Abstract
Cytochrome c oxidase (COX), the terminal enzyme of the mitochondrial electron transport chain, is regulated by isozyme expression, allosteric effectors such as the ATP/ADP ratio, and reversible phosphorylation. Of particular interest is the ‘allosteric ATP-inhibition’, which has been hypothesized to keep the mitochondrial membrane potential at low healthy values (< 140 mV), thus preventing the formation of superoxide radical anions, which have been implicated in multiple degenerative diseases. It has been proposed that the ‘allosteric ATPinhibition’ is switched on by the protein kinase A-dependent phosphorylation of COX. The goal of this study was to identify the phosphorylation site(s) involved in the ‘allosteric ATPinhibition’ of COX. We report the mass spectrometric identification of four new phosphorylation sites in bovine heart COX. The identified phosphorylation sites include Tyr-218 in subunit II, Ser-1 in subunit Va, Ser-2 in subunit Vb, and Ser-1 in subunit VIIc. With the exeption of for Ser- 2 in subunit Vb, the identified phosphorylation sites were found in enzyme samples with and without ‘allosteric ATP inhibition’, making Ser-2 of subunit Vb a candidate site enabling allosteric regulation. We therefore hypothesize that additional phosphorylation(s) may be required for the ‘allosteric ATP-inhibition’, and that these sites may be easily dephosphorylated or difficult to identify by mass spectrometry.
Keywords: Cytochrome c oxidase, enzyme kinetics, mass spectrometry, protein phosphorylation, allosteric ATP-inhibition.
1 Introduction
Main functions of the mitochondrial oxidative phosphorylation (OxPhos) system in mammals are the synthesis of ATP, the production of ROS (reactive oxygen species) as signaling molecules, and the generation of heat. The demand for these activities varies continuously and between different tissues/organs. Endogenous and neuronal signals turn on signaling pathways resulting in phosphorylation of target proteins, which are partly localized in mitochondria [1–5]. Some protein kinases have been shown to translocate into the mitochondria after their activation [6–8] and to bind or phosphorylate mitochondrial proteins, including cytochrome c oxidase (COX) [9–12].
COX is the terminal enzyme of the respiratory chain, consisting of 3 mitochondriaencoded catalytic and 10 nuclear-encoded regulatory subunits. It has been suggested to be the rate-limiting enzyme of the electron transport chain in vivo [13–16]. In addition, COX appears to be a key regulatory site of OxPhos, since it is regulated by three ways: 1) by expression of developmental-, tissue-, and species-specific subunit isoforms [17–19], while no subunit isoforms have been found for the other complexes; 2) by allosteric interactions, e.g., via ADP/ATP [20,21], 3,5-diiodothyronine [22], palmitate [23], and calcium [24]; and 3) by reversible phosphorylation of the subunits [25–28]. Based on consensus sequences for protein kinase A (PKA) [29], 53 potential phosphorylation sites for serine or threonine are present in the bovine heart enzyme (11 in subunit I, 10 in II, 3 in III, 6 in IV, 3 in Va, 3 in Vb, 4 in VIa, 3 in VIb, 2 in VIc, 2 in VIIa, 1 in VIIb, 1 in VIIc and 4 in VIII). Although various phosphorylation sites have been identified in COX by mass spectrometry [12,20,27,28], in only a few cases could a functional role be ascribed.
The activity of COX is regulated by various “allosteric” effectors, which bind and act on sites different from the binding sites for the substrates dioxygen and cytochrome c. A specific binding site for ADP or ATP, depending on the free ATP/ADP ratio, was identified at the matrix domain of subunit IV which at high ATP/ADP ratios (half-maximal at ATP/ADP = 28 [22]) changes the kinetics from a hyperbolic into an inhibited sigmoidal curve [21]. This ‘allosteric ATP-inhibition’ of COX is graphically visualized when the polarographically measured oxygen consumption is plotted against increasing cytochrome c concentrations. The ‘allosteric ATPinhibition’ is independent of the mitochondrial membrane potential (ΔΨm) [30] and keeps ΔΨm at low ‘healthy’ values (< 140 mV) [31], thus preventing the formation of ROS, which increase exponentially at ΔΨm values above 140 mV. ROS have been implicated in various degenerative diseases [32]. The ‘allosteric ATP-inhibition’ was found to be reversibly switched on and off by phosphorylation [25,27, 33, 34] and is suggested to be switched off under conditions of stress [12].
The goal of the present investigation was to identify candidate phosphorylation site(s) responsible for the ‘allosteric ATP-inhibition’ of COX. To overcome methodological problems related to the hydrophobic nature of the COX complex chemical protein cleavage with cyanogen bromide (CNBr) was combined with an enzymatic trypsin digestion to generate phosphopeptides of an appropriate length and hydrophilic nature. This enabled their TiO2-based enrichment and identification by mass-spectrometry (MS). We identified 4 new phosphorylation sites in bovine heart COX by MS: subunit II-Tyr218, Va-Ser1, Vb-Ser2, and VIIc-Ser1. In addition, we confirmed two previously described phosphorylation sites, pSer126 of subunit II [35] and pSer4 of subunit Va [27]. The phosphorylated serine 2 of subunit Vb was exclusively identified in COX with ‘allosteric ATP-inhibition’ while the other 3 new phosphorylation sites were found in samples with and without it. Vb-Ser2 cannot be the only site responsible for 'allosteric ATPinhibition' since it is located at the matrix side, while the 'allosteric ATP-inhibition' was induced by phosphorylation from the cytosolic side [33]. The quantification was possible only in case of pSer126, but we found no differences for enzyme kinetics with and without ‘allosteric ATPinhibition’. We therefore conclude that the phosphorylation site(s) responsible for 'allosteric ATP-inhibition', yet unknown, is sensitive to dephosphorylation and is difficult to map in COX samples with allosteric regulation.
2 Materials and methods
2.1 Isolation of mitochondria
Mitochondria were isolated from either fresh or frozen bovine hearts. The hearts were obtained from the slaughterhouse about two hours after animal death, transported on ice and either directly used for preparation of mitochondria or frozen at −80 °C. Frozen hearts were thawed overnight on ice, cut into cubes, minced in a meat grinder, and mixed in a commercial blender for 3×10 s at maximal speed in 3 volumes of a buffer containing 250 mM sucrose, 20 mM HEPES, pH 7.4, 2 mM EGTA. Mitochondria were also isolated in the presence of two phosphatase inhibitors: 25 mM NaF and 10 nM okadaic acid. However, the presence or absence of these two phosphatase inhibitors did not change the (variable) extent of ‘allosteric ATP-inhibition’ (data not shown). The pH was readjusted during homogenization with 2 M Tris. The homogenate was centrifuged for 10 min at 650×g, and mitochondria were collected by centrifugation for 15 min at 16000×g and washed once with the same buffer.
2.2 Isolation of COX and Western analysis
COX was purified from mitochondria either by a standard procedure [36] or by blue native polyacrylamide gel electrophoresis (BN-PAGE) [37,38]. Mitochondria were dissolved in lauryl-maltoside in the presence of 6-aminohexanoic acid and Coomassie blue G-250. Western blots of COX, isolated by standard procedure or by BN-PAGE, were performed as described [27].
2.3 Measurement of COX activity
The kinetics of COX were measured polarographically at 25 °C in a volume of 0.5 ml using an Oxygraph System (Hansatech, Norfolk, U.K.). The isolated enzyme was dissolved in 50 mM potassium phosphate, pH 7.4, 2 mM EGTA, 5 mM MgSO4, and 1% Tween 20. Oxygen consumption was measured with increasing concentrations of cytochrome c (0.2 – 50 µM) in the presence of 15 mM ascorbate and either 5 mM ADP, or 5 mM ATP and an ATP-regenerating system consisting of 10 mM phosphoenolyruvate and 200 U/ml pyruvate kinase.
2.4 Protein cleavage with CNBr and trypsin
Prior to chemical cleavage, cysteines were alkylated with iodoacetamide and Coomassie blue was removed from the gel bands by alternating treatment with digestion buffer (50 mM ammonium bicarbonate) and a 1:1 mixture of digestion buffer and acetonitrile (ACN) until the protein bands were colorless. Depending of the gel slice, 10 to 20 µl of CNBr-solution (10% CNBr in 70% formic acid (FA)) was added, enough to rehydrate the dried slice. After 2 h in dark, the reaction was stopped by adding a 5-fold volume of ultrapure water and incubating for 30 min. The supernatant was collected and the CNBr-cleaved peptides were extracted in two steps with 50% and 100% ACN and the pooled extracts were lyophilized overnight to remove residual volatile CNBr and most of the FA. Remaining CNBr-solutions were deactivated according to Lunn and Sansone [39]. The samples were used directly for a 2 h tryptic digestion (50 °C) or for phosphopeptide enrichment using TiO2 [27,40]. In the latter case the flow-through of the TiO2-enrichment step was cleaned by reversed phase chromatography as previously described [27], dried, and used for a tryptic digestion before an additional TiO2-based phosphopeptide enrichment step was performed. The lyophilizates were solubilized for 5 min in 5 µl of 5 M urea, diluted with 70 µl digestion buffer and digested for 2 h at 50 °C after the addition of 5 µl trypsin solution (62.5 ng/µl, NB sequencing grade, modified from porcine pancreas, Serva Electrophoresis).
2.5 Enrichment of phosphopeptides and sample cleanup for MS analysis
The CNBr- and/or CNBr/trypsin-cleavages were performed prior to TiO2-based enrichment of phosphopeptides as described before [27,40] with few modifications. The samples were mixed with a 5-fold volume of TiO2-loading solution (10% lactic acid, 5% TFA, 80% ACN, 5% ultrapure water) to which the TiO2-material was added simultaneously. After 30 min binding at room temperature and strong agitation the material was sedimented and the phosphopeptide-depleted supernatant was collected, dried, and prepared for MS analysis as described below. The TiO2-material was aspirated after addition of 50 µl loading solution and loaded on a 200 µl tip (Eppendorf, Germany) equipped with a C8 membrane plug (3 M Empore™ C8 extraction disc, IVA Analysentechnik, Germany). A 20 ml syringe equipped with a Chromabond® adapter (for PP-columns, Machery-Nagel, Germany) was used to press the solutions through the column material. The flow-through was pooled with the phosphopeptide-depleted supernatant. The TiO2-material was washed with 50 µl of TiO2 loading solution and 50 µl of washing solution (1% TFA, 80% ACN, 19% ultrapure water) to reduce the amount of non-specifically-bound peptides. Phosphopeptide elution was performed using 20 µl of the alkaline elution solution (60 µl of 24.5% NH4OH (J. T. Baker) and 940 µl ultrapure water), which was slowly pressed through the column. The elution step was repeated after resuspension of the TiO2-material. Finally, peptides that remained on the membrane plug were eluted with 20 µl of 30% ACN, pooled with the eluates, immediately acidified with 4 µl FA, and dried under vacuum.
Prior to MS analysis the TiO2-enriched fractions were desalted as previously described [27,40], using reversed-phase Poros medium R3 (Biosystems, USA) packed 2 mm high in a 200 µl-tip equipped with a C8 membrane plug, R2-material (Biosystems) was used to desalt the phosphopeptide-depleted supernatants.
2.6 NanoLC-ESI-MS/MS analysis and identification of phosphopeptides
For nanoLC-ESIMS/ MS analysis 15 µl of the TiO2-fractions were injected into a U3000 HPLC system (Dionex LC Packings). For peptide separation a solvent system consisting of solvent A (0.1% (v/v) FA) and solvent B (0.1% (v/v) FA, 84% (v/v) ACN) was used with a gradient of 5 to 30% solvent B in 34 min, 30 to 95% solvent B in 5 min, held at 95% solvent B for 5 min before the column was conditioned for the next run for 15 min with 5% solvent B.
The HPLC-system was coupled to a nano-ESI-source with distal coated silicaTips™ (FS360-20-10-D-20, New Objective Inc., Woburn, USA) and to an HCTultra PTM Discovery System™ (Bruker Daltonics) ion-trap mass spectrometer. Peptides were analyzed in the positive mode after ionisation with 1.4 kV source voltage. For tandem mass spectrometric (MS/MS) analysis of TiO2-flow through peptides, ten single spectra were summed to desalt in the MS mode by the MS control software (Esquiere Control V. 6.1; Bruker Daltonics) and the three most abundant ions were selected for alternating CID- and ETD-MS/MS experiments before the masses were put into an exclusion list for 1.2 min. For ETD experiments the accumulation of fluoranthene radical anions was set manually to 5 to 10 ms for efficient fragmentation. The TiO2- eluates were analyzed using the same MS settings. Neutral loss experiments were performed with initial CID fragmentation and additional MS3 experiments in case of potential neutral losses of phosphoric acid for singly-, doubly-, and tripl-charged precursor ions (−98, −49, and −32.7 m/z), as well as ETD fragmentation experiments of the precursor ion.
MS data were processed with DataAnalysis V. 4.0 (Bruker Daltonics) and mgf- or xml-files were stored in the Proteinscape V. 1.3 database (Bruker Daltonics) and used for database searches against the bovine NCBI-nr decoy sub-database (77,870 sequences, update June 01, 2011) with the Mascot® V. 2.2 search algorithm. The following search parameters were selected: CNBr or CNBr combined with trypsin peptide cleavage, peptide mass accuracy of 0.6 Da (monoisotopic), fragment mass accuracy of 0.6 Da (monoisotopic), variable modification due to carbamidomethylation of cysteine, formylation of the peptide N-terminus, homoserine lactone at C-terminal methionine, phosphorylation of serine, threonine, and tyrosine, and a maximum of three missed cleavage sites. The spectra quality was monitored manually by theoretical fragmentation using the MS-Product software tool. The charge states of all abundant fragment ions were determined from their isotopic pattern and compared to theoretical ions.
3 Results
In this study multiple samples of whole COX or isolated COX subunits from SDS-PAGE have been used for MS analysis. The isolated bovine heart enzyme and the mitochondria, used to isolate COX by BN-PAGE, partly exhibited ‘allosteric ATP-inhibition’, while this inhibition was lacking in other samples. The ‘allosteric ATP-inhibition’ was analyzed polarographically by measuring oxygen consumption via titrations with increasing cytochrome c concentrations, a) in the presence of ATP and b) in the presence of ADP. Allosteric responsiveness is indicated by a sigmoidal inhibition curve in the presence of ATP, while a hyperbolic curve is seen in the presence of ADP [21]. In the absence of the ‘allosteric ATP-inhibition’ both curves are hyperbolic and almost identical.
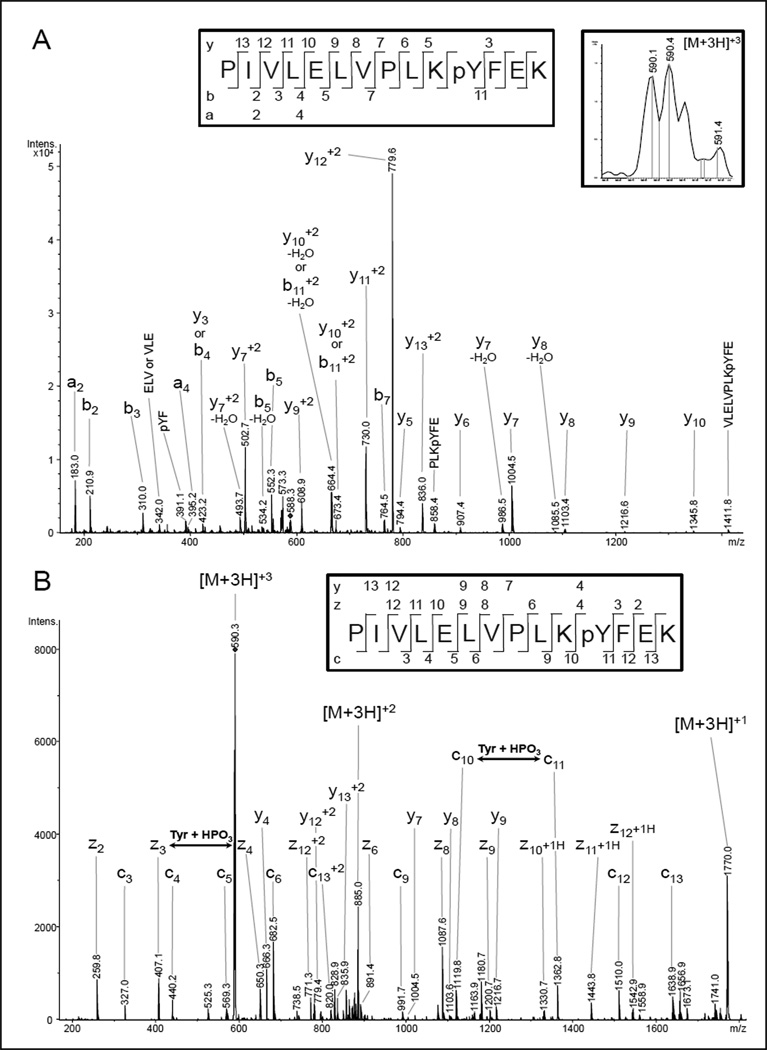
We applied a direct combination of CNBr cleavage followed by tryptic digestion and TiO2 enrichment to isolate and identify phosphorylated peptides by MS in subunits of COX. In 9 out of 12 cow heart COX samples from different animals two phosphorylation sites were identified in COX subunit II: the previously described site at Ser-126 [35] and a new site at Tyr- 218 (Fig. 1). These sites are phosphorylated independently of the presence or absence of ‘allosteric ATP-inhibition’. A combination of CID and ETD experiments enable the unambiguous determination of the phosphosites with Mascot® scores of up to 72 for pTyr-218 in single experiments. Tyr-218 was unambiguously identified in the peptide PIVLELVPLKpYFEK (590.1 m/z, triply charged) with high coverage in the y- and b-ion series (CID fragment-ion pattern) with y5 and b11 distinctly assigning the phosphorylation site (Fig. 1A). ETD experiments result in a complete determination of the amino acid sequence with the N-terminal c10 and c11 ions as well as the C-terminal z3 and z4 ions for the phosphorylated tyrosine (Fig. 1B). The pSer- 126 and pTyr-218 peptides were initially identified with the ion-trap mass spectrometer after TiO2 enrichment. It was also possible to identify them directly without further enrichment, in a targeted proteomic approach via multiple reaction monitoring (MRM) triggered CID-MS/MS experiments (enhanced product ion experiments) in a triple-quadrupole/linear ion trap hybrid mass-spectrometer (4000 Qtrap, AB Sciex). However, with a pure MRM-assay (without additional enhanced product ion experiments) to quantify the phosphorylated peptide with Ser- 126 of subunit II, no differences were found for enzymes with or without ATP-inhibition kinetics (data not shown).
Fig 1.
NanoLC-ESI-MS/MS analysis of CcO subunit II. Phosphotyrosine-218 in COX subunit II was localized in a CID experiment (A) and unequivocally confirmed with an additional ETD experiment (B) of the triply-charged precursor ion (590.1 m/z) of PIVLELVPLKpYFEK.
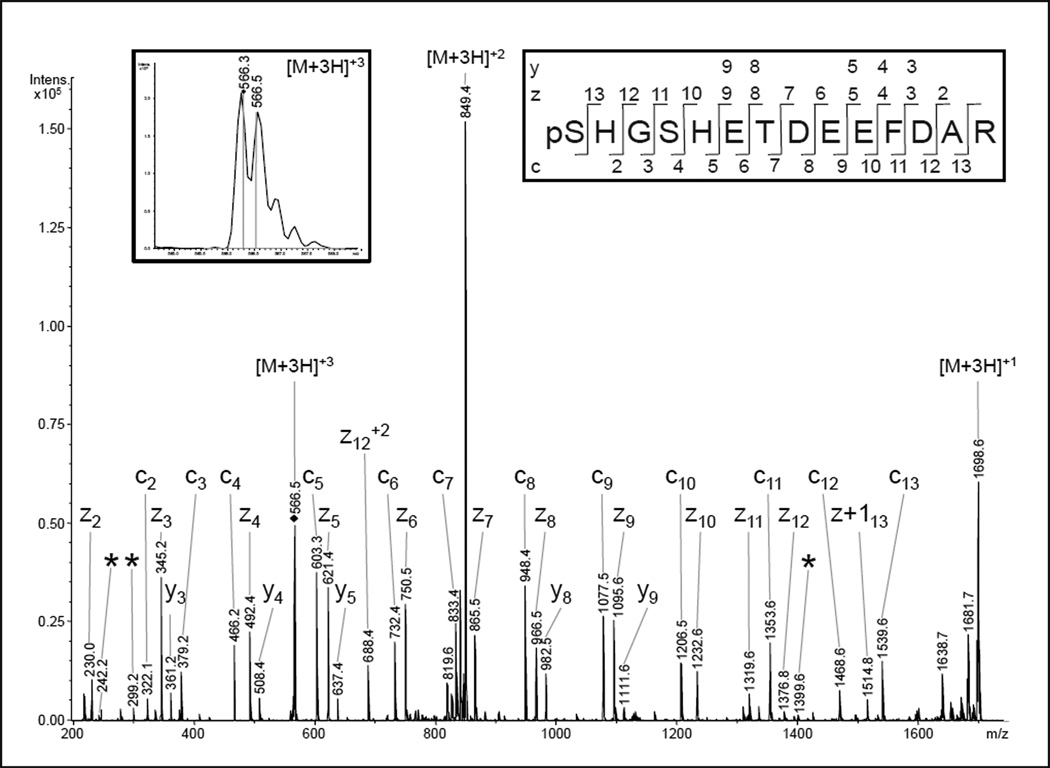
Phosphorylation of Ser-4 in COX subunit Va was previously identified at the Philipps- University Marburg [27] and was confirmed in cow heart COX preparations at Wayne State University (WSU). For all WSU preparations the peptide of subunit Va was found to contain a mixture of phosphorylations at Ser-1 and Ser-4 (Fig. 2). The identification with the Mascot® search algorithm results in a score of 94 for pSer-1 and 72 for pSer-4 (ETD-fragment ion spectrum) (Fig. 2) and 39 or 38 for the corresponding CID-MS/MS spectrum of the triply charged (566.2 m/z) ion of (p)SHG(p)SHETDEEFDAR (not shown). The result of the ETD experiment suggests that the pSer-1 variant is correctly assigned, but in comparison with pSer-4 specific fragment ions from previous identifications [27] and a theoretical fragmentation, a mixture of both peptide types is likely. The explanation for the pSer-1 preference of the search results is a higher abundance of the c2 and c3 ions and the corresponding z11 and z12 ions discriminating pSer-1 and pSer-4. The pSer-4 specific fragments show minor intensities but are present in all samples from WSU when a phosphorylation of this peptide is detected. A doubly phosphorylated peptide was never identified suggesting that a single phosphorylation of the Ser-1-Ser-4 spanning epitope renders the other serine unrecognizable for the corresponding kinase. An alternative explanation may be a methodological problem due to the enrichment method. The loss of multiple phosphorylated peptides due to their strong affinity to the TiO2-material is a known shortcoming [41,42]. On the other hand, Palumbo and co-workers [43] described an intramolecular gas phase phosphate group transfer occurring for protonated phosphotyrosine-, phosphothreonine-, and phosphoserine-containing peptides during CID fragmentation experiments. However, we also observed these pSer1 and pSer4-specific masses in ETD experiments which are not effected by this phenomenon.
Fig 2.
NanoLC-ESI-MS/MS analysis of COX subunit Va. A phosphorylation at serine-4 was previously described for a peptide of the same sequence [27]. The presented ETD spectrum of the triply-charged ion (566.3 m/z) was matched to the peptide pSHGSHETDEEFDAR by the Mascot® search algorithm with scores of 94 for a phosphorylation at serine-1 and 72 in case of pSer4. The discrimination between these phosphorylation sites should be possible, as ions from the four N-terminal amino acids as well as the ions enabling the assignment of pSer1 were also present (c2, c3, z11, z12, z13). However, in comparison with the fragmentation behavior of the previously-identified pSer-4 peptide and the calculated fragment ion masses, three pSer4-specific fragments of low relative abundance were also present (*). Therefore, a HPLC co-elution and ion trap co-isolation of both phosphopeptides is likely and the spectrum presumably represents a mixed fragment ion pattern.
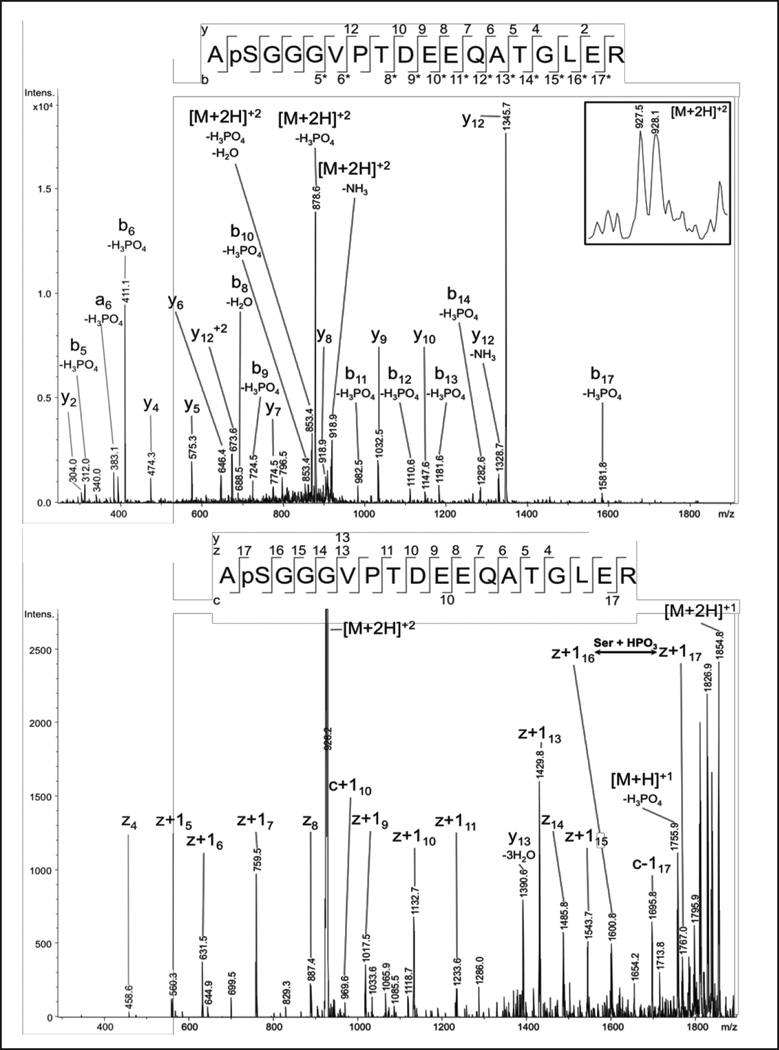
In subunit Vb of one COX preparation from Marburg a new phosphorylation site was found to be localized at Ser-2 of the mature protein, by mass-spectrometry after TiO2 enrichment, with Mascot® scores of up to 82 (Fig. 3). The N-terminal peptide ApSGGGVPTDEEQATGLER is identified to be doubly charged (927.5 m/z). From the CID experiments no assignment of the first five amino acids is possible including pSer-2, but the C-terminal part including numerous b-ions with CID-typical neutral losses of phosphoric acid can be assigned. In addition, the z-ion series of the ETD fragmentation clearly assigns the amino acid sequence including the unambiguous identification of pSer-2 by the z16(+1H) and z17(+1H) ions. Phosphorylation of Ser- 2 in COX subunit Vb was also found in two COX preparations from WSU. All three preparations exhibited the allosteric ATP-inhibition.
Fig 3.
NanoLC-ESI-MS/MS analysis of COX subunit Vb. The analysis of the doubly charged precursor ion (927.5 m/z) resulted in the identification of pSer2 in the peptide ApSGGGVPTDEEQATGLER with Mascot® scores of up to 82. The CID experiment (A) of the N-terminal peptide of the mature protein produced typical neutral losses of phosphoric acid from the precursor ion and nearly all b-type ions. Together with the y-ions the C-terminal part of the peptide is well defined. In contrast to the CID-fragmentation behavior, the z-ions of the ETD experiment also define the N-terminal part, including the phosphorylation site, with site-specific ions z16(+1H) and z17(+1H).
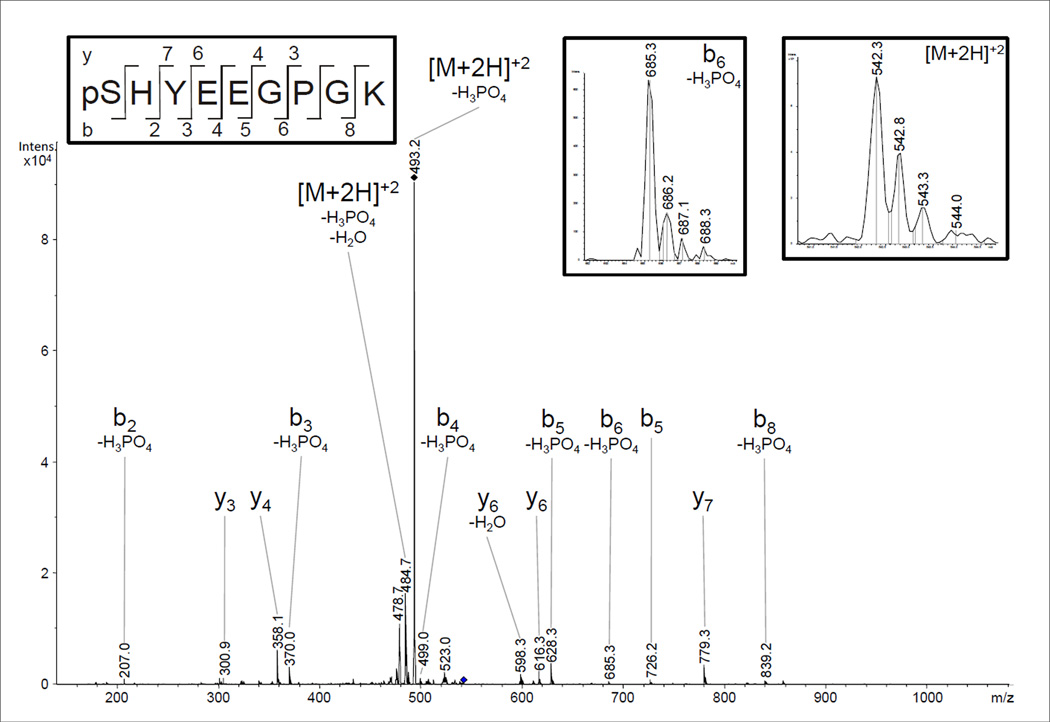
A further phosphorylation site was identified in subunit VIIc with Mascot® scores of up to 52 in 7 biological replicates after CNBr-cleavage and TiO2 enrichment. It was found in samples with and without allosteric ATP-inhibition, isolated by standard methods or by BN-PAGE. The CID-spectrum of pSHYEEGPGK (542.3 m/z, doubly charged) shown in Fig. 4 proofs phosphorylation of Ser-1.
Fig 4.
NanoLC-ESI-MS/MS analysis of COX subunit VIIc. The N-terminal phosphorylation of the mature protein was identified with Mascot® scores of up to 52 in CID experiments on the doubly-charged ion (m/z 542.3) from the peptide pSHYEEGPGK. The spectrum shows neutral loss of H3PO4, and the amino acid sequence-defining ions were well resolved (b6-H3PO4 ion).
In previous studies the ‘allosteric ATP inhibition’ of COX was suggested to be due to phosphorylation of Ser-441 in COX subunit I [30]. Within our study, we isolated and identified the Ser-441-containing peptide PRRYSDYPDAYTM after CNBr cleavage and MS detection. This non-phosphorylated peptide is prominent in all nanoLC-ESI-MS/MS experiments, but it was not detectable in the phosphorylated form in any of the samples.
4 Discussion
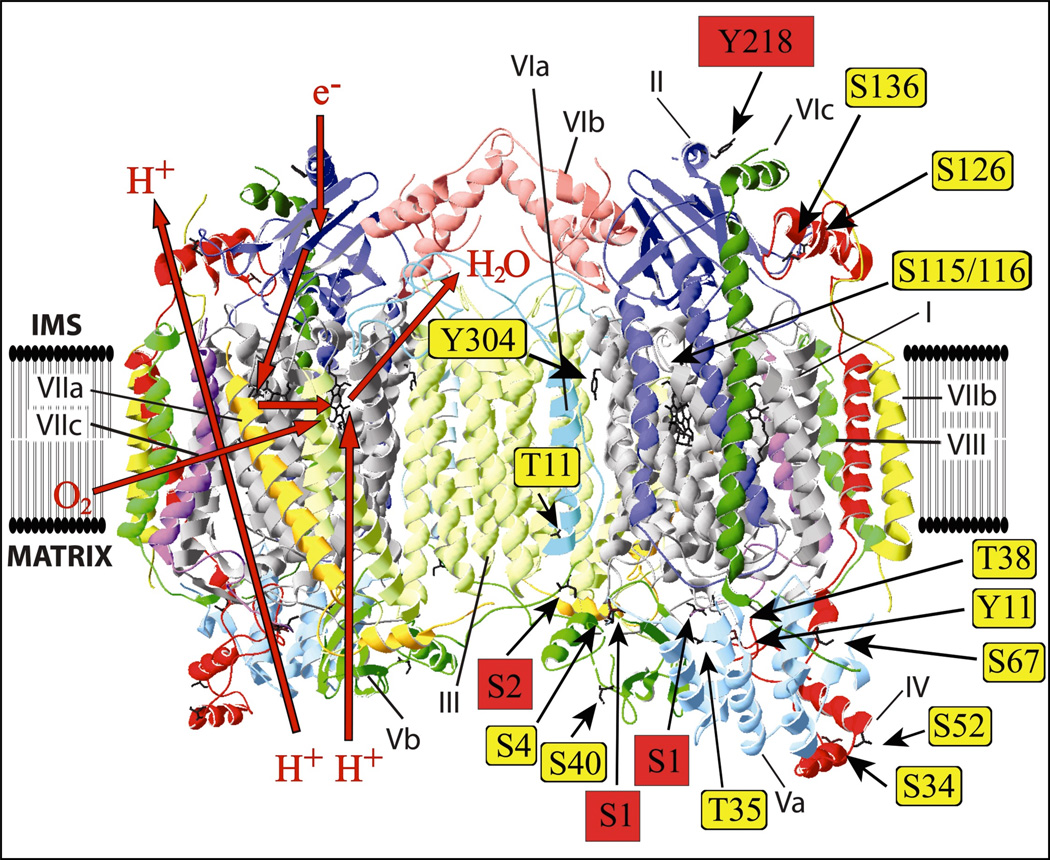
Previously, 14 phosphorylation sites were identified in mammalian COX. In rabbit heart: I-Ser115, I-Ser116, IV-Thr52 and Vb-Ser40 [26]; in bovine liver: I-Tyr304 [44]; in bovine heart: II-Ser126 [35]; IV-Tyr11 [45]; IV-Ser34, Va-Ser4 and Va-Thr35 [27]; VIa-H-Thr11 [46]; in HeLa cells IV-Ser67, IV-Ser136, and Va-Thr38 [47]. Fig. 5 shows the location of these sites based on the crystal structure of the bovine heart enzyme [48]. The four newly-identified phosphorylation sites of this study are highlighted in red. The large number of known phosphorylation sites in mammalian COX suggests multiple regulatory pathways acting on COX, but only very few could be related to a physiological function.
Fig 5.
Identified phosphorylation sites in the crystal structure of bovine heart COX [48]. Crystal structure data of cow heart COX [48] were used and processed with the program Swiss- PDBViewer 4.0.3. Identified phosphorylated amino acids in mammals are represented as sticks. The four newly-identified phosphorylated amino acids are highlighted in red. The red arrows on the left side represent the pathways of hydrogen ions, electrons, dioxygen and water.
Most MS identifications of this study were found in COX preparations with and without ‘allosteric ATP-inhibition’. Interestingly, three phosphorylation sites for subunit Vb-Ser2 were found only in enzyme preparations with ‘allosteric ATP-inhibition’ (not shown). Since it is suggested that the ‘allosteric ATP-inhibition’ involves cooperativity between the two binding sites for cytochrome c in the dimeric enzyme complex (Fig. 5), it is possible that the phosphate group at Ser-2 in subunit Vb, located at the interface between the two monomers on the matrix side, participates in the dimerization of COX and tightens the interaction between the monomers. It is possible that this phosphorylation is necessary but not sufficient for the ‘allosteric ATPinhibition’, since it can be switched on in vitro by PKA phosphorylation of the reconstituted COX from the cytosolic side [33]. It was postulated that subunit I-Ser441 is responsible for the ‘allosteric ATP-inhibition’ of COX [27,33], since it represents the only consensus sequence for PKA phosphorylations [29] on the cytosolic side of subunit I. The lack of phosphorylation of the corresponding peptide (PRRYSDYPDAYTM) of subunit I containing Ser441 does not exclude this site from switching on the allosteric regulation, for example., it could be dephosphorylated during isolation of the peptide for MS. In fact, the ‘allosteric ATP-inhibition’ is easily lost either in vivo or during isolation of the enzyme [31]. Alternatively, different and independent phosphorylation may enable allosteric regulation of COX by ATP/ADP and Ser-2 of subunit Vb may be one of them.
In the crystal structure of bovine heart COX [46] the phosphorylation of Thr11 in the membrane region of subunit VIa-H was identified, and it was suggested that it participates in the dimeric structure of the 13 subunit enzyme complex. This is another subunit that bridges the two monomers in the dimeric structure. Recently, Acin-Perez and colleagues identified a new phosphorylation site in mouse COX subunit IV-1 at Ser-58 [49]. This site is close to the postulated binding site for ATP at the matrix side of subunit IV [17], and it was concluded that it prevents the ‘allosteric ATP-inhibition’ of COX. However, the kinetics presented do not show sigmoidal tendencies in the presence of ATP [49]. In bovine liver COX phosphorylation of Tyr- 11 in subunit IV has been identified [44]. This phosphorylation site is also located on the matrix side close to the postulated binding site for ATP [17] and could modify the ‘allosteric ATPinhibition’ of COX.
In addition to the 18 phosphorylation sites identified thus far, many more potential sites exist in COX (53 predicted sites for serine/threonine [29]). For their identification, new conditions of sample preparation for MS analysis which can prevent artificial dephosphorylation are required. Methods allowing for reliable quantification of the identified phosphosites need to be established since in the same tissue COX complexes have been shown to be differently phosphorylated, for example., subunit Va-Ser1 and Va-Ser4, which only occur separately. The large number of possible phosphorylation patterns of COX suggests a highly complex and variable regulation of COX. In addition to mechanistic aspects, the complex regulation of COX by reversible phosphorylation is also of great interest in clinical research where understanding its role in multiple human diseases remains a challenge [50].
Acknowledgements
The technical assistance of Petra Weber is gratefully acknowledged and we also want to thank Martin Eisenacher for the generation of PRIDE XML versions of the data sets with ProCon (http://www.medizinisches-proteom-center.de/software). This work was supported by the Deutsche Forschungsgemeinschaft (Ka 192/40-1) and by grant GM089900 (MH) from the National Institutes of Health. We thank Dr. Jeffrey Doan for comments on the manuscript.
Abbreviations
- COX
cytochrome c oxidase
- ROS
reactive oxygen species
- OxPhos
oxidative phosphorylation
- BN-PAGE
blue native polyacrylamide gel electrophoresis
- CNBr
cyanogen bromide
- FA
formic acid
- ESI
electrospray ionization
- MS/MS
tandem mass spectrometry
- CID
collision induced dissociation
- ETD
electron transfer dissociation
- MRM
multiple reaction monitoring
- WSU
Wayne State University.
References
- 1.Goldenthal MJ, Marín-García J. Mitochondrial signaling pathways: A receiver/integrator organelle. Mol. Cell. Biochem. 2004;262:1–16. doi: 10.1023/b:mcbi.0000038228.85494.3b. [DOI] [PubMed] [Google Scholar]
- 2.Horbinski C, Chu CT. Kinase signaling cascades in the mitochondrion: a matter of life or death. Free Rad. Biol. Med. 2005;38:2–11. doi: 10.1016/j.freeradbiomed.2004.09.030. [DOI] [PubMed] [Google Scholar]
- 3.Boneh A. Regulation of mitochondrial oxidative phosphorylation by second messenger-mediated signal transduction mechanisms. Cell. Mol. Life Sci. 2006;63:1236–1248. doi: 10.1007/s00018-005-5585-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pagliarini DJ, Dixon JE. Mitochondrial modulation: reversible phosphorylation takes center stage? Trends Biochem. Sci. 2006;31:26–34. doi: 10.1016/j.tibs.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 5.Vogt S, Rhiel A, Koch V, Kadenbach B. Regulation of oxidative phosphorylation by inhibition of its enzyme complexes via reversible phosphorylation. Curr. Enzyme Inhib. 2007;3:189–206. [Google Scholar]
- 6.Bijur GN, Jope RS. Rapid accumulation of Akt in mitochondria following phosphatidylinositol 3-kinase activation. J. Neurochem. 2003;87:1427–1435. doi: 10.1046/j.1471-4159.2003.02113.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Murphy E, Steenbergen C. Mechanisms Underlying Acute Protection From Cardiac Ischemia-Reperfusion Injury. Physiol. Rev. 2008;88:581–609. doi: 10.1152/physrev.00024.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miura T, Tanno M, Sato T. Mitochondrial kinase signalling pathways in myocardial protection from ischaemia/reperfusion-induced necrosis. Cardiovasc Res. 2010;88:7–15. doi: 10.1093/cvr/cvq206. [DOI] [PubMed] [Google Scholar]
- 9.Boerner JL, Demory ML, Silva C, Parsons SJ. Phosphorylation of Y845 on the epidermal growth factor receptor mediates binding to the mitochondrial protein cytochrome c oxidase subunit II. Mol. Cell. Biol. 2004;24:7059–7071. doi: 10.1128/MCB.24.16.7059-7071.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ogbi M, Chew CS, Pohl J, Stuchlik O, et al. Cytochrome c oxidase subunit IV as a marker of protein kinase Cε function in neonatal cardiac myocytes: implications for cytochrome c oxidase activity. Biochem. J. 2004;382:923–932. doi: 10.1042/BJ20040468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ogbi M, Johnson JA. Protein kinase Cε interacts with cytochrome c oxidase subunit IV and enhances cytochrome c oxidase activity in neonatal cardiac myocyte preconditioning. Biochem. J. 2006;393:191–199. doi: 10.1042/BJ20050757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kadenbach B, Ramzan R, Wen L, Vogt S. New extension of the Mitchell Theory for oxidative phosphorylation in mitochondria of living organisms. Biochim. Biophys. Acta. 2010;1800:205–212. doi: 10.1016/j.bbagen.2009.04.019. [DOI] [PubMed] [Google Scholar]
- 13.Villani G, Attardi G. In vivo control of respiration by cytochrome c oxidase in wild-type and mitochondrial DNA mutation-carrying human cells. Proc. Natl. Acad. Sci. USA. 1997;94:1166–1171. doi: 10.1073/pnas.94.4.1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Villani G, Attardi G. In vivo measurements of respiration control by cytochrome c oxidase and in situ analysis of oxidative phosphorylation. Methods Cell Biol. 2001;65:119–131. doi: 10.1016/s0091-679x(01)65007-6. [DOI] [PubMed] [Google Scholar]
- 15.Piccoli C, Scrima R, Boffoli D, Capitanio N. Control by cytochrome c oxidase of the callular oxidative phosphorylation system depends on the mitochondrial energy state. Biochem. J. 2006;396:573–583. doi: 10.1042/BJ20060077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dalmonte ME, Forte E, Genova ML, Giuffrè A, et al. Control of respiration by cytochrome c oxidase in intact cells: role of the membrane potential. J. Biol. Chem. 2009;284:32331–32335. doi: 10.1074/jbc.M109.050146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hüttemann M, Kadenbach B, Grossman LI. Mammalian subunit IV isoforms of cytochrome c oxidase. Gene. 2001;267:111–123. doi: 10.1016/s0378-1119(01)00385-7. [DOI] [PubMed] [Google Scholar]
- 18.Hüttemann M, Jaradat S, Grossman LI. Cytochrome c oxidase of mammals contains a testes-specific isoform of subunit VIb – the counterpart to testes-specific cytochrome c ? Mol. Reprod. Dev. 2003;66:8–16. doi: 10.1002/mrd.10327. [DOI] [PubMed] [Google Scholar]
- 19.Hüttemann M, Schmidt TR, Grossman LI. A third isoform of cytochrome c oxidase subunit VIII is present in mammals. Gene. 2003;312:95–102. doi: 10.1016/s0378-1119(03)00604-8. [DOI] [PubMed] [Google Scholar]
- 20.Frank V, Kadenbach B. Regulation of the H+/e−-stoichiometry of cytochrome c oxidase from bovine heart by intraliposomal ATP/ADP ratios. FEBS Lett. 1996;382:121–124. doi: 10.1016/0014-5793(96)00096-8. [DOI] [PubMed] [Google Scholar]
- 21.Arnold S, Kadenbach B. Cell respiration is controlled by ATP an allosteric inhibitor of cytochrome c oxidase. Eur. J. Biochem. 1997;249:350–354. doi: 10.1111/j.1432-1033.1997.t01-1-00350.x. [DOI] [PubMed] [Google Scholar]
- 22.Arnold S, Goglia F, Kadenbach B. 3,5-diiodothyronine binds to subunit Va of cytochrome c oxidase and abolishes the allosteric inhibition of respiration by ATP. Eur. J. Biochem. 1998;252:325–330. doi: 10.1046/j.1432-1327.1998.2520325.x. [DOI] [PubMed] [Google Scholar]
- 23.Lee I, Kadenbach B. Palmitate decreases proton pumping of liver-type cytochrome c oxidase. Eur. J. Biochem. 2001;268:6329–6334. doi: 10.1046/j.0014-2956.2001.02602.x. [DOI] [PubMed] [Google Scholar]
- 24.Kirichenko AV, Pfitzner U, Ludwig B, Soares CM, et al. Cytochrome c oxidase as a calcium binding protein. Studies on the role of a conserved aspartate in helices XI-XII cytoplasmic loop in cation binding. Biochemistry. 2005;44:12391–12401. doi: 10.1021/bi050376v. [DOI] [PubMed] [Google Scholar]
- 25.Lee I, Bender E, Kadenbach B. Control of mitochondrial membrane potential and ROS formation by reversible phosphorylation of cytochrome c oxidase. Mol. Cell. Biochem. 2002;234/235:63–70. [PubMed] [Google Scholar]
- 26.Fang JK, Prabu SK, Sepuri NB, Raza H, et al. Site specific phosphorylation of cytochrome c oxidase subunits I IVi1 and Vb in rabbit hearts subjected to ischemia/reperfusion. FEBS Lett. 2007;581:1302–1310. doi: 10.1016/j.febslet.2007.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Helling S, Vogt S, Rhiel A, Ramzan R, et al. Phosphorylation and kinetics of mammalian cytochrome c oxidase. Mol. Cell. Proteomics. 2008;7:1714–1724. doi: 10.1074/mcp.M800137-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hüttemann M, Lee I, Samavati L, Yu H, Doan JW. Regulation of mitochondrial oxidative phosphorylation through cell signaling. Biochim. Biophys. Acta. 2007;1773:1701–1720. doi: 10.1016/j.bbamcr.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 29.Pearson RB, Kemp BE. Protein kinase phosphorylation site sequences and consensus specificity motifs: tabulations. Methods Enzymol. 1991;200:62–81. doi: 10.1016/0076-6879(91)00127-i. [DOI] [PubMed] [Google Scholar]
- 30.Arnold S, Kadenbach B. The intramitochondrial ATP/ADP-ratio controls cytochrome c oxidase activity allosterically. FEBS Lett. 1999;443:105–108. doi: 10.1016/s0014-5793(98)01694-9. [DOI] [PubMed] [Google Scholar]
- 31.Ramzan R, Staniek K, Kadenbach B, Vogt S. Mitochondrial respiration and membrane potential are regulated by the allosteric ATP-inhibition of cytochrome c oxidase. Biochim. Biophys. Acta. 2010;1797:1672–1680. doi: 10.1016/j.bbabio.2010.06.005. [DOI] [PubMed] [Google Scholar]
- 32.Valko M, Leibfritz D, Moncola J, Cronin MTD, et al. Free radicals and antioxidants in normal physiological functions and human disease. Int. J. Biochem. Cell Biol. 2007;39:44–84. doi: 10.1016/j.biocel.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 33.Lee I, Bender E, Arnold S, Kadenbach B. Minireview-Hypothesis. New control of mitochondrial membrane potential and ROS-formation. Biol. Chem. 2001;382:1629–1636. doi: 10.1515/BC.2001.198. [DOI] [PubMed] [Google Scholar]
- 34.Bender E, Kadenbach B. The allosteric ATP-inhibition of cytochrome c oxidase is reversibly switched on by cAMP-dependent phosphorylation. FEBS Lett. 2000;466:130–134. doi: 10.1016/s0014-5793(99)01773-1. [DOI] [PubMed] [Google Scholar]
- 35.Hüttemann M, Helling S, Sanderson TH, Sinkler C, et al. Regulation of mitochondrial respiration and apoptosis through cell signaling: cytochrome c oxidase and cytochrome c in ischemia/reperfusion injury and inflammation. Biochim. Biophys. Acta. 2011 Jul 13; doi: 10.1016/j.bbabio.2011.07.001. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kadenbach B, Stroh A, Ungibauer M, Kuhn-Nentwig L, et al. Isozymes of cytochrome c oxidase: characterization and isolation from different tissues. Methods Enzymol. 1986;126:32–45. doi: 10.1016/s0076-6879(86)26006-1. [DOI] [PubMed] [Google Scholar]
- 37.Schägger H, von Jagow G. Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal. Biochem. 1991;199:223–231. doi: 10.1016/0003-2697(91)90094-a. [DOI] [PubMed] [Google Scholar]
- 38.Wittig I, Braun HP, Schägger H. Blue native PAGE. Nature Protocols. 2006;1:418–428. doi: 10.1038/nprot.2006.62. [DOI] [PubMed] [Google Scholar]
- 39.Lunn G, Sansone EB. Destruction of cyanogen bromide and inorganic cyanides. Anal. Biochem. 1985;147:245–250. doi: 10.1016/0003-2697(85)90034-x. [DOI] [PubMed] [Google Scholar]
- 40.Thingholm TE, Jorgensen TJ, Jensen ON, Larsen MR. Highly selective enrichment of phosphorylated peptides using titanium dioxide. Nature Protoc. 2006;1:1929–1935. doi: 10.1038/nprot.2006.185. [DOI] [PubMed] [Google Scholar]
- 41.Helling S, Shinde S, Brosseron F, Schnabel A, et al. Ultratrace Enrichment of Tyrosine Phosphorylated Peptides on an Imprinted Polymer. Anal. Chem. 2011;83:1862–1865. doi: 10.1021/ac103086v. [DOI] [PubMed] [Google Scholar]
- 42.Thingholm TE, Jensen ON, Robinson PJ, Larsen MR. SIMAC (Sequential Elution from IMAC), a Phosphoproteomics Strategy for the Rapid Separation of Monophosphorylated from Multiply Phosphorylated Peptides. Mol. Cell. Proteomics. 2008;7:661–671. doi: 10.1074/mcp.M700362-MCP200. [DOI] [PubMed] [Google Scholar]
- 43.Palumbo AM, Reid GE. Evaluation of Gas-Phase Rearrangement and Competing Fragmentation Reactions on Protein Phosphorylation Site Assignment Using Collision Induced Dissociation-MS/MS and MS. Anal. Chem. 2008;80:9735–9747. doi: 10.1021/ac801768s. [DOI] [PubMed] [Google Scholar]
- 44.Lee I, Salomon AR, Ficarro S, Mathes I, et al. cAMP-dependent tyrosine phosphorylation of subunit I inhibits cytochrome c oxidase activity. J. Biol. Chem. 2005;280:6094–6100. doi: 10.1074/jbc.M411335200. [DOI] [PubMed] [Google Scholar]
- 45.Lee I, Salomon AR, Yu K, Samavati L, et al. Isolation of regulatory-competent, phosphorylated cytochrome c oxidase. Methods Enzymol. 2009;457:193–210. doi: 10.1016/S0076-6879(09)05011-3. [DOI] [PubMed] [Google Scholar]
- 46.Tsukihara T, Shimokata K, Katayama Y, Shimada H, et al. The low-spin heme of cytochrome c oxidase as the driving element of the protonpumping process. Proc. Natl. Acad. Sci. U.S A. 2003;100:15304–15305. doi: 10.1073/pnas.2635097100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Olsen JV, Vermeulen M, Santamaria A, Kumar C, et al. Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci. Signal. 2010;3:ra3. doi: 10.1126/scisignal.2000475. [DOI] [PubMed] [Google Scholar]
- 48.Tsukihara T, Aoyama H, Yamashita E, Tomizaki T, et al. The whole structure of the 13-subunit oxidized cytochrome c oxidase at 2.8 A. Science. 1996;272:1136–1144. doi: 10.1126/science.272.5265.1136. [DOI] [PubMed] [Google Scholar]
- 49.Acin-Perez R, Gatti DL, Bai Y, Manfredi G. Protein Phosphorylation and Prevention of Cytochrome Oxidase Inhibition by ATP: Coupled Mechanisms of Energy Metabolism Regulation. Cell Metab. 2011;13:712–719. doi: 10.1016/j.cmet.2011.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hüttemann M, Lee I, Pecinova A, Pecina P, et al. Regulation of oxidative phosphorylation, the mitochondrial membrane potential, and their role in human disease. J. Bioenerg. Biomembr. 2008;40:445–456. doi: 10.1007/s10863-008-9169-3. [DOI] [PubMed] [Google Scholar]