Abstract
The genetic code, formerly thought to be frozen, is now known to be in a state of evolution. This was first shown in 1979 by Barrell et al. (G. Barrell, A. T. Bankier, and J. Drouin, Nature [London] 282:189-194, 1979), who found that the universal codons AUA (isoleucine) and UGA (stop) coded for methionine and tryptophan, respectively, in human mitochondria. Subsequent studies have shown that UGA codes for tryptophan in Mycoplasma spp. and in all nonplant mitochondria that have been examined. Universal stop codons UAA and UAG code for glutamine in ciliated protozoa (except Euplotes octacarinatus) and in a green alga, Acetabularia. E. octacarinatus uses UAA for stop and UGA for cysteine. Candida species, which are yeasts, use CUG (leucine) for serine. Other departures from the universal code, all in nonplant mitochondria, are CUN (leucine) for threonine (in yeasts), AAA (lysine) for asparagine (in platyhelminths and echinoderms), UAA (stop) for tyrosine (in planaria), and AGR (arginine) for serine (in several animal orders) and for stop (in vertebrates). We propose that the changes are typically preceded by loss of a codon from all coding sequences in an organism or organelle, often as a result of directional mutation pressure, accompanied by loss of the tRNA that translates the codon. The codon reappears later by conversion of another codon and emergence of a tRNA that translates the reappeared codon with a different assignment. Changes in release factors also contribute to these revised assignments. We also discuss the use of UGA (stop) as a selenocysteine codon and the early history of the code.
Full text
PDF

















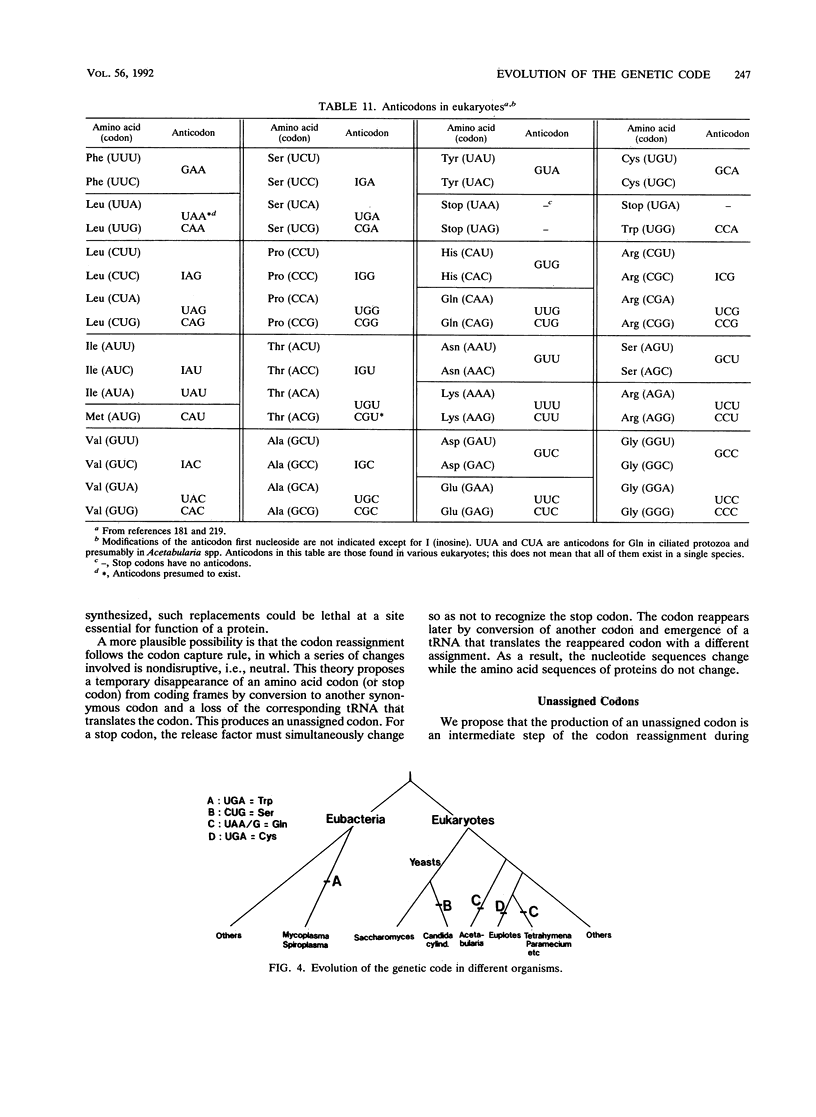





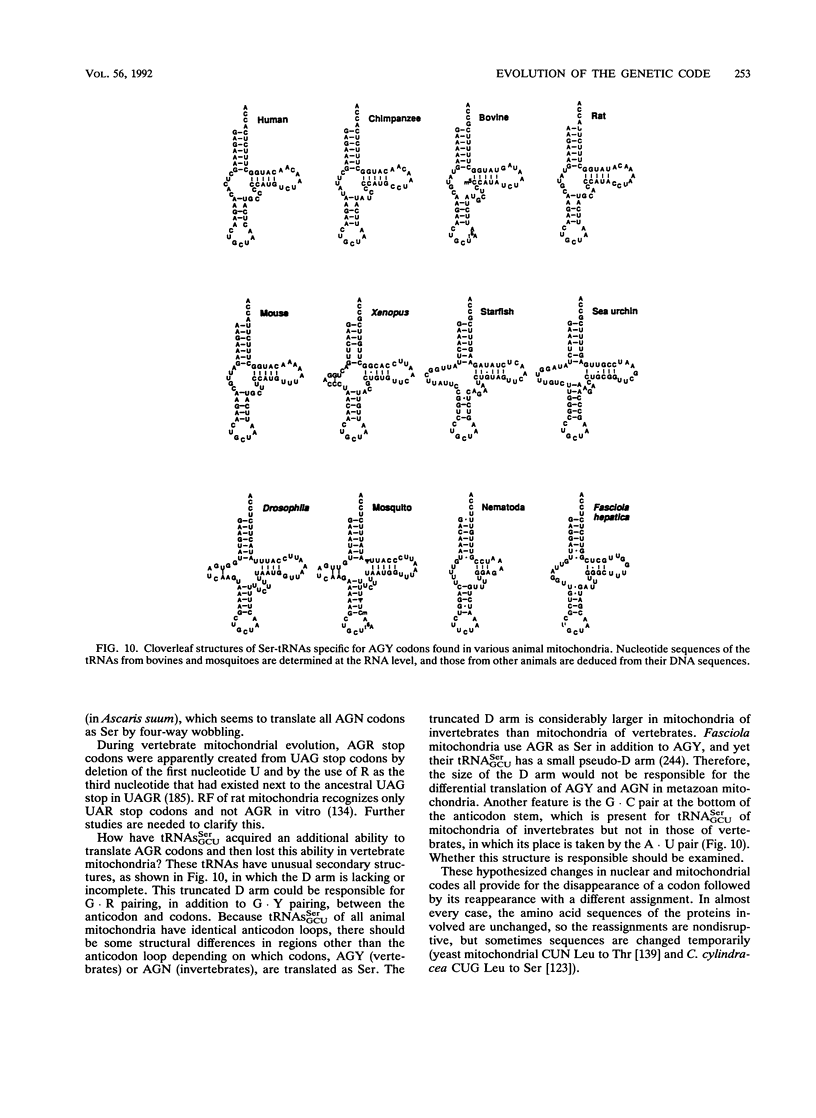











Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ainley W. M., Macreadie I. G., Butow R. A. var1 Gene on the mitochondrial genome of Torulopsis glabrata. J Mol Biol. 1985 Aug 20;184(4):565–576. doi: 10.1016/0022-2836(85)90303-1. [DOI] [PubMed] [Google Scholar]
- Akiyama M., Maki H., Sekiguchi M., Horiuchi T. A specific role of MutT protein: to prevent dG.dA mispairing in DNA replication. Proc Natl Acad Sci U S A. 1989 Jun;86(11):3949–3952. doi: 10.1073/pnas.86.11.3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andachi Y., Yamao F., Iwami M., Muto A., Osawa S. Occurrence of unmodified adenine and uracil at the first position of anticodon in threonine tRNAs in Mycoplasma capricolum. Proc Natl Acad Sci U S A. 1987 Nov;84(21):7398–7402. doi: 10.1073/pnas.84.21.7398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andachi Y., Yamao F., Muto A., Osawa S. Codon recognition patterns as deduced from sequences of the complete set of transfer RNA species in Mycoplasma capricolum. Resemblance to mitochondria. J Mol Biol. 1989 Sep 5;209(1):37–54. doi: 10.1016/0022-2836(89)90168-x. [DOI] [PubMed] [Google Scholar]
- Anderson S., Bankier A. T., Barrell B. G., de Bruijn M. H., Coulson A. R., Drouin J., Eperon I. C., Nierlich D. P., Roe B. A., Sanger F. Sequence and organization of the human mitochondrial genome. Nature. 1981 Apr 9;290(5806):457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- Anderson S., de Bruijn M. H., Coulson A. R., Eperon I. C., Sanger F., Young I. G. Complete sequence of bovine mitochondrial DNA. Conserved features of the mammalian mitochondrial genome. J Mol Biol. 1982 Apr 25;156(4):683–717. doi: 10.1016/0022-2836(82)90137-1. [DOI] [PubMed] [Google Scholar]
- Anfinsen C. B., Corley L. G. An active variant of staphylococcal nuclease containing norleucine in place of methionine. J Biol Chem. 1969 Oct 10;244(19):5149–5152. [PubMed] [Google Scholar]
- Aota S., Ikemura T. Diversity in G + C content at the third position of codons in vertebrate genes and its cause. Nucleic Acids Res. 1986 Aug 26;14(16):6345–6355. doi: 10.1093/nar/14.16.6345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asakawa S., Kumazawa Y., Araki T., Himeno H., Miura K., Watanabe K. Strand-specific nucleotide composition bias in echinoderm and vertebrate mitochondrial genomes. J Mol Evol. 1991 Jun;32(6):511–520. doi: 10.1007/BF02102653. [DOI] [PubMed] [Google Scholar]
- BOROSOOK H., DEASY C. L., HAAGEN-SMIT A. J., KEIGHLEY G., LOWY P. H. The uptake in vitro of C14-labeled glycine, l-leucine, and l-lysine by different components of guinea pig liver homogenate. J Biol Chem. 1950 Jun;184(2):529–543. [PubMed] [Google Scholar]
- Barahona I., Soares H., Cyrne L., Penque D., Denoulet P., Rodrigues-Pousada C. Sequence of one alpha- and two beta-tubulin genes of Tetrahymena pyriformis. Structural and functional relationships with other eukaryotic tubulin genes. J Mol Biol. 1988 Aug 5;202(3):365–382. doi: 10.1016/0022-2836(88)90271-9. [DOI] [PubMed] [Google Scholar]
- Baron C., Heider J., Böck A. Mutagenesis of selC, the gene for the selenocysteine-inserting tRNA-species in E. coli: effects on in vivo function. Nucleic Acids Res. 1990 Dec 11;18(23):6761–6766. doi: 10.1093/nar/18.23.6761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrell B. G., Anderson S., Bankier A. T., de Bruijn M. H., Chen E., Coulson A. R., Drouin J., Eperon I. C., Nierlich D. P., Roe B. A. Different pattern of codon recognition by mammalian mitochondrial tRNAs. Proc Natl Acad Sci U S A. 1980 Jun;77(6):3164–3166. doi: 10.1073/pnas.77.6.3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrell B. G., Bankier A. T., Drouin J. A different genetic code in human mitochondria. Nature. 1979 Nov 8;282(5735):189–194. doi: 10.1038/282189a0. [DOI] [PubMed] [Google Scholar]
- Bernardi G., Bernardi G. Compositional constraints and genome evolution. J Mol Evol. 1986;24(1-2):1–11. doi: 10.1007/BF02099946. [DOI] [PubMed] [Google Scholar]
- Bernardi G., Olofsson B., Filipski J., Zerial M., Salinas J., Cuny G., Meunier-Rotival M., Rodier F. The mosaic genome of warm-blooded vertebrates. Science. 1985 May 24;228(4702):953–958. doi: 10.1126/science.4001930. [DOI] [PubMed] [Google Scholar]
- Bernardi G. The isochore organization of the human genome. Annu Rev Genet. 1989;23:637–661. doi: 10.1146/annurev.ge.23.120189.003225. [DOI] [PubMed] [Google Scholar]
- Berry M. J., Banu L., Chen Y. Y., Mandel S. J., Kieffer J. D., Harney J. W., Larsen P. R. Recognition of UGA as a selenocysteine codon in type I deiodinase requires sequences in the 3' untranslated region. Nature. 1991 Sep 19;353(6341):273–276. doi: 10.1038/353273a0. [DOI] [PubMed] [Google Scholar]
- Berry M. J., Banu L., Larsen P. R. Type I iodothyronine deiodinase is a selenocysteine-containing enzyme. Nature. 1991 Jan 31;349(6308):438–440. doi: 10.1038/349438a0. [DOI] [PubMed] [Google Scholar]
- Bibb M. J., Van Etten R. A., Wright C. T., Walberg M. W., Clayton D. A. Sequence and gene organization of mouse mitochondrial DNA. Cell. 1981 Oct;26(2 Pt 2):167–180. doi: 10.1016/0092-8674(81)90300-7. [DOI] [PubMed] [Google Scholar]
- Brown C. M., Stockwell P. A., Trotman C. N., Tate W. P. Sequence analysis suggests that tetra-nucleotides signal the termination of protein synthesis in eukaryotes. Nucleic Acids Res. 1990 Nov 11;18(21):6339–6345. doi: 10.1093/nar/18.21.6339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown C. M., Stockwell P. A., Trotman C. N., Tate W. P. The signal for the termination of protein synthesis in procaryotes. Nucleic Acids Res. 1990 Apr 25;18(8):2079–2086. doi: 10.1093/nar/18.8.2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning K. S., RajBhandary U. L. Cytochrome oxidase subunit III gene in Neurospora crassa mitochondria. Location and sequence. J Biol Chem. 1982 May 10;257(9):5253–5256. [PubMed] [Google Scholar]
- Burger G., Scriven C., Machleidt W., Werner S. Subunit 1 of cytochrome oxidase from Neurospora crassa: nucleotide sequence of the coding gene and partial amino acid sequence of the protein. EMBO J. 1982;1(11):1385–1391. doi: 10.1002/j.1460-2075.1982.tb01327.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böck A., Forchhammer K., Heider J., Leinfelder W., Sawers G., Veprek B., Zinoni F. Selenocysteine: the 21st amino acid. Mol Microbiol. 1991 Mar;5(3):515–520. doi: 10.1111/j.1365-2958.1991.tb00722.x. [DOI] [PubMed] [Google Scholar]
- Böck A., Stadtman T. C. Selenocysteine, a highly specific component of certain enzymes, is incorporated by a UGA-directed co-translational mechanism. Biofactors. 1988 Oct;1(3):245–250. [PubMed] [Google Scholar]
- COWIE D. B., COHEN G. N., BOLTON E. T., DE ROBICHON-SZULMAJSTER H. Amino acid analog incorporation into bacterial proteins. Biochim Biophys Acta. 1959 Jul;34:39–46. doi: 10.1016/0006-3002(59)90230-6. [DOI] [PubMed] [Google Scholar]
- Canaday J., Dirheimer G., Martin R. P. Yeast mitochondrial methionine initiator tRNA: characterization and nucleotide sequence. Nucleic Acids Res. 1980 Apr 11;8(7):1445–1457. doi: 10.1093/nar/8.7.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantatore P., Roberti M., Rainaldi G., Gadaleta M. N., Saccone C. The complete nucleotide sequence, gene organization, and genetic code of the mitochondrial genome of Paracentrotus lividus. J Biol Chem. 1989 Jul 5;264(19):10965–10975. [PubMed] [Google Scholar]
- Caron F., Meyer E. Does Paramecium primaurelia use a different genetic code in its macronucleus? Nature. 1985 Mar 14;314(6007):185–188. doi: 10.1038/314185a0. [DOI] [PubMed] [Google Scholar]
- Cerretti D. P., Dean D., Davis G. R., Bedwell D. M., Nomura M. The spc ribosomal protein operon of Escherichia coli: sequence and cotranscription of the ribosomal protein genes and a protein export gene. Nucleic Acids Res. 1983 May 11;11(9):2599–2616. doi: 10.1093/nar/11.9.2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers I., Frampton J., Goldfarb P., Affara N., McBain W., Harrison P. R. The structure of the mouse glutathione peroxidase gene: the selenocysteine in the active site is encoded by the 'termination' codon, TGA. EMBO J. 1986 Jun;5(6):1221–1227. doi: 10.1002/j.1460-2075.1986.tb04350.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevalier C., Saillard C., Bové J. M. Organization and nucleotide sequences of the Spiroplasma citri genes for ribosomal protein S2, elongation factor Ts, spiralin, phosphofructokinase, pyruvate kinase, and an unidentified protein. J Bacteriol. 1990 May;172(5):2693–2703. doi: 10.1128/jb.172.5.2693-2703.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark-Walker G. D., McArthur C. R., Sriprakash K. S. Location of transcriptional control signals and transfer RNA sequences in Torulopsis glabrata mitochondrial DNA. EMBO J. 1985 Feb;4(2):465–473. doi: 10.1002/j.1460-2075.1985.tb03652.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clary D. O., Wolstenholme D. R. The mitochondrial DNA molecular of Drosophila yakuba: nucleotide sequence, gene organization, and genetic code. J Mol Evol. 1985;22(3):252–271. doi: 10.1007/BF02099755. [DOI] [PubMed] [Google Scholar]
- Cone J. E., Del Río R. M., Davis J. N., Stadtman T. C. Chemical characterization of the selenoprotein component of clostridial glycine reductase: identification of selenocysteine as the organoselenium moiety. Proc Natl Acad Sci U S A. 1976 Aug;73(8):2659–2663. doi: 10.1073/pnas.73.8.2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox E. C., Yanofsky C. Altered base ratios in the DNA of an Escherichia coli mutator strain. Proc Natl Acad Sci U S A. 1967 Nov;58(5):1895–1902. doi: 10.1073/pnas.58.5.1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crick F. H., Brenner S., Klug A., Pieczenik G. A speculation on the origin of protein synthesis. Orig Life. 1976 Dec;7(4):389–397. doi: 10.1007/BF00927934. [DOI] [PubMed] [Google Scholar]
- Crick F. H. Codon--anticodon pairing: the wobble hypothesis. J Mol Biol. 1966 Aug;19(2):548–555. doi: 10.1016/s0022-2836(66)80022-0. [DOI] [PubMed] [Google Scholar]
- Crick F. H., Griffith J. S., Orgel L. E. CODES WITHOUT COMMAS. Proc Natl Acad Sci U S A. 1957 May 15;43(5):416–421. doi: 10.1073/pnas.43.5.416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crick F. H. The origin of the genetic code. J Mol Biol. 1968 Dec;38(3):367–379. doi: 10.1016/0022-2836(68)90392-6. [DOI] [PubMed] [Google Scholar]
- Cronin J. R., Pizzarello S. Amino acids in meteorites. Adv Space Res. 1983;3(9):5–18. doi: 10.1016/0273-1177(83)90036-4. [DOI] [PubMed] [Google Scholar]
- DOUNCE A. L. [Duplicating mechanism for peptide chain and nucleic acid synthesis]. Enzymologia. 1952 Sep 1;15(5):251–258. [PubMed] [Google Scholar]
- Desjardins P., Morais R. Sequence and gene organization of the chicken mitochondrial genome. A novel gene order in higher vertebrates. J Mol Biol. 1990 Apr 20;212(4):599–634. doi: 10.1016/0022-2836(90)90225-B. [DOI] [PubMed] [Google Scholar]
- Diamond A., Dudock B., Hatfield D. Structure and properties of a bovine liver UGA suppressor serine tRNA with a tryptophan anticodon. Cell. 1981 Aug;25(2):497–506. doi: 10.1016/0092-8674(81)90068-4. [DOI] [PubMed] [Google Scholar]
- Dubin D. T., HsuChen C. C. Sequence and structure of a methionine transfer RNA from mosquito mitochondria. Nucleic Acids Res. 1984 May 25;12(10):4185–4189. doi: 10.1093/nar/12.10.4185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudler R., Schmidhauser C., Parish R. W., Wettenhall R. E., Schmidt T. A mycoplasma high-affinity transport system and the in vitro invasiveness of mouse sarcoma cells. EMBO J. 1988 Dec 1;7(12):3963–3970. doi: 10.1002/j.1460-2075.1988.tb03283.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eigen M., Lindemann B. F., Tietze M., Winkler-Oswatitsch R., Dress A., von Haeseler A. How old is the genetic code? Statistical geometry of tRNA provides an answer. Science. 1989 May 12;244(4905):673–679. doi: 10.1126/science.2497522. [DOI] [PubMed] [Google Scholar]
- Elliott M. S., Trewyn R. W. Inosine biosynthesis in transfer RNA by an enzymatic insertion of hypoxanthine. J Biol Chem. 1984 Feb 25;259(4):2407–2410. [PubMed] [Google Scholar]
- Fiers W., Contreras R., Duerinck F., Haegeman G., Iserentant D., Merregaert J., Min Jou W., Molemans F., Raeymaekers A., Van den Berghe A. Complete nucleotide sequence of bacteriophage MS2 RNA: primary and secondary structure of the replicase gene. Nature. 1976 Apr 8;260(5551):500–507. doi: 10.1038/260500a0. [DOI] [PubMed] [Google Scholar]
- Flohe L., Günzler W. A., Schock H. H. Glutathione peroxidase: a selenoenzyme. FEBS Lett. 1973 May 15;32(1):132–134. doi: 10.1016/0014-5793(73)80755-0. [DOI] [PubMed] [Google Scholar]
- Forchhammer K., Böck A. Selenocysteine synthase from Escherichia coli. Analysis of the reaction sequence. J Biol Chem. 1991 Apr 5;266(10):6324–6328. [PubMed] [Google Scholar]
- Forstrom J. W., Zakowski J. J., Tappel A. L. Identification of the catalytic site of rat liver glutathione peroxidase as selenocysteine. Biochemistry. 1978 Jun 27;17(13):2639–2644. doi: 10.1021/bi00606a028. [DOI] [PubMed] [Google Scholar]
- Fox T. D. Natural variation in the genetic code. Annu Rev Genet. 1987;21:67–91. doi: 10.1146/annurev.ge.21.120187.000435. [DOI] [PubMed] [Google Scholar]
- Gadaleta G., Pepe G., De Candia G., Quagliariello C., Sbisà E., Saccone C. The complete nucleotide sequence of the Rattus norvegicus mitochondrial genome: cryptic signals revealed by comparative analysis between vertebrates. J Mol Evol. 1989 Jun;28(6):497–516. doi: 10.1007/BF02602930. [DOI] [PubMed] [Google Scholar]
- Garesse R. Drosophila melanogaster mitochondrial DNA: gene organization and evolutionary considerations. Genetics. 1988 Apr;118(4):649–663. doi: 10.1093/genetics/118.4.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garey J. R., Wolstenholme D. R. Platyhelminth mitochondrial DNA: evidence for early evolutionary origin of a tRNA(serAGN) that contains a dihydrouridine arm replacement loop, and of serine-specifying AGA and AGG codons. J Mol Evol. 1989 May;28(5):374–387. doi: 10.1007/BF02603072. [DOI] [PubMed] [Google Scholar]
- Grisi E., Brown T. A., Waring R. B., Scazzocchio C., Davies R. W. Nucleotide sequence of a region of the mitochondrial genome of Aspergillus nidulans including the gene for ATPase subunit 6. Nucleic Acids Res. 1982 Jun 11;10(11):3531–3539. doi: 10.1093/nar/10.11.3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivell L. A. Molecular evolution. Deciphering divergent codes. Nature. 1986 Nov 13;324(6093):109–110. doi: 10.1038/324109a0. [DOI] [PubMed] [Google Scholar]
- Gupta R. Halobacterium volcanii tRNAs. Identification of 41 tRNAs covering all amino acids, and the sequences of 33 class I tRNAs. J Biol Chem. 1984 Aug 10;259(15):9461–9471. [PubMed] [Google Scholar]
- Günzler W. A., Steffens G. J., Grossmann A., Kim S. M., Otting F., Wendel A., Flohé L. The amino-acid sequence of bovine glutathione peroxidase. Hoppe Seylers Z Physiol Chem. 1984 Feb;365(2):195–212. doi: 10.1515/bchm2.1984.365.1.195. [DOI] [PubMed] [Google Scholar]
- Hanyu N., Kuchino Y., Nishimura S., Beier H. Dramatic events in ciliate evolution: alteration of UAA and UAG termination codons to glutamine codons due to anticodon mutations in two Tetrahymena tRNAs. EMBO J. 1986 Jun;5(6):1307–1311. doi: 10.1002/j.1460-2075.1986.tb04360.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper D. S., Jahn C. L. Differential use of termination codons in ciliated protozoa. Proc Natl Acad Sci U S A. 1989 May;86(9):3252–3256. doi: 10.1073/pnas.86.9.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmanis M. G., Stadtman T. C. Isolation of a selenium-containing thiolase from Clostridium kluyveri: identification of the selenium moiety as selenomethionine. Proc Natl Acad Sci U S A. 1982 Aug;79(16):4912–4916. doi: 10.1073/pnas.79.16.4912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatfield D., Diamond A., Dudock B. Opal suppressor serine tRNAs from bovine liver form phosphoseryl-tRNA. Proc Natl Acad Sci U S A. 1982 Oct;79(20):6215–6219. doi: 10.1073/pnas.79.20.6215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckman J. E., Sarnoff J., Alzner-DeWeerd B., Yin S., RajBhandary U. L. Novel features in the genetic code and codon reading patterns in Neurospora crassa mitochondria based on sequences of six mitochondrial tRNAs. Proc Natl Acad Sci U S A. 1980 Jun;77(6):3159–3163. doi: 10.1073/pnas.77.6.3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heider J., Leinfelder W., Böck A. Occurrence and functional compatibility within Enterobacteriaceae of a tRNA species which inserts selenocysteine into protein. Nucleic Acids Res. 1989 Apr 11;17(7):2529–2540. doi: 10.1093/nar/17.7.2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helftenbein E., Müller E. Both alpha-tubulin genes are transcriptionally active in Stylonychia lemnae. Curr Genet. 1988 May;13(5):425–432. doi: 10.1007/BF00365664. [DOI] [PubMed] [Google Scholar]
- Helftenbein E. Nucleotide sequence of a macronuclear DNA molecule coding for alpha-tubulin from the ciliate Stylonychia lemnae. Special codon usage: TAA is not a translation termination codon. Nucleic Acids Res. 1985 Jan 25;13(2):415–433. doi: 10.1093/nar/13.2.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendriks L., Van Broeckhoven C., Vandenberghe A., Van de Peer Y., De Wachter R. Primary and secondary structure of the 18S ribosomal RNA of the bird spider Eurypelma californica and evolutionary relationships among eukaryotic phyla. Eur J Biochem. 1988 Oct 15;177(1):15–20. doi: 10.1111/j.1432-1033.1988.tb14339.x. [DOI] [PubMed] [Google Scholar]
- Hensgens L. A., Brakenhoff J., De Vries B. F., Sloof P., Tromp M. C., Van Boom J. H., Benne R. The sequence of the gene for cytochrome c oxidase subunit I, a frameshift containing gene for cytochrome c oxidase subunit II and seven unassigned reading frames in Trypanosoma brucei mitochondrial maxi-circle DNA. Nucleic Acids Res. 1984 Oct 11;12(19):7327–7344. doi: 10.1093/nar/12.19.7327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hensgens L. A., Grivell L. A., Borst P., Bos J. L. Nucleotide sequence of the mitochondrial structural gene for subunit 9 of yeast ATPase complex. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1663–1667. doi: 10.1073/pnas.76.4.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrick G., Hunter D., Williams K., Kotter K. Alternative processing during development of a macronuclear chromosome family in Oxytricha fallax. Genes Dev. 1987 Dec;1(10):1047–1058. doi: 10.1101/gad.1.10.1047. [DOI] [PubMed] [Google Scholar]
- Hill K. E., Lloyd R. S., Yang J. G., Read R., Burk R. F. The cDNA for rat selenoprotein P contains 10 TGA codons in the open reading frame. J Biol Chem. 1991 Jun 5;266(16):10050–10053. [PubMed] [Google Scholar]
- Himeno H., Masaki H., Kawai T., Ohta T., Kumagai I., Miura K., Watanabe K. Unusual genetic codes and a novel gene structure for tRNA(AGYSer) in starfish mitochondrial DNA. Gene. 1987;56(2-3):219–230. doi: 10.1016/0378-1119(87)90139-9. [DOI] [PubMed] [Google Scholar]
- Hiratsuka J., Shimada H., Whittier R., Ishibashi T., Sakamoto M., Mori M., Kondo C., Honji Y., Sun C. R., Meng B. Y. The complete sequence of the rice (Oryza sativa) chloroplast genome: intermolecular recombination between distinct tRNA genes accounts for a major plastid DNA inversion during the evolution of the cereals. Mol Gen Genet. 1989 Jun;217(2-3):185–194. doi: 10.1007/BF02464880. [DOI] [PubMed] [Google Scholar]
- Holmquist G. P. Evolution of chromosome bands: molecular ecology of noncoding DNA. J Mol Evol. 1989 Jun;28(6):469–486. doi: 10.1007/BF02602928. [DOI] [PubMed] [Google Scholar]
- Holmquist R., Jukes T. H., Pangburn S. Evolution of transfer RNA. J Mol Biol. 1973 Jun 25;78(1):91–116. doi: 10.1016/0022-2836(73)90430-0. [DOI] [PubMed] [Google Scholar]
- Hori H., Osawa S. Evolutionary change in 5S RNA secondary structure and a phylogenic tree of 54 5S RNA species. Proc Natl Acad Sci U S A. 1979 Jan;76(1):381–385. doi: 10.1073/pnas.76.1.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori H., Osawa S. Origin and evolution of organisms as deduced from 5S ribosomal RNA sequences. Mol Biol Evol. 1987 Sep;4(5):445–472. doi: 10.1093/oxfordjournals.molbev.a040455. [DOI] [PubMed] [Google Scholar]
- Horowitz S., Gorovsky M. A. An unusual genetic code in nuclear genes of Tetrahymena. Proc Natl Acad Sci U S A. 1985 Apr;82(8):2452–2455. doi: 10.1073/pnas.82.8.2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HsuChen C. C., Cleaves G. R., Dubin D. T. A major lysine tRNA with a CUU anticodon in insect mitochondria. Nucleic Acids Res. 1983 Dec 20;11(24):8659–8662. doi: 10.1093/nar/11.24.8659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HsuChen C. C., Dubin D. T. A cluster of four transfer RNA genes in mosquito mitochondrial DNA. Biochem Int. 1984 Mar;8(3):385–391. [PubMed] [Google Scholar]
- HsuChen C. C., Kotin R. M., Dubin D. T. Sequences of the coding and flanking regions of the large ribosomal subunit RNA gene of mosquito mitochondria. Nucleic Acids Res. 1984 Oct 25;12(20):7771–7785. doi: 10.1093/nar/12.20.7771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudspeth M. E., Ainley W. M., Shumard D. S., Butow R. A., Grossman L. I. Location and structure of the var1 gene on yeast mitochondrial DNA: nucleotide sequence of the 40.0 allele. Cell. 1982 Sep;30(2):617–626. doi: 10.1016/0092-8674(82)90258-6. [DOI] [PubMed] [Google Scholar]
- Ikeda R., Ohama T., Muto A., Osawa S. Nucleotide sequences of nine tRNA genes from Micrococcus luteus. Nucleic Acids Res. 1990 Dec 11;18(23):7154–7154. doi: 10.1093/nar/18.23.7154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda R., Ohama T., Muto A., Osawa S. Nucleotide sequences of two tRNA gene clusters from Micrococcus luteus. Nucleic Acids Res. 1990 Dec 11;18(23):7155–7155. doi: 10.1093/nar/18.23.7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikemura T., Aota S. Global variation in G+C content along vertebrate genome DNA. Possible correlation with chromosome band structures. J Mol Biol. 1988 Sep 5;203(1):1–13. doi: 10.1016/0022-2836(88)90086-1. [DOI] [PubMed] [Google Scholar]
- Ikemura T. Correlation between the abundance of Escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes. J Mol Biol. 1981 Feb 15;146(1):1–21. doi: 10.1016/0022-2836(81)90363-6. [DOI] [PubMed] [Google Scholar]
- Ikemura T. Correlation between the abundance of Escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes: a proposal for a synonymous codon choice that is optimal for the E. coli translational system. J Mol Biol. 1981 Sep 25;151(3):389–409. doi: 10.1016/0022-2836(81)90003-6. [DOI] [PubMed] [Google Scholar]
- Ikemura T. Correlation between the abundance of yeast transfer RNAs and the occurrence of the respective codons in protein genes. Differences in synonymous codon choice patterns of yeast and Escherichia coli with reference to the abundance of isoaccepting transfer RNAs. J Mol Biol. 1982 Jul 15;158(4):573–597. doi: 10.1016/0022-2836(82)90250-9. [DOI] [PubMed] [Google Scholar]
- Inamine J. M., Denny T. P., Loechel S., Schaper U., Huang C. H., Bott K. F., Hu P. C. Nucleotide sequence of the P1 attachment-protein gene of Mycoplasma pneumoniae. Gene. 1988 Apr 29;64(2):217–229. doi: 10.1016/0378-1119(88)90337-x. [DOI] [PubMed] [Google Scholar]
- Inamine J. M., Ho K. C., Loechel S., Hu P. C. Evidence that UGA is read as a tryptophan codon rather than as a stop codon by Mycoplasma pneumoniae, Mycoplasma genitalium, and Mycoplasma gallisepticum. J Bacteriol. 1990 Jan;172(1):504–506. doi: 10.1128/jb.172.1.504-506.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwabe N., Kuma K., Hasegawa M., Osawa S., Miyata T. Evolutionary relationship of archaebacteria, eubacteria, and eukaryotes inferred from phylogenetic trees of duplicated genes. Proc Natl Acad Sci U S A. 1989 Dec;86(23):9355–9359. doi: 10.1073/pnas.86.23.9355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs H. T., Asakawa S., Araki T., Miura K., Smith M. J., Watanabe K. Conserved tRNA gene cluster in starfish mitochondrial DNA. Curr Genet. 1989 Mar;15(3):193–206. doi: 10.1007/BF00435506. [DOI] [PubMed] [Google Scholar]
- Jacobs H. T., Elliott D. J., Math V. B., Farquharson A. Nucleotide sequence and gene organization of sea urchin mitochondrial DNA. J Mol Biol. 1988 Jul 20;202(2):185–217. doi: 10.1016/0022-2836(88)90452-4. [DOI] [PubMed] [Google Scholar]
- Jukes T. H. A change in the genetic code in Mycoplasma capricolum. J Mol Evol. 1985;22(4):361–362. doi: 10.1007/BF02115692. [DOI] [PubMed] [Google Scholar]
- Jukes T. H. Amino acid codes in mitochondria as possible clues to primitive codes. J Mol Evol. 1981;18(1):15–17. doi: 10.1007/BF01733206. [DOI] [PubMed] [Google Scholar]
- Jukes T. H., Bessho Y., Ohama T., Osawa S. Release factors and the genetic code. Nature. 1991 Aug 15;352(6336):575–575. doi: 10.1038/352575a0. [DOI] [PubMed] [Google Scholar]
- Jukes T. H., Bhushan V. Silent nucleotide substitutions and G + C content of some mitochondrial and bacterial genes. J Mol Evol. 1986;24(1-2):39–44. doi: 10.1007/BF02099949. [DOI] [PubMed] [Google Scholar]
- Jukes T. H. Evolution of the amino acid code: inferences from mitochondrial codes. J Mol Evol. 1983;19(3-4):219–225. doi: 10.1007/BF02099969. [DOI] [PubMed] [Google Scholar]
- Jukes T. H. Genetic code 1990. Outlook. Experientia. 1990 Dec 1;46(11-12):1149–1157. doi: 10.1007/BF01936925. [DOI] [PubMed] [Google Scholar]
- Jukes T. H., Holmquist R., Moise H. Amino acid composition of proteins: Selection against the genetic code. Science. 1975 Jul 4;189(4196):50–51. doi: 10.1126/science.237322. [DOI] [PubMed] [Google Scholar]
- Jukes T. H., Osawa S., Muto A. Divergence and directional mutation pressures. Nature. 1987 Feb 19;325(6106):668–668. doi: 10.1038/325668b0. [DOI] [PubMed] [Google Scholar]
- Jukes T. H., Osawa S., Muto A., Lehman N. Evolution of anticodons: variations in the genetic code. Cold Spring Harb Symp Quant Biol. 1987;52:769–776. doi: 10.1101/sqb.1987.052.01.086. [DOI] [PubMed] [Google Scholar]
- Jukes T. H., Osawa S. The genetic code in mitochondria and chloroplasts. Experientia. 1990 Dec 1;46(11-12):1117–1126. doi: 10.1007/BF01936921. [DOI] [PubMed] [Google Scholar]
- Jukes T. H. Possibilities for the evolution of the genetic code from a preceding form. Nature. 1973 Nov 2;246(5427):22–26. doi: 10.1038/246022a0. [DOI] [PubMed] [Google Scholar]
- Jukes T. H. The genetic code. II. Am Sci. 1965 Dec;53(4):477–487. [PubMed] [Google Scholar]
- Jungck J. R. The genetic code as a periodic table. J Mol Evol. 1978 Aug 2;11(3):211–224. doi: 10.1007/BF01734482. [DOI] [PubMed] [Google Scholar]
- Kagawa Y., Nojima H., Nukiwa N., Ishizuka M., Nakajima T., Yasuhara T., Tanaka T., Oshima T. High guanine plus cytosine content in the third letter of codons of an extreme thermophile. DNA sequence of the isopropylmalate dehydrogenase of Thermus thermophilus. J Biol Chem. 1984 Mar 10;259(5):2956–2960. [PubMed] [Google Scholar]
- Kano A., Andachi Y., Ohama T., Osawa S. Novel anticodon composition of transfer RNAs in Micrococcus luteus, a bacterium with a high genomic G + C content. Correlation with codon usage. J Mol Biol. 1991 Sep 20;221(2):387–401. doi: 10.1016/0022-2836(91)80061-x. [DOI] [PubMed] [Google Scholar]
- Kawaguchi Y., Honda H., Taniguchi-Morimura J., Iwasaki S. The codon CUG is read as serine in an asporogenic yeast Candida cylindracea. Nature. 1989 Sep 14;341(6238):164–166. doi: 10.1038/341164a0. [DOI] [PubMed] [Google Scholar]
- King J. L., Jukes T. H. Non-Darwinian evolution. Science. 1969 May 16;164(3881):788–798. doi: 10.1126/science.164.3881.788. [DOI] [PubMed] [Google Scholar]
- Komine Y., Adachi T., Inokuchi H., Ozeki H. Genomic organization and physical mapping of the transfer RNA genes in Escherichia coli K12. J Mol Biol. 1990 Apr 20;212(4):579–598. doi: 10.1016/0022-2836(90)90224-A. [DOI] [PubMed] [Google Scholar]
- Kondo K., Sone H., Yoshida H., Toida T., Kanatani K., Hong Y. M., Nishino N., Tanaka J. Cloning and sequence analysis of the arginine deiminase gene from Mycoplasma arginini. Mol Gen Genet. 1990 Mar;221(1):81–86. doi: 10.1007/BF00280371. [DOI] [PubMed] [Google Scholar]
- Kuchino Y., Hanyu N., Tashiro F., Nishimura S. Tetrahymena thermophila glutamine tRNA and its gene that corresponds to UAA termination codon. Proc Natl Acad Sci U S A. 1985 Jul;82(14):4758–4762. doi: 10.1073/pnas.82.14.4758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köchel H. G., Lazarus C. M., Basak N., Küntzel H. Mitochondrial tRNA gene clusters in Aspergillus nidulans: organization and nucleotide sequence. Cell. 1981 Feb;23(2):625–633. doi: 10.1016/0092-8674(81)90158-6. [DOI] [PubMed] [Google Scholar]
- Lagerkvist U. "Two out of three": an alternative method for codon reading. Proc Natl Acad Sci U S A. 1978 Apr;75(4):1759–1762. doi: 10.1073/pnas.75.4.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagerkvist U. Unorthodox codon reading and the evolution of the genetic code. Cell. 1981 Feb;23(2):305–306. doi: 10.1016/0092-8674(81)90124-0. [DOI] [PubMed] [Google Scholar]
- Lee B. J., Rajagopalan M., Kim Y. S., You K. H., Jacobson K. B., Hatfield D. Selenocysteine tRNA[Ser]Sec gene is ubiquitous within the animal kingdom. Mol Cell Biol. 1990 May;10(5):1940–1949. doi: 10.1128/mcb.10.5.1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B. J., Worland P. J., Davis J. N., Stadtman T. C., Hatfield D. L. Identification of a selenocysteyl-tRNA(Ser) in mammalian cells that recognizes the nonsense codon, UGA. J Biol Chem. 1989 Jun 15;264(17):9724–9727. [PubMed] [Google Scholar]
- Lee C. C., Timms K. M., Trotman C. N., Tate W. P. Isolation of a rat mitochondrial release factor. Accommodation of the changed genetic code for termination. J Biol Chem. 1987 Mar 15;262(8):3548–3552. [PubMed] [Google Scholar]
- Lehman N., Jukes T. H. Genetic code development by stop codon takeover. J Theor Biol. 1988 Nov 21;135(2):203–214. doi: 10.1016/s0022-5193(88)80074-2. [DOI] [PubMed] [Google Scholar]
- Leinfelder W., Forchhammer K., Veprek B., Zehelein E., Böck A. In vitro synthesis of selenocysteinyl-tRNA(UCA) from seryl-tRNA(UCA): involvement and characterization of the selD gene product. Proc Natl Acad Sci U S A. 1990 Jan;87(2):543–547. doi: 10.1073/pnas.87.2.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leinfelder W., Zehelein E., Mandrand-Berthelot M. A., Böck A. Gene for a novel tRNA species that accepts L-serine and cotranslationally inserts selenocysteine. Nature. 1988 Feb 25;331(6158):723–725. doi: 10.1038/331723a0. [DOI] [PubMed] [Google Scholar]
- Li M., Tzagoloff A. Assembly of the mitochondrial membrane system: sequences of yeast mitochondrial valine and an unusual threonine tRNA gene. Cell. 1979 Sep;18(1):47–53. doi: 10.1016/0092-8674(79)90352-0. [DOI] [PubMed] [Google Scholar]
- Loftfield R. B., Vanderjagt D. The frequency of errors in protein biosynthesis. Biochem J. 1972 Aug;128(5):1353–1356. doi: 10.1042/bj1281353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MILLER S. L. A production of amino acids under possible primitive earth conditions. Science. 1953 May 15;117(3046):528–529. doi: 10.1126/science.117.3046.528. [DOI] [PubMed] [Google Scholar]
- Macino G., Coruzzi G., Nobrega F. G., Li M., Tzagoloff A. Use of the UGA terminator as a tryptophan codon in yeast mitochondria. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3784–3785. doi: 10.1073/pnas.76.8.3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macino G., Tzagoloff A. Assembly of the mitochondrial membrane system: partial sequence of a mitochondrial ATPase gene in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1979 Jan;76(1):131–135. doi: 10.1073/pnas.76.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin R. P., Schneller J. M., Stahl A. J., Dirheimer G. Import of nuclear deoxyribonucleic acid coded lysine-accepting transfer ribonucleic acid (anticodon C-U-U) into yeast mitochondria. Biochemistry. 1979 Oct 16;18(21):4600–4605. doi: 10.1021/bi00588a021. [DOI] [PubMed] [Google Scholar]
- Martin R. P., Sibler A. P., Gehrke C. W., Kuo K., Edmonds C. G., McCloskey J. A., Dirheimer G. 5-[[(carboxymethyl)amino]methyl]uridine is found in the anticodon of yeast mitochondrial tRNAs recognizing two-codon families ending in a purine. Biochemistry. 1990 Jan 30;29(4):956–959. doi: 10.1021/bi00456a016. [DOI] [PubMed] [Google Scholar]
- Maréchal-Drouard L., Guillemaut P., Cosset A., Arbogast M., Weber F., Weil J. H., Dietrich A. Transfer RNAs of potato (Solanum tuberosum) mitochondria have different genetic origins. Nucleic Acids Res. 1990 Jul 11;18(13):3689–3696. doi: 10.1093/nar/18.13.3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maréchal L., Guillemaut P., Grienenberger J. M., Jeannin G., Weil J. H. Sequence and codon recognition of bean mitochondria and chloroplast tRNAsTrp: evidence for a high degree of homology. Nucleic Acids Res. 1985 Jun 25;13(12):4411–4416. doi: 10.1093/nar/13.12.4411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon N. K., Peck H. D., Jr, Gall J. L., Przybyla A. E. Cloning and sequencing of the genes encoding the large and small subunits of the periplasmic (NiFeSe) hydrogenase of Desulfovibrio baculatus. J Bacteriol. 1987 Dec;169(12):5401–5407. doi: 10.1128/jb.169.12.5401-5407.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer F., Schmidt H. J., Plümper E., Hasilik A., Mersmann G., Meyer H. E., Engström A., Heckmann K. UGA is translated as cysteine in pheromone 3 of Euplotes octocarinatus. Proc Natl Acad Sci U S A. 1991 May 1;88(9):3758–3761. doi: 10.1073/pnas.88.9.3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizutani T., Kurata H., Yamada K. Study of mammalian selenocysteyl-tRNA synthesis with [75Se]HSe. FEBS Lett. 1991 Sep 2;289(1):59–63. doi: 10.1016/0014-5793(91)80908-l. [DOI] [PubMed] [Google Scholar]
- Motchnik P. A., Tappel A. L. Multiple selenocysteine content of selenoprotein P in rats. J Inorg Biochem. 1990 Nov;40(3):265–269. doi: 10.1016/0162-0134(90)80060-b. [DOI] [PubMed] [Google Scholar]
- Motchnik P. A., Tappel A. L. Rat plasma selenoprotein P properties and purification. Biochim Biophys Acta. 1989 Oct 13;993(1):27–35. doi: 10.1016/0304-4165(89)90138-4. [DOI] [PubMed] [Google Scholar]
- Muramatsu T., Nishikawa K., Nemoto F., Kuchino Y., Nishimura S., Miyazawa T., Yokoyama S. Codon and amino-acid specificities of a transfer RNA are both converted by a single post-transcriptional modification. Nature. 1988 Nov 10;336(6195):179–181. doi: 10.1038/336179a0. [DOI] [PubMed] [Google Scholar]
- Muramatsu T., Yokoyama S., Horie N., Matsuda A., Ueda T., Yamaizumi Z., Kuchino Y., Nishimura S., Miyazawa T. A novel lysine-substituted nucleoside in the first position of the anticodon of minor isoleucine tRNA from Escherichia coli. J Biol Chem. 1988 Jul 5;263(19):9261–9267. doi: 10.1351/pac198961030573. [DOI] [PubMed] [Google Scholar]
- Muto A., Andachi Y., Yuzawa H., Yamao F., Osawa S. The organization and evolution of transfer RNA genes in Mycoplasma capricolum. Nucleic Acids Res. 1990 Sep 11;18(17):5037–5043. doi: 10.1093/nar/18.17.5037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muto A., Kawauchi Y., Yamao F., Osawa S. Preferential use of A- and U-rich codons for Mycoplasma capricolum ribosomal proteins S8 and L6. Nucleic Acids Res. 1984 Nov 12;12(21):8209–8217. doi: 10.1093/nar/12.21.8209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muto A., Osawa S. The guanine and cytosine content of genomic DNA and bacterial evolution. Proc Natl Acad Sci U S A. 1987 Jan;84(1):166–169. doi: 10.1073/pnas.84.1.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NIRENBERG M. W., MATTHAEI J. H. The dependence of cell-free protein synthesis in E. coli upon naturally occurring or synthetic polyribonucleotides. Proc Natl Acad Sci U S A. 1961 Oct 15;47:1588–1602. doi: 10.1073/pnas.47.10.1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nellen W., Gallwitz D. Actin genes and actin messenger RNA in Acanthamoeba castellanii. Nucleotide sequence of the split actin gene I. J Mol Biol. 1982 Jul 25;159(1):1–18. doi: 10.1016/0022-2836(82)90028-6. [DOI] [PubMed] [Google Scholar]
- Netzker R., Köchel H. G., Basak N., Küntzel H. Nucleotide sequence of Aspergillus nidulans mitochondrial genes coding for ATPase subunit 6, cytochrome oxidase subunit 3, seven unidentified proteins, four tRNAs and L-rRNA. Nucleic Acids Res. 1982 Aug 11;10(15):4783–4794. doi: 10.1093/nar/10.15.4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nghiem Y., Cabrera M., Cupples C. G., Miller J. H. The mutY gene: a mutator locus in Escherichia coli that generates G.C----T.A transversions. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2709–2713. doi: 10.1073/pnas.85.8.2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura S. Structure, biosynthesis, and function of queuosine in transfer RNA. Prog Nucleic Acid Res Mol Biol. 1983;28:49–73. doi: 10.1016/s0079-6603(08)60082-3. [DOI] [PubMed] [Google Scholar]
- Nomoto M., Imai N., Saiga H., Matsui T., Mita T. Characterization of two types of histone H2B genes from macronuclei of Tetrahymena thermophila. Nucleic Acids Res. 1987 Jul 24;15(14):5681–5697. doi: 10.1093/nar/15.14.5681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oba T., Andachi Y., Muto A., Osawa S. CGG: an unassigned or nonsense codon in Mycoplasma capricolum. Proc Natl Acad Sci U S A. 1991 Feb 1;88(3):921–925. doi: 10.1073/pnas.88.3.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oba T., Andachi Y., Muto A., Osawa S. Translation in vitro of codon UGA as tryptophan in Mycoplasma capricolum. Biochimie. 1991 Jul-Aug;73(7-8):1109–1112. doi: 10.1016/0300-9084(91)90153-r. [DOI] [PubMed] [Google Scholar]
- Oh S. H., Ganther H. E., Hoekstra W. G. Selenium as a component of glutathione periodase isolated from ovine erythrocytes. Biochemistry. 1974 Apr 23;13(9):1825–1829. doi: 10.1021/bi00706a008. [DOI] [PubMed] [Google Scholar]
- Ohama T., Muto A., Osawa S. Role of GC-biased mutation pressure on synonymous codon choice in Micrococcus luteus, a bacterium with a high genomic GC-content. Nucleic Acids Res. 1990 Mar 25;18(6):1565–1569. doi: 10.1093/nar/18.6.1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohama T., Muto A., Osawa S. Spectinomycin operon of Micrococcus luteus: evolutionary implications of organization and novel codon usage. J Mol Evol. 1989 Nov;29(5):381–395. doi: 10.1007/BF02602908. [DOI] [PubMed] [Google Scholar]
- Ohama T., Osawa S., Watanabe K., Jukes T. H. Evolution of the mitochondrial genetic code. IV. AAA as an asparagine codon in some animal mitochondria. J Mol Evol. 1990 Apr;30(4):329–332. doi: 10.1007/BF02101887. [DOI] [PubMed] [Google Scholar]
- Ohama T., Yamao F., Muto A., Osawa S. Organization and codon usage of the streptomycin operon in Micrococcus luteus, a bacterium with a high genomic G + C content. J Bacteriol. 1987 Oct;169(10):4770–4777. doi: 10.1128/jb.169.10.4770-4777.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkubo S., Muto A., Kawauchi Y., Yamao F., Osawa S. The ribosomal protein gene cluster of Mycoplasma capricolum. Mol Gen Genet. 1987 Dec;210(2):314–322. doi: 10.1007/BF00325700. [DOI] [PubMed] [Google Scholar]
- Ohyama K., Fukuzawa H., Kohchi T., Sano T., Sano S., Shirai H., Umesono K., Shiki Y., Takeuchi M., Chang Z. Structure and organization of Marchantia polymorpha chloroplast genome. I. Cloning and gene identification. J Mol Biol. 1988 Sep 20;203(2):281–298. doi: 10.1016/0022-2836(88)90001-0. [DOI] [PubMed] [Google Scholar]
- Old J. M., Jones D. S. The recognition of methionine analogues by Escherichia coli methionyl-transfer ribonucleic acid synthetase. Biochem Soc Trans. 1975;3(5):659–660. doi: 10.1042/bst0030659. [DOI] [PubMed] [Google Scholar]
- Osawa S., Collins D., Ohama T., Jukes T. H., Watanabe K. Evolution of the mitochondrial genetic code. III. Reassignment of CUN codons from leucine to threonine during evolution of yeast mitochondria. J Mol Evol. 1990 Apr;30(4):322–328. doi: 10.1007/BF02101886. [DOI] [PubMed] [Google Scholar]
- Osawa S., Jukes T. H. Codon reassignment (codon capture) in evolution. J Mol Evol. 1989 Apr;28(4):271–278. doi: 10.1007/BF02103422. [DOI] [PubMed] [Google Scholar]
- Osawa S., Jukes T. H. Evolution of the genetic code as affected by anticodon content. Trends Genet. 1988 Jul;4(7):191–198. doi: 10.1016/0168-9525(88)90075-3. [DOI] [PubMed] [Google Scholar]
- Osawa S., Muto A., Jukes T. H., Ohama T. Evolutionary changes in the genetic code. Proc Biol Sci. 1990 Jul 23;241(1300):19–28. doi: 10.1098/rspb.1990.0060. [DOI] [PubMed] [Google Scholar]
- Osawa S., Muto A., Ohama T., Andachi Y., Tanaka R., Yamao F. Prokaryotic genetic code. Experientia. 1990 Dec 1;46(11-12):1097–1106. doi: 10.1007/BF01936919. [DOI] [PubMed] [Google Scholar]
- Osawa S., Ohama T., Jukes T. H., Watanabe K. Evolution of the mitochondrial genetic code. I. Origin of AGR serine and stop codons in metazoan mitochondria. J Mol Evol. 1989 Sep;29(3):202–207. doi: 10.1007/BF02100203. [DOI] [PubMed] [Google Scholar]
- Osawa S., Ohama T., Jukes T. H., Watanabe K., Yokoyama S. Evolution of the mitochondrial genetic code. II. Reassignment of codon AUA from isoleucine to methionine. J Mol Evol. 1989 Nov;29(5):373–380. doi: 10.1007/BF02602907. [DOI] [PubMed] [Google Scholar]
- Osawa S., Ohama T., Yamao F., Muto A., Jukes T. H., Ozeki H., Umesono K. Directional mutation pressure and transfer RNA in choice of the third nucleotide of synonymous two-codon sets. Proc Natl Acad Sci U S A. 1988 Feb;85(4):1124–1128. doi: 10.1073/pnas.85.4.1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozeki H., Ohyama K., Inokuchi H., Fukuzawa H., Kohchi T., Sano T., Nakahigashi K., Umesono K. Genetic system of chloroplasts. Cold Spring Harb Symp Quant Biol. 1987;52:791–804. doi: 10.1101/sqb.1987.052.01.088. [DOI] [PubMed] [Google Scholar]
- PATTERSON E. L., MILSTREY R., STOKSTAD E. L. Effect of selenium in preventing exudative diathesis in chicks. Proc Soc Exp Biol Med. 1957 Aug-Sep;95(4):617–620. doi: 10.3181/00379727-95-23307. [DOI] [PubMed] [Google Scholar]
- PINSENT J. The need for selenite and molybdate in the formation of formic dehydrogenase by members of the coli-aerogenes group of bacteria. Biochem J. 1954 May;57(1):10–16. doi: 10.1042/bj0570010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pape L. K., Tzagoloff A. Cloning and characterization of the gene for the yeast cytoplasmic threonyl-tRNA synthetase. Nucleic Acids Res. 1985 Sep 11;13(17):6171–6183. doi: 10.1093/nar/13.17.6171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prat A., Katinka M., Caron F., Meyer E. Nucleotide sequence of the Paramecium primaurelia G surface protein. A huge protein with a highly periodic structure. J Mol Biol. 1986 May 5;189(1):47–60. doi: 10.1016/0022-2836(86)90380-3. [DOI] [PubMed] [Google Scholar]
- Preer J. R., Jr, Preer L. B., Rudman B. M., Barnett A. J. Deviation from the universal code shown by the gene for surface protein 51A in Paramecium. Nature. 1985 Mar 14;314(6007):188–190. doi: 10.1038/314188a0. [DOI] [PubMed] [Google Scholar]
- Pritchard A. E., Sable C. L., Venuti S. E., Cummings D. J. Analysis of NADH dehydrogenase proteins, ATPase subunit 9, cytochrome b, and ribosomal protein L14 encoded in the mitochondrial DNA of Paramecium. Nucleic Acids Res. 1990 Jan 11;18(1):163–171. doi: 10.1093/nar/18.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard A. E., Seilhamer J. J., Mahalingam R., Sable C. L., Venuti S. E., Cummings D. J. Nucleotide sequence of the mitochondrial genome of Paramecium. Nucleic Acids Res. 1990 Jan 11;18(1):173–180. doi: 10.1093/nar/18.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read R., Bellew T., Yang J. G., Hill K. E., Palmer I. S., Burk R. F. Selenium and amino acid composition of selenoprotein P, the major selenoprotein in rat serum. J Biol Chem. 1990 Oct 15;265(29):17899–17905. [PubMed] [Google Scholar]
- Renbaum P., Abrahamove D., Fainsod A., Wilson G. G., Rottem S., Razin A. Cloning, characterization, and expression in Escherichia coli of the gene coding for the CpG DNA methylase from Spiroplasma sp. strain MQ1(M.SssI). Nucleic Acids Res. 1990 Mar 11;18(5):1145–1152. doi: 10.1093/nar/18.5.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roe B. A., Ma D. P., Wilson R. K., Wong J. F. The complete nucleotide sequence of the Xenopus laevis mitochondrial genome. J Biol Chem. 1985 Aug 15;260(17):9759–9774. [PubMed] [Google Scholar]
- Rotruck J. T., Pope A. L., Ganther H. E., Swanson A. B., Hafeman D. G., Hoekstra W. G. Selenium: biochemical role as a component of glutathione peroxidase. Science. 1973 Feb 9;179(4073):588–590. doi: 10.1126/science.179.4073.588. [DOI] [PubMed] [Google Scholar]
- SUEOKA N. On the genetic basis of variation and heterogeneity of DNA base composition. Proc Natl Acad Sci U S A. 1962 Apr 15;48:582–592. doi: 10.1073/pnas.48.4.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safran M., Farwell A. P., Leonard J. L. Evidence that type II 5'-deiodinase is not a selenoprotein. J Biol Chem. 1991 Jul 25;266(21):13477–13480. [PubMed] [Google Scholar]
- Samuelsson T., Guindy Y. S., Lustig F., Borén T., Lagerkvist U. Apparent lack of discrimination in the reading of certain codons in Mycoplasma mycoides. Proc Natl Acad Sci U S A. 1987 May;84(10):3166–3170. doi: 10.1073/pnas.84.10.3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Brownlee G. G., Barrell B. G. A two-dimensional fractionation procedure for radioactive nucleotides. J Mol Biol. 1965 Sep;13(2):373–398. doi: 10.1016/s0022-2836(65)80104-8. [DOI] [PubMed] [Google Scholar]
- Schneider S. U., Leible M. B., Yang X. P. Strong homology between the small subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase of two species of Acetabularia and the occurrence of unusual codon usage. Mol Gen Genet. 1989 Sep;218(3):445–452. doi: 10.1007/BF00332408. [DOI] [PubMed] [Google Scholar]
- Schön A., Kannangara C. G., Gough S., Söll D. Protein biosynthesis in organelles requires misaminoacylation of tRNA. Nature. 1988 Jan 14;331(6152):187–190. doi: 10.1038/331187a0. [DOI] [PubMed] [Google Scholar]
- Sebald W., Wachter E., Tzagoloff A. Identification of amino acid substitutions in the dicyclohexylcarbodiimide-binding subunit of the mitochondrial ATPase complex from oligomycin-resistant mutants of Saccharomyces cerevisiae. Eur J Biochem. 1979 Oct 15;100(2):599–607. doi: 10.1111/j.1432-1033.1979.tb04207.x. [DOI] [PubMed] [Google Scholar]
- Seilhamer J. J., Cummings D. J. Altered genetic code in Paramecium mitochondria: possible evolutionary trends. Mol Gen Genet. 1982;187(2):236–239. doi: 10.1007/BF00331123. [DOI] [PubMed] [Google Scholar]
- Sharp P. M., Li W. H. An evolutionary perspective on synonymous codon usage in unicellular organisms. J Mol Evol. 1986;24(1-2):28–38. doi: 10.1007/BF02099948. [DOI] [PubMed] [Google Scholar]
- Shimayama T., Himeno H., Sasuga J., Yokobori S., Ueda T., Watanabe K. The genetic code of a squid mitochondrial gene. Nucleic Acids Symp Ser. 1990;(22):73–74. [PubMed] [Google Scholar]
- Shinozaki K., Ohme M., Tanaka M., Wakasugi T., Hayashida N., Matsubayashi T., Zaita N., Chunwongse J., Obokata J., Yamaguchi-Shinozaki K. The complete nucleotide sequence of the tobacco chloroplast genome: its gene organization and expression. EMBO J. 1986 Sep;5(9):2043–2049. doi: 10.1002/j.1460-2075.1986.tb04464.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuber A. P., Orr E. C., Recny M. A., Schendel P. F., May H. D., Schauer N. L., Ferry J. G. Cloning, expression, and nucleotide sequence of the formate dehydrogenase genes from Methanobacterium formicicum. J Biol Chem. 1986 Oct 5;261(28):12942–12947. [PubMed] [Google Scholar]
- Sibler A. P., Dirheimer G., Martin R. P. Codon reading patterns in Saccharomyces cerevisiae mitochondria based on sequences of mitochondrial tRNAs. FEBS Lett. 1986 Jan 1;194(1):131–138. doi: 10.1016/0014-5793(86)80064-3. [DOI] [PubMed] [Google Scholar]
- Sibler A. P., Dirheimer G., Martin R. P. Nucleotide sequence of a yeast mitochondrial threonine-tRNA able to decode the C-U-N leucine codons. FEBS Lett. 1981 Sep 28;132(2):344–348. doi: 10.1016/0014-5793(81)81194-5. [DOI] [PubMed] [Google Scholar]
- Smith M. J., Banfield D. K., Doteval K., Gorski S., Kowbel D. J. Nucleotide sequence of nine protein-coding genes and 22 tRNAs in the mitochondrial DNA of the sea star Pisaster ochraceus. J Mol Evol. 1990 Sep;31(3):195–204. doi: 10.1007/BF02109496. [DOI] [PubMed] [Google Scholar]
- Stadtman T. C. Selenium biochemistry. Annu Rev Biochem. 1990;59:111–127. doi: 10.1146/annurev.bi.59.070190.000551. [DOI] [PubMed] [Google Scholar]
- Stadtman T. C. Selenium-dependent enzymes. Annu Rev Biochem. 1980;49:93–110. doi: 10.1146/annurev.bi.49.070180.000521. [DOI] [PubMed] [Google Scholar]
- Stadtman T. C. Some selenium-dependent biochemical processes. Adv Enzymol Relat Areas Mol Biol. 1979;48:1–28. doi: 10.1002/9780470122938.ch1. [DOI] [PubMed] [Google Scholar]
- Stadtman T. C. Specific occurrence of selenium in enzymes and amino acid tRNAs. FASEB J. 1987 Nov;1(5):375–379. doi: 10.1096/fasebj.1.5.2445614. [DOI] [PubMed] [Google Scholar]
- Sueoka N. CORRELATION BETWEEN BASE COMPOSITION OF DEOXYRIBONUCLEIC ACID AND AMINO ACID COMPOSITION OF PROTEIN. Proc Natl Acad Sci U S A. 1961 Aug;47(8):1141–1149. doi: 10.1073/pnas.47.8.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sueoka N. Directional mutation pressure and neutral molecular evolution. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2653–2657. doi: 10.1073/pnas.85.8.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukenaga Y., Ishida K., Takeda T., Takagi K. cDNA sequence coding for human glutathione peroxidase. Nucleic Acids Res. 1987 Sep 11;15(17):7178–7178. doi: 10.1093/nar/15.17.7178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka R., Muto A., Osawa S. Nucleotide sequence of tryptophan tRNA gene in Acholeplasma laidlawii. Nucleic Acids Res. 1989 Jul 25;17(14):5842–5842. doi: 10.1093/nar/17.14.5842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vold B. S. Structure and organization of genes for transfer ribonucleic acid in Bacillus subtilis. Microbiol Rev. 1985 Mar;49(1):71–80. doi: 10.1128/mr.49.1.71-80.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WILKINS M. H. F., STOKES A. R., WILSON H. R. Molecular structure of deoxypentose nucleic acids. Nature. 1953 Apr 25;171(4356):738–740. doi: 10.1038/171738a0. [DOI] [PubMed] [Google Scholar]
- Wada K., Aota S., Tsuchiya R., Ishibashi F., Gojobori T., Ikemura T. Codon usage tabulated from the GenBank genetic sequence data. Nucleic Acids Res. 1990 Apr 25;18 (Suppl):2367–2411. doi: 10.1093/nar/18.suppl.2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waring R. B., Davies R. W., Lee S., Grisi E., Berks M. M., Scazzocchio C. The mosaic organization of the apocytochrome b gene of Aspergillus nidulans revealed by DNA sequencing. Cell. 1981 Nov;27(1 Pt 2):4–11. doi: 10.1016/0092-8674(81)90354-8. [DOI] [PubMed] [Google Scholar]
- Warrick H. M., Spudich J. A. Codon preference in Dictyostelium discoideum. Nucleic Acids Res. 1988 Jul 25;16(14A):6617–6635. doi: 10.1093/nar/16.14.6617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber A. L., Miller S. L. Reasons for the occurrence of the twenty coded protein amino acids. J Mol Evol. 1981;17(5):273–284. doi: 10.1007/BF01795749. [DOI] [PubMed] [Google Scholar]
- Weber F., Dietrich A., Weil J. H., Maréchal-Drouard L. A potato mitochondrial isoleucine tRNA is coded for by a mitochondrial gene possessing a methionine anticodon. Nucleic Acids Res. 1990 Sep 11;18(17):5027–5030. doi: 10.1093/nar/18.17.5027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss W. A., Friedberg E. C. Normal yeast tRNA(CAGGln) can suppress amber codons and is encoded by an essential gene. J Mol Biol. 1986 Dec 20;192(4):725–735. doi: 10.1016/0022-2836(86)90024-0. [DOI] [PubMed] [Google Scholar]
- Williams K. R., Herrick G. Expression of the gene encoded by a family of macronuclear chromosomes generated by alternative DNA processing in Oxytricha fallax. Nucleic Acids Res. 1991 Sep 11;19(17):4717–4724. doi: 10.1093/nar/19.17.4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. R., Dugre D. H., Dugre S. A., Kondo M., Saxinger W. C. On the fundamental nature and evolution of the genetic code. Cold Spring Harb Symp Quant Biol. 1966;31:723–736. doi: 10.1101/sqb.1966.031.01.093. [DOI] [PubMed] [Google Scholar]
- Wolstenholme D. R., Macfarlane J. L., Okimoto R., Clary D. O., Wahleithner J. A. Bizarre tRNAs inferred from DNA sequences of mitochondrial genomes of nematode worms. Proc Natl Acad Sci U S A. 1987 Mar;84(5):1324–1328. doi: 10.1073/pnas.84.5.1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong J. T., Cedergren R. Natural selection versus primitive gene structure as determinant of codon usage. Eur J Biochem. 1986 Aug 15;159(1):175–180. doi: 10.1111/j.1432-1033.1986.tb09849.x. [DOI] [PubMed] [Google Scholar]
- Wong J. T. The evolution of a universal genetic code. Proc Natl Acad Sci U S A. 1976 Jul;73(7):2336–2340. doi: 10.1073/pnas.73.7.2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamao F., Andachi Y., Muto A., Ikemura T., Osawa S. Levels of tRNAs in bacterial cells as affected by amino acid usage in proteins. Nucleic Acids Res. 1991 Nov 25;19(22):6119–6122. doi: 10.1093/nar/19.22.6119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamao F., Iwagami S., Azumi Y., Muto A., Osawa S., Fujita N., Ishihama A. Evolutionary dynamics of tryptophan tRNAs in Mycoplasma capricolum. Mol Gen Genet. 1988 May;212(2):364–369. doi: 10.1007/BF00334708. [DOI] [PubMed] [Google Scholar]
- Yamao F., Muto A., Kawauchi Y., Iwami M., Iwagami S., Azumi Y., Osawa S. UGA is read as tryptophan in Mycoplasma capricolum. Proc Natl Acad Sci U S A. 1985 Apr;82(8):2306–2309. doi: 10.1073/pnas.82.8.2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama S., Watanabe T., Murao K., Ishikura H., Yamaizumi Z., Nishimura S., Miyazawa T. Molecular mechanism of codon recognition by tRNA species with modified uridine in the first position of the anticodon. Proc Natl Acad Sci U S A. 1985 Aug;82(15):4905–4909. doi: 10.1073/pnas.82.15.4905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinoni F., Birkmann A., Stadtman T. C., Böck A. Nucleotide sequence and expression of the selenocysteine-containing polypeptide of formate dehydrogenase (formate-hydrogen-lyase-linked) from Escherichia coli. Proc Natl Acad Sci U S A. 1986 Jul;83(13):4650–4654. doi: 10.1073/pnas.83.13.4650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinoni F., Heider J., Böck A. Features of the formate dehydrogenase mRNA necessary for decoding of the UGA codon as selenocysteine. Proc Natl Acad Sci U S A. 1990 Jun;87(12):4660–4664. doi: 10.1073/pnas.87.12.4660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruijn M. H. Drosophila melanogaster mitochondrial DNA, a novel organization and genetic code. Nature. 1983 Jul 21;304(5923):234–241. doi: 10.1038/304234a0. [DOI] [PubMed] [Google Scholar]
- van der Feltz M. J., Shivji M. K., Allen P. B., Heisterkamp N., Groffen J., Wiedemann L. M. Nucleotide sequence of both reciprocal translocation junction regions in a patient with Ph positive acute lymphoblastic leukaemia, with a breakpoint within the first intron of the BCR gene. Nucleic Acids Res. 1989 Jan 11;17(1):1–10. doi: 10.1093/nar/17.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]


