Abstract
Aims
Muscle ring finger (MuRF) proteins have been implicated in the transmission of mechanical forces to nuclear cell signaling pathways through their association with the sarcomere. We recently reported that MuRF1, but not MurF2, regulates pathologic cardiac hypertrophy in vivo. This was surprising given that MuRF1 and MuRF2 interact with each other and many of the same sarcomeric proteins experimentally.
Methods and Results
Mice missing all four MuRF1 and MuRF2 alleles [MuRF1/MuRF2 double null (DN)] were born with a massive spontaneous hypertrophic cardiomyopathy and heart failure; mice that were null for one of the genes, but heterozygous for the other (i.e. MuRF1−/−//MuRF2+/− or MuRF1+/−//MuRF2−/−) were phenotypically identical to wild type mice. Microarray analysis of genes differentially expressed between MuRF1/MuRF2 DN, mice missing 3 of the four alleles, and wild type mice revealed a significant enrichment of genes regulated by the E2F transcription factor family. Over 85% of the differentially expressed genes had E2F promoter regions (E2f:DP; p<0.001). Western analysis of E2F revealed no differences between MuRF1/MuRF2 DN hearts and wild type hearts; however, chromatin IP studies revealed that MuRF1/MuRF2 DN hearts had significantly less binding of E2F1 in the promoter regions of genes previously defined to be regulated by E2F1 (p21, Brip1, and PDK4, p<0.01).
Conclusion(s)
These studies suggest that MuRF1 and MuRF2 play a redundant role in regulating developmental physiologic hypertrophy, by regulating E2F transcription factors essential for normal cardiac development by supporting E2F localization to the nucleus, but not through a process that degrades the transcription factor.
Keywords: MuRF1, MuRF2, ubiquitin ligase, E2F, cardiac hypertrophy
Introduction
The muscle ring finger (MuRF) proteins are muscle-specific proteins implicated in the regulation of cardiac development and contractility 1–3. Both MuRF1 and MuRF2 are found at the M-line of the sarcomere 3–5 and affect activity of the transcription factor serum response factor 6, 7. This has been hypothesized to occur in response to biomechanical stimuli, suggesting that MuRF proteins have the ability to recognize stress and respond appropriately 6. We recently found that MuRF1, but not MuRF2, regulates the development of pathologic cardiac hypertrophy in adult mice 7. The involvement of only MuRF1 in this process was surprising given that MuRF1 and MuRF2 are known to homo- and hetero-dimerize 8, and have been found to redundantly target myofibrillar proteins in yeast two hybrid studies 9. We hypothesized that MuRF1 and MuRF2 may have redundant function during cardiac development, and therefore created mice lacking either 3 (MuRF1−/−//MuRF2+/− or MuRF1+/−//MuRF2−/−) or 4 (MuRF1−/−//MuRF2−/−; MuRF1/MuRF2 DN) of the MuRF1 and MuRF2 alleles and compared them functionally. We then analyzed these hearts for redundancies by gene expression profiling.
Materials and Methods
Generation of MuRF1−/− // MuRF2 −/− mice
All mouse experiments were approved by the Institutional Animal Care and Use Committee (IACUC) review board at the University of North Carolina and were in compliance with the rules governing animal use as published by the National Institutes of Health. We generated MuRF1−/− // MuRF2 −/−(MuRF1/MuRF2 double null (DN)) mice using mice previously characterized by our laboratory (129S/C57BL6 background) 7. Briefly, we mated MuRF1 −/− and MuRF2 −/−mice to generate double heterozygous mice (MuRF1+/− //MuRF2+/−). Double heterozygous mice were then mated to generate double null and wild type controls on the same background. Equal numbers of both sexes were included in the studies and equal numbers of male and female offspring were identified. The significant decrease in expected MuRF1/MuRF2 DN mice (only 4 out of 268 pups born/Chi2=0.0000255) resulted in a limited number of MuRF1/MuRF2 DN breeding pairs. For this reason, we chose to breed the double null mice to either MuRF1+/−//MuRF2−/− or MuRF1−/−//MuRF2+/− mice, resulting in the production of mice lacking 3 of the 4 MuRF1 and MuRF2 alleles (i.e. MuRF1−/−MuRF2+/− or MuRF1+/−MuRF2−/−) in addition to MuRF1/MuRF2 DN and wild type mice. At the time of harvest, mice were exposed to isoflurane in an enclosed environment until respiration stopped; cervical dislocation was then performed and then tissues were immediately removed for analysis.
Experimental design
Histological analysis was performed on MuRF1/MuRF2 DN mice either within 12 hours post-mortem (for those mice that died post-natally) or at 12 weeks of age (for surviving mice). Both experimental groups (i.e. those that died post-natally and those that lived) contained an even distribution of male and female mice. Hearts were dissected from the body and perfused with 4% paraformaldehyde. Paraffin sections were stained with H&E, Masson’s Trichrome, or Lectin as previously described7.
Echocardiography was performed on conscious mice by both M-mode and two-dimensional imaging using the Vevo 770 ultrasound system as previously described 7 at 12 weeks of age.
Real time PCR analysis of gene expression
For gene expression studies, a two-step reaction was used to determine mRNA expression of fetal genes associated with cardiac hypertrophy as previously described 7.
RNA extraction and microarray processing
Total RNA was isolated from 12-week-old mouse cardiac apices using the All Prep DNA/RNA/Protein isolation kit (Qiagen, Inc., Valencia CA) was verified for integrity using the BioAnalyzer 2100 (Agilent Technologies, Inc., Santa Clara, CA). RNA samples labeled with cyanine-5 CTP in a T-7 transcription reaction using the Agilent Low Input Linear RNA Amplification/Labeling System were hybridized to 4×44K microarray slides (Agilent GPL4134) in the presence of equimolar concentrations of cyanine-3 CTP-labeled mouse reference RNA 10. Slides were hybridized, washed, and scanned on an Axon 4000b microarray scanner and data were processed using Feature Extraction (version 9.1.3.1, Agilent). Post-processing included Loess- 11, 12 and median-centered normalization using Genespring GX (version 10.0.1 Build 81217; Agilent). The Database for Annotation, Visualization and Integrated Discovery (DAVID) 13, 14 identified significantly enriched functional clusters (high classification stringency, group enrichment score of >1.3, p<0.05) using multiple annotation libraries from lists of differentially expressed genes, using the genes represented on the microarray as background (see Supplemental Table 1 for DAVID annotation libraries used). Complete, MIAME-compliant datasets were deposited with the Gene Expression Omnibus of the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/geo/) 15, and are accessible through GEO Series accession number GSE14512.
Chromatin IP (ChIP) analysis of E2F1
Chromatin IP (ChIP) from heart tissues was based on the Farnham protocol (http://farnham.genomecenter.ucdavis.edu/). p21, Brip1, and PDK4 promoter regions were investigated because these were genes differentially expressed in the microarray analyses for which E2F1 regulation had been published in the peer-reviewed literature and primers had been described in the mouse. PCR primers were designed to amplify a 96, 148, and 128 bp region of the p21, Brip1, and PDK4 promoters, respectively. The sequences of the PCR primers we used to amplify the p21 locus 16 were 5′-TGT ATG TGG CTC TGC TGG TG-3′(forward) and 5′-CCT CCC CTC TGG GAA TCT AA-3′ (reverse). The sequences of the PCR primers we used to amplify the Brip locus 17 were 5′-CTG TGT GAT TGG CTG ACT GG-3′(forward) and 5′-TACAGCCACTCCTCCCTCTC-3′ (reverse). The sequences of the PCR primers we used to amplify the PDK4 locus 18 were 5′-TCC TAG CGA CCT GGG ATC TA -3′(forward) and 5′-CTA GAA AGG CCT GGC ACA GT-3′ (reverse). The amplified DNA products were analyzed by real time qPCR using SYBR® Green and confirmed by product melting curve analysis.
Results
MuRF1/MuRF2 DN mice exhibit a spontaneous cardiac hypertrophy from birth
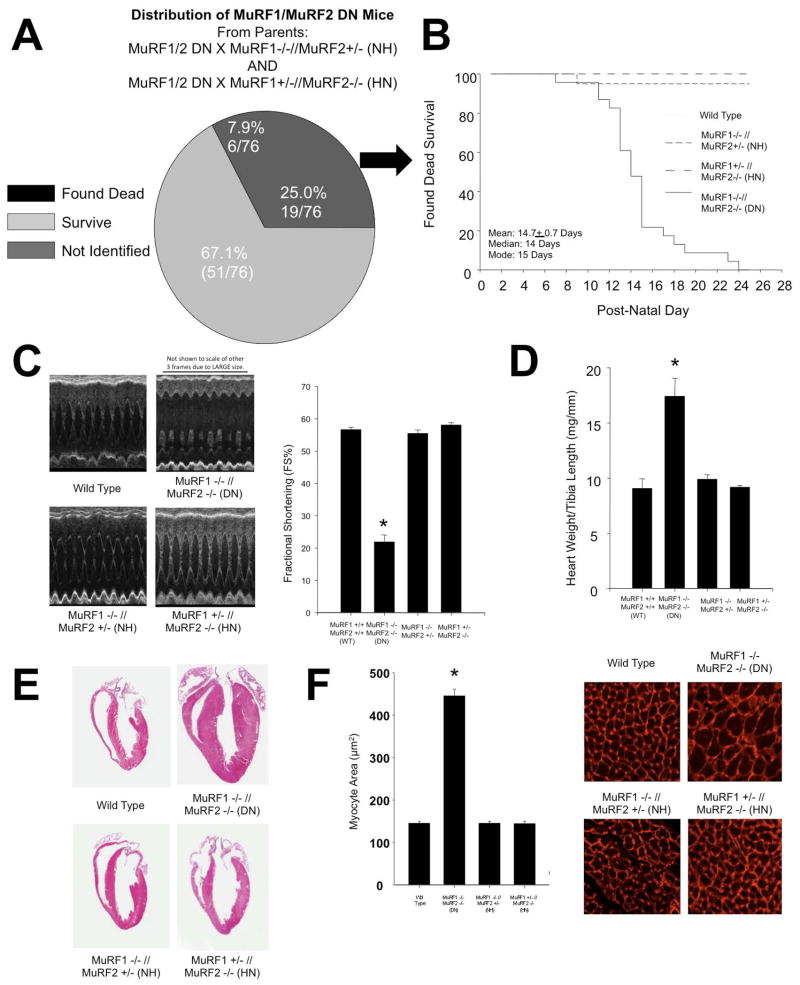
We previously demonstrated that mice lacking either MuRF1 (MuRF1−/−) or MuRF2 (MuRF2−/−) do not exhibit a cardiac phenotype at baseline 7. To extend these previous studies and to test for functional redundancy, we examined the cardiac phenotype of mice lacking 3 or 4 of the MuRF1/MuRF2 alleles. Breeding MuRF1 +/− // MuRF2 +/− (to generate MuRF1/MuRF2 DN mice) led to very few viable MuRF1/MuRF2 DN mice. We therefore bred the limited number of surviving MuRF1/MuRF2 DN mice to either MuRF1+/−//MuRF2−/− or MuRF1−/−//MuRF2+/− mice. This breeding strategy afforded us the benefit of examining not only the phenotype of MuRF1/MuRF2 DN mice but also the phenotype of mice lacking 3 of the 4 MuRF1 and MuRF2 alleles. These breedings resulted in the birth of 76 mice lacking 3 of the 4 MuRF1/MuRF2 alleles (i.e. MuRF1−/−MuRF2+/− or MuRF1+/−MuRF2−/−) and 25 MuRF1/MuRF2 DN mice. Of the 25 MuRF1/MuRF2 DN mice that were born alive, 19 died between birth and post-natal day 23, while 6 survived to adulthood. Using Mendelian principals to assume that the non-MuRF1/MuRF2 DN mice represented ~50% of the population, of the predicted 76 MuRF1/MuRF2 DN mice that should have been born, 51 (67.1%) were unaccounted for by live births, as shown in Figure 1A. We propose that these mice died in utero due to cardiac defects as post-mortem histological analysis consistently identified extensive cardiac hypertrophy and the presence of fibrosis (illustrated in Supplemental Figure 1B, see blue extracellular matrix), with the majority of the thoracic cavity taken up by the enlarged heart (Supplemental Figure 1A). In contrast, MuRF1/MuRF2 DN mice that did not die did not exhibit any fibrosis anywhere in their hearts (see Figure 1A, right column vs. adjacent middle column). Of the 25% MuRF1/MuRF2 DN mice found dead after birth, the average age of death was 14.7+0.7 days (Figure 1B).
Figure 1. MuRF1 −/− // MuRF2 −/− (DN) mice die in utero and peri-natally and exhibit a spontaneous cardiac hypertrophy.
A. Analysis of the fate of MuRF1/MuRF2 DN pups born from breedings of MuRF1/MuRF2 DN mice and either MuRF1−/−MuRF2+/−or MuRF1+/−MuRF2−/− mice (N=76 estimated from the 76 non-MuRF1/MuRF2 DN mice). B. Graphical representation of the average day of death of mice with varying genotypes (N=22). C. Echocardiograms from 12-week-old mice of varying genotype. MuRF1/MuRF2 DN mice exhibit a pronounced decrease in cardiac function. D. Cardiac hypertrophy was determined by measuring the ratio of increased heart weight/tibia length. E. Paraffin cross sections through the hearts of mice with varying genotypes. F. Measurement of myocyte cross sectional area. A One-Way ANOVA was performed to determine significance, followed by a multiple comparison procedures (Holm-Sidak method) to determine significance between groups. *p<0.001
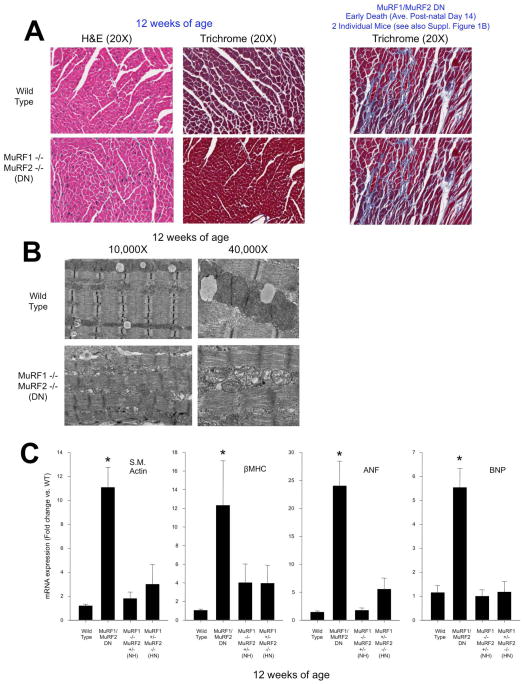
MuRF1/MuRF2 DN mice that survived into adulthood displayed normal activity and breeding, despite a significant impairment in cardiac function determined by echocardiography (see Figure 1C, Table 1), which did not progress with age (data not shown). The earliest time point we investigated after birth (6 weeks of age) identified significant defects in MuRF1/MuRF2 DN cardiac function (fractional shortening=20.5+3.4% vs. other three groups FS=55.6+0.8%; ejection fraction=42.0+6.2 vs. 87.3+0.6%; N=3 & 5, respectively, p<0.01). At 12 weeks of age, MuRF1/MuRF2 DN mice displayed significant cardiac dysfunction, as measured by a dramatic decrease in the percentage of fractional shortening (FS) compared to wild type mice (21.9+2.2% vs. 56.6+0.7%, respectively). Mice in which 3 of the 4 MuRF1/MuRF2 alleles were missing (MuRF1 −/− //MuRF2 +/− or MuRF1+/−//MuRF2−/−) showed no deficit in FS% (55.4+1.1% and 58.1+0.8%, respectively) (Figure 1C). MuRF1/MuRF2 DN mice also developed cardiac hypertrophy as indicated by increased heart weight/tibia length (Figure 1D), increased gross histological size (Figure 1E), and increased cardiomyocyte cross sectional area (Figure 1F). Fibrosis and other gross defects were not detected in 12-week-old MuRF1/MuRF2 DN mice histologically (Figure 2A), which was in sharp contrast to the widespread fibrosis found in most MuRF1/MuRF2 DN mice that died within the first several weeks of life (Supplemental Figure 1B). It is interesting to note that MuRF1 and MuRF2 are found in cardiomyocytes, so the fibrotic response of the fibroblasts in these early deaths is likely due to indirect and/or developmental effects of changes seen in the cardiomyocytes. The cause of death in these mice that died in the first 2 weeks of life appeared to be heart failure, as indicated by significant decrease in cardiac function and edematous lungs. Interestingly, it is during this same window of time (i.e. post-natal days 5 through 14) that mouse cardiomyocytes undergo a dramatic period of hypertrophic growth 19. This may suggest that MuRF1 and MuRF2 may play a key regulatory role during this vital physiologic developmental hypertrophy growth process as it is during this same time frame that most MuRF1/MuRF2 DN post-natal deaths occur (average day 14.0±0.7). Transmission electron microscopy indicated that hearts from the MuRF1/MuRF2 DN mice that died on average day 14 exhibited disrupted sarcomeres, with both Z disc and M line defects, in addition to gross abnormalities in mitochondria (Figure 2B). In contrast, no such defects and fewer mitochondria were identified in skeletal muscle (gastrocnemius) of the same MuRF1/MuRF2 DN mice by TEM (Supplemental Figure 1C). These findings suggest that overlap of MuRF1 and MuRF2 function in sarcomeric stability may be less important in skeletal muscle development compared to the developing heart.
Table 1.
Echocardiography of Dimensions and Function in MuRF1/MuRF2 DN, wild type, MuRF1−/−MuRF2+/− (NH), and MuRF2+/−MuRF2−/− (HN) mice at 12 weeks of age. Data represents an approximate even distribution of male and female mice. *p<0.001 vs. MuRF1/MuRF2 wild type, MuRF1+/−MuRF2−/−, and MuRF1−/−MuRF2+/−. DN=double null; WT=wild type.
| MuRF1+/+ MuRF2+/+ (WT) (N=5) | MuRF1/ MuRF2 DN (N=5) | MuRF1 −/− MuRF2+/− (NH) (N=5) | MuRF1 +/− MuRF2 −/− (HN) (N=5) | |
|---|---|---|---|---|
| BW (g) | 26.5±0.9 | 23.3±3.5 | 27.1±1.15 | 23.0±2.6 |
| HW/TL (mg/mm) | 9.1±0.9 | 17.4±1.6* | 10.9±0.4 | 9.2±0.2 |
| LV mass index (mg) | 110.3±7.3 | 206.2±20.2* | 104.3±5.4 | 103.0±13.0 |
| LVMass/BW (mg/g) | 4.2±0.3 | 8.88±0.47* | 4.2±0.2 | 4.5±0.6 |
| HR (bpm) | 609.0±29.3 | 644.6±24.3 | 643.6±14.3 | 612.2±22.6 |
| IVSTD (mm) | 0.99±0.02 | 1.14±0.03* | 1.04±0.02 | 1.01±0.01 |
| IVSTS (mm) | 1.57±0.13 | 1.47±0.01 | 1.69±0.07 | 1.70±0.05 |
| PWTD (mm) | 0.98±0.02 | 1.08±0.03* | 1.01±0.01 | 0.97±0.01 |
| PWTS (mm) | 1.67±0.04 | 1.20±0.03* | 1.68±0.08 | 1.52±0.03 |
| LVEDD (mm) | 2.94±0.13 | 3.98±0.24* | 2.90±0.20 | 2.84±0.20 |
| LVESD (mm) | 1.24±0.06 | 3.23±0.30* | 1.21±0.06 | 1.20±0.06 |
| EF (%) | 88.0±0.6 | 44.3±3.9* | 89.2±0.4 | 83.4±4.3 |
Transthoracic echocardiography on unanesthetized mice. BW, body weight; HW, heart weight; TL, tibia length; HR, heart rate; LV mass index [(ExLVD3d-LVED3d) × 1.055]; ExLVD, external left ventricular diameter; bpm, heart beats per minute; IVSTD, interventricular septal thickness in diastole; IVSTS, interventricular septal thickness in systole; PWTD, posterior wall thickness in diastole; PWTS, posterior wall thickness in systole; LVEDD, left ventricular enddiastolic dimension; LVESD, left ventricular end-systolic dimension; FS, fractional shortening, calculated as (LVEDD-LVESD)/LVEDD × 100; ND, not determined.
Figure 2. MuRF1/MuRF2 DN hearts exhibit significant increases in fetal gene expression and disrupted sarcomere organization and mitochondria.
A. Histological analysis of 12-week-old hearts. B. Electron microscopic analysis of wild type and MuRF1/MuRF2 DN hearts. C. Markers of cardiac hypertrophy are increased in 12-week-old MuRF1/MuRF2 DN hearts. Data in all panels represent at least five mice/group. A One-Way ANOVA was performed to determine significance, followed by a multiple comparison procedures (Holm-Sidak method) to determine significance between groups. *p<0.001
MuR F1/MuRF2 DN hearts have discrete increases in hypertrophic “fetal gene” expression
Our results suggest that only 1 of the 4 MuRF1 and MuRF2 alleles is necessary to maintain a phenotype comparable to wild type mice during cardiac and skeletal muscle development. This led us to propose that MuRF1 and MuRF2 have redundant functions in developmental cardiac growth, a finding that contrasts sharply with our observations that MuRF1, but not MuRF2, regulates pathologic cardiac hypertrophy in adult mice in vivo 7. To determine whether the cardiac hypertrophy seen in the MuRF1/MuRF2 DN mice was associated with “fetal gene” expression and pathologic hypertrophy, we assayed for the characteristic upregulation of skeletal muscle actin, βMHC (myosin heavy chain), ANF (atrial natriuretic factor), and BNP (brain natriuretic peptide) in MuRF1/MuRF2 DN, MuRF1−/−//MuRF2+/− (Null Het), MuRF1+/−//MuRF2−/− (Het Null) and wild type mice. All four “fetal gene” mRNA levels were significantly increased in the MuRF1/MuRF2 DN mice compared to mice missing 3 of the 4 MuRF1/MuRF2 alleles or to wild type mice (Figure 2C). This suggested that MuRF1/MuRF2 DN hearts underwent either pathologic cardiac hypertrophy and/or exhibited an inhibition of cardiac development.
Global analysis of differential gene expression
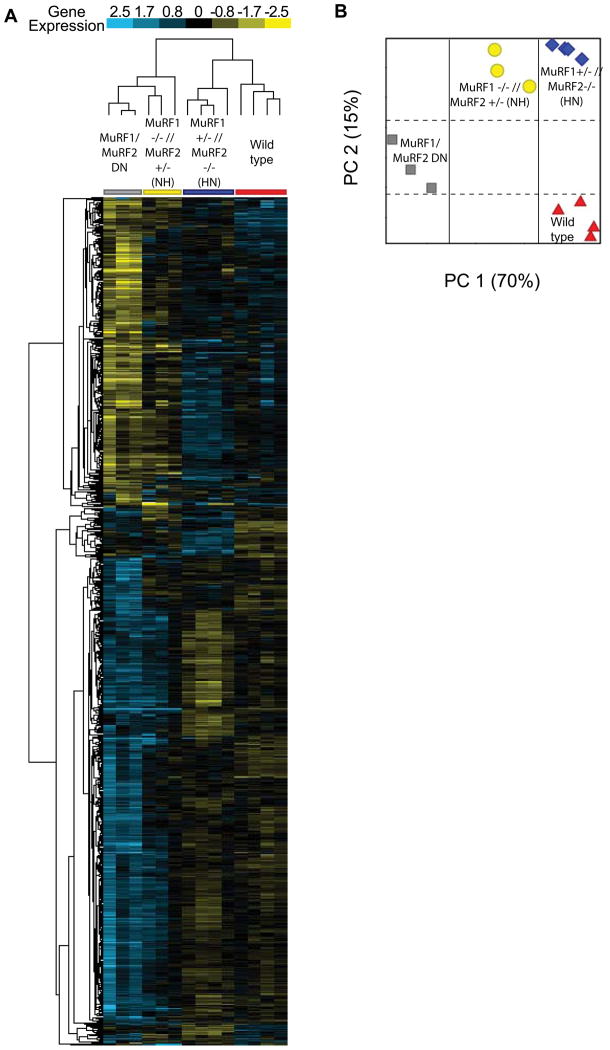
Our physiologic, histologic, and Real Time PCR analysis of fetal genes demonstrated the functional redundancy of MuRF1 and MuRF2: the absence of 3 of the 4 MuRF1/MuRF2 alleles resulted in normal cardiac phenotype, whereas the loss of all 4 alleles led to a dramatic spontaneous cardiac hypertrophy. To investigate the redundancy of MuRF1 and MuRF2 at the level of the transcriptome, we used microarray analysis to identify differentially expressed genes in 12-week-old MuRF1/MuRF2 DN, MuRF1 −/− // MuRF2 +/− (Null Het), MuRF1 +/− // MuRF2 −/− (Het Null), and strain-matched wild type mice (Figure 3; Supplemental Table 2). Based on the functional redundancy of MuRF1 and MuRF2, we anticipated few differences between the wild type mice, and the 2 groups of mice lacking 3 of the 4 alleles. However, an unsupervised cluster analysis of differentially expressed genes between all 4 genotypes revealed a very different story. According to this analysis, MuRF1 +/− // MuRF2 −/− mice share a similar cardiac gene expression profile with WT mice (right 2 groups, Figure 3A), whereas the cardiac gene expression profile of the MuRF1 −/− // MuRF2 +/− mice align more closely with the MuRF1/MuRF2 DN mice (left 2 groups, Figure 3A). This latter finding was unexpected since both sets of mice lacking three of the 4 MuRF1 and MuRF2 alleles were functionally identical to wild type mice.
Figure 3.
Cluster analysis of differentially expressed genes in 12-week-old MuRF1/MuRF2 DN, MuRF1 −/− // MuRF2 +/− (NH), MuRF1 +/− // MuRF2 −/− (HN), and strain-matched wild type (WT) hearts. Represented are differentially expressed genes with a fold change > 2.0, p value cut-off < 0.05, analyzed by One-Way ANOVA and post-hoc by Tukey HSD and Benjamini-Hochberg multiple testing correction. A. An unsupervised clustering analysis was then performed on genes differentially expressed statistically. B. A principal components analysis was then performed on the 14 individual data sets included in the analysis, which determined similarities between wild type and MuRF1 +/− // MuRF2 −/− (HN) hearts by principal component 1, accounting for 70% of the differences between the data sets.
To better delineate the relationships between the four groups of mice, we performed a principle components analysis (PCA) of the four genotypes using the clustered gene set (Figure 3B). A PCA is a linear transformation used to reduce the dimensions of a microarray (containing thousands of genes) to a new coordinate system (x, y) by retaining characteristics of the data that contribute most to the variance. This method for characterizing or mathematically representing the relationship between individual components of multiple complex data sets allows the determination of structure between variables, in the present case between genotypes. The first principle component (PC1, which accounts for 70% of the total variance across all of the samples) could not distinguish the cardiac gene expression patterns in wild type and MuRF1 +/− // MuRF2 −/− mice (right column, Figure 3B). This analysis also identified that the cardiac gene expression profile of MuRF1 −/− // MuRF2 +/− and MuRF1/MuRF2 DN mice was substantially removed from the other 2 groups, consistent with the unsupervised clustering. Interestingly, the second principal component (PC2, 15% of the total variance) demonstrated similarities between the cardiac gene expression profile of the MuRF1 −/− // MuRF2 +/− and the MuRF1 +/− // MuRF2 −/− mice that were distinct from any similarities shared with either wild type or DN mice. Consistent with this analysis, we identified only a few genes unique to MuRF1 −/− // MuRF2 +/− (11) and MuRF1 +/−// MuRF2 −/− mice (33) (Supplemental Figure 4, Supplemental Table 3).
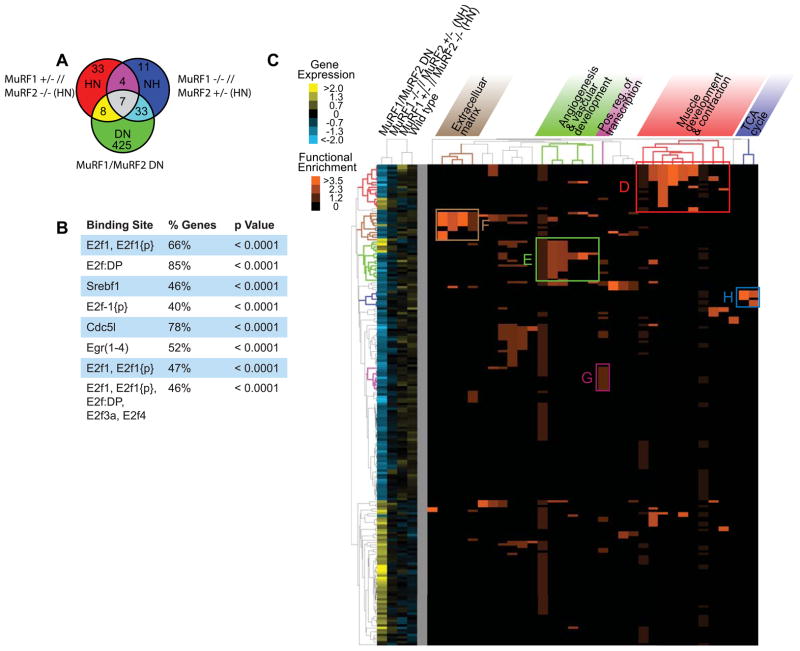
We next compared the change in gene expression in the MuRF1/MuRF2 DN hearts compared to all other groups. First, we identified all the genes differentially expressed from wild type hearts, represented as the 3 circles in Figure 4A. We then compared these differentially expressed genes between the MuRF1/MuRF2 DN, MuRF1 −/− // MuRF2 +/− (Null Het), and MuRF1 +/− // MuRF2 −/− (Het Null) and identified 425 differentially expressed genes unique to the DN mice (Figure 4A). Analysis of the upstream promoter regions of the 425 differentially expressed genes by TRANSFAC analysis using Gather (http://gather.genome.duke.edu/) identified 8 transcription factor binding sites significantly enriched in the promoter region of these genes (Figure 4B). Strikingly, 5 of the 8 binding sites belonged to the E2F family of transcription factors, and were present in 40–85% of the genes with TRANSFAC annotations (Figure 4B). The number of genes with predicted E2F promoter binding sites was evenly distributed between up- and down-regulated genes (data not shown), suggesting redundant regulation of E2F transcription activity may be mediated, in part, by MuRF1 and MuRF2. TRANSFAC analysis also identified SREBf1, Cdc51, and Egr(1–4) promoter binding sites in 46–78% of the genes differentially expressed in MuRF1/MuRF2 DN mouse hearts (Figure 4B). With E2F1 so richly represented in this analysis in the MuRF1/MuRF1 DN hearts, we performed Western blot analysis for E2F1 on all four genotypes investigated in this study. At the protein level, E2F1 levels were not different between the four MuRF1/MuRF2 genotype groups (Supplemental Figure 2A and 2B).
Figure 4.
Analysis of genes differentially expressed only in MuRF1/MuRF2 DN hearts. Represented are differentially expressed genes with a fold change > 2.0, p value cut-off < 0.05, analyzed by One-Way ANOVA and post-hoc by Tukey HSD and Benjamini-Hochberg multiple testing correction. A. A Venn diagram representation of the genes analyzed for common transcription factor binding sites by TRANSFAC analysis and enriched functional groups by DAVID analysis. Genes differentially expressed in the MuRF1/MuRF2 DN, MuRF1 +/− // MuRF2 −/− (HN), and MuRF1 −/− //MuRF2 +/− (NH) compared to wild type mice were compared. The 425 genes unique to the MuRF1/MuRF2 DN mice were investigated further. B. TRANSFAC analysis using Gather of the putative binding sites in the promoter region of the 425 genes differentially expressed in MuRF1/MuRF2 DN as a percentage the annotated genes. C. A cluster analysis of the higher ordered DAVID analysis of the annotated genes revealed enriched functional groups in: D. Muscle Development and Contraction; E. Angiogenesis and vascular development; F. Extracellular matrix; G. TCA cycle; and H. Positive regulation of transcription. The functional enrichment of the functional categories by the heat map in orange (see insert for key).
Genes differentially expressed in MuRF1/MuRF2 DN mice are involved in amino acid metabolism and ligase activity
A higher order analysis of the differentially expressed genes using DAVID Bioinformatics Resources 13 was next used to perform a functional annotation of genes differentially expressed exclusively in the MuRF1/MuRF2 DN compared to all other groups (Figure 4C, Supplemental Figure 3). The functional groups differentially expressed only in the MuRF1/MuRF2 DN mice in relation to the other 3 groups included in this study were (Figure 4C, Supplemental Figure 3): 1) muscle development and contraction (Figure 4D); 2) angiogenesis and vascular development (Figure 4E); 3) extracellular matrix (Figure 4F); 4) positive regulation of transcription (Figure 4G); and 5) the TCA cycle (Figure 4H). Detailed functional enrichment analysis of these groups can be found in the supplemental results section and Supplemental Figure 3. Upon extensive literature searches, we identified that, although not previously reported in mice lacking MuRF1 and MuRF2 20, several of these differentially expressed genes have been associated with both fetal cardiac development and cardiac hypertrophy (see supplemental results section). The fact that we had identified genes associated with cardiac hypertrophy was no surprise, given the extreme phenotype in the MuRF1/MuRF2 DN. However, we were intrigued in the differences in gene expression associated with the genotype of these mice (i.e. MuRF1−/−//MuRF2−/−) as opposed to the phenotype (i.e. cardiac hypertrophy).
MuRF1−/−//MuRF2−/− hearts have decreased E2F1 occupancy at promoter regions of multiple E2F1-regulated genes
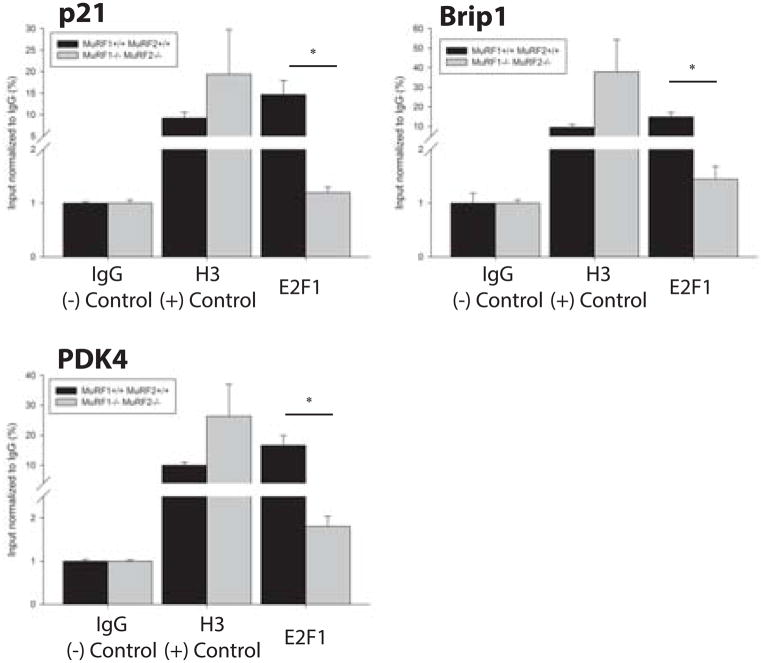
Since no differences in E2F1 protein levels were identified between MuRF1−/−//MuRF2−/− (DN), MuRF1−/−//MuRF2+/− (NH), MuRF1+/−//MuRF2−/− (HN), and wild type mice (Supplementary Figure 2A and 2B), we hypothesized that E2F1 transcriptional activity was regulated by MuRF1/MuRF2 expression in the heart. To test this hypothesis, we performed chromatin immunoprecipitation (ChIP) using cardiac tissue from MuRF1−/−//MuRF2−/− (DN) and their age-matched wild type controls to measure the occupancy of E2F1 in the promoter regions of three genes, p21, Brip1, and PDK4, which we found to have significantly different expression levels between MuRF1−/−//MuRF2−/− (DN) and wild type hearts in microarray analyses. Primers to E2F1 binding sites in the promoter regions of p21, Brip1, and PDK4, previously identified by Radhakrishnan et al. 16(15048076), Eelen et al. 17 (18345034), and Hsieh et al. 18 (18667418), respectively, were designed to flank the E2F1 binding sites for each of these genes in order to amplify DNA isolated by ChIP. A significantly decreased amount of E2F1 occupancy in these promoters was identified in MuRF1−/−//MuRF2−/− (DN) hearts compared to wild type for each of the E2F1 target genes (Figure 5), including a role for MuRF1 and MuRF1. Given that the total E2F1 protein levels between these hearts does not differ, we propose that MuRF1 and MuRF2 redundantly prevent E2F1 from going in to the nucleus or they enhance E2F1 removal from the nucleus by its ubiquitin ligase activities, to decrease its overall apparent interaction with the promoter regions of E2F1 regulated genes.
Figure 5. E2F1 occupancy in the promoter regions of E2F1 regulated genes, p21, Brip1, and PDK4, is significantly decreased in MuRF1−/−//MuRF2−/− (DN) hearts compared to wild type.
Fresh cardiac tissue was cut into 1mm pieces and crosslinked with formaldehyde in order to establish protein-DNA complexes. Crosslinked tissue was lyzed, chromatin was extracted, and sonication was performed as described in the Materials and Methods. The resulting chromatin was immunoprecipitated using antibodies specific to E2F1, histone H3 (positive control), or IgG (negative control). Following immunoprecipitation, protein-DNA complexes were decrosslinked and DNA was extracted. Immunoprecipitated DNA was submitted to RT-PCR analysis using primers flanking the E2F1 binding sites within the promoter regions of p21, Brip1, and PDK4. CT-values were compared to input CT values to generate %input values for each experimental group and final data was normalized to negative control (IgG). Resulting data show that the amount of immunoprecipitated DNA corresponding to the E2F1 binding regions of p21, Brip1, and PDK4 gene promoters was significantly decreased in MuRF1−/−//MuRF2−/− hearts (gray bars) compared to wild types (black bars). At Student’s t-test was used to determine the significance between genotypes. *p<0.01
Discussion
In this study, we identified for the first time that MuRF1 and MuRF2 play a necessary but functionally redundant role in cardiac development in vivo. We found that MuRF1/MuRF2 DN mice are born with an extreme cardiac hypertrophy with a high rate of lethality peri-natally within the first few weeks of life. However, in mice lacking 3 of the 4 MuRF1 and MuRF2 alleles the cardiac phenotype was identical to that found in wild type mice. By microarray analysis of cardiac gene expression, we identified that MuRF1−/−//MuRF2−/− hearts had a number of differentially expressed many genes, which we identified having E2F1 promoter regions using a bioinformatics analysis (TRANSFAC). Despite no differences in E2F1 protein levels in MuRF1/MuRF2 DN mice (Supplemental Figure 5), we did identify differences in E2F1 protein located on the promoter regions of E2F1-regulated genes using ChIP assays (Figure 5). In this context, the present studies indicate that MuRF1 and MuRF2 may have overlapping function confined to the unique cardiac hypertrophy that occurs during development, regulating key cellular processes controlled by the transcription factor E2F1. Additionally, MuRF1 and MuRF2 had hypertrophy-independent effects, based on principal components analysis, which were mediated by other transcriptions, as presented in Supplemental Figure 4 and discussed in the online data supplement.
We identified that genes differentially expressed in MuRF1/MuRF2 DN compared to the other three groups of mice had an overwhelming representation of E2F transcription factor sites (Figure 4). Likewise, analysis of the differentially expressed genes in the MuRF1/MuRF2 DN and MuRF1−/−//MuRF2+/− groups (Supplemental Figure 4) identified that 85% of the genes differentially expressed had E2F transcription factor sites in their promoter region. However, the finding of 85% of the genes having E2F binding sites was not included in our results since the p value was >0.05; regardless, it does point to a consistent trend in our data sets. The identification of E2F1 transcription factor sites is intriguing because E2F1 in non-cardiac cells is known to be regulated by other ubiquitin ligases, including the ubiquitin ligase MDM2 21, 22 and Cullin proteins. In association with ROC1 and SKP, Cullins-1, 2, 3, and 5 also ubiquitinate E2F1 in non-cardiac cells 23. In cardiomyocytes, E2F1 has the ability to drive quiescent cells into S phase in both neonatal and adult cardiomyocytes 24, 25. E2F1 also regulates cardiomyocyte size 26, induces cardiomyocyte cell proliferation without stimulating apoptosis 27, and activates autophagy and the intrinsic death pathway 28. Given the prominent role of E2F transcription factors in cardiac cell cycle, cell proliferation, cell death, and glucose oxidation, it is not surprising that they have a prominent role in cardiac development. The role of E2F in the cardiac hypertrophy in the present model, however, cannot be definitely interpreted as the cause of the hypertrophy. Further studies will be necessary to determine if changes in E2F are directly causing the phenotype, or are an indirect effect of the phenotype itself.
If MuRF1 and MuRF2 are redundantly regulating E2F1 to cause the present phenotype, the mechanism is likely complex given the number of proteins involved in the E2F complex. Mechanistically, the regulation of E2F1 transcription factors by MuRF1 and MuRF2 may not be intuitive, given MuRF proteins role as a ubiquitin ligase that has been reported to poly-ubiquitinate and degrade several proteins 2, 29. However, MuRF1, as well as other ubiquitin ligases regulates proteins by several non-canonical ubiquitination pathways, including mono-ubiquitination 30, 31. One way in which MuRF1 and MuRF2 may support E2F1 to go to the nuclear may be by supporting its nuclear localization or inhibiting its nuclear export 30, 31, although this was not tested directly. Alternatively, we speculate that a number of proteins are inhibitors of the E2F1 protein; one of the most well-studied group of proteins which limit E2F1 transcriptional activity are pocket proteins, including retinoblastoma susceptibility gene (Rb), p107, and p130—all of which are expressed in the heart 32. Since the regulation of these pocket proteins in relation to their ability to inhibit E2F1 has been found to be important for normal cardiac development and function 32, MuRF1/MuRF2 could be exacting its activation of E2F1 transcriptional activity by targeting pocket proteins for ubiquitin-dependent proteasomal degradation. Our data are consistent with MuRF1 and MuRF2 redundantly promoting the recruitment of E2F1 to its binding sites within the promoter regions of E2F1-regulated genes in the heart, since the absence of both MuRF1 and MuRF2 leads to reduced E2F1 promoter occupancy by ChIP analysis. This may be through mechanisms that directly support E2F1 stability, or through mechanisms that promote the degradation of E2F1 inhibitors, resulting in more E2F1 at nuclear promoter sites of E2F1-regulated genes.
Mice lacking E2F transcription factors have a cardiac phenotype and embryonic lethality similar to that seen in our MuRF1/MuRF2 DN mice. For example, mice lacking E2F3 are born at approximately 25% of the expected ratio, and the small proportion that survive to adulthood exhibit growth retardation dependent on the dosage of E2F3 33. Crossing the E2F3 null mice with mice having Retinoblastoma (Rb) mutations improves E2F3 null mouse survival, suggesting a role of Rb in this process, since E2F-RB dimers occur in vivo 33. Compound E2F3 +/− // E2F1 +/− heterozygous mice similarly develop congestive heart failure, suggesting that the dosage of free activating E2F is critical to normal cardiac development and function 33.
A recent study identified that mice lacking MuRF1 and MuRF2 created had a spontaneous cardiac and quadriceps muscle hypertrophy, a decrease in body weight, and chronically activated protein synthesis 20. They additionally identified that Carp, FHL2, and SQSTM1/p62 were increased in MuRF1/MuRF2 DN hearts, but not in wild type, MuRF1 −/−, or MuRF2 −/− mice, indicating that these may be redundant targets of MuRF1 and MuRF2 and underlie the abnormal phenotype20. In the present study, we additionally demonstrate the redundancy of MuRF1 and MuRF2 by showing that mice missing 3 of the 4 alleles (MuRF1 −/− // MuRF2 +/− and MuRF1 +/− // MuRF2 −/−) do not differ from strain-matched wild type control mice. We additionally demonstrate the MuRF1 and MuRF2 redundantly regulate E2F1 by microarray, TRASFAC analysis, and confirmation by ChIP assays of E2F1 binding to p21, Brip1, and PDK4 promoter regions.
While this previous study identified a 74% post-natal mortality rate 20, the present study estimated that approximately 67.1% of the MuRF1/MuRF2 DN mice died in utero, with an additional 25% dying at day 14, and the remaining 6.7% surviving past the peri-natal period. We also found that 4 of the 11 mice that we let survive into adulthood (i.e. not killed for our studies) died at 3.3, 2.4, 2.1, and 0.7 months of age. In contrast, this previous study of MuRF1/MuRF2 dKO mice all survived to 18 months, which is distinctly different than our findings20. This may be due, in part, to the greater number of animals we evaluated overall. Of these adult MuRF1/MuRF2 DN mice in the current study, one has survived 25 months indicating the longevity of some MuRF1/MuRF2 DN mice if they survive the post-natal death, which peaks at ~day 14. The present study emphasizes the developmental nature of the defect in the hearts missing both MuRF1 and MuRF2, and suggests that the importance of the redundant affects of these proteins predominantly is due to developmental cardiac defects, a common cause of spontaneous abortion 34.
We identified comparable increases in the MuRF1/MuRF2 DN heart masses compared to wild type hearts and hearts missing 3 of the 4 MuRF1/MuRF2 alleles. However, we did not identify a skeletal muscle hypertrophy in the gastrocnemius or tibialis anterior normalized for tibia length. Interestingly, though, the soleus muscle was significantly decreased in the MuRF1/MuRF2 DN mice compared to all the other genotypes tested. This is consistent with the decreased body weight we identified in the MuRF1/MuRF2 DN at 12 weeks of age (Supplemental Table 4) compared to all other groups. While Witt, et al. identified a similar age-dependent decrease in body weight of the MuRF1/MuRF2 DN mice, they identified hypertrophy in the quadriceps muscle at 18 months of age. There are many factors that may account for these differences, including the differences in age, the background of the mice, and the targeting strategies of the MuRF1 and MuRF2 alleles. However, the predominant and profound changes in the cardiac phenotype are consistent between the two studies. The mild changes in skeletal muscle suggest less of a redundant role of MuRF1 and MuRF2 in vivo.
Despite their muscle specificity, mice lacking all four MuRF1 and MuRF2 alleles that died at ~14 days of age, exhibited a diffuse fibrosis throughout their hearts, while those MuRF1/MuRF2 DN mice that survive did not have any detectable fibrosis (Figure 1A). The observed fibrosis may be related to the associated premature death that was not seen mice that survived this 2 weeks period and had normal life spans (up to 25 months of age). Since MuRF1 and MuRF2 are found in muscle, the effects of MuRF1 and MuRF2 deficiency on fibroblast activation and secretion of fibrosis in the mice that died prematurely after birth is due to indirect effects. The activation of fibrosis in cardiomyocyte-specific gene expression has been described in multiple models 35–37 and speaks to the extensive cross talk between cardiomyocytes and fibroblasts by inflammatory mediators 38 that control cardiac function. A pivotal role of cardiomyocyte TGF-β, for example, has been implicated in the fibrosis seen in pressure overload induced cardiac hypertrophy 39. Reparative fibrosis, in the context of cardiomyocyte death, occurs to replace the dying cell and occurs in intermuscular spaces focally 40. The more diffuse fibrosis that can occur, as described in the MuRF1/MuRF2 DN mice that die at ~day 14, is regulated by cardiac fibroblasts that control the synthesis of collagens and enzymes that degrade them (MMPs)40. It is unclear how the relatively small subset of mice that survived the initial day 14 premature death associated with fibrosis avoided the development of the diffuse fibrosis activated by indirect stimulation of fibroblasts. And it is unexpected that this fibrotic response is absent in the surviving mice with the severe cardiac dysfunction as fibrosis itself can contribute to cardiac dysfunction. Since there is a strong relationship between myocardial extracellular matrix remodeling, increased fibrosis, and decreased ventricular function 41, we hypothesize that the pathophysiology of the acute death seen at ~day 14 is compounded by mechanisms seen in the surviving mice (i.e. E2F1 mediated mechanisms) in addition to indirect fibrotic mechanisms that contribute to the decreased function observed after birth, heart failure, and sudden death not previously reported in this MuRF1/MuRF2 DN mouse model 20.
Supplementary Material
Acknowledgments
Funding
This work was supported by the National Institutes of Health (R37HL065619 to C.P., R01HL104129 to M.W.); the American Heart Association Scientist Development Grant (to M.W.); and the Leducq Foundation (to C.P., M.W.).
The authors wish to thank Janice Weaver of the University of North Carolina Animal Histopathology Core Facility and Vicky Madden of the University of North Carolina Microscopy Services Laboratory for their assistance with histological and TEM preparations and analysis.
Non-standard Abbreviations and Acronyms
- MuRF1
Muscle Ring Finger-1
- MuRF2
Muscle Ring Finger-2
- DN
MuRF1/MuRF2 double null
- S.M. actin
skeletal muscle actin
- ANF
atrial natriuretic factor
- βMHC
β myosin heavy chain
- BNP
brain natriuretic peptide
- CDKN1(p21)
cyclin-dependent kinase inhibitor 1A
- BRCA1
Breast cancer 2, early onset
- Brip1
interacting protein C-terminal helicase 1
- PDK4
pyruvate dehydrogenase kinase, isozyme 4
Footnotes
All work was performed at the University of North Carolina at Chapel Hill
Conflict of Interest
The authors have declared that no conflict of interest exists (M.W., J.R., K.W., J.S.,P.L., E.H., D.G., C.P.).
References
- 1.Gregorio CC, Perry CN, McElhinny AS. Functional properties of the titin/connectin-associated proteins, the muscle-specific RING finger proteins (MURFs), in striated muscle. J Muscle Res Cell Motil. 2005;26:389–400. doi: 10.1007/s10974-005-9021-x. [DOI] [PubMed] [Google Scholar]
- 2.Kedar V, McDonough H, Arya R, Li HH, Rockman HA, Patterson C. Muscle-specific RING finger 1 is a bona fide ubiquitin ligase that degrades cardiac troponin I. Proc Natl Acad Sci U S A. 2004;101:18135–18140. doi: 10.1073/pnas.0404341102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McElhinny AS, Kakinuma K, Sorimachi H, Labeit S, Gregorio CC. Muscle-specific RING finger-1 interacts with titin to regulate sarcomeric M-line and thick filament structure and may have nuclear functions via its interaction with glucocorticoid modulatory element binding protein-1. J Cell Biol. 2002;157:125–136. doi: 10.1083/jcb.200108089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoshijima M. Mechanical stress-strain sensors embedded in cardiac cytoskeleton: Z disk, titin, and associated structures. Am J Physiol Heart Circ Physiol. 2006;290:H1313–1325. doi: 10.1152/ajpheart.00816.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McElhinny AS, Perry CN, Witt CC, Labeit S, Gregorio CC. Muscle-specific RING finger-2 (MURF-2) is important for microtubule, intermediate filament and sarcomeric M-line maintenance in striated muscle development. J Cell Sci. 2004;117:3175–3188. doi: 10.1242/jcs.01158. [DOI] [PubMed] [Google Scholar]
- 6.Lange S, Xiang F, Yakovenko A, Vihola A, Hackman P, Rostkova E, Kristensen J, Brandmeier B, Franzen G, Hedberg B, Gunnarsson LG, Hughes SM, Marchand S, Sejersen T, Richard I, Edstrom L, Ehler E, Udd B, Gautel M. The kinase domain of titin controls muscle gene expression and protein turnover. Science. 2005;308:1599–1603. doi: 10.1126/science.1110463. [DOI] [PubMed] [Google Scholar]
- 7.Willis MS, Ike C, Li L, Wang DZ, Glass DJ, Patterson C. Muscle ring finger 1, but not muscle ring finger 2, regulates cardiac hypertrophy in vivo. Circ Res. 2007;100:456–459. doi: 10.1161/01.RES.0000259559.48597.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Centner T, Yano J, Kimura E, McElhinny AS, Pelin K, Witt CC, Bang ML, Trombitas K, Granzier H, Gregorio CC, Sorimachi H, Labeit S. Identification of muscle specific ring finger proteins as potential regulators of the titin kinase domain. J Mol Biol. 2001;306:717–726. doi: 10.1006/jmbi.2001.4448. [DOI] [PubMed] [Google Scholar]
- 9.Witt SH, Granzier H, Witt CC, Labeit S. MURF-1 and MURF-2 target a specific subset of myofibrillar proteins redundantly: towards understanding MURF-dependent muscle ubiquitination. J Mol Biol. 2005;350:713–722. doi: 10.1016/j.jmb.2005.05.021. [DOI] [PubMed] [Google Scholar]
- 10.He XR, Zhang C, Patterson C. Universal mouse reference RNA derived from neonatal mice. Biotechniques. 2004;37:464–468. doi: 10.2144/04373RT02. [DOI] [PubMed] [Google Scholar]
- 11.Riva A, Carpentier AS, Torresani B, Henaut A. Comments on selected fundamental aspects of microarray analysis. Comput Biol Chem. 2005;29:319–336. doi: 10.1016/j.compbiolchem.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 12.Smyth GK, Speed T. Normalization of cDNA microarray data. Methods. 2003;31:265–273. doi: 10.1016/s1046-2023(03)00155-5. [DOI] [PubMed] [Google Scholar]
- 13.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 14.Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- 15.Brazma A, Hingamp P, Quackenbush J, Sherlock G, Spellman P, Stoeckert C, Aach J, Ansorge W, Ball CA, Causton HC, Gaasterland T, Glenisson P, Holstege FC, Kim IF, Markowitz V, Matese JC, Parkinson H, Robinson A, Sarkans U, Schulze-Kremer S, Stewart J, Taylor R, Vilo J, Vingron M. Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nat Genet. 2001;29:365–371. doi: 10.1038/ng1201-365. [DOI] [PubMed] [Google Scholar]
- 16.Radhakrishnan SK, Feliciano CS, Najmabadi F, Haegebarth A, Kandel ES, Tyner AL, Gartel AL. Constitutive expression of E2F-1 leads to p21-dependent cell cycle arrest in S phase of the cell cycle. Oncogene. 2004;23:4173–4176. doi: 10.1038/sj.onc.1207571. [DOI] [PubMed] [Google Scholar]
- 17.Eelen G, Vanden Bempt I, Verlinden L, Drijkoningen M, Smeets A, Neven P, Christiaens MR, Marchal K, Bouillon R, Verstuyf A. Expression of the BRCA1-interacting protein Brip1/BACH1/FANCJ is driven by E2F and correlates with human breast cancer malignancy. Oncogene. 2008;27:4233–4241. doi: 10.1038/onc.2008.51. [DOI] [PubMed] [Google Scholar]
- 18.Hsieh MC, Das D, Sambandam N, Zhang MQ, Nahle Z. Regulation of the PDK4 isozyme by the Rb-E2F1 complex. J Biol Chem. 2008;283:27410–27417. doi: 10.1074/jbc.M802418200. [DOI] [PubMed] [Google Scholar]
- 19.Leu M, Ehler E, Perriard JC. Characterisation of postnatal growth of the murine heart. Anat Embryol (Berl) 2001;204:217–224. doi: 10.1007/s004290100206. [DOI] [PubMed] [Google Scholar]
- 20.Witt CC, Witt SH, Lerche S, Labeit D, Back W, Labeit S. Cooperative control of striated muscle mass and metabolism by MuRF1 and MuRF2. EMBO J. 2008;27:350–360. doi: 10.1038/sj.emboj.7601952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kowalik TF, DeGregori J, Leone G, Jakoi L, Nevins JR. E2F1-specific induction of apoptosis and p53 accumulation, which is blocked by Mdm2. Cell Growth Differ. 1998;9:113–118. [PubMed] [Google Scholar]
- 22.O’Connor DJ, Lam EW, Griffin S, Zhong S, Leighton LC, Burbidge SA, Lu X. Physical and functional interactions between p53 and cell cycle co-operating transcription factors, E2F1 and DP1. EMBO J. 1995;14:6184–6192. doi: 10.1002/j.1460-2075.1995.tb00309.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ohta T, Xiong Y. Phosphorylation- and Skp1-independent in vitro ubiquitination of E2F1 by multiple ROC-cullin ligases. Cancer Res. 2001;61:1347–1353. [PubMed] [Google Scholar]
- 24.Agah R, Kirshenbaum LA, Abdellatif M, Truong LD, Chakraborty S, Michael LH, Schneider MD. Adenoviral delivery of E2F-1 directs cell cycle reentry and p53-independent apoptosis in postmitotic adult myocardium in vivo. J Clin Invest. 1997;100:2722–2728. doi: 10.1172/JCI119817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kirshenbaum LA, Abdellatif M, Chakraborty S, Schneider MD. Human E2F-1 reactivates cell cycle progression in ventricular myocytes and represses cardiac gene transcription. Dev Biol. 1996;179:402–411. doi: 10.1006/dbio.1996.0270. [DOI] [PubMed] [Google Scholar]
- 26.Angelis E, Garcia A, Chan SS, Schenke-Layland K, Ren S, Goodfellow SJ, Jordan MC, Roos KP, White RJ, MacLellan WR. A cyclin D2-Rb pathway regulates cardiac myocyte size and RNA polymerase III after biomechanical stress in adult myocardium. Circ Res. 2008;102:1222–1229. doi: 10.1161/CIRCRESAHA.107.163550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ebelt H, Liu Z, Muller-Werdan U, Werdan K, Braun T. Making omelets without breaking eggs: E2F-mediated induction of cardiomyoycte cell proliferation without stimulation of apoptosis. Cell Cycle. 2006;5:2436–2439. doi: 10.4161/cc.5.21.3402. [DOI] [PubMed] [Google Scholar]
- 28.Yurkova N, Shaw J, Blackie K, Weidman D, Jayas R, Flynn B, Kirshenbaum LA. The cell cycle factor E2F-1 activates Bnip3 and the intrinsic death pathway in ventricular myocytes. Circ Res. 2008;102:472–479. doi: 10.1161/CIRCRESAHA.107.164731. [DOI] [PubMed] [Google Scholar]
- 29.Li HH, Du J, Fan YN, Zhang ML, Liu DP, Li L, Lockyer P, Kang EY, Patterson C, Willis MS. The Ubiquitin Ligase MuRF1 Protects Against Cardiac Ischemia/Reperfusion Injury by Its Proteasome-Dependent Degradation of Phospho-c-Jun. Am J Pathol. 2011;178:1043–1058. doi: 10.1016/j.ajpath.2010.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rodriguez JE, Li L, Schisler JC, Patterson C, Willis MS. The cardiac ubiquitin ligase muscle ring finger-1 (MuRF1) ubiquitinates and degrades PPAR-alpha to regulate fatty acid and glucose metabolism. Circulation. 2009;120:S854–855. [Google Scholar]
- 31.Rodríguez JE, Li L, Willis MS. Muscle ring finger-1 regulates cardiac fatty acid and glucose metabolism via its interaction with PPARalpha. FASEB J. 2010;24:38.33. [Google Scholar]
- 32.Ahuja P, Sdek P, MacLellan WR. Cardiac myocyte cell cycle control in development, disease, and regeneration. Physiol Rev. 2007;87:521–544. doi: 10.1152/physrev.00032.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cloud JE, Rogers C, Reza TL, Ziebold U, Stone JR, Picard MH, Caron AM, Bronson RT, Lees JA. Mutant mouse models reveal the relative roles of E2F1 and E2F3 in vivo. Mol Cell Biol. 2002;22:2663–2672. doi: 10.1128/MCB.22.8.2663-2672.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tennstedt C, Chaoui R, Korner H, Dietel M. Spectrum of congenital heart defects and extracardiac malformations associated with chromosomal abnormalities: results of a seven year necropsy study. Heart. 1999;82:34–39. doi: 10.1136/hrt.82.1.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yoon PO, Lee MA, Cha H, Jeong MH, Kim J, Jang SP, Choi BY, Jeong D, Yang DK, Hajjar RJ, Park WJ. The opposing effects of CCN2 and CCN5 on the development of cardiac hypertrophy and fibrosis. J Mol Cell Cardiol. 2010;49:294–303. doi: 10.1016/j.yjmcc.2010.04.010. [DOI] [PubMed] [Google Scholar]
- 36.Panek AN, Posch MG, Alenina N, Ghadge SK, Erdmann B, Popova E, Perrot A, Geier C, Dietz R, Morano I, Bader M, Ozcelik C. Connective tissue growth factor overexpression in cardiomyocytes promotes cardiac hypertrophy and protection against pressure overload. PLoS One. 2009;4:e6743. doi: 10.1371/journal.pone.0006743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jacoby JJ, Kalinowski A, Liu MG, Zhang SS, Gao Q, Chai GX, Ji L, Iwamoto Y, Li E, Schneider M, Russell KS, Fu XY. Cardiomyocyte-restricted knockout of STAT3 results in higher sensitivity to inflammation, cardiac fibrosis, and heart failure with advanced age. Proc Natl Acad Sci U S A. 2003;100:12929–12934. doi: 10.1073/pnas.2134694100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takeda N, Manabe I. Cellular Interplay between Cardiomyocytes and Nonmyocytes in Cardiac Remodeling. Int J Inflam. 2011;2011:535241. doi: 10.4061/2011/535241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koitabashi N, Danner T, Zaiman AL, Pinto YM, Rowell J, Mankowski J, Zhang D, Nakamura T, Takimoto E, Kass DA. Pivotal role of cardiomyocyte TGF-beta signaling in the murine pathological response to sustained pressure overload. J Clin Invest. 2011;121:2301–2312. doi: 10.1172/JCI44824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Manabe I, Shindo T, Nagai R. Gene expression in fibroblasts and fibrosis: involvement in cardiac hypertrophy. Circ Res. 2002;91:1103–1113. doi: 10.1161/01.res.0000046452.67724.b8. [DOI] [PubMed] [Google Scholar]
- 41.Segura AM, Frazier OH, Buja LM. Fibrosis and heart failure. Heart Fail Rev. 2012 doi: 10.1007/s10741-012-9365-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.