008). That these broad phylum level changes have not been found consistently (Ley, 2010; Ravussin et al., 2012), may indicate that the important events involve lower taxonomic levels within these phyla.
Dietary composition can alter the microbiota, independent of the calories consumed. In a comparison of ad-libitum fed or caloric-restricted mice receiving either a DIO diet or normal chow, the dietary composition had a greater influence on shaping the microbiota than caloric intake or host obese status (Ravussin et al., 2012). However, whether the metabolic phenotype was transferrable to germ-free recipients was not assessed, so it could not be determined whether the dietary composition or the increased calories select for an obesogenic microbiota. That excessive caloric intake in ob/ob mice receiving a normal chow and in mice fed ad-libitum on a DIO diet selects for an obesogenic microbiota argues that the increased caloric intake, not dietary composition, is the driving force.
In the large intestine, complex nutrients that have not been absorbed by the host are fermented to short-chain fatty acids (SCFA), which contribute a small but significant number of calories to the host (Macfarlane and Macfarlane, 2012), estimated to account for up to 10% of caloric intake in humans (Bergman, 1990) (Figure 2). The total production of SCFA, and thus calories contributed by microbiota, depend on both dietary and microbiota composition. The majority of SCFA are produced from carbohydrates, but a smaller fraction can be produced from protein degradation, which yields SCFA plus other metabolic end-products, such as amines, phenols, thiols, and hydrogen sulfide (Macfarlane and Macfarlane, 2012). Leptin-deficient ob/ob mice harbor a microbiota that produce more SCFA than wild type mice (wt) and extract more calories from the diet (Turnbaugh et al., 2006). A similar increase in microbial energy harvest was observed in lean humans switched from consuming 2,400 to 3,400 calories/day (Jumpertz et al., 2011). These findings show that in a GIO murine model and a DIO human simulation, the intestinal microbiota can adapt to an enriched diet and contribute additional calories to the host (Figure 1).
Metabolic disturbances can occur that are not caused by either abnormal host gene status or dietary changes, but instead are driven by an aberrant microbiota (microbe-induced obesity, MIO). For example, the microbiota may become altered as a result of an abnormal founding population, as occurs with Caesarian section (Biasucci et al., 2010; Dominguez-Bello et al., 2010; Huurre et al., 2008; Pandey et al., 2012). Epidemiologic studies indicate an association of C-section with the development of childhood obesity (Blustein et al., 2013; Huh et al., 2012; Li et al., 2013). Similarly, the microbiota can be altered by antibiotic exposure (Antonopoulos et al., 2009; Dethlefsen et al., 2008; Ubeda and Pamer, 2012). In particular, antibiotic exposure in early life, just when host adipocyte populations are developing (Greenwood and Hirsch, 1974), has been associated with the development of adiposity in humans (Ajslev et al., 2011; Trasande et al., 2013). In a murine model of MIO, early life sub-therapeutic antibiotic treatment (STAT) changed microbiome composition and increased fat mass (Cho et al., 2012). STAT increased the abundance of microbial genes involved in SCFA production, as well as increased acetate, butyrate, and propionate concentrations in the intestine. Thus, if SCFA are markers of increased energy extraction, one driving force behind MIO has been identified.
SCFAs also provide important microbial signals to the host, in part via the G protein-coupled receptors GPR41 and GPR43 (FFAR2 and FFAR1, respectively). The GPR41 receptor on enteroendocrine cells sense SCFAs and induce secretion of peptide YY (Ichimura et al., 2009), which both slows intestinal transit time and increases satiety. GPR41-deficient mice are leaner than their wt counterparts (Table 1), due to decreased PYY and increased intestinal transit rate (Samuel et al., 2008). However, germ-free GPR41 −/− mice exhibited no weight differences from germ-free wt mice, indicating that the effects are dependent on the microbiota. Absorbed SCFA also provide metabolic signals to distant tissues. SCFA bind GPR41 on adipocytes, increasing adipogenesis, inhibiting lipolysis, and inducing leptin secretion (Ichimura et al., 2009). The major product of microbial fermentation, acetate, is a substrate for hepatic cholesterol and triglyceride synthesis (Wong et al., 2006). Early life STAT increased intestinal acetate, as well as the downstream (via the portal circulation) hepatic expression of genes involved in fatty acid metabolism and lipogenesis (Cho et al., 2012).
Table 1.
Metabolic consequences of specified host and dietary interactions with the microbiome
| Host state | Phenotype with microbiota | Phenotype in GF mice or with reduced microbiotaa | Major microbial changes | Transferb | Reference |
|---|---|---|---|---|---|
| ob −/− | ↑ weight and adiposity | Resistance to GIOa | ↑ Firmicutes ↓ Bacteroidetes |
Yes | Ley PNAS 2005, Turnbaugh, Nature 2006 Cani, Diabetes 2008 |
| High calorie diet | ↑ weight and adiposity | Resistance to DIO | ↑ Firmicutes ↓ Bacteroidetes |
Yes | Turnbaugh, Cell Host and Microbe 2008 |
| GPR41 −/− | Lean | No difference from controls | Unknown | Unknown | Samuel, PNAS 2008 |
| Pregnancy | ↑ weight and adiposity | Unknown | ↑ lactic acid bacteria ↓ butyrate producers |
Yes | Koren, Cell 2012 |
| TLR5 −/− | Obese, insulin resistant, hyperphagic | No difference from controlsa | Changes at the OTU level, no changes at the phylum level | Yes | Vijay-Kumar, Science 2010 |
| Inflammasomes Asc−/−, Casp1 −/−, NLP3 −/−, IL18 −/− | ↑ NASH | No difference from controlsa | ↑ Prevotellaceae and Porphyromonadaceae | Yes | Henao-Mejia, Nature 2012 |
| Lymphotoxin deficientc Ltbr −/− Lta −/−, Ltb −/− | Resistant to DIO | Unknown | ↑ SFB & Cytophaga ↓ Erysipelotrichi |
Yes, but transient, obese wt → ltbr−/− | Uphadyay, Nat Immunol 2012 |
| Roux-en-Y Gastric bypass | Lean | Unknown | ↑ Gammaproteobacteria ↑ Akkermansia |
Yes, | Liou, Science Translational Medicine 2013 |
Reduction in microbiota by high-dose broad-spectrum antibiotics
Transfer of the phenotype to conventional mice has been accomplished
Resistance to DIO also achieved by deleting IL23a and RORγt, elements downstream of the lymphotoxin pathway
The extent of total energy extracted from the diet depends on the structure of the intestinal tract, the composition of the microbiota, and the composition of the diet. The dynamic metabolic relationships can be understood through considering continuous flow fermentation systems, in which the population of microbes is maintained by balancing nutrient infusion and waste removal. The evolution of a conserved and stable microbial consortium requires overcoming a considerable set of challenges. These concepts were first compiled by Hungate in his classic work: The Rumen and Its Microbes (Hungate, 1966). Because of the constant downward movement of intestinal contents, a stable microbiota can only be achieved if the net microbial growth equals the flow rate of the system. Over-nutrition can disrupt the microbial ecosystem and can have severe consequences for both host and microbiota. When a ruminant (i.e. cow or sheep) is rapidly switched from a grass-based diet to a grain-based diet, the sudden influx of starch in the digestive tract leads to an overgrowth of lactate-producing Streptococcus bovis (Hungate et al., 1952). Lactic acid accumulates in the rumen faster than can be utilized by other ruminant microbes (Nagaraja and Titgemeyer, 2007). The excess lactate decreases the pH of the rumen below 5, resulting in acute systemic acidosis, which can lead to death within 24 hours. This is an extreme example, since ruminants are substantially different from mice and humans in the fundamental digestive tract organization. Ruminants are foregut fermenters, while humans and mice are hindgut fermenters, which may have evolved as an adaptation enabling wider variations in diet. For mice and humans, most of the microbiota only receive nutrients after the host has extracted many of the simple molecules, and thus hindgut fermenters are not vulnerable to fatal indigestion from nutrient excess.
Major anatomical rearrangements of the gastrointestinal tract of mice and humans produce large effects on energy extraction and on the intestinal microbiota, in part due to changing factors related to nutrient flux and pH (Aron-Wisnewsky et al., 2012). In obese subjects undergoing Roux-en-Y gastric bypass (RYGB) surgery, metabolic improvements can be observed in the first week after the surgery, preceding significant weight loss (Ahn et al., 2011; Rubino et al., 2004). These weight-independent metabolic improvements may be mediated by rapid changes in microbiota composition since major reconstruction of the gastrointestinal tract as part of gastric bypass significantly alters the microbiota shortly after the surgery (Graessler et al., 2012; Li et al., 2011). In a recent study by Liou et. al., RYGB had a more substantial effect on the microbiome than sham surgery or caloric restriction, even though mice receiving caloric restriction also were significantly leaner than the sham-surgery mice (Liou et al., 2013), demonstrating that gut structure had greater impact on the microbiota than obesity status. RYBG was associated with increased populations of Proteobacteria (Escherichia) and Verrucomicrobia (Akkermansia), and a reduction in Firmicutes compared to sham-surgery and weight-restriction. Mice that underwent gastric bypass extracted significantly less energy from the diet than mice that underwent sham-surgery or caloric restricted mice, which may have been mediated by a combination of gut restructuring and changes in the intestinal microbiota. Germ-free mice colonized with microbiota from RYGB mice weighed significantly less than mice colonized with microbiota from sham surgery mice, showing that the RYGB-altered microbiota play an active role in weight loss and metabolic status. Both the RYGB cecal donors and their recipient mice had decreased total SCFA compared to sham donors and recipients, with major reductions in acetate levels and a relative increase in propionate levels, providing further evidence that SCFAs influence obesity.
2. INFLAMMATION AND IMMUNITY
The interaction between diet, genes, and the intestinal microbiota in the context of obesity is complex (Parks et al., 2013), since each of these factors has the potential to modulate inflammation and immunity (Kau et al., 2011). Obesity is a low-grade inflammatory disorder and the role of the microbiota via specific immune pathways has been studied with the aid of experiments involving germ-free and knockout mice (Table 1).
LPS
High fat diet can increase absorption of lipopolysaccharide (LPS), a component of gram-negative bacterial cell walls, either by incorporation into chylomicrons or by increasing intestinal permeability (Cani et al., 2007; Cani et al., 2008). LPS is a potent inflammatory mediator that signals in a CD14/TLR4-dependent manner, and infusion of LPS alone can increase weight gain, adiposity, insulin resistance, and liver triglycerides similar to a high fat diet. High dose antibiotic treatment or deletion of CD14 reduces inflammatory cytokine expression and ameliorates weight gain on HFD, demonstrating the important interaction of microbiota and inflammatory signaling cascades. Prebiotic fibers fermented to SCFA by the intestinal microbiota can block LPS-mediated metabolic consequences by increasing colonic expression of tight junction proteins zona occludens 1 and claudin 3, improving gut barrier function and reducing systemic LPS (Neyrinck et al., 2012) (Figure 2).
Akkermansia muciniphila, a microbial specialist that can derive its carbon and energy sources from mucus lining the intestinal tract, increases with prebiotic treatment (Everard et al., 2011). In addition to the metabolic and inflammatory changes previously discussed, HFD reduces A. muciniphila levels and thins the mucus layer (Everard et al., 2013). Introduction of A. muciniphila by daily oral gavage to mice fed HFD restores the mucus layer, decreases circulating LPS levels, decreases fat mass gain, and increases glucose tolerance compared to mice gavaged with PBS. These changes in metabolism and inflammation were not produced by killed A. muciniphila or by the “probiotic” strain Lactobacillus plantarum, indicating that live A. muciniphila has specialist actions that improve the gut barrier and reduce the effects of DIO.
TLR5
Deletion of the innate immunity receptor TLR5, which senses bacterial flagellin, induces inflammation, which if mild, causes a metabolic syndrome including hyperphagia-dependent weight gain, insulin resistance, and increased adiposity, blood pressure, and cholesterolemia (Vijay-Kumar et al., 2010). These changes were linked with increased adipocyte pro-inflammatory cytokines IL-1β and INF-γ. An antibiotic regimen that reduced microbial density 100-fold abrogated the metabolic syndrome in TLR5 −/− mice, and conversely, germ-free mice that were colonized with TLR5 −/− microbiota showed many of the disease phenotypes. Together these results indicate the sufficiency of the altered microbiota to yield the pathologies that accompany TLR5 deficiency. However, some TLR5 −/− mice have an opposing phenotype of severe colitis leading to weight loss, not gain (Vijay-Kumar et al., 2007). In a separate study, TLR5 −/− mice housed at two different animal facilities did not show an obesity phenotype or increased basal intestinal inflammation (Letran et al., 2011). The circulating microbes in separate facilities can lead to animals with differential microbiota colonization, altering host immune phenotypes (Ivanov et al., 2009). Since the genetic background was the same for the TLR5 −/− mice, the local microbiota in each animal facility most likely accounted for differences in the immune and metabolic phenotypes. Thus, in this model, the colonizing microbiota characteristics might determine inflammation extent and which pathway dominates.
Lymphotoxin mediates diet-induced obesity through the microbiota
Lymphotoxin, secreted by Th1 lymphocytes, and implicated in obesity by GWAS, is involved in the control of mucosal intestinal microbiota (Upadhyay et al., 2012). Lymphotoxin-receptor-deficient (ltbr −/−) mice have reduced colonic IL-22 and RegIII antimicrobial peptide expression, and elevated levels of segmented filamentous bacteria (SFB), consistent with diminished immune control at the mucosal surface (Upadhyay et al., 2012). Addition of HFD further shifts both immune responses and metabolism, including increasing colonic IL-23 expression and increasing in weight and adiposity. When any of the components in the axis are eliminated, including HFD, lymphotoxin α or β subunits, lymphotoxin receptor (ltbr −/−), IL-23, or RORγt (Th17) cells, mice become DIO-resistant (Figure 2, inset). The dietary changes and the differences in ltbr status account for 52% and 18% of the variation in the microbiome, respectively. Several expected microbiome shifts associated with introduction of a high fat diet, such as an increase in Firmicutes and a loss in diversity, were only observed in ltbr +/− mice, but not in ltbr −/− mice. GF wt mice conventionalized with microbiota from HFD-fed ltbr −/− mice transiently resist DIO, but the phenotype gradually reverts, due to their (wt) immune development. When HFD-fed ltbr −/− mice are co-housed with DIO-susceptible ltbr +/− mice, the ltbr −/− mice gain more weight, indicating the dominance of the transferred microbiota selected by HFD in the presence of a functional lymphotoxin pathway.
Inflammasomes and NASH
Multi-protein complexes that sense pathogen-associated molecular patterns (PAMPs), known as inflammasomes, aid in mucosal defenses by activating inflammatory cytokine precursors pro-IL-1β and pro-IL-18. Inflammasomes have been implicated in the progression from non-alcoholic fatty liver disease to non-alcoholic steatohepatitis (NASH) (Henao-Mejia et al., 2012). Apoptosis-associated speck-like protein containing a CARD (Asc) is a key adaptor protein of the inflammasome complex. Asc-deficient mice show higher levels of TLR4 and TLR9 agonists in the portal circulation and higher hepatic TNFα expression than wt mice, indicating that microbial products translocate to the liver and induce inflammation. When given a methionine-choline deficient diet known to induce NASH, mice lacking Asc or other components of the inflammasome complex (caspase 1, NLRP3) or downstream signaling (IL-18 −/−) develop NASH at a higher incidence than wt mice. The enhanced NASH phenotype in asc −/− mice was ablated with deletion of either TLR4, TLR9, MyD88:Trif, or TNFα, demonstrating the importance of innate immune pathways. However, the presence of the intestinal microbiota is also necessary because high dose broad-spectrum antibiotics in asc −/− mice reduces NASH incidence. When wt mice were co-housed with asc −/− mice, wt mice developed NASH more frequently, which was associated with increased Prevotellaceae, showing the importance of the altered microbiome in disease risk.
All told, these models of DIO and GIO illustrate the importance of immune control of intestinal microbiota, whose relaxation or activation can result in pathologic metabolic consequences.
3. METABOLIC IMPACT OF MICROBIOME PERTURBATION
Systems approaches
There are long-standing immunologic and metabolic relationships between the host and microbiota, and there can be consequences following natural and artificial perturbations. Microbial diversity and stability are important when considering the complex intra-community interactions that lead to emergent properties of the system. Well-evolved ecosystems often have functional redundancy in which several species can perform the same task. This concept explains how the taxa that constitute the mammalian gut microbiome often are highly diverse and variable from person to person, yet share high degree functional capacity (HMPConsortium, 2012; Kurokawa et al., 2007; Turnbaugh et al., 2009). However, ecosystems can be disrupted. In the case of forests, it might be fires, landslides, or droughts (Paine et al., 1998). For human microbiota, it might be famine, or surfeit, antibiotic treatments, or even parturition. Each of these events has the potential to change the energy flow in tightly regulated systems and consequences, including loss in diversity, vary according to the system’s status and the extent of functional redundancy. As diversity declines, loss of function may not be immediately apparent until the last redundant member able to perform a function is gone, however, communities with lower diversity are less resilient when subject to further perturbations (Kau et al., 2011). For perturbations of host-associated microbiota, timing is important, because the effects and their consequences may differ according to stage of life.
Despite the obvious differences in geography, diet, and lifestyle, there are strong similarities in the age-specific features of the microbial composition in persons living in the United States, Malawi, and Venezuela (Yatsunenko et al., 2012). Regardless of origin, babies begin life with low diversity, which progressively increases over the first years of life, and for all, the outline of the adult microbiome is essentially established by the age of three. Not surprising considering the long-standing differences in diet reflected in the three populations, the biochemical pathways represented in the metagenomes varied substantially. The study subjects differed substantially in the overall diversity in adulthood; the average adult in the US had from 225 to 400 fewer operational taxonomic units (OTUs) (or approximately 15 to 25 percent fewer) than did the Malawians or Venezuelan Amerindians. This observation is consistent with our earlier hypothesis of a ‘disappearing microbiota’ due to the changes represented by modernity (Blaser and Falkow, 2009), and may be the harbinger of functional losses as well.
Pregnancy
Parturition is a major biological perturbation, yet a critical event in the animal life cycle. The energetics are complex since the needs of the mother and her offspring must be balanced, and in mammals, the mother’s role extends far beyond birth. Nature’s challenge is to provide a sufficient endowment to the next generation while safeguarding the mother’s own health, for the essential tasks ahead. Multiple adaptations in the maternal immune system allow tolerance of the growing fetus (Erlebacher, 2013; Thellin and Heinen, 2003). The GI microbiota composition shifts remarkably over the course of pregnancy and continuing beyond (Koren et al., 2012). Its progressive nature implies that an optimum exists, timed to the events of parturition, and consistent with a model of balanced outcome.
There are temporal shifts in both the diversity within a single microbial community, known as α-diversity, which can be measured by phylogenetic diversity, species richness, and Shannon’s index, and in the diversity shared among different communities, known as β-diversity, which, for example, can be measured by UniFrac distances, changes in microbial abundance, and the Jaccard index, (Lozupone and Knight, 2008). The microbiota in late pregnancy has reduced α-diversity, but higher β-diversity compared to non-pregnant women or early pregnancy (Koren et al., 2012). In the 3rd trimester, there is greater representation of lactic acid bacteria (Lactobacillus, Streptococcus, and Enterococcus), which are highly prevalent in the infant gut, whereas butyrate-producing bacteria (Faecalibacterium, Blautia, and Ruminococcus), which dominate the gut in adulthood, are enriched in early pregnancy. Since mammals have evolved to provide a portion of their microbes to their offspring, shifts occurring during pregnancy may reflect selection that increases the probability of offspring survival. The greater representation of lactic acid bacteria may be seen as an adaptation to prepare the mother for transfer of these organisms in the birth and peri-natal period to her offspring to take maximum advantage of the main energy source for the child, lactose in its mother’s milk. The increased beta-diversity reflects the variation in the different pathways by which nature accomplishes its goals of intergenerational transfer.
When germ-free mice were conventionalized with fecal microbiota from women in their first (T1) or third (T3) trimester of pregnancy, those receiving the T3 microbiota had increased intestinal cytokines, gained more weight, and were more glucose-intolerant than those receiving the T1 microbiota. This result implies that the T3 microbiota induces an alternative metabolic state in its host, associated with greater energy storage. The findings in mice mimic those in pregnant women who also gain weight and develop insulin-resistance, but vary in the degree of these phenotypes. The trimester-specific differences provide evidence for a microbiota-driven energy optimum to maximize maternal fitness and caloric transfer to the next generation. This concept is consistent with the notion of a co-evolved microbiota that optimizes host survival through effects on its progeny.
Perturbation
Perturbation, an important constraint in biological systems, may be random, episodic, or programed, like pregnancy (see above) or hibernation (see below). Well-evolved ecosystems have long histories of perturbation, in part documented by and captured within their existing population structure (Paine et al., 1998). Ecosystem stability reflects both its resistance to change and its resilience following perturbation (Costello et al., 2012; Shade et al., 2012). Changes in diet in omnivorous hosts substantially perturb the gut microbiome (e.g. adding high fat (Turnbaugh et al., 2008), fiber (Cox et al., 2013), and protein (Faith et al., 2011)), and changes in the mode of birth delivery are associated with alterations in the patterns established for vertical microbiome transmission, comparing vaginal and Caesarian sections (Dominguez-Bello et al., 2010). Direct exposure to anti-bacterial agents has the greatest potential for perturbation, and occurs frequently in the US. Based on recent CDC data (Hicks et al., 2013), more than 250 million courses of antibiotics were prescribed in the US in 2010, which represents 880 courses of antibiotics per 1000 population. Short antibiotic courses in adults cause profound but incompletely understood changes in community structure, with variable resilience (Dethlefsen et al., 2008). Perturbation during early life, when the gut community structure is emerging (Yatsunenko et al., 2012), may be most important, since it is occurring during a developmentally sensitive host window (Cho et al., 2012). The CDC data indicate that the average child in the US is receiving about 3 antibiotic courses in first 2 years of life and nearly 11 by age 10 (Hicks et al., 2013). To understand the impact of these perturbations, ecological theory can guide us through the ground-rules governing ecosystem responses.
Within ecosystems, there are r- or K-selected species, which have evolved alternate strategies to cope with environmental disruption (Reznick et al., 2002). r-selected species evolved to have a rapid growth rate to monopolize resources when competition is low following a large ecological disruption, whereas K-selected species have evolved for efficiency to persist when the carrying capacity of the environment is low (Figure 3). In the intestine, large perturbations can result from antibiotic exposure, extreme changes in diet, surgical interventions, and acute gastrointestinal illnesses. Carrying capacity can be limited by availability of nutrients, electron acceptors (e.g. oxygen), or even physical niches. A weed is an example of an r-selected species with high proliferative potential. Weeds often are normal members of the ecosystem that live at the fringe in microhabitats, since competitors lock up the necessary resources for its expansion, but after a large perturbation, weeds can grow out of control. Whether native or introduced, the r-selected weeds are usually considered as invaders because of their unbalanced residency in the environment. Members of the phylum Proteobacteria or Verrucomicrobia usually represent a minor population within the intestinal microbiome, but can dominate following antibiotic therapy (Dubourg et al., 2013; Ubeda and Pamer, 2012), gastric bypass (Graessler et al., 2012; Liou et al., 2013), or dietary shifts (Parks et al., 2013), and thus may be examples of r-selected species dominating an environment following a perturbation. The gradual shift from facultative anaerobic bacteria (Proteobacteria and Bacilli) to strict anaerobes (Bacteroidia, Clostridia, and Erysipelotrichi) in the developing infant gut (Pantoja-Feliciano et al., 2013) may be driven by a competitive advantage of the K-species strict anaerobes to maintain populations when oxygen is limited.
After an ecosystem is perturbed, exogenous organisms are more likely to successfully invade. Recovery from perturbation requires a full complement of species or of functional groups. In the gut, there appears to be enormous functional redundancy (HMPConsortium, 2012; Turnbaugh et al., 2009). However, there could be keystone species, defined as those that have disproportional influence on the behavior of the system as a whole, that when lost or gained has a profound influence on health. For example, on the plains, wolves have a very small population footprint, but their predatory presence affects much larger communities of grazers, and even larger populations of plants that the grazers eat. An important consequence of perturbations is loss of keystone species, in which small populations can oscillate down to zero, and thus are lost.
Very successful organisms, whether they are r- or K-selected, often have bi-stable states. Bi-stability is an ancient adaptation to long-term niches that are subject to significant and frequent perturbations. Unpredictable food supplies, which constrain most animal populations, are an example of a typical perturbation. The alternative forms in bi-stable states allow optimization in changing milieus. When conditions change, rapid growers like Clostridium species, can form spores to transmit to the next susceptible host. At the cost of not adding to the population of proliferating cells that stably colonize a given host, exchanging metabolic signals with the host and/or other microbiota, Clostridium sp. diverts its metabolism to the spore form that is most likely to survive transmission. There is fitness cost in diverting energy to formation of the seeds that spores represent, but there also is great fitness value, since host life span is finite. Such dynamics affect the partitioning of energy in the gut lumen, with downstream effects on both microbiota and host. Unlike their animal hosts, bacteria have the ability to bloom on a very short time-scale with logarithmic expansion of their share of the energy pie, ultimately affecting transmission at critical junctures. This characteristic is a factor, intrinsic to any well-evolved ecology, subject to competition by other organisms operating with parallel strategies, and to host constraints, which are obligatorily obeyed.
Energy source and perturbation: the case of hibernation
Throughout the biosphere, there is vigorous competition for energy. In well-evolved mature stable ecosystems, carbon metabolism is tightly regulated (Mackelprang et al., 2011), implying that checks exist against one species gaining a disproportionate share of the available nutrients. Hibernation is a massive physiological shift that is programmed that affects both central metabolism and the immune system (Bouma et al., 2010; Carey et al., 2003; Humphries et al., 2003). That energy storage and utilization is central to hibernation physiology provides an excellent model system to examine microbial characteristics associated with an annual perturbation cycle. Some colonizers of the mammalian colon have the ability to obtain their energy, not from the food substrate passing through the intestine, but rather from digesting host-derived molecules (Berry et al., 2013). One important example is Akkermansia muciniphila, a member of the Verrucomicrobia phylum, which can use mucin as its sole carbon and nitrogen source (Derrien et al., 2004). That Akkermansia utilizes mucins for energy, which are virtually unlimited in supply in the vertebrate gut, represents an apparent paradox. With a readily available and apparently inexhaustible relatively exclusive food supply, why are Verrucomicrobia not always the dominant organism in the gut ecosystem? Similarly, what are the consequences to the host of an organism that has the ability to deplete its mucus, an important component of its barrier against potentially pathogenic microbiota and their toxic products?
Verrucomicrobia abundance is not constant in the intestinal microbiota; proportions increase with disruption and dysbiosis, e.g. antibiotic treatment (Dubourg et al., 2013), or increase with an influx of fermentable fiber (Tachon et al., 2012). The change in microbiota in hibernating ground squirrels provides an excellent paradigm (Carey et al., 2013) with broad implications. Squirrels store energy in the summer, and hibernate in the winter. In the late winter, essentially all of the energy available to the microbiota is host-derived. Hibernation, rather than specific host, diet, or age, is the major determinant of the gut microbial community composition. During the seasons of feeding, Firmicutes, which are specialists in dietary carbohydrates (Flint et al., 2012), dominate, but after several months of fasting, their representation falls, replaced by Verrucomicrobia, as well as by Bacteroidetes, which can switch their expression of genes related to diet or host-related energy sources (Martens et al., 2008; Salyers et al., 1977).
This programmed seasonal restructuring in relation to hibernation and its consequent dietary limitation is useful to model the energy relationships between the major gut microbiota taxa classes and the effects on host pathophysiology (Figure 4). In a steady-state in most mammals (and in times of feeding for hibernators), the populations of (r-selected) organisms that utilize host-derived substrates are held in check by competing organisms that use the plentiful dietary food sources. Change in the mucus layer is minimized. Dietary utilizers also break down complex macronutrients into small monomers or ferment them to SCFA, which are required nutrients for other microbial consortia. Use of SCFA by these transactional species is a benefit to dietary utilizers because it is removing a waste product that could detrimentally lower pH. However, in perturbed states (and in times of fasting for hibernators), the organisms that use dietary sources are disrupted and fail to check the growth of the host substrate-users and fail provide cross-feeding metabolites. The resulting overgrowth of organisms that utilize host substrates depletes the mucus layer and diminishes its barrier function. Similarly, butyrate, produced by fermentation of dietary carbohydrates, and known to enhance tight junctions (Wong et al., 2006), is reduced during fasting. These changes are manifested as an increase in the cellularity of the lamina propria and a reduction in villus length (Kurtz and Carey, 2007). To the pathologist, this would be ‘colitis,’ except that in the hibernator, it is the physiologic consequence of its lifestyle. The digestion of mucus and the loss of butyrate resulting from perturbations that affect the microbes requiring dietary energy sources are examples of secondary effects on the interface between the microbiota and the host. These can result in differential signaling to epithelial and immune cells, with both immune and metabolic consequences (Canani, 2011).
The dynamics of the interactions suggest that there are boundary conditions that govern the microbial population structure in each gut locality. However, in total, the conserved nature of the relationships of the major phyla to one another, and their fluctuation with perturbation suggests the operation of a Nash equilibrium (Blaser and Kirschner, 2007), a game theory construct in which good behavior by players is rewarded, and ‘cheaters’ achieve inferior results (Nash, 1951). Within the boundary conditions of the gut, with competing organisms, and alternative energy substrates that affect host viability, the development of a Nash equilibrium can explain the conserved dynamic changes observed between the microbe phyla and the host. The overall conservation of the mammalian colonic microbial community structure is an important observation that indicates shared solutions between hosts and their resident microbes that have persisted for > 50 million years of evolution (Ley et al., 2008a). This time-scale implies that important boundary conditions have selected against marked divergence from a common ancestry. In contrast, the presence of alternative macro-level population structures in mammals (Arumugam et al., 2011) could be a condition inconsistent with a conserved Nash equilibrium.
Conclusions
Host phenotypes in obesity are dependent on the interactions between the diet, the resident microbiota, and host immunity. Evolutionary and ecological theories ultimately govern, but the principal actors include the differential microbial energetics and the host cells that monitor the microbial denizens. We are just beginning to explore a complex and ultimately crucial biological system, all the more important, because it appears to be changing out of proportion to evolutionary time.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahn SM, Pomp A, Rubino F. Metabolic surgery for type 2 diabetes. Ann NY Acad Sci. 2011;1212:E37–E45. doi: 10.1111/j.1749-6632.2011.05984.x. [DOI] [PubMed] [Google Scholar]
- Ajslev TA, Andersen CS, Gamborg M, Sørensen TIA, Jess T. Childhood overweight after establishment of the gut microbiota: the role of delivery mode, pre-pregnancy weight and early administration of antibiotics. Int J Obes. 2011;35:522–529. doi: 10.1038/ijo.2011.27. [DOI] [PubMed] [Google Scholar]
- Antonopoulos DA, Huse SM, Morrison HG, Schmidt TM, Sogin ML, Young VB. Reproducible Community Dynamics of the Gastrointestinal Microbiota following Antibiotic Perturbation. Infection and Immunity. 2009;77:2367–2375. doi: 10.1128/IAI.01520-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aron-Wisnewsky J, Doré J, Clement K. The importance of the gut microbiota after bariatric surgery. Nature Rev Gastroenterol & Hepatol. 2012;9:590–598. doi: 10.1038/nrgastro.2012.161. [DOI] [PubMed] [Google Scholar]
- Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, Fernandes GR, Tap J, Bruls T, Batto JM, et al. Enterotypes of the human gut microbiome. Nature. 2011;473:174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bäckhed F, Ding H, Wang T, Hooper LV, Koh GY, Nagy A, Semenkovich CF, Gordon JI. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci USA. 2004;101:15718–15723. doi: 10.1073/pnas.0407076101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergman EN. Energy contributions of volatile fatty acids from the gastrointestinal tract in various species. Physiol Rev. 1990;70:567–590. doi: 10.1152/physrev.1990.70.2.567. [DOI] [PubMed] [Google Scholar]
- Bernbom N, Licht TR, Brogren CH, Jelle B, Johansen AH, Badiola I, Vogensen FK, Nørrung B. Effects of Lactococcus lactis on Composition of Intestinal Microbiota: Role of Nisin. Appl Environ Microbiol. 2006;72:239–244. doi: 10.1128/AEM.72.1.239-244.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry D, Stecher B, Schintlmeister A, Reichert J, Brugiroux S, Wild B, Wanek W, Richter A, Rauch I, Decker T, et al. Host-compound foraging by intestinal microbiota revealed by single-cell stable isotope probing. Proc Natl Acad Sci USA. 2013;110:4720–4725. doi: 10.1073/pnas.1219247110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biasucci G, Rubini M, Riboni S, Morelli L, Bessi E, Retetangos C. Mode of delivery affects the bacterial community in the newborn gut. Early Hum Dev. 2010;86:13–15. doi: 10.1016/j.earlhumdev.2010.01.004. [DOI] [PubMed] [Google Scholar]
- Blaser MJ, Falkow S. What are the consequences of the disappearing human microbiota? Nat Rev Micro. 2009;7:887–894. doi: 10.1038/nrmicro2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaser MJ, Kirschner D. The equilibria that allow bacterial persistence in human hosts. Nature. 2007;449:843–849. doi: 10.1038/nature06198. [DOI] [PubMed] [Google Scholar]
- Blustein J, Attina T, Liu M, Ryan AM, Cox LM, Blaser MJ, Trasande L. Association of caesarean delivery with child adiposity from age 6 weeks to 15 years. Int J Obes. 2013 doi: 10.1038/ijo.2013.49. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouma HR, Carey HV, Kroese FGM. Hibernation: the immune system at rest? J Leukocyte Biol. 2010;88:619–624. doi: 10.1189/jlb.0310174. [DOI] [PubMed] [Google Scholar]
- Canani RB. Potential beneficial effects of butyrate in intestinal and extraintestinal diseases. World J Gastroenterol. 2011;17:1519. doi: 10.3748/wjg.v17.i12.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cani PD, Amar J, Iglesias MA, Poggi M, Knauf C, Bastelica D, Neyrinck AM, Fava F, Tuohy KM, Chabo C, et al. Metabolic Endotoxemia Initiates Obesity and Insulin Resistance. Diabetes. 2007;56:1761–1772. doi: 10.2337/db06-1491. [DOI] [PubMed] [Google Scholar]
- Cani PD, Bibiloni R, Knauf C, Waget A, Neyrinck AM, Delzenne NM, Burcelin R. Changes in Gut Microbiota Control Metabolic Endotoxemia-Induced Inflammation in High-Fat Diet-Induced Obesity and Diabetes in Mice. Diabetes. 2008;57:1470–1481. doi: 10.2337/db07-1403. [DOI] [PubMed] [Google Scholar]
- Carey HV, Andrews MT, Martin SL. Mammalian hibernation: cellular and molecular responses to depressed metabolism and low temperature. Physiol Rev. 2003;83:1153–1181. doi: 10.1152/physrev.00008.2003. [DOI] [PubMed] [Google Scholar]
- Carey HV, Walters WA, Knight R. Seasonal restructuring of the ground squirrel gut microbiota over the annual hibernation cycle. Am J Physiol-Reg I. 2013;304:R33–R42. doi: 10.1152/ajpregu.00387.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho I, Blaser MJ. The human microbiome: at the interface of health and disease. Nat Rev Genet. 2012;13:260–270. doi: 10.1038/nrg3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho I, Yamanishi S, Cox L, Methe BA, Zavadil J, Li K, Gao Z, Mahana D, Raju K, Teitler I, et al. Antibiotics in early life alter the murine colonic microbiome and adiposity. Nature. 2012;488:621–626. doi: 10.1038/nature11400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coates ME, Fuller R, Harrison GF, Lev M, Suffolk SF. A comparison of the growth of chicks in the Gustafsson germ-free apparatus and in a conventional environment, with and without dietary supplements of penicillin. Br J Nutr. 1963;17:141–150. doi: 10.1079/bjn19630015. [DOI] [PubMed] [Google Scholar]
- Costello EK, Stagaman K, Dethlefsen L, Bohannan BJM, Relman DA. The Application of Ecological Theory Toward an Understanding of the Human Microbiome. Science. 2012;336:1255–1262. doi: 10.1126/science.1224203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox LM, Cho I, Young SA, Anderson WHK, Waters BJ, Hung SC, Gao Z, Mahana D, Bihan M, Alekseyenko AV, et al. The nonfermentable dietary fiber hydroxypropyl methylcellulose modulates intestinal microbiota. FASEB J. 2013;27:692–702. doi: 10.1096/fj.12-219477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien M, Vaughan EE, Plugge CM, de Vos WM. Akkermansia muciniphila gen. nov., sp. nov., a human intestinal mucin-degrading bacterium. Int J Syst Evol Micr. 2004;54:1469–1476. doi: 10.1099/ijs.0.02873-0. [DOI] [PubMed] [Google Scholar]
- Dethlefsen L, Huse S, Sogin ML, Relman DA. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 2008;6:e280. doi: 10.1371/journal.pbio.0060280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez-Bello MG, Blaser MJ, Ley RE, Knight R. Development of the Human Gastrointestinal Microbiota and Insights From High-Throughput Sequencing. Gastroenterology. 2011;140:1713–1719. doi: 10.1053/j.gastro.2011.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, Knight R. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci USA. 2010;107:11971–11975. doi: 10.1073/pnas.1002601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubourg G, Lagier JC, Armougom F, Robert C, Audoly G, Papazian L, Raoult D. High-level colonisation of the human gut by Verrucomicrobia following broad-spectrum antibiotic treatment. Int Journal Antimicrob Ag. 2013;41:149–155. doi: 10.1016/j.ijantimicag.2012.10.012. [DOI] [PubMed] [Google Scholar]
- Erlebacher A. Immunology of the Maternal-Fetal Interface. Annu Rev Immunol. 2013;31:387–411. doi: 10.1146/annurev-immunol-032712-100003. [DOI] [PubMed] [Google Scholar]
- Everard A, Belzer C, Geurts L, Ouwerkerk JP, Druart C, Bindels LB, Guiot Y, Derrien M, Muccioli GG, Delzenne NM, et al. Crosstalk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc Natl Acad Sci USA. 2013:1–6. doi: 10.1073/pnas.1219451110. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everard A, Lazarevic V, Derrien M, Girard M, Muccioli GG, Muccioli GM, Neyrinck AM, Possemiers S, Van Holle A, François P, et al. Responses of gut microbiota and glucose and lipid metabolism to prebiotics in genetic obese and diet-induced leptin-resistant mice. Diabetes. 2011;60:2775–2786. doi: 10.2337/db11-0227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faith JJ, McNulty NP, Rey FE, Gordon JI. Predicting a Human Gut Microbiotaís Response to Diet in Gnotobiotic Mice. Science. 2011;333:101–104. doi: 10.1126/science.1206025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flegal KM, Carroll MD, Kit BK, Ogden CL. Prevalence of obesity and trends in the distribution of body mass index among us adults, 1999–2010. JAMA. 2012;307:491–497. doi: 10.1001/jama.2012.39. [DOI] [PubMed] [Google Scholar]
- Flint HJ, Scott KP, Duncan SH, Louis P, Forano E. Microbial degradation of complex carbohydrates in the gut. Gut Microbes. 2012;3:289–306. doi: 10.4161/gmic.19897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graessler J, Qin Y, Zhong H, Zhang J, Licinio J, Wong M-L, Xu A, Chavakis T, Bornstein AB, Ehrhart-Bornstein M, et al. Metagenomic sequencing of the human gut microbiome before and after bariatric surgery in obese patients with type 2 diabetes: correlation with inflammatory and metabolic parameters. Pharmacogenomics J. 2012:1–9. doi: 10.1038/tpj.2012.43. [DOI] [PubMed] [Google Scholar]
- Greenblum S, Turnbaugh PJ, Borenstein E. Metagenomic systems biology of the human gut microbiome reveals topological shifts associated with obesity and inflammatory bowel disease. Proc Natl Acad Sci USA. 2012;109:594–599. doi: 10.1073/pnas.1116053109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwood MR, Hirsch J. Postnatal development of adipocyte cellularity in the normal rat. J Lipid Res. 1974;15:474–483. [PubMed] [Google Scholar]
- Haslam DW, James WPT. Obesity. Lancet. 2005;366:1197–1209. doi: 10.1016/S0140-6736(05)67483-1. [DOI] [PubMed] [Google Scholar]
- Henao-Mejia J, Elinav E, Jin C, Hao L, Mehal WZ, Strowig T, Thaiss CA, Kau AL, Eisenbarth SC, Jurczak MJ, et al. Inflammasome-mediated dysbiosis regulates progression of NAFLD and obesity. Nature. 2012;482:179–185. doi: 10.1038/nature10809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks LA, Taylor TH, Hunkler RJ. U.S. Outpatient Antibiotic Prescribing, 2010. New Engl J Med. 2013;368:1461–1462. doi: 10.1056/NEJMc1212055. [DOI] [PubMed] [Google Scholar]
- HMPConsortium. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollopeter G, Erickson JC, Palmiter RD. Role of neuropeptide Y in diet-, chemical-and genetic-induced obesity of mice. Int J Obes. 1998;22:506–512. doi: 10.1038/sj.ijo.0800615. [DOI] [PubMed] [Google Scholar]
- Holmes E, Li JV, Marchesi JR, Nicholson JK. Gut Microbiota Composition and Activity in Relation to Host Metabolic Phenotype and Disease Risk. Cell Metabolism. 2012;16:559–564. doi: 10.1016/j.cmet.2012.10.007. [DOI] [PubMed] [Google Scholar]
- Huh SY, Rifas-Shiman SL, Zera CA, Edwards JWR, Oken E, Weiss ST, Gillman MW. Delivery by caesarean section and risk of obesity in preschool age children: a prospective cohort study. Arch Dis Childhood. 2012;97:610–616. doi: 10.1136/archdischild-2011-301141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphries MM, Thomas DW, Kramer DL. The role of energy availability in Mammalian hibernation: a cost-benefit approach. Physiol Biochem Zool. 2003;76:165–179. doi: 10.1086/367950. [DOI] [PubMed] [Google Scholar]
- Hungate RE. The Rumen and Its Microbes. New York: Academic Press; 1966. [Google Scholar]
- Hungate RE, Dougherty RW, Bryant MP, Cello RM. Microbiological and physiological changes associated with acute indigestion in sheep. Cornell Vet. 1952;42:423–449. [PubMed] [Google Scholar]
- Huurre A, Kalliomäki M, Rautava S, Rinne M, Salminen S, Isolauri E. Mode of Delivery – Effects on Gut Microbiota and Humoral Immunity. Neonatology. 2008;93:236–240. doi: 10.1159/000111102. [DOI] [PubMed] [Google Scholar]
- Ichimura A, Hirasawa A, Hara T, Tsujimoto G. Free fatty acid receptors act as nutrient sensors to regulate energy homeostasis. Prostaglandins Other Lipid Mediat. 2009;89:82–88. doi: 10.1016/j.prostaglandins.2009.05.003. [DOI] [PubMed] [Google Scholar]
- Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, et al. Induction of Intestinal Th17 Cells by Segmented Filamentous Bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumpertz R, Le DS, Turnbaugh PJ, Trinidad C, Bogardus C, Gordon JI, Krakoff J. Energy-balance studies reveal associations between gut microbes, caloric load, and nutrient absorption in humans. Am J Clin Nutr. 2011;94:58–65. doi: 10.3945/ajcn.110.010132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kau AL, Ahern PP, Griffin NW, Goodman AL, Gordon JI. Human nutrition, the gut microbiome and the immune system. Nature. 2011;474:327–336. doi: 10.1038/nature10213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren O, Goodrich Julia K, Cullender Tyler C, Spor A, Laitinen K, Kling Bäckhed H, Gonzalez A, Werner Jeffrey J, Angenent Largus T, Knight R, et al. Host Remodeling of the Gut Microbiome and Metabolic Changes during Pregnancy. Cell. 2012;150:470–480. doi: 10.1016/j.cell.2012.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurokawa K, Itoh T, Kuwahara T, Oshima K, Toh H, Toyoda A, Takami H, Morita H, Sharma VK, Srivastava TP, et al. Comparative Metagenomics Revealed Commonly Enriched Gene Sets in Human Gut Microbiomes. DNA Res. 2007;14:169–181. doi: 10.1093/dnares/dsm018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz CC, Carey HV. Seasonal changes in the intestinal immune system of hibernating ground squirrels. Dev Comp Immunol. 2007;31:415–428. doi: 10.1016/j.dci.2006.07.003. [DOI] [PubMed] [Google Scholar]
- Letran SE, Lee SJ, Atif SM, Flores-Langarica A, Uematsu S, Akira S, Cunningham AF, McSorley SJ. TLR5-Deficient Mice Lack Basal Inflammatory and Metabolic Defects but Exhibit Impaired CD4 T Cell Responses to a Flagellated Pathogen. J Immunol. 2011;186:5406–5412. doi: 10.4049/jimmunol.1003576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE. Obesity and the human microbiome. Curr Opin Gastroenterol. 2010;26:5–11. doi: 10.1097/MOG.0b013e328333d751. [DOI] [PubMed] [Google Scholar]
- Ley RE, Bäckhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proc Natl Acad Sci USA. 2005;102:11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE, Hamady M, Lozupone CA, Turnbaugh PJ, Ramey RR, Bircher JS, Schlegel ML, Tucker TA, Schrenzel MD, Knight R, et al. Evolution of Mammals and Their Gut Microbes. Science. 2008a;320:1647–1651. doi: 10.1126/science.1155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE, Lozupone CA, Hamady M, Knight R, Gordon JI. Worlds within worlds: evolution of the vertebrate gut microbiota. Nature Rev Micro. 2008b;6:776–788. doi: 10.1038/nrmicro1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- Li H, Ye R, Pei L, Ren A, Zheng X, Liu J. Caesarean delivery, caesarean delivery on maternal request and childhood overweight: a Chinese birth cohort study of 181 380 children. Pediatr Obes. 2013 doi: 10.1111/j.2047-6310.2013.00151.x. [DOI] [PubMed] [Google Scholar]
- Li JV, Ashrafian H, Bueter M, Kinross J, Sands C, Le Roux CW, Bloom SR, Darzi A, Athanasiou T, Marchesi JR, et al. Metabolic surgery profoundly influences gut microbial-host metabolic cross-talk. Gut. 2011;60:1214–1223. doi: 10.1136/gut.2010.234708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liou AP, Paziuk M, Luevano JM, Machineni S, Turnbaugh PJ, Kaplan LM. Conserved Shifts in the Gut Microbiota Due to Gastric Bypass Reduce Host Weight and Adiposity. Sci Trans Med. 2013;5:178ra141–178ra141. doi: 10.1126/scitranslmed.3005687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone CA, Knight R. Species divergence and the measurement of microbial diversity. FEMS Microbiol Rev. 2008;32:557–578. doi: 10.1111/j.1574-6976.2008.00111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macfarlane GT, Macfarlane S. Bacteria, Colonic Fermentation, and Gastrointestinal Health. J AOAC Int. 2012;95:50–60. doi: 10.5740/jaoacint.sge_macfarlane. [DOI] [PubMed] [Google Scholar]
- Mackelprang R, Waldrop MP, DeAngelis KM, David MM, Chavarria KL, Blazewicz SJ, Rubin EM, Jansson JK. Metagenomic analysis of a permafrost microbial community reveals a rapid response to thaw. Nature. 2011;480:368–371. doi: 10.1038/nature10576. [DOI] [PubMed] [Google Scholar]
- Martens EC, Chiang HC, Gordon JI. Mucosal Glycan Foraging Enhances Fitness and Transmission of a Saccharolytic Human Gut Bacterial Symbiont. Cell Host Microbe. 2008;4:447–457. doi: 10.1016/j.chom.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaraja TG, Titgemeyer EC. Ruminal Acidosis in Beef Cattle: The Current Microbiological and Nutritional Outlook1,2. J Dairy Sci. 2007;90:E17–E38. doi: 10.3168/jds.2006-478. [DOI] [PubMed] [Google Scholar]
- Nash J. Non-cooperative games. Ann Math. 1951;54:286–295. [Google Scholar]
- Neyrinck AM, Van Hee VF, Piront N, De Backer F, Toussaint O, Cani PD, Delzenne NM. Wheat-derived arabinoxylan oligosaccharides with prebiotic effect increase satietogenic gut peptides and reduce metabolic endotoxemia in diet-induced obese mice. Nutr Diabetes. 2012;2:e28. doi: 10.1038/nutd.2011.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paine RT, Tegner MJ, Johnson EA. Compounded perturbations yield ecological surprises. Ecosystems. 1998;1:535–545. [Google Scholar]
- Pandey PK, Verma P, Kumar H, Bavdekar A, Patole MS, Shouche YS. Comparative analysis of fecal microflora of healthy full-term Indian infants born with different methods of delivery (vaginal vs cesarean): Acinetobacter sp. prevalence in vaginally born infants. J Bioscience. 2012;37:989–998. doi: 10.1007/s12038-012-9268-5. [DOI] [PubMed] [Google Scholar]
- Pantoja-Feliciano IG, Clemente JC, Costello EK, Perez ME, Blaser MJ, Knight R, Dominguez-Bello MG. Biphasic assembly of the murine intestinal microbiota during early development. ISME J. 2013:1–4. doi: 10.1038/ismej.2013.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks BW, Nam E, Org E, Kostem E, Norheim F, Hui ST, Pan C, Civelek M, Rau CD, Bennett BJ, et al. Genetic Control of Obesity and Gut Microbiota Composition in Response to High-Fat, High-Sucrose Diet in Mice. Cell Metabolism. 2013;17:141–152. doi: 10.1016/j.cmet.2012.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne AN, Chassard C, Lacroix C. Gut microbial adaptation to dietary consumption of fructose, artificial sweeteners and sugar alcohols: implications for host–microbe interactions contributing to obesity. Obes Rev. 2012;13:799–809. doi: 10.1111/j.1467-789X.2012.01009.x. [DOI] [PubMed] [Google Scholar]
- Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravussin Y, Koren O, Spor A, LeDuc C, Gutman R, Stombaugh J, Knight R, Ley RE, Leibel RL. Responses of Gut Microbiota to Diet Composition and Weight Loss in Lean and Obese Mice. Obesity. 2012;20:738–747. doi: 10.1038/oby.2011.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reznick D, Bryant MJ, Bashey F. r-and K-selection revisited: the role of population regulation in life-history evolution. Ecology. 2002;83:1509–1520. [Google Scholar]
- Rubino F, Gagner M, Gentileschi P, Kini S, Fukuyama S, Feng J, Diamond E. The Early Effect of the Roux-en-Y Gastric Bypass on Hormones Involved in Body Weight Regulation and Glucose Metabolism. Ann Surg. 2004;240:236–242. doi: 10.1097/01.sla.0000133117.12646.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salyers AA, Vercellotti JR, West SE, Wilkins TD. Fermentation of mucin and plant polysaccharides by strains of Bacteroides from the human colon. Appl Environ Microbiol. 1977;33:319–322. doi: 10.1128/aem.33.2.319-322.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel BS, Shaito A, Motoike T, Rey FE, Backhed F, Manchester JK, Hammer RE, Williams SC, Crowley J, Yanagisawa M, et al. Effects of the gut microbiota on host adiposity are modulated by the short-chain fatty-acid binding G protein-coupled receptor, Gpr41. Proc Natl Acad Sci USA. 2008;105:16767–16772. doi: 10.1073/pnas.0808567105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage DC. Microbial ecology of the gastrointestinal tract. Annu Rev Microbiol. 1977;31:107–133. doi: 10.1146/annurev.mi.31.100177.000543. [DOI] [PubMed] [Google Scholar]
- Shade A, Peter H, Allison SD, Baho DL, Berga M, Bürgmann H, Huber DH, Langenheder S, Lennon JT, Martiny JBH, et al. Fundamentals of microbial community resistance and resilience. Front Microbiol. 2012;3:417. doi: 10.3389/fmicb.2012.00417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachon S, Zhou J, Keenan M, Martin R, Marco ML. The intestinal microbiota in aged mice is modulated by dietary resistant starch and correlated with improvements in host responses. FEMS Microbiol Ecol. 2012;83:299–309. doi: 10.1111/j.1574-6941.2012.01475.x. [DOI] [PubMed] [Google Scholar]
- Thellin O, Heinen E. Pregnancy and the immune system: between tolerance and rejection. Toxicology. 2003;185:179–184. doi: 10.1016/s0300-483x(02)00607-8. [DOI] [PubMed] [Google Scholar]
- Trasande L, Blustein J, Liu M, Corwin E, Cox LM, Blaser MJ. Infant antibiotic exposures and early-life body mass. Int J Obes. 2013;37:16–23. doi: 10.1038/ijo.2012.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh P, Backhed F, Fulton L, Gordon J. Diet-Induced Obesity Is Linked to Marked but Reversible Alterations in the Mouse Distal Gut Microbiome. Cell Host Microbe. 2008;3:213–223. doi: 10.1016/j.chom.2008.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1131. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- Ubeda C, Pamer EG. Antibiotics, microbiota, and immune defense. Trends Immunol. 2012;33:459–466. doi: 10.1016/j.it.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhyay V, Poroyko V, Kim T-j, Devkota S, Fu S, Liu D, Tumanov AV, Koroleva EP, Deng L, Nagler C, et al. Lymphotoxin regulates commensal responses to enable diet-induced obesity. Nat Immunol. 2012;13:947–953. doi: 10.1038/ni.2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijay-Kumar M, Aitken JD, Carvalho FA, Cullender TC, Mwangi S, Srinivasan S, Sitaraman SV, Knight R, Ley RE, Gewirtz AT. Metabolic Syndrome and Altered Gut Microbiota in Mice Lacking Toll-Like Receptor 5. Science. 2010;328:228–231. doi: 10.1126/science.1179721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijay-Kumar M, Sanders CJ, Taylor RT, Kumar A, Aitken JD, Sitaraman SV, Neish AS, Uematsu S, Akira S, Williams IR, et al. Deletion of TLR5 results in spontaneous colitis in mice. J Clin Invest. 2007;117:3909–3921. doi: 10.1172/JCI33084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong JMW, de Souza R, Kendall CWC, Emam A, Jenkins DJA. Colonic health: fermentation and short chain fatty acids. J Clin Gastroenterol. 2006;40:235–243. doi: 10.1097/00004836-200603000-00015. [DOI] [PubMed] [Google Scholar]
- Wright J, Kennedy-Stephenson J, Wang C, McDowell M, Johnson C. Trends in intake of energy and macronutrients–United States, 1971–2000. JAMA. 2004;291:1193–1194. [Google Scholar]
- Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]