Superresolution fluorescence microscopy shows that bacterial MreB forms filamentous structures of length up to 3.4 µm, which run at various angles underneath the cell membrane with a speed of 85 nm/s. Movement depends on ATPase activity. Thus MreB is a true orthologue of actin, whose extended filament architecture drives cells.
Abstract
The maintenance of rod-cell shape in many bacteria depends on actin-like MreB proteins and several membrane proteins that interact with MreB. Using superresolution microscopy, we show that at 50-nm resolution, Bacillus subtilis MreB forms filamentous structures of length up to 3.4 μm underneath the cell membrane, which run at angles diverging up to 40° relative to the cell circumference. MreB from Escherichia coli forms at least 1.4-μm-long filaments. MreB filaments move along various tracks with a maximal speed of 85 nm/s, and the loss of ATPase activity leads to the formation of extended and static filaments. Suboptimal growth conditions lead to formation of patch-like structures rather than extended filaments. Coexpression of wild-type MreB with MreB mutated in the subunit interface leads to formation of shorter MreB filaments and a strong effect on cell shape, revealing a link between filament length and cell morphology. Thus MreB has an extended-filament architecture with the potential to position membrane proteins over long distances, whose localization in turn may affect the shape of the cell wall.
INTRODUCTION
The discovery of cytoskeletal elements in bacteria has dramatically changed the view of the cell biology of all cells because these proteins must have developed very early in evolution rather than more recently with the evolution of eukaryotes. It is generally accepted that FtsZ (found in bacteria and Archaea) is an orthologue of tubulin and thus a genuine cytoskeletal protein. FtsZ forms a polymeric ring at the cell center in almost all bacteria, as well as in a large part of the Archaea, and initiates cell division by recruiting all further downstream proteins that build up the septum (Adams and Errington, 2009). Further, coiled-coil–rich proteins (e.g., crescentin) affect cell curvature and form extended intermediate-filament (IF)–like structures in several bacterial species, possibly representing a divergent evolution of IF proteins (Cabeen and Jacobs-Wagner, 2010). In addition, MreB and ParM have a structure that can be superimposed with that of actin with little deviation (van den Ent et al., 2001, 2002). These three proteins share the actin fold with additional proteins, such as FtsA and sugar kinases; the latter are not believed to form filaments that play a role in vivo. Like actin, MreB and ParM form magnesium- and nucleotide-dependent filaments in vitro and in vivo (FtsA has been shown to form filaments in vitro but not yet in vivo; Szwedziak et al., 2012) but have a different architecture from that of actin (van den Ent et al., 2001; Graumann, 2007; Mayer and Amann, 2009). In contrast to actin, MreB can form straight double filaments or sheets of filaments, although under certain salt conditions, helical MreB filaments have been observed, whereas ParM forms double-helical filaments with opposite handedness to those of actin (Popp et al., 2008, 2010; Salje et al., 2009). MreB is an essential protein in many species and localizes underneath the bacterial cell membrane in many rod-shaped bacteria through direct interaction with the membrane (Dempwolff et al., 2011; Salje et al., 2011). The depletion of MreB leads to a loss of rod shape, until the round cells lyse, and point mutations in the ATPase-binding pocket have a dominant-negative effect on cell morphology (Kruse et al., 2003; Defeu Soufo and Graumann, 2006). In addition, MreB affects the positioning of intracellular protein complexes and even cell surface structures in many bacterial species (Gitai et al., 2004; Cowles and Gitai, 2010; Defeu Soufo and Graumann, 2005). Diffraction-limited epifluorescence, total internal reflection fluorescence (TIRF), and confocal fluorescence microscopy techniques have been used to interpret MreB structures as helical structures that show a high turnover and thus remodeling on a time scale of tens of seconds (Jones et al., 2001; Carballido-Lopez and Errington, 2003; Defeu Soufo and Graumann, 2004; Graumann, 2007; Kim et al., 2006). MreB was therefore termed a cytoskeletal element. Recently, the view that MreB forms extended filamentous structures was challenged, and a passive motion mechanism was proposed (Dominguez-Escobar et al., 2011; Garner et al., 2011; van Teeffelen et al., 2011). Several reports concluded that MreB does not form extended filaments but forms “patch” structures (i.e., filaments of a size smaller than the resolution of light microscopy) that passively move along with the putative cell wall extension machinery from one side of the cell to the other in a circumferential motion. Such short structures would connect cytosolic proteins with membrane proteins but would not be able to connect membrane proteins on a longer range. These ideas suggest that the fundamental aspect of the formation of extended filamentous structures that affect cell shape and other dynamic subcellular processes may be dissimilar between MreB and actin. Using superresolution microscopy, we show that MreB forms filaments ranging from shorter (250 nm) to longer (greater than a half-turn around the cell periphery) sizes, which have the potential to confer long-range interactions underneath the cell membrane. We show that mutations in the ATP-binding pocket, as well as in the subunit interface, alter filament dynamics or length, accompanied by alterations in cell morphology, showing that filament architecture of MreB is crucial for proper cell shape maintenance.
Bacterial cell shape is dictated by the peptidoglycan wall (Cabeen and Jacobs-Wagner, 2005). In addition to MreB, several proteins are essential for the generation and maintenance of rod cell morphology in bacteria. RodA and MreC are essential for viability, and their individual depletion leads to the formation of round cells (Matsuzawa et al., 1973; Iwaya et al., 1978; Henriques et al., 1998; Lee and Stewart, 2003; Kruse et al., 2005; Bendezu and de Boer, 2008). This is also true for the depletion of MreD, but the cells are able to survive as cocci, although growth is highly retarded. MreB (or its paralogue in Bacillus subtilis, Mbl) interacts with MreC and MreD (Kruse et al., 2005; Defeu Soufo and Graumann, 2006), and also with penicillin-binding proteins, components of the machinery that extends the cell wall during bacterial growth (Divakaruni et al., 2005; Kawai et al., 2009). In an alternative (but not mutually exclusive) model to the passenger model described earlier, the localization of MreB assemblies might direct the positioning of membrane proteins, thus organizing the synthesis of the wall into a regular pattern.
RESULTS
B. subtilis MreB and Mbl form dynamic filaments of length up to 3.4 μm underneath the cell membrane in vivo
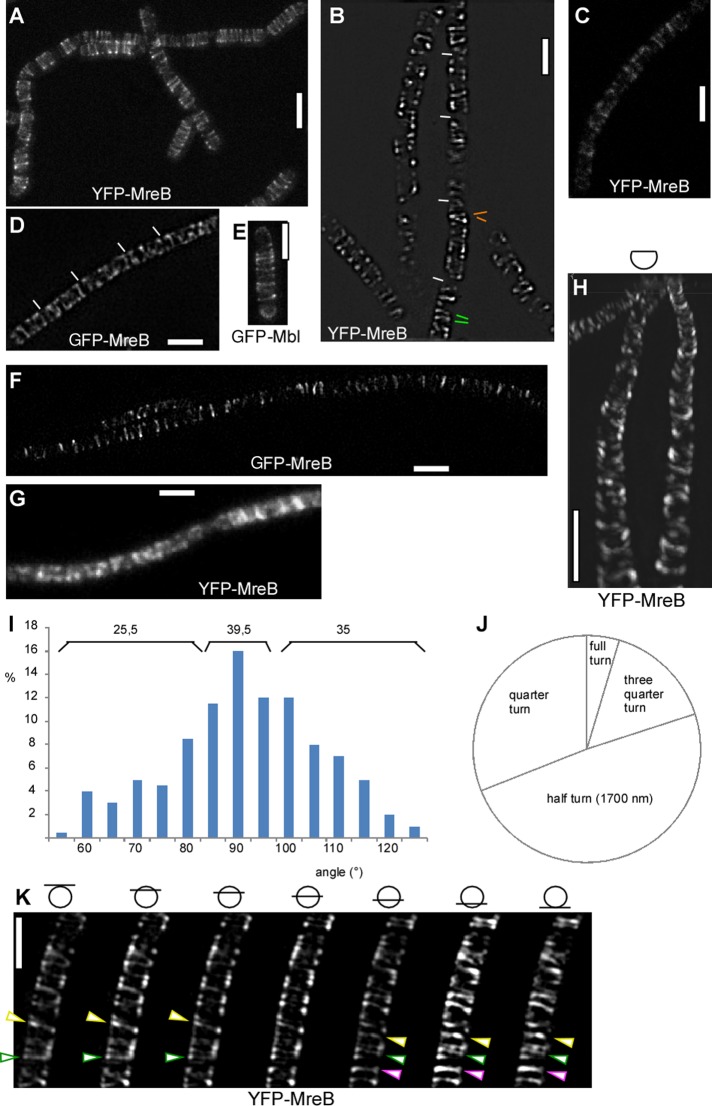
We addressed the fundamental question of whether MreB forms extended filamentous structures in a bacterium, like its orthologue actin in eukaryotes, or builds up small assemblies that would only be capable of conferring interactions between cytosolic and membrane proteins but no interactions along the membrane. We therefore used stimulated emission depletion (STED) and structured illumination (SIM) superresolution microscopy to visualize MreB filaments at highest possible resolution in live bacterial cells. We imaged exponentially growing B. subtilis cells, which express a green fluorescent protein (GFP)–MreB fusion from the original gene locus, which can functionally replace the wild-type copy (Defeu Soufo and Graumann, 2004), or a yellow fluorescent protein (YFP)–MreB fusion from an ectopic site on the chromosome (Defeu Soufo and Graumann, 2006), driven by the xylose-inducible promoter. We also used a published strain in which the original mreB gene is deleted and GFP-MreB is expressed from the amylase locus (a kind gift of Rut Carballido-Lopez, French National Institute for Agricultural Research, Jouy-en-Josas, France; Dominguez-Escobar et al., 2011). In addition, we generated and visualized a GFP-Mbl fusion, which is expressed under the control of the original mbl promoter from the native gene locus and is also expressed as the sole source of Mbl in the cell. Qualitatively, STED and SIM images showed similar results. We reached a resolution of 125 nm with N-SIM (Nikon) and SIM (Zeiss), 80 nm with STED, and 42 nm with gated (G-) STED (Leica). YFP-MreB or GFP-MreB (as a sole source of the protein) formed distinct filaments, the shortest of which were diffraction limited (50 nm) and a large number of which were between 0.8 and 1.7 μm (the latter fraction extended around an entire half-circumference, cell width being 1.1 μm) or possibly longer (Figure 1, A–D). Acquisition of 15 images for three-dimensional (3D) N-SIM took 1.2 s, or 500 ms using nine Zeiss SIM images (30-ms exposure time), whereas acquisition of YFP-MreB by STED took <1 s for a single cell of 3-μm length (1000 Hz, four line scans). Average movement of MreB structures (or possibly extension of MreB filaments) has been described as occurring at a speed of 22 nm/s (Garner et al., 2011) or 50 nm/s (Dominguez-Escobar et al., 2011) and maximal movement of 90 nm/s (see later discussion), such that negligible change in the position of MreB filaments would occur within the acquisition time, showing that MreB forms true filaments and that the length of the imaged structures is a very good estimate of their actual size in vivo. GFP-Mbl formed filaments indistinguishable from those of GFP-MreB (Figure 1E). Based on the fact that MreB and Mbl colocalize and interact (Carballido-Lopez et al., 2006; Defeu Soufo and Graumann, 2006), these experiments argue against an overproduction artifact for filament formation of MreB paralogues. As a further verification that filament formation is not based on overexpression of MreB, we induced YFP-MreB fusion with 0.005% xylose rather than with 0.5%. Even at this very low concentration of inducer, YFP-MreB still formed visible filaments (Figure 1C). A fusion expressing functional GFP-MreB from the amylase locus as sole source of the protein used in a different study (Dominguez-Escobar et al., 2011) also showed similar filamentous structures (Figure 1F). When imaged with TIRF microscopy, cells grown under similar conditions as in Figure 1, A–F, showed a more patch–like pattern of fluorescence (Figure 1G), likely because TIRF microscopy only excites fluorescence within the upper 100–200 nm of the cell's curved surface.
FIGURE 1:
Superresolution microscopy of MreB in exponentially growing bacterial cells. (A) STED image of YFP-MreB (80-nm resolution). (B) N-SIM image of YFP-MreB (125-nm resolution). Green bars show neighboring filaments with different angles; orange bars show opposite angles relative to 90°. (C) G-STED image of YFP-MreB expressed at very low concentration of inducer (0.005% xylose). (D) SIM (Zeiss) image of GFP-MreB expressed from original locus. (E) STED image of GFP-Mbl (original promoter). (F) STED image of GFP-MreB expressed from an ectopic site on the chromosome as sole source of the protein (Formstone and Errington, 2005; Dominguez-Escobar et al., 2011). (G) TIRF image of YFP-MreB (corresponding to A, B, and D). (H) N-SIM 3D reconstruction of the upper two-thirds of a chain of cells. (I) Angular distribution of GFP-MreB filaments in B. subtilis. Numbers above the bars show percentage of filaments with 90° (±5°) relative to the longitudinal axis or with angles with >10° difference relative to the diametral axis of the cell. (J) Relative contributions of filaments of different sizes. (K) SIM (Zeiss) Z-stack with planes taken every 150 nm. Bars show the position of focal planes relative to the cell body. Green triangles indicate filament making a full helical turn; yellow triangle, filament making a quarter turn (not completed full turn); and purple triangle, filament making a half-turn. White bars, 2 μm.
MreB filaments showed a highly variable angular distribution, from circumferential to a tilt of 40°. About 40% of the filaments were circumferential, with a variation of ±5°, and 60% had an angle of 10° or more (Figure 1I). Measurements from STED and SIM experiments yielded indistinguishable angular distributions. Circumferential filaments could be adjacent to highly tilted filaments, and even filaments of opposite tilt were found to be present next to each other (Figure 1B) within the same cell. These data show that the orientation of MreB filaments is surprisingly irregular and much less well organized than previously deduced from conventional fluorescence microscopy.
Using N-SIM or Zeiss SIM, we performed 3D reconstructions of MreB filaments. Figure 1K (also see Supplemental Movie S1) shows that many GFP-MreB filaments extended for more than a half-circumference. Figure 1H shows a 3D reconstruction of the upper two-thirds of the cell diameter, revealing the extended nature of a majority of filaments. The number of such longer filaments was highly variable between cells, and on average, we found 1.2 long filaments (>1.7 μm) and 3.1 medium-length filaments (1.7 μm) per cell (of length 3 ± 0.2 μm), in addition to 2.5 shorter filaments. From complete 3D reconstructions, we measured filament length and categorized filaments into four length classes: <850 nm (quarter-turn), up to 1.7 μm (half-turn), three-quarter turn, and full turn (3.4 μm; Figure 1J). Two-thirds of all filaments were longer than one-quarter turn, with half-turns making up almost half of all filaments. Thus MreB filaments showed clear discontinuities; long filaments were frequently neighbored by short filaments, revealing that at 125-nm resolution, MreB does not form a continuous structure. MreB filaments are long enough, however, that they could confer long-range interactions between membrane proteins or MreB-associated cytosolic proteins and thus are able of coupling molecules from opposing sides of the cell's circumference.
YFP-MreB filaments move along various tracks underneath the cell membrane
Using SIM, we performed time-lapse experiments, capturing a set of 3D (N-SIM) or two-dimensional (2D; Zeiss SIM) images every 5 s. Figure 2A and Supplemental Movies S2–S4 show that YFP-MreB filaments undergo considerable remodeling. We found that most filaments moved at various angles relative to the circumference (reflecting the angular degrees shown in Figure 1I) from one side of the cell to the other, whereas many filaments moved along a circumferential path. Frequently, both ends of short filaments moved at an identical speed, which is hard to reconcile with a treadmilling-like mode of movement; identical on- or off-binding rates would need to be present at both ends. On the other hand, we found filaments of size of 1.7 μm or longer, which underwent apparent shrinkage, although it cannot be excluded that the entire long filament moved, because the leading edge cannot be seen in the time lapse. We also found many cases in which filaments reversed their direction between 5-s intervals (Figure 2 and Supplemental Movies S2–S4), in agreement with the observed reversal of MreB “patches” (Garner et al., 2011). This could most clearly be seen for short filaments, which reversed in the same direction from which they had entered the field of view (Figure 2B). Reversal was also seen for longer filaments, where only one end could be seen, indicating that backward movement can also occur for extended filaments of 1.7 μm or maybe longer (this would require 3D time-lapse experiments, which are not yet feasible with superresolution techniques). Neighboring filaments could be observed to have the opposite direction of movement (Supplemental Movies S2 and S4); we were not able to detect a pattern of movement, that is, a preferred directionality, as was proposed based on conventional epifluorescence microscopy (Defeu Soufo and Graumann, 2004). Thus, at the present resolution, we can state that MreB filaments appear to follow random paths of directionality, which can be rapidly reversed. Our study reveals a novel mode of filament dynamics of MreB, namely the fusion of filaments. Figure 2 shows an example of two filaments meeting from opposite directions, and, after a short period of an apparent stop, resuming with movement in one direction. Although fusion was a rare event (<2% of all filaments followed by time-lapse SIM), this suggests that filaments take certain tracks, even though these seem to have a highly relaxed arrangement. Of note, we never observed a case of filaments crossing each other, in full agreement with the finding that MreB has intrinsic membrane affinity (Dempwolff et al., 2011) and directly binds to the membrane via an amphipathic helix (Salje et al., 2011).
FIGURE 2:
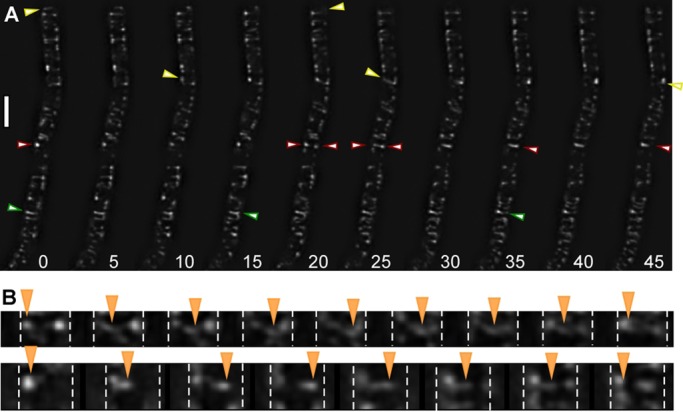
Dynamics of YFP-MreB filaments. (A) Time-lapse experiments with images captured every 5 s, using N-SIM. Yellow triangles indicate filaments moving from left to right; green triangle, filament reversing its direction; red triangles, filaments moving in from opposite directions and fusing, with the left-moving filament prevailing in its direction. (B) Two examples of filaments reversing their direction of movement, indicated by orange triangles; images captured every 5 s. Dashed white lines mark the lateral sides of the cells. The filament in the top moves right at an angle, whereas the filament in the bottom moves close to 90°. Both filaments move back at the same angle as they enter the field of view. White bar, 2 μm.
The speed of filament movement was highly variable. We observed maximal extension/movement of 1.7 μm in 3.75 frames (12 visually fast filaments tracked), that is, 1700 nm in 18.75 s, or 90 nm/s. When the same cells were imaged twice, however (Supplemental Movie S3 was taken ∼5 min after Supplemental Movie S2), movement of MreB filaments was reduced to about half of the speed measured in the first experiment (52 nm/s for the fastest six filaments). When cells were imaged 5 min after they had been transferred from a shaking flask into a closed tube before mounting to the slide for imaging, speed of movement was lower by a factor of three to four than that of the fastest filaments seen under optimal growth conditions (data not shown). Thus light stress (and possible additional heat stress) and oxygen limitation already affect the speed of movement of MreB filaments. It was reported that MreB filaments dissipate after a sudden step down in the energy source supplied for the cells (Defeu Soufo and Graumann, 2004) and that addition of antibiotics affecting cell wall synthesis slows movement of MreB filaments (Dominguez-Escobar et al., 2011; Garner et al., 2011; van Teeffelen et al., 2011). Our experiments reveal that extreme care must be taken to interpret the effects of compounds or mutations leading to cell wall stress, which can be sensed by σ-factor and two-component signal transduction systems (Eiamphungporn and Helmann, 2008; Rietkotter et al., 2008), on the movement of MreB filaments, which are clearly very sensitive to different kinds of stresses.
Irregular YFP-MreB structures in cells growing in a transition phase
When cells were grown under suboptimal conditions (e.g., in the absence of aeration through shaking, doubling time of >3 h), only focal or spot-like structures of length <80 nm were visible using STED microscopy, as well as few filamentous structures, which were regularly positioned (Figure 3A). This agrees with our previous finding that MreB filaments dissipate upon nutrient downshift within few minutes to form spot-like structures (Defeu Soufo and Graumann, 2004). We analyzed how the rapidly moving filaments are generated from the short structures, and shifted nonshaking cells to optimal aeration conditions. On resumption of faster growth, YFP-MreB filaments slowly increased in length but were highly disordered (Figure 3, B–D). During the transition, filaments could even extend longitudinally along the lateral membrane, a condition never seen under optimal growth conditions (Figure 3C). Eventually, filaments assumed a more regular localization pattern as growth became maximal (Figure 3E). We were not able to determine whether abnormally positioned filaments gradually find a more regular circular/helical pattern or are completely dissolved and more and more normally oriented filaments appear, but given the high remodeling capacity of MreB filaments, the latter scenario is more likely. To test whether the ability of MreB to form filaments is correlated with the amount of protein, we performed Western blot analysis. At 1 h after arrest of strong aeration of cultures (Supplemental Figure S1B), GFP-MreB or YFP-MreB filaments had a spotty appearance similar to that shown in Figure 3A. The amount of GFP-MreB expressed from the original locus remained rather constant, whereas that of YFP-MreB expressed from the amylase locus was equal or slightly lower in different experiments (Supplemental Figure S1A). The shift from anaerobic to aeration condition restored the filamentous phenotype, and no differences in expression levels were detected for both protein fusions (Supplemental Figure S1A). Thus MreB levels do not show considerable differences between optimal or suboptimal growth conditions. It is interesting that the large distribution of angles of MreB filaments under optimal growth conditions is even much greater under conditions of growth transitions (Figure 3), although it is unlikely that the structure of the cell wall is dramatically different during the transition phase.
FIGURE 3:
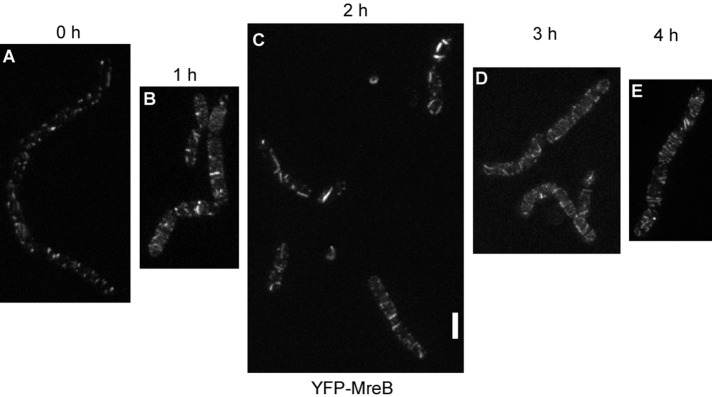
STED microscopy of cells grown in culture in the absence of shaking (A) and successive changes in the pattern of localization of YFP-MreB after commencement of shaking (B–E). White bar, 2 μm; images are scaled equally.
We also analyzed the dynamics of MreB structures formed under suboptimal growth conditions. Time-lapse microscopy with cells grown under nonshaking conditions for 1 h (Supplemental Figure S1B) revealed that the patchy structures formed under these conditions do not move but are statically located underneath the cell membrane (Supplemental Movie S5). Thus MreB becomes immobile under suboptimal growth/stress conditions, in strong contrast to optimal aeration conditions.
E. coli MreB forms moving filaments
We sought to extend the analysis of MreB structures at high resolution to the model bacterium from Gram-negative bacteria, E. coli. We used an MreB-sandwich–red fluorescent protein (RFP) fusion reported to be functional (Bendezu et al., 2009). Although we found ∼5% of the population having aberrant cell morphology, most cells possessed normal rod shape (Figure 4B), indicating that the fusion protein can largely complement for the missing wild-type protein. SIM showed that EcMreB also forms filaments in growing cells (Figure 4, A and B). It was not possible to obtain a 3D image, due to the fast bleaching and low quantum efficiency of RFP. From the 2D SIM analysis, a majority of filaments were diffraction limited (i.e., length <125 nm), and cells contained between one and five filaments of length at least 1.5 μm (half-turn). Thus EcMreB filaments appear to be somewhat shorter than those seen in B. subtilis. Time-lapse SIM revealed highly dynamic movement of short and long filaments (Supplemental Movies S6 and S7). Filaments moved with a speed of 65 nm/s, and very few filaments were static. Similar to B. subtilis, a majority of filaments observed (60%) moved in an angular rather than circular pattern (40%). We conclude that in two model bacteria, MreB forms dynamic filaments of sufficient length to confer long-range connectivity underneath the cell membrane, which show similar patterns of movement but different lengths between species.
FIGURE 4:
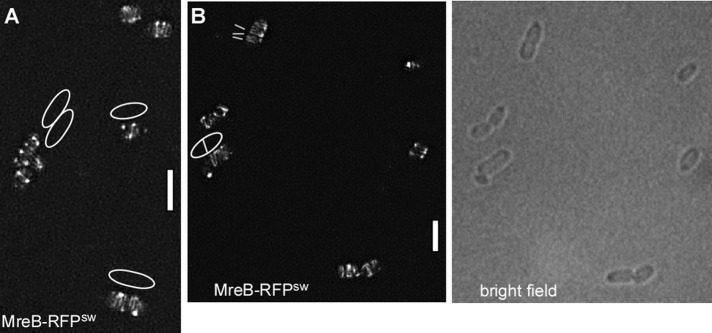
E. coli MreB forms filaments. (A, B) 3D SIM (Zeiss) images of E. coli cells expressing a sandwich MreB-RFP fusion (the RFP is integrated into a loop within the MreB molecule), EcMreB-RFPsw. (A) White lines indicate the orientation of filaments; outlines of cells are shown that are difficult to deduce from the fluorescence signal. (B) Outlines of cells seen by bright-field illumination. White bars, 2 μm.
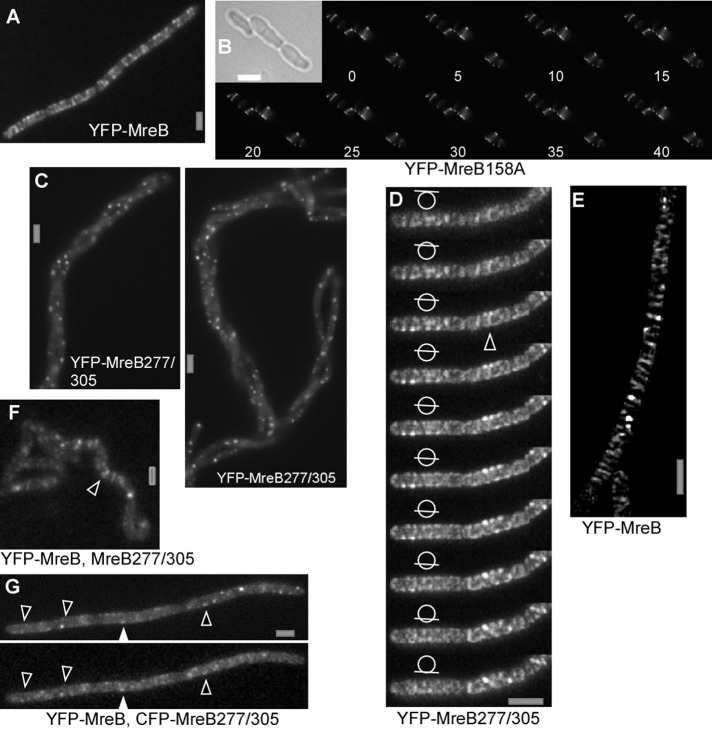
A B. subtilis MreB ATPase mutant forms static filaments, and mutations in the polymer interface affect filament length and cell morphology
It has been reported that the D158A mutation in MreB reduces ATPase activity and leads to a dominant-negative effect on cell morphology in E. coli and in B. subtilis (Kruse et al., 2003; Defeu Soufo and Graumann, 2006). The localization and dynamics of wild-type (wt) MreB or of Mbl filaments are affected through the expression of MreBD158A (Defeu Soufo and Graumann, 2006). We visualized D158A mutant YFP-MreB in B. subtilis using SIM. Like wt MreB, ATPase mutant MreB formed filaments, although these were brighter and less abundant (Figure 5B) than those of wt protein (Figure 5A). Dynamics of MreBD158A was analyzed using time-lapse SIM. Figure 5B and Supplemental Movies S8 and S9 show that unlike wt MreB, only few (<5%) mutant filaments showed movement between 5-s intervals for the entire acquisition (up to 40 exposures). Thus mutant filaments do not show dynamic turnover or movement, whereas cell morphology is strongly altered (Figure 5B). These experiments show that ATPase activity is required for the movement of MreB filaments, unlike previously reports (Garner et al., 2011), and in turn are necessary for proper cell shape maintenance. The basis for the discrepancy between our data and those of Garner et al. (2011) is unknown.
FIGURE 5:
Fluorescence microscopy of exponentially growing chains of B. subtilis cells. (A) Epifluorescence of YFP-MreB. (B) Time-lapse SIM (Zeiss; 5-s intervals) of cells expressing D158A mutant YFP-MreB; inset, outlines of cells by bright-field light. (C) Expression of double mutant YFP-MreB277/305 for 8 h, epifluorescence. (D) YFP-MreB277/305 expressed for 3 h; SIM (Zeiss) stack with planes indicated by bars in circle; triangle shows an example of an extended (<750 nm) filament. (E) SIM (Zeiss) image of cells expressing YFP-MreB under identical conditions as in D. (F) Localization of YFP-MreB 4 h after expression of double mutant MreB. Note that the helical phenotype has not fully arisen at this time, but the localization of proteins is easier to see because they are still mostly flat. White triangle indicates clear, extended filament. (G) Colocalization of CFP-MreB and YFP-MreB277/305 (examples indicated by triangles) 4 h after expression of double-mutant MreB (epifluorescence). White bars, 2 μm.
Our experiments are neither proof nor disproof of treadmilling, however, and do not exclude the possibility that MreB filaments could be moved by the cell wall synthesis machinery (Dominguez-Escobar et al., 2011; Garner et al., 2011; van Teeffelen et al., 2011): for example, ATPase mutant MreB could form thicker filament bundles than those formed by wt MreB (in case it forms filament bundles in vivo), or mutant filaments could be much longer, and thus movement may be blocked indirectly. To test this, we measured the width of MreB filaments using gated-STED, a technique that allows a resolution of ≤50 nm. We measured 125 filaments from wt cells and 105 from MreBD158A-expressing cells at 2 h after addition of inducer to obtain similar conditions. Figure 6 shows a zoom on two filaments in a cell, one of which is 42 nm wide (the width of the peak at half-maximal intensity is used for size measurements) and the other 80 nm, verifying that filaments can be visualized with high resolution, and showing that some MreB filaments are indeed much wider than actin filaments. On average, wt MreB filaments had a width of 75.6 ± 17.7 nm and mutant MreB filaments 83.2 ± 19.5 nm, revealing that both types of filaments do not vary greatly in their diameter. The change in cell morphology becomes visible ∼3–4 h after induction of MreBD158A. The average width of 75 mutant filaments measured did not differ significantly from that after 2 h. These findings favor the idea that mutant MreB filaments cannot be moved efficiently because they lack sufficient turnover rather than being too heavy to be moved. The experiments also argue against the idea that mutant MreB slows movement by laterally adding to wt MreB filaments, because a similar extent of induction of wt YFP-MreB does not slow filament movement. It is possible, however, that movement is slowed because of an increase in length of the filaments, based on slower subunit exchange. These aspects need to be investigated in future analysis.
FIGURE 6:
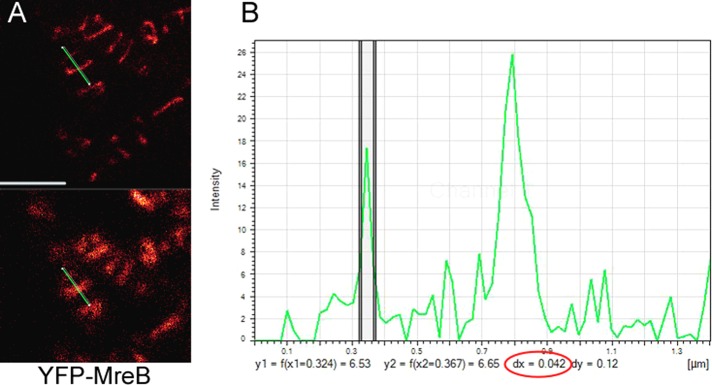
Measurement of the thickness of YFP-MreB filaments in exponentially growing B. subtilis cells. (A) G-STED image (top) and conventional confocal image (bottom). The green bar indicates the line scan through a thin and a much thicker filament. (B) Intensity plot of the line scan. The y axis shows fluorescence intensity, and the x axis shows size in micrometers. The two vertical lines indicate the half-maximum intensity of the filament, which has a diameter of 42 nm (number circled in red); the thicker filament is on the right and has a size of 85 nm.
It was recently shown that several mutations in the ATP-binding pocket of MreB affect cell shape maintenance (Dye et al., 2011). Of interest, a single mutation at the interaction surface of MreBH affects the architecture of filaments in vivo (Defeu Soufo and Graumann, 2010). To extend the analysis of requirements for protofilament assembly of MreB, we introduced several single and double mutations into the known subunit contact sites between MreB monomers. When combined (but not single), two mutations, A277Q and K305D, generated a striking phenotype: cells became bent and helical, and eventually (after six to eight doubling times) coiled around other chains of cells in liquid culture (Figure 5C and Supplemental Figure S2A). In an attempt to understand this interesting phenotype, we purified wt and double-mutant MreB and performed dynamic light scattering analysis as a measure of the ability to form filaments. Supplemental Figure S3 shows that wt MreB, as well as MreB277/305, rapidly induced filament formation when magnesium was added. At higher magnesium concentration, double-mutant MreB scattered light more strongly than wt MreB. For both versions of MreB, addition of 10 mM or more of magnesium reversed the speed of scattering (data not shown). Although we cannot explain the enhanced formation of filaments at 5 mM magnesium for the double-mutant MreB, it is clear that the initial nucleation of filaments is similar for the mutant version and wt protein. Of note, this analysis does not address the stability of the rapidly formed MreB filaments nor their length. To analyze filament formation in vivo, we next localized the double-mutant YFP-MreB expressed from an ectopic site on the chromosome by SIM. Three hours after induction, the protein formed few filaments and mostly resolution-limited structures, most likely short filaments of <125 nm (Figure 5D and Z-stack seen in Supplemental Movie S10), much shorter than those formed by wt MreB (Figure 5E). In time-lapse SIM, soon after induction, extended as well as diffraction-limited filaments of YFP-MreB277/305 moved with similar speed as wt filaments (Supplemental Movie S11). After prolonged induction (>2 h), YFP-MreB277/305 formed only diffraction-limited filaments (Supplemental Figure S2B), which no longer showed any movement (data not shown). Therefore the double mutation affects filament formation of MreB in that filament length decreases. Owing to technical limitations, it was not possible to obtain SIM or STED localization of both wt MreB and double-mutant MreB. When coexpressed, YFP-MreB277/305 largely colocalized with cyan fluorescent protein (CFP)–MreB in short or diffraction-limited structures (Figure 5G and Supplemental Figure S4), and wt MreB also formed fewer and mainly shorter filaments that were static when it was coexpressed with MreB277/305 (Figure 5F and Supplemental Movie S12). These data show that the expression of double-mutant MreB affects the formation as well as the dynamics of wt MreB filaments. Thus our experiments establish that the alteration of contact sites between MreB monomers affect the size of filaments, leading to shorter filament length, which in turn affects cell morphology.
DISCUSSION
Several models have been proposed for the distinct structures observed for MreB in live cells: 1) a continuous cytoskeletal structure extending from pole to pole, 2) shorter helical filaments forming a discontinuous cytoskeleton, 3) filaments of <250 nm, and 4) patches not containing filaments at all, rotating passively around a 90° angle underneath the cell membrane (Jones et al., 2001; Defeu Soufo and Graumann, 2005; Dominguez-Escobar et al., 2011; Garner et al., 2011; van Teeffelen et al., 2011). All models entail a membrane association of MreB, which has been shown to be an intrinsic property of MreB (Dempwolff et al., 2011) due to an amphipathic helix within MreB (Salje et al., 2011), but the models differ greatly in what can be achieved by MreB: in models 1 and 2, MreB can confer long-range connection/interactions underneath the membrane and confer mechanical functions as well, which would not be the case for model 3. In model 4, MreB would only be able to confer interactions in Z (between cytosol and membrane/cell wall), but not in X or Y (along the membrane). Using two different superresolution fluorescence microscopy techniques and functional fluorescent protein fusions to MreB and its paralogue Mbl, we show that model 2 is correct: MreB forms a discontinuous structure that is highly dynamic, including filaments as long as 3.4 μm. Thus MreB can confer positional as well as mechanical properties within cells, shown here for two bacterial model organisms. With an average width of 75 nm as determined by G-STED, MreB filaments must consist of lateral bundles of protofilaments, which could set up considerable force against the membrane. Using superresolution techniques, we show unexpected properties of MreB: even neighboring filaments can have strikingly different angles relative to the 90° circumference of the cell; filaments can fuse with each other and change their direction of movement but never cross each other. The latter finding concurs with the idea of direct membrane binding of MreB proteins. If MreB filaments were moved by cell wall–synthesizing enzymes, the paths of the enzymes must by highly variable within the wall, as evidenced by our time-lapse experiments, which is an intriguing concept.
When ATPase activity of MreB was compromised (i.e., a mutation was introduced that generates this effect in actin), filaments became much more static, having a drastic effect on cell morphology. This finding does not argue against a possible motor within the cell wall that has been proposed to move MreB structures (Dominguez-Escobar et al., 2011; Garner et al., 2011; van Teeffelen et al., 2011) nor is it in favor of treadmilling: possibly, MreB filaments have to undergo a sufficient amount of internal turnover to be short and flexible enough to be driven by an external motor. The idea that MreB may have a more cytomotive than cytoskeletal function must be stringently tested, and if true should be reflected in no longer calling MreB a cytoskeletal element. We show that MreB filaments are very sensitive to different perturbations of cell growth, revealing that care must be taken in interpreting effects of cell envelope stress on the movement of MreB filaments. In any event, filament turnover is clearly a prerequisite for the movement and function of MreB, now confirmed by high-resolution live-cell microscopy.
It was recently shown that an N-terminal YFP fusion to E. coli MreB (which is not fully functional) generates structures visible by cryo–electron microscopy, which are not seen when untagged or an RFP sandwich fusion of MreB is imaged (Swulius and Jensen, 2012). We wonder whether an N-terminal fusion might artificially generate thick bundles of MreB filaments, whereas functional fusions and wild-type MreB might form very thin filaments that are difficult to image in fixed cells. In addition, because MreB filaments are directly associated with the cell membrane, they might be masked by large, membrane-embedded proteins and thereby might not be distinguishable from other membrane proteins, many of which extend into the cytosol much more deeply than MreB filaments (e.g., complex 1 or ATP synthetase). In any case, our experiments demonstrate that a functional sandwich FP-MreB forms 1.4-μm or longer filaments in E. coli cells, which move at various angles relative to the cell circumference. Thus MreB also forms extended filamentous structures in a second model bacterium.
Our investigations also provide evidence that the nature of MreB filaments is pivotal for a regular cell shape: two mutations in the interaction interface of MreB subunits give rise to a dominant-negative effect on the formation of short, wild-type MreB filaments in vivo accompanied by a change in cell morphology. This reveals that intact interaction surfaces are required for the formation of extended filaments, which in turn are essential for the function of MreB.
Several reports provided evidence that the bacterial peptidoglycan sacculus is synthesized in a helical pattern (Tilby, 1977; Mendelson et al., 1984; Wang et al., 2012). It is interesting to see that the formation of shorter-than-normal MreB filaments drives cells into a “superhelical” pattern. These findings suggest that MreB filaments may be important to force cell wall synthesis into a “less helical” pattern, such that rod shape is established from an otherwise helical/randomly shaped cell, through the activity of extended MreB filaments. These findings support the view that the bacterial MreB cytoskeleton also has a mechanical function (Defeu Soufo and Graumann, 2010; Wang et al., 2010), in addition to a function in the positioning of the cell wall synthesis machinery (Divakaruni et al., 2005; Kawai et al., 2009). It is intriguing that the helical “supercoil” phenotype obtained through the MreB double mutant can also be generated by a lyt rodA double mutation (Rogers and Thurman, 1978), reinforcing the idea that cell wall–synthetic enzymes and the MreB cytoskeleton are tightly connected and in concert responsible for the shape of the cell.
Taken together, our work yields unprecedented insight into the nature of filaments formed by MreB, which have the potential to establish long-range interactions between enzymes involved in cell wall synthesis. Our finding that MreB forms dynamic filaments in vivo, as does actin, strongly supports the idea that early in evolution, metabolic enzymes evolved into a common ancestor of a filament-forming protein (Barry and Gitai, 2011) that now performs a multitude of structural and organizational tasks in all domains of life (Jockusch and Graumann, 2011).
MATERIALS AND METHODS
Growth conditions
B. subtilis strains were grown at 30°C in LB (lysogeny broth) medium or on LB plates containing the appropriate antibiotic. For microscopy analysis, strains were inoculated in S750 minimal medium (Jaacks et al., 1989), complemented with 1% casamino acid. For induction of xylose or hyperspank promotors, glucose was exchanged for fructose in S750 medium and 0.5% (wt/vol) xylose or for 1 mM isopropyl-β-d-thiogalactoside (IPTG), respectively. Cells were grown under shaking condition (200 rpm) to mid–exponential phase at 30˚C.
Construction of plasmids and strains
To construct a B. subtilis strain in which the transcription of gfp-mbl at the original gene locus is controlled by the endogenous mbl promoter, 250 base pairs upstream of the mbl open reading frame were amplified from chromosomal DNA of PY79 (Table 1) using the primers 2390 and 2391 (Supplemental Table S1) and inserted between BglII and KpnI sites in pJS13 (Supplemental Table S2), resulting in pJS152 (Supplemental Table S2), such that the xylose promoter is replaced by the mbl promoter. B. subtilis PY79-competent cells were transformed with the obtained plasmid selecting for chloramphenicol (5 μg/ml) resistance to generate the strain JS158 (Table 1). Mutant YFP-MreB with mutations A277Q and K305D in the contact surfaces between MreB monomers, termed YFP-MreB277/305, was generated by site-directed PCR mutagenesis. Therefore primers 2515, 2516, 2563, and 2564 (Supplemental Table S1) and pJS24 or pJS64 (Supplemental Table S2), respectively, as DNA templates, were used to generate pCR4 and pCR5 (Supplemental Table S2). Transformation of PY79 with pCR4, selecting for spectinomycin (100 μg/ml) resistance generated CR1 (Table 1). To introduce the mreBA277Q, K305D mutant allele in the B. subtilis strain expressing YFP-MreB, the fragment corresponding to mreB277/305 was amplified by PCR with the primers 2962 and 2616 (Supplemental Table S1) and with pCR4 as DNA template. The fragment was digested with NheI and SphI and cloned into the corresponding sites of pDP150 (Kearns and Losick, 2005; Supplemental Table S2), generating pCR6 (Supplemental Table S2). JS36 (Defeu Soufo and Graumann, 2006; Table 1) was transformed with pCR6, selecting for erythromycin (1 μg/ml), lincomycin (25 μg/ml), and spectinomycin (100 μg/ml) resistance, to generate CR2 (Table 1). For dual visualization of YFP-MreB277/305 and CFP-MreB in B. subtilis, the fragment corresponding to cfp-mreB was amplified by PCR with the primers 2861 and 2616 (Supplemental Table S1) and chromosomal DNA from JS41 (Table 1) as DNA template and subsequently cloned into pDP150 to generate pCR7 (Supplemental Table S2). PY79 was transformed with pCR7, selecting for erythromycin and lincomycin resistance, to generate CR3 (Table 1). Transformation of CR3 with pCR4, selecting for erythromycin, lincomycin, and spectinomycin resistance, generated CR4 (Table 1).
TABLE 1:
Strains.
| Name | Parent | Genotype | Source or reference |
|---|---|---|---|
| PY79 | B. subtilis | Wild type | |
| JS12 | B. subtilis PY79 | Pxyl-gfp-mreB (cmr) | Defeu Soufo and Graumann (2004) |
| JS36 | B. subtilis PY79 | Pxyl-yfp-mreB::amyE (spcr) | Defeu Soufo and Graumann (2006) |
| JS158 | B. subtilis PY79 | Pmbl-gfp-mbl (cmr) | This work |
| 3723 | B. subtilis 168 | Pxyl-gfp-mreB::amyE, trpC2 Ωneo3427 ∆mreB (kan, spcr) | Dominguez-Escobar et al. (2011) |
| FB72 | E. coli DY329 | mreB’-rfp-’mreB (cmr) | Bendezu et al. (2009) |
| JS51 | B. subtilis PY79 | Pxyl-yfp-mreBD158A::amyE (spcr) | Defeu Soufo and Graumann (2006) |
| CR1 | B. subtilis PY79 | Pxyl-yfp-mreBA277Q, K305D::amyE (spcr) | This work |
| CR2 | B. subtilis PY79 | Phyperspank-mreBA277Q, K305D::thrC, Pxyl-yfp-mreB::amyE (mls, spcr) | This work |
| JS41 | B. subtilis PY79 | Pxyl-cfp-mreB (cmr) | Defeu Soufo and Graumann (2006) |
| CR3 | B. subtilis PY79 | Phyperspank-cfp-mreB::thrC (mlsr) | This work |
| CR4 | B. subtilis PY79 | Phyperspank-cfp-mreB::thrC, Pxyl-yfp-A277Q, K305D::amyE (mlsr, spcr) | This work |
Resistance (r) gene abbreviations: cm, chloramphenicol; kan, kanamycin; mls, macrolide-lincosamide-streptogramin; spc, spectinomycin.
Image acquisition
For microscopy analysis, exponentially growing B. subtiliscells were mounted on agarose pads containing S750 medium on object slides. Epifluorescence and TIRF microscopy were performed using a Zeiss Axio Observer Z1 microscope (Carl Zeiss, Jena, Germany) with a 1.45 numerical aperture objective and a Photometrics Cascade charge-coupled device camera (Photometrics, Tucson, AZ). The electronic data processing was conducted with MetaMorph 6.3 software (Meta Imaging Software, Molecular Devices, Sunnyvale, CA). Subsequent image analysis was performed using ImageJ (National Institutes of Health, Bethesda, MD). Superresolution microscopy was performed using SIM (Zeiss and Nikon) or STED (Leica) technique. STED was performed using a Leica STED-CW confocal laser scanning microscope in Mannheim, Germany, and G-STED using a Leica G-STED SP8 microscope in Marburg, Germany. Images were acquired with three or four line scans at 1000 Hz. SIM was performed using a Zeiss Elyra System (Munich, Germany) or an N-SIM (Nikon) System (Heidelberg, Germany), using the respective software. For N-SIM, only 3D acquisitions were performed (15 images), and for Zeiss SIM, 2D acquisition was chosen for time-lapse microscopy and for the acquisition of Z-stacks and 3D for single-plane acquisitions. GFP-MreB and GFP-Mbl were acquired using 488-nm laser excitation, and MreB-RFPsw using 561-nm excitation.
Protein expression and purification
E. coli BL21 (λDE3) was transformed with pJS64 and pCR5 (Supplemental Table S2), respectively. Cultures were grown at 37°C in 400 μl of LB supplemented with streptomycin (50 μg/ml) to an OD600 of 0.6–0.8. Protein expression was induced by addition of IPTG to a final concentration of 1 mM and incubation for additional 4 h at 30°C. Cells were collected by centrifugation and stored at −80°C until use. Cells were disrupted with French press in appropriate buffer (100 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, pH 7.5), and lysates were cleared by centrifugation. Proteins were purified using Strep-tag affinity purification kit (IBA GmbH, Göttingen, Germany) and dialyzed against low-salt polymerization/storage buffer (5 mM Tris-HCl, 0.1 mM CaCl2, 0.2 mM ATP, pH 7.5).
Light scattering assay
Proteins were diluted with low-salt polymerization buffer to a final concentration of 5 μM. Polymerization reactions were measured at 315-nm excitation and 318-nm emission using a fluorescence spectrometer (LS55; PerkinElmer, Rodgau, Germany) with 2.5-nm slit width for excitation and emission. Reactions were carried out at 25°C with 100-μl samples in quartz cuvettes (Quartz SUPRASIL Ultra-micro, light path 10 mm; PerkinElmer). The time interval of measurement was 5 s, with an integration time of 2 s. After 100- to 150-s equilibration time, MgCl2 to a final concentration of 2.5 and 5 mM, respectively, was added to the reaction sample to trigger the polymerization, and the record of scattered light was continued for an additional 15 min. Data were analyzed using Excel (Microsoft, Redmond, WA).
Supplementary Material
Acknowledgments
We are very grateful to the superresolution teams of Leica (Nathalie Garin, Ulf Schwarz, and Manfred Jung), Zeiss (Sven Poppelreuther and Jacques Paysan), and Nikon (Klaus Nettesheim and Kim-Miriam Beer) for their time and help with using their microscopes. We are grateful to the reviewers for their very thoughtful and constructive comments. This work was supported by the Deutsche Forschungsgemeinschaft (SPP1474, FOR 929).
Abbreviations used:
- 3D
three dimensional
- ATP
adenosine triphosphate
- a.u.
arbitrary unit
- CFP
cyan fluorescent protein
- FP
fluorescent protein
- FtsZ
gene product of ftsZ: filamentous temperature-sensitive Z
- GFP
green fluorescent protein
- IPTG
isopropyl β-d-1-thiogalactopyranoside
- Mbl
gene product of mbl: MreB-like
- MgCl2
magnesium chloride
- MreB
gene product of mreB: murein cluster e B
- MreC
gene product of mreC: murein cluster e C
- MreD
gene product of mreD: murein cluster e D
- SIM
structured illumination
- STED
stimulated emission depletion
- Strep
streptavidin-tag
- TIRF
total internal reflection fluorescence
- wt
wild type
- YFP
yellow fluorescent protein
- Xyl
xylose
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E12-10-0728) on June 19, 2013.
REFERENCES
- Adams DW, Errington J. Bacterial cell division: assembly, maintenance and disassembly of the Z ring. Nat Rev Microbiol. 2009;7:642–653. doi: 10.1038/nrmicro2198. [DOI] [PubMed] [Google Scholar]
- Barry RM, Gitai Z. Self-assembling enzymes and the origins of the cytoskeleton. Curr Opin Microbiol. 2011;14:704–711. doi: 10.1016/j.mib.2011.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendezu FO, de Boer PA. Conditional lethality, division defects, membrane involution, and endocytosis in mre and mrd shape mutants of Escherichia coli. J Bacteriol. 2008;190:1792–1811. doi: 10.1128/JB.01322-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendezu FO, Hale CA, Bernhardt TG, de Boer PA. RodZ (YfgA) is required for proper assembly of the MreB actin cytoskeleton and cell shape in E. coli. EMBO J. 2009;28:193–204. doi: 10.1038/emboj.2008.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabeen MT, Jacobs-Wagner C. Bacterial cell shape. Nat Rev Microbiol. 2005;3:601–610. doi: 10.1038/nrmicro1205. [DOI] [PubMed] [Google Scholar]
- Cabeen MT, Jacobs-Wagner C. The bacterial cytoskeleton. Annu Rev Genet. 2010;44:365–392. doi: 10.1146/annurev-genet-102108-134845. [DOI] [PubMed] [Google Scholar]
- Carballido-Lopez R, Errington J. The bacterial cytoskeleton: in vivo dynamics of the actin-like protein Mbl of Bacillus subtilis. Dev Cell. 2003;4:19–28. doi: 10.1016/s1534-5807(02)00403-3. [DOI] [PubMed] [Google Scholar]
- Carballido-Lopez R, Formstone A, Li Y, Ehrlich SD, Noirot P, Errington J. Actin homolog MreBH governs cell morphogenesis by localization of the cell wall hydrolase LytE. Dev Cell. 2006;11:399–409. doi: 10.1016/j.devcel.2006.07.017. [DOI] [PubMed] [Google Scholar]
- Cowles KN, Gitai Z. Surface association and the MreB cytoskeleton regulate pilus production, localization and function in Pseudomonas aeruginosa. Mol Microbiol. 2010;76:1411–1426. doi: 10.1111/j.1365-2958.2010.07132.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Defeu Soufo HJ, Graumann PL. Dynamic movement of actin-like proteins within bacterial cells. EMBO Rep. 2004;5:789–794. doi: 10.1038/sj.embor.7400209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Defeu Soufo HJ, Graumann PL. Bacillus subtilis actin-like protein MreB influences the positioning of the replication machinery and requires membrane proteins MreC/D and other actin-like proteins for proper localization. BMC Cell Biol. 2005;6:10. doi: 10.1186/1471-2121-6-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Defeu Soufo HJ, Graumann PL. Dynamic localization and interaction with other Bacillus subtilis actin-like proteins are important for the function of MreB. Mol Microbiol. 2006;62:1340–1356. doi: 10.1111/j.1365-2958.2006.05457.x. [DOI] [PubMed] [Google Scholar]
- Defeu Soufo HJ, Graumann PL. Bacillus subtilis MreB paralogues have different filament architectures and lead to shape remodelling of a heterologous cell system. Mol Microbiol. 2010;78:1145–1158. doi: 10.1111/j.1365-2958.2010.07395.x. [DOI] [PubMed] [Google Scholar]
- Dempwolff F, Reimold C, Reth M, Graumann PL. Bacillus subtilis MreB orthologs self-organize into filamentous structures underneath the cell membrane in a heterologous cell system. PLoS One. 2011;6:e27035. doi: 10.1371/journal.pone.0027035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Divakaruni AV, Loo RR, Xie Y, Loo JA, Gober JW. The cell-shape protein MreC interacts with extracytoplasmic proteins including cell wall assembly complexes in Caulobacter crescentus. Proc Natl Acad Sci USA. 2005;102:18602–18607. doi: 10.1073/pnas.0507937102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez-Escobar J, Chastanet A, Crevenna AH, Fromion V, Wedlich-Soldner R, Carballido-Lopez R. Processive movement of MreB-associated cell wall biosynthetic complexes in bacteria. Science. 2011;333:225–228. doi: 10.1126/science.1203466. [DOI] [PubMed] [Google Scholar]
- Dye NA, Pincus Z, Fisher IC, Shapiro L, Theriot JA. Mutations in the nucleotide binding pocket of MreB can alter cell curvature and polar morphology in Caulobacter. Mol Microbiol. 2011;81:368–394. doi: 10.1111/j.1365-2958.2011.07698.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiamphungporn W, Helmann JD. The Bacillus subtilis sigma(M) regulon and its contribution to cell envelope stress responses. Mol Microbiol. 2008;67:830–848. doi: 10.1111/j.1365-2958.2007.06090.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Formstone A, Errington J. A magnesium-dependent mreB null mutant: implications for the role of mreB in Bacillus subtilis. Mol Microbiol. 2005;55:1646–1657. doi: 10.1111/j.1365-2958.2005.04506.x. [DOI] [PubMed] [Google Scholar]
- Garner EC, Bernard R, Wang W, Zhuang X, Rudner DZ, Mitchison T. Coupled, circumferential motions of the cell wall synthesis machinery and MreB filaments in B. subtilis. Science. 2011;333:222–225. doi: 10.1126/science.1203285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitai Z, Dye N, Shapiro L. An actin-like gene can determine cell polarity in bacteria. Proc Natl Acad Sci USA. 2004;101:8643–8648. doi: 10.1073/pnas.0402638101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graumann PL. Cytoskeletal elements in bacteria. Annu Rev Microbiol. 2007;61:589–618. doi: 10.1146/annurev.micro.61.080706.093236. [DOI] [PubMed] [Google Scholar]
- Henriques AO, Glaser P, Piggot PJ, Moran CP., Jr Control of cell shape and elongation by the rodA gene in Bacillus subtilis. Mol Microbiol. 1998;28:235–247. doi: 10.1046/j.1365-2958.1998.00766.x. [DOI] [PubMed] [Google Scholar]
- Iwaya M, Jones CW, Khorana J, Strominger JL. Mapping of the mecillinam-resistant, round morphological mutants of Escherichia coli. J Bacteriol. 1978;133:196–202. doi: 10.1128/jb.133.1.196-202.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaacks KJ, Healy J, Losick R, Grossman AD. Identification and characterization of genes controlled by the sporulation regulatory gene spo0H in Bacillus subtilis. J Bacteriol. 1989;171:4121–4129. doi: 10.1128/jb.171.8.4121-4129.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jockusch BM, Graumann PL. The long journey: actin on the road to pro- and eukaryotic cells. Rev Physiol Biochem Pharmacol. 2011;161:67–85. doi: 10.1007/112_2011_1. [DOI] [PubMed] [Google Scholar]
- Jones LJ, Carballido-Lopez R, Errington J. Control of cell shape in bacteria: helical, actin-like filaments in Bacillus subtilis. Cell. 2001;104:913–922. doi: 10.1016/s0092-8674(01)00287-2. [DOI] [PubMed] [Google Scholar]
- Kawai Y, Daniel RA, Errington J. Regulation of cell wall morphogenesis in Bacillus subtilis by recruitment of PBP1 to the MreB helix. Mol Microbiol. 2009;71:1131–1144. doi: 10.1111/j.1365-2958.2009.06601.x. [DOI] [PubMed] [Google Scholar]
- Kearns DB, Losick R. Cell population heterogeneity during growth of Bacillus subtilis. Genes Dev. 2005;19:3083–3094. doi: 10.1101/gad.1373905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SY, Gitai Z, Kinkhabwala A, Shapiro L, Moerner WE. Single molecules of the bacterial actin MreB undergo directed treadmilling motion in Caulobacter crescentus. Proc Natl Acad Sci USA. 2006;103:10929–10934. doi: 10.1073/pnas.0604503103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruse T, Bork-Jensen J, Gerdes K. The morphogenetic MreBCD proteins of Escherichia coli form an essential membrane-bound complex. Mol Microbiol. 2005;55:78–89. doi: 10.1111/j.1365-2958.2004.04367.x. [DOI] [PubMed] [Google Scholar]
- Kruse T, Moller-Jensen J, Lobner-Olesen A, Gerdes K. Dysfunctional MreB inhibits chromosome segregation in Escherichia coli. EMBO J. 2003;22:5283–5292. doi: 10.1093/emboj/cdg504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JC, Stewart GC. Essential nature of the mreC determinant of Bacillus subtilis. J Bacteriol. 2003;185:4490–4498. doi: 10.1128/JB.185.15.4490-4498.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzawa H, Hayakawa K, Sato T, Imahori K. Characterization and genetic analysis of a mutant of Escherichia coli K-12 with rounded morphology. J Bacteriol. 1973;115:436–442. doi: 10.1128/jb.115.1.436-442.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer JA, Amann KJ. Assembly properties of the Bacillus subtilis actin, MreB. Cell Motil Cytoskeleton. 2009;66:109–118. doi: 10.1002/cm.20332. [DOI] [PubMed] [Google Scholar]
- Mendelson NH, Favre D, Thwaites JJ. Twisted states of Bacillus subtilis macrofibers reflect structural states of the cell wall. Proc Natl Acad Sci USA. 1984;81:3562–3566. doi: 10.1073/pnas.81.11.3562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popp D, Narita A, Maeda K, Fujisawa T, Ghoshdastider U, Iwasa M, Maeda Y, Robinson RC. Filament structure, organization, and dynamics in MreB sheets. J Biol Chem. 2010;285:15858–15865. doi: 10.1074/jbc.M109.095901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popp D, Narita A, Oda T, Fujisawa T, Matsuo H, Nitanai Y, Iwasa M, Maeda K, Onishi H, Maeda Y. Molecular structure of the ParM polymer and the mechanism leading to its nucleotide-driven dynamic instability. EMBO J. 2008;27:570–579. doi: 10.1038/sj.emboj.7601978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rietkotter E, Hoyer D, Mascher T. Bacitracin sensing in Bacillus subtilis. Mol Microbiol. 2008;68:768–785. doi: 10.1111/j.1365-2958.2008.06194.x. [DOI] [PubMed] [Google Scholar]
- Rogers HJ, Thurman PF. Double mutants of Bacillus subtilis growing as helices. J Bacteriol. 1978;133:1508–1509. doi: 10.1128/jb.133.3.1508-1509.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salje J, van den Ent F, de Boer P, Lowe J. Direct membrane binding by bacterial actin MreB. Mol Cell. 2011;43:478–487. doi: 10.1016/j.molcel.2011.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salje J, Zuber B, Lowe J. Electron cryomicroscopy of E. coli reveals filament bundles involved in plasmid DNA segregation. Science. 2009;323:509–512. doi: 10.1126/science.1164346. [DOI] [PubMed] [Google Scholar]
- Swulius MT, Jensen GJ. The helical MreB cytoskeleton in E. coli MC1000/pLE7 is an artifact of the N-terminal YFP tag. J Bacteriol. 2012;194:6382–6386. doi: 10.1128/JB.00505-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szwedziak P, Wang Q, Freund SM, Lowe J. FtsA forms actin-like protofilaments. EMBO J. 2012;31:2249–22460. doi: 10.1038/emboj.2012.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilby MJ. Helical shape and wall synthesis in a bacterium. Nature. 1977;266:450–452. doi: 10.1038/266450a0. [DOI] [PubMed] [Google Scholar]
- van den Ent F, Amos LA, Lowe J. Prokaryotic origin of the actin cytoskeleton. Nature. 2001;413:39–44. doi: 10.1038/35092500. [DOI] [PubMed] [Google Scholar]
- van den Ent F, Moller-Jensen J, Amos LA, Gerdes K, Lowe J. F-actin-like filaments formed by plasmid segregation protein ParM. EMBO J. 2002;21:6935–6943. doi: 10.1093/emboj/cdf672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Teeffelen S, Wang S, Furchtgott L, Huang KC, Wingreen NS, Shaevitz JW, Gitai Z. The bacterial actin MreB rotates, and rotation depends on cell-wall assembly. Proc Natl Acad Sci USA. 2011;108:15822–15827. doi: 10.1073/pnas.1108999108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Arellano-Santoyo H, Combs PA, Shaevitz JW. Actin-like cytoskeleton filaments contribute to cell mechanics in bacteria. Proc Natl Acad Sci USA. 2010;107:9182–9185. doi: 10.1073/pnas.0911517107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Furchtgott L, Huang KC, Shaevitz JW. Helical insertion of peptidoglycan produces chiral ordering of the bacterial cell wall. Proc Natl Acad Sci USA. 2012;109:E595–E604. doi: 10.1073/pnas.1117132109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.