Abstract
Malaria is holoendemic in the lowlands of Papua New Guinea (PNG), and interactions among Plasmodium species may influence prevalence of mixed infections. Previously, field samples from a cross-sectional survey in Dreikikir, East Sepik Province, analyzed by blood smear and polymerase chain reaction (PCR), showed that mixed infections were common and randomly distributed in this malaria endemic region. To evaluate further whether Plasmodium species distribution is random, blood smear– and PCR/sequence-specific oligonucleotide probe hybridization–based analyses of cross-sectional survey samples were conducted in 2 additional malaria holoendemic regions of northern PNG. Despite ecologic, species prevalence, and transmission season differences in these new surveys, all 4 Plasmodium species were found to be randomly distributed in each area; random distribution patterns also were observed when study populations were divided into age groups. These findings provide consistent evidence that Plasmodium species infections occur independently of one another in PNG malaria holoendemic sites. This independent occurrence suggests that age-dependent, acquired malaria immunity has limited influence on the distribution pattern of Plasmodium species infections in endemic human populations; infection by 1 human malaria parasite species does not reduce susceptibility to infection by others; and malaria vaccines would exhibit limited protection against blood-stage infection by heterologous Plasmodium species.
INTRODUCTION
Human malaria is caused by 4 species of the protozoan parasite Plasmodium: P. falciparum, P. vivax, P. malariae, and P. ovale. In malaria endemic regions, individuals often are infected with >1 Plasmodium species (mixed species infections).1–9 Various epidemiologic studies involving species co-occurrence recorded fewer mixed infections than expected by chance, suggesting in-host competitive interactions whereby one species has eliminated, or reduced the density of, another species to an undetectable level.6–9 These interactions may be mediated by innate host factors that regulate parasite density,10 parasite metabolism, or factors contributing to the acquisition of heterologous (or cross-species) immunity.11,12 If heterologous immunity reduces the prevalence of mixed species infections, as has been proposed,11–13 a malaria vaccine may not need to be specific for each species of Plasmodium to be effective.11
To determine whether interactions occur in mixed Plasmodium species infections, we previously analyzed blood samples from >2,000 individuals living in Dreikikir, East Sepik Province, a region of Papua New Guinea (PNG) endemic for all 4 Plasmodium species infections.5,14 To assess the prevalence and complexity of infections with greater sensitivity and specificity, we also analyzed a subset of samples by nested polymerase chain reaction (PCR).5,15 Using a model in which acquisition of a species infection is assumed to be an independent event, and uninfected individuals are included in calculating expected frequencies for all infection assemblages,16 our findings showed that all 4 Plasmodium species were randomly distributed in blood-stage infections, whether diagnosed by blood smear or PCR. This random distribution was true in the overall population and when the population was divided into 2 age groups.5 These results suggest that Plasmodium species blood-stage infections occur independently of one another and that age-dependent, acquired immunity against malaria may have limited influence on the distribution pattern of parasite species.
Our previous study5 was limited because sampling for malaria infections occurred at a single time point in 1 area, did not consider parasite density, and did not include children <5 years old. In the present study, we analyzed blood samples from 2,731 individuals collected during cross-sectional surveys in 2 different malaria holoendemic areas at different time points during 2 different transmission seasons and determined prevalence of single infections, mixed species infections, or no infection by blood smear and PCR/sequence-specific oligonucleotide probe hybridization (SSOPH)–based diagnostic assays. These results were analyzed further in the context of parasitemia within the 2 different endemic communities.
MATERIALS AND METHODS
Study areas, populations, and blood sample collection
This study was conducted in the Wosera (grassland/marsh, Screw River floodplain; East Sepik Province) and the Liksul (coastal rainforest; Madang Province) areas. For each location, rainfall is heaviest between November and June (wet season) and is lighter between July and October (dry season).17–19 Entomologic inoculation rates (defined as the average number of infectious bites/person/night) for these areas are observed to vary (0.15–0.70).17,20–22 Earlier blood smear surveys reported overall Plasmodium infection prevalence in these areas as 60% and 37.5%.18,19 In the Wosera, malariometric indices have shown slight irregular changes over time, but, in general, there is no clear-cut seasonal transmission pattern.18 In the Liksul area, malaria transmission has been characterized as perennial, with slightly higher transmission during the wet season.19,23 We have observed that molecular polymorphisms associated with chloroquine-resistant P. falciparum are prevalent in both study sites but with a significantly higher prevalence in the Liksul area (Fisher’s exact test, P < 0.0001).24
Blood samples were obtained as a result of convenience sampling from all individuals volunteering to participate in malaria prevalence surveys. The 1,759 samples from the Wosera (July 1998–January 1999) represent 95.2% of the 6 villages surveyed (combined population, N = 1,848) and included individuals <1–85 years old. The 972 samples from the Liksul area (May–July 2000) represent 78.3% of the 3 villages surveyed (combined population, N = 1,242) and included individuals 1–82 years old. Overall, age was not used as a selection criterion. Blood samples were collected in potassium-ethylene diaminetetraacetic acid (EDTA)–coated Vacutainer tubes, and stored at −20°C until DNA extraction could be performed. Ethical approval for this study and the procedures for oral informed consent were obtained from the Medical Research and Advisory Council of PNG and the Institutional Review Board for Human Investigation of Case Western Reserve University and the University Hospitals of Cleveland, Ohio.
Blood smear examination
Blood smear results were obtained retrospectively in both of the malaria prevalence surveys conducted for this study. Malaria blood smear surveys are performed on all blood samples collected for infectious disease epidemiology in collaboration with the Papua New Guinea Institute of Medical Research (PNGIMR). In compliance with PNG Ministry of Health guidelines, treatment of asymptomatic Plasmodium infection is not recommended so that development or maintenance of immunity to clinical malaria is not compromised. If any study volunteers showed signs of clinical malaria, they were transported immediately to the nearest health care center for treatment by local medical personnel. Thick and thin smears were stained with 4% Giemsa and examined by PNGIMR-trained microscopists under oil immersion (100×). Parasite species and species-specific densities were identified and recorded while counting the number of microscope fields inclusive of 200 leukocytes. Based on an average leukocyte count of 8,000/μl,18 the same blood volume would contain approximately 125,000 erythrocytes. If 1 parasitized erythrocyte were observed during this evaluation, the lower limit of parasite density would be 0.001% (1/125,000). These results were subjected to a re-analysis of 10% of the slides to confirm accuracy.
DNA extraction
DNA was extracted from whole blood (200 μl) from study subjects using QIAamp 96 spin blood kits (QIAGEN, Valencia, CA).
Polymerase chain reaction amplification
Amplification of small subunit (ssu) rDNA was performed after nested PCR strategies, using 2 sets of Plasmodium genus–specific primers (Research Genetics, Huntsville, AL). The nest 1 primers were upstream 5′-TTC AGA TGT CAG AGG TGA AAT TCT-3′ and downstream 5′-AAT TAG CAG GTT AAG ATC TCG TTC-3′. The nest 2 primers were upstream 5′-ACG ATC AGA TAC CGT CGT AAT CTT-3′ and downstream 5′-GAA CCC AAA GAC TTT GAT TTC TCA T-3′. All amplification reactions were performed in PCR reaction mixtures described previously,5 using Peltier Thermal Cycler, PTC-225 (MJ Research, Watertown, MA). Nest 1 conditions were 92°C 2 minutes (1×); 92°C 30 seconds, 63°C 2 minutes (35×); and 63°C 5 minutes (1×). A 3-μl aliquot of the nest 1 reaction was used as a template in the nest 2 reaction. Nest 2 conditions were 92°C 2 minutes (1×); 92°C 30 seconds, 70°C 1 minute (40×); and 70°C 5 minutes (1×). To evaluate overall amplification efficiency, PCR products (145–159 bp) from nest 2 reactions were electrophoresed on 2% agarose gels, stained with SYBR Gold (Molecular Probes, Eugene, OR), and visualized on a Storm 860 using ImageQuant software (Molecular Dynamics, Sunnyvale, CA).
Sequence-specific oligonucleotide probe hybridization assay
PCR products were prepared for dot blotting by heating 10 μl of amplicon solution to 95°C for 2 minutes, followed by addition of an equal volume (10 μl) of 20× standard saline citrate (3 M sodium chloride, 0.3 M Na3-citrate, pH 7.0). This solution (2 μl) was spotted onto Hybond N+ filters (Amersham Pharmacia Biotech, Piscataway, NJ). After drying, the nylon filters were bathed first in denaturing buffer (0.4 N NaOH) for 10 minutes and second in neutralizing buffer (1.8 M NaCl, 0.1 M NaH2PO4, 0.01 M EDTA) for 1 minute. The filters were air-dried and cross-linked by UV light exposure in a UV Stratalinker 2400 (Stratagene, La Jolla, CA). Filters prepared in this manner were incubated in 50-ml conical tubes in 10 ml of hybridization buffer (0.75 M NaCl, 0.075 M Na3-citrate, 0.1% sodium dodecylsulfate, 5% liquid block [Amersham Pharmacia Biotech, Piscataway, NJ], 30% dextran sulfate) for 1 hour before adding labeled sequence-specific oligonucleotide probes (SSOPs). Species-specific, fluorescein isothiocyanate-labeled SSOPs (Table 1) were added to the 50-ml conical tubes containing individual filters and hybridization buffer for overnight incubation at 35°C. After hybridization, the filters were twice washed in 5× standard saline citrate at room temperature for 5 minutes each, followed by 2 × 15-minute high-stringency washes optimized for specific hybridization of each individual SSOP (Table 1). Detection was performed using the enhanced chemifluorescence signal amplification kit (Amersham Pharmacia Biotech, Piscataway, NJ) following the supplier’s recommended protocol. Fluorescence was detected using the Storm 860 (Molecular Dynamics, Sunnyvale, CA).
Table 1.
Sequence-specific oligonucleotide probes for Plasmodium species detection
| Species* | Probes† | Washing buffer/temperature |
|---|---|---|
| Pf | 5′-AGG TGA CTT TTA GAT TG-3′ | 1.0× SSC, 0.1% (w/v) SDS/37°C |
| Pv | 5′-CTC TTC GGA GTT TAT-3′ | 0.6× SSC, 0.1% (w/v) SDS/37°C |
| Pm | 5′-TAT ATA TGA GTG TTT C-3′ | 0.6× SSC, 0.1% (w/v) SDS/37°C |
| Po | 5′-AAA TTT CTT AGA TTG C-3′ | 1.0× SSC, 0.1% (w/v) SDS/37°C |
Pf = Plasmodium falciparum; Pv = P. vivax; Pm = P. malariae; Po = P. ovale.
Probe sequences based on previously reported ssu rDNA sequences in GenBank accession numbers AF145334, AF145335, AF145336, and AF145337.
Statistical analyses
For each study area, we determined the prevalence of individual species infections by enumerating the positive samples and dividing by the total study population. The multiple-kind lottery (MKL) model16 was used to calculate the expected numbers of parasite species infection assemblages in each population.5 This model assumes that (1) acquisition of infection is an independent event, and (2) one possible outcome of exposure is no acquisition of infection. Infection is designated as p1, and the reciprocal, absence of infection, is designated as 1-p1 or q1. Extending this to the 4 Plasmodium species, p1, p2, p3, and p4 would represent infection by the individual species, P. falciparum, P. vivax, P. malariae, and P. ovale, whereas q1, q2, q3, and q4 would represent absence of infection by the individual species. The expected frequency of infection by 1 species alone (e.g., P. falciparum only) is the product p1q2q3q4, infection by 2 species (e.g., P. falciparum and P. vivax) is the product p1p2q3q4. Chi-square values were calculated using heterogeneity tests to compare observed with expected values. We used linear regression to analyze the effect of age on parasite density following natural log transformation of parasitemia. All analyses were performed using Microsoft Excel (Microsoft Corporation, Redmond, WA), StatView 5.0.1 (SAS Institute, Inc, Cary, NC), or SAS 8.2 (SAS Institute, Inc, Cary, NC).
RESULTS
Specificity of polymerase chain reaction/sequence-specific oligonucleotide probe hybridization–based diagnostic assay
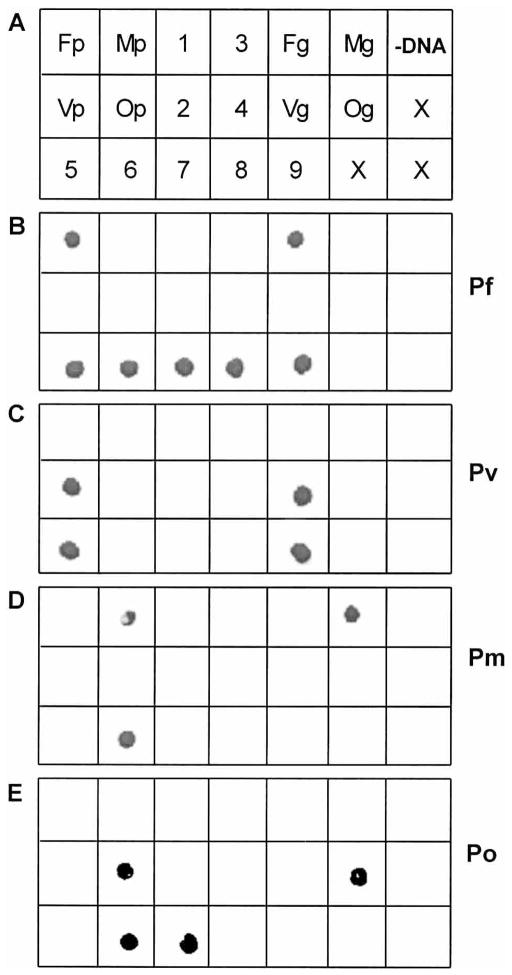
Genomic DNA preparations of P. falciparum, P. vivax, P. malariae, and P. ovale and cloned, species-specific, plasmid-based approximately 1,200-bp ssu rDNA templates5 were used as positive controls to assess the specificity of the PCR/SSOPH assay. In addition, genomic DNA preparations from 5 individuals (samples 5–9, Figure 1) found to be positive for various patterns of Plasmodium species infections by the previously used nested PCR assay were subjected to PCR/SSOPH analysis.5,15 Negative control reactions included genomic DNA preparations from 4 individuals never exposed to malaria infection (samples 1–4, Figure 1) and PCR buffer to which no DNA was added (−DNA, Figure 1). Nest 1 PCR amplification produced amplicons of 491–500 bp, and nest 2 PCR amplification produced amplicons of 145–159 bp, depending on species-specific template length differences (data not shown). Under optimized conditions, all probes produced species-specific hybridization results across all positive (plasmid—Fp, Vp, Mp, Op, or genomic—Fg, Vg, Mg, Og) controls (Figure 1). Probe hybridization results for the 5 malaria-endemic individuals were identical to results obtained previously (Figure 1).5 No evidence of species-specific probe hybridization was observed for any of the negative controls.
Figure 1.
Specificity of polymerase chain reaction (PCR)/sequence-specific oligonucleotide probe hybridization–based diagnostic assay. A) DNA templates from which PCR products were amplified before dot blotting onto nylon membranes. Positive and negative controls appear in rows 1 and 2, and malaria-infected Papua New Guinea study subjects appear in row 3. Species-specific, cloned, plasmid-purified, approximately 1,200 bp ssu rDNA templates from Plasmodium falciparum (Fp), P. vivax (Vp), P. malariae (Mp), and P. ovale (Op); negative control reactions from genomic DNA of 4 individuals (1–4) never exposed to malaria infection; genomic DNA preparations from P. falciparum (Fg), P. vivax (Vg), P. malariae (Mg), and P. ovale (Og); PCR buffer to which no DNA was added (-DNA). Genomic DNA preparations from 5 individuals (5–9) found positive for various of Plasmodium species assemblages by the nested PCR assay; X represents no sample added. B–E) Four separate nylon membranes hybridized with P. falciparum–specific, P. vivax–specific, P. malariae–specific, and P. ovale–specific probes.
Plasmodium species infection assessed by blood smear
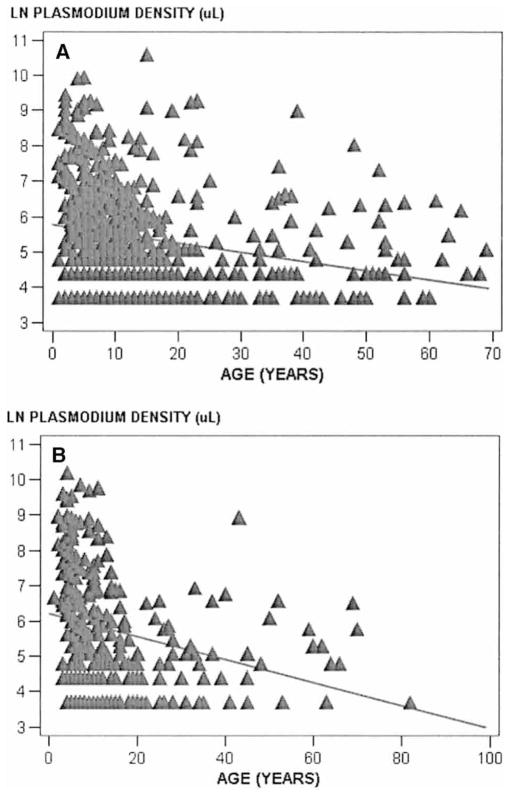
Overall, parasite densities were observed to decrease with increased age in each study population (Wosera, F = 40.1, P < 0.0001; Liksul, F = 23.3, P < 0.0001) (Figure 2). No consistent relationships were observed to suggest that high parasitemia of 1 Plasmodium species suppressed parasitemia of other Plasmodium species (data not shown).
Figure 2.
Linear regression of overall Plasmodium species blood smear density for the Wosera (A) and Liksul (B). The data show natural log-transformed combined parasite density for Plasmodium falciparum, P. vivax, P. malariae, and P. ovale. The diagonal shows the regression line for the relationship between parasitemia and age.
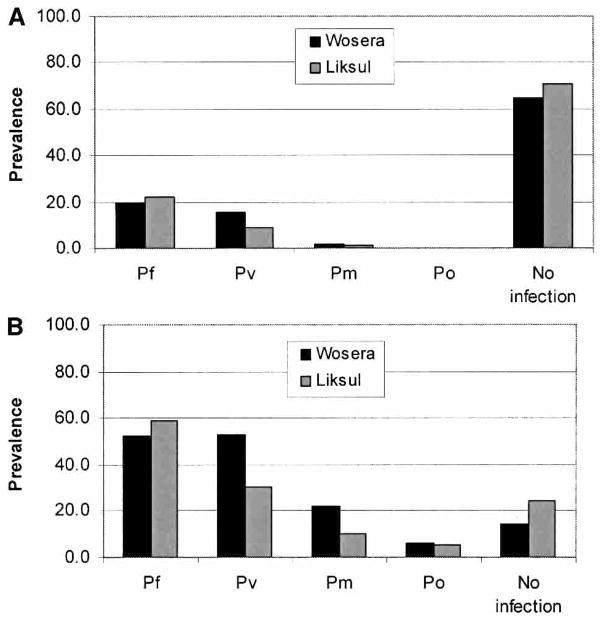
Results from blood smear analysis are summarized in Figure 3A. Overall, prevalence of Plasmodium species infection was 35.1% in the Wosera and 29.6% in the Liksul areas. The prevalence of P. falciparum infections was 20% and 22.3%; P. vivax, 15.8% and 9%; and P. malariae, 2% and 1.1%, in the Wosera and Liksul. Prevalence of P. ovale infections was 0.1% in Liksul, and no P. ovale infections were detected in the Wosera. Further analyses were performed to assess the prevalence of various single-species and mixed-species infections in these areas. Prevalence of all single-species infections was 32.4% in the Wosera and 26.7% in Liksul. Prevalence of all mixed-species infections was 2.7% in the Wosera and 2.9% in Liksul.
Figure 3.
Prevalence of Plasmodium species infections in 2 malaria holoendemic areas in Papua New Guinea. Diagnosis was done by blood smear (A) and by polymerase chain reaction/sequence-specific oligonucleotide probe hybridization (B). Pf = Plasmodium falciparum; Pv = P. vivax; Pm = P. malariae; Po = P. ovale; No infection = individuals judged not to be infected by the diagnostic assay.
To evaluate whether prevalence of infection assemblages (i.e., no infection, single-species infections, or mixed-species infections) within the study populations deviated from an independent random distribution pattern (null hypothesis), the MKL model16 was applied. Observed and expected numbers of infections were not significantly different in either of the study areas (Wosera, chi-square15df = 10.04, 0.90 ≤ P ≤ 0.80; and Liksul, chi-square15df = 4.78, P = 0.994). If interactions occurred, departures from the expected values would have been observed.
Plasmodium species infection prevalence assessed by polymerase chain reaction/sequence-specific oligonucleotide probe hybridization
From each study population, randomly selected sample subsets from the Wosera (n = 340) and Liksul (n = 330) areas were analyzed by the PCR/SSOPH-based diagnostic assay. Using this assay, the prevalence of Plasmodium species infections was 85.9% in the Wosera and 75.5% in Liksul, which were approximately 2.5-fold greater than that detected by blood smear analysis. Prevalence of P. falciparum infections was 52.6% and 59.1%; P. vivax, 53.2% and 30.3%; and P. malariae, 21.8% and 10% in the Wosera and Liksul (Figure 3B). Overall, P. falciparum, P. vivax, and P. malariae infections were 2.6-fold to 10.9-fold greater compared with respective blood smear analyses. All 21 of the P. ovale infections in the Wosera were detected using the PCR/SSOPH assay; 1 of the 17 P. ovale infections in Liksul was detected by PCR/SSOPH and blood smear analyses. The prevalence of P. ovale infections detected by PCR/SSOPH for the Wosera and Liksul were 6.2% and 5.2% (Figure 3B). Further analysis of these data revealed prevalences of 46.2% and 50% for all combined single species infections and 39.7% and 25.5% for all combined mixed-species infections in the Wosera and Liksul. Three individuals (prevalence 0.9%) in the Wosera and no individuals in Liksul were found to be infected by all 4 species using the PCR/SSOPH assay (Table 2). Overall, the prevalence of mixed-species infections diagnosed by this assay was 8.8-fold to 14.7-fold higher in these populations than by blood smear analysis. Observed and expected numbers of infection assemblages (single species, mixed species, not infected) were not significantly different in the Wosera (chi-square15df = 12.63, P = 0.63) (Table 2) or in Liksul (chi-square15df = 9.41, P = 0.86) (Table 2).
Table 2.
Number of Plasmodium infections in overall populations detected by PCR/SSOPH assay
| Parasite assemblage* | Wosera (n = 340)
|
Liksul (n = 330)
|
||||
|---|---|---|---|---|---|---|
| Observed | Expected | chi-square | Observed | Expected | chi-square | |
| Pf | 65 | 61.44 | 0.21 | 119 | 116.02 | 0.08 |
| Pv | 68 | 62.91 | 0.41 | 36 | 34.92 | 0.03 |
| Pm | 20 | 15.37 | 1.39 | 8 | 8.92 | 0.10 |
| Po | 4 | 3.64 | 0.04 | 2 | 4.36 | 1.28 |
| Pf + Pv | 70 | 69.95 | 0.00 | 46 | 50.44 | 0.39 |
| Pf + Pm | 15 | 17.09 | 0.26 | 12 | 12.89 | 0.06 |
| Pf + Po | 4 | 4.04 | 0.00 | 7 | 6.30 | 0.08 |
| Pv + Pm | 15 | 17.50 | 0.36 | 6 | 3.88 | 1.16 |
| Pv + Po | 3 | 4.14 | 0.31 | 1 | 1.90 | 0.42 |
| Pm + Po | 3 | 1.01 | 3.90 | 0 | 0.48 | 0.48 |
| Pf + Pv + Pm | 18 | 19.46 | 0.11 | 5 | 5.60 | 0.07 |
| Pf + Pv + Po | 4 | 4.60 | 0.08 | 5 | 2.74 | 1.86 |
| Pf + Pm + Po | 0 | 1.13 | 1.13 | 1 | 0.70 | 0.13 |
| Pv + Pm + Po | 0 | 1.15 | 1.15 | 1 | 0.21 | 2.96 |
| Pf + Pv + Pm + Po | 3 | 1.28 | 2.31 | 0 | 0.30 | 0.30 |
| Not infected | 48 | 55.27 | 0.96 | 81 | 80.32 | 0.01 |
| chi-square (15 df) | 12.63 | 9.41 | ||||
| P value | 0.63 | 0.86 | ||||
Pf = Plasmodium falciparum; Pv = P. vivax; Pm = P. malariae; Po = P. ovale.
Distribution of Plasmodium species infection prevalence in age-group categories
Further analyses of the PCR/SSOPH diagnostic assay data were performed to determine whether the random distribution of parasite species was altered when the overall populations were categorized into age groups. For this analysis, subjects were categorized into 3 age groups, 2–4, 5–9, and ≥10 years, in the Wosera and Liksul. A comparison of observed and expected numbers of infections in all age groups showed an independent random distribution of Plasmodium species in the Wosera (for 2–4 years, chi-square15df = 5.03, P = 0.99; for 5–9 years, chi-square15df = 9.43, P = 0.85; and for ≥10 years, chi-square15df = 17.01; P = 0.32) (Table 3) and in Liksul (for 2–4 years, chi-square15df = 2.82, P > 0.995; for 5–9 years, chi-square15df = 5.08, P = 0.99; for ≥10 years, chi-square15df = 11.48; P = 0.72) (Note: Total numbers of individuals from the Wosera [N = 332] and Liksul [N = 329] were reduced from the totals shown in Table 2 from omission of children <2 years old and individuals not reporting age) (Table 3). Observed and expected numbers of infections were not significantly different when analyses were performed using blood smears to determine prevalence of Plasmodium species infections (data not shown).
Table 3.
Number of Plasmodium infections in each population by age group detected by PCR/SSOPH assay
| Age group | Parasite assemblage* | Wosera (n =332)
|
Liksul (n =329)
|
||||
|---|---|---|---|---|---|---|---|
| Observed | Expected | chi-square | Observed | Expected | chi-square | ||
| 2–4 yr | Pf | 8 | 8.31 | 0.01 | 12 | 12.82 | 0.05 |
| Pv | 7 | 6.67 | 0.02 | 0 | 0.95 | 0.95 | |
| Pm | 0 | 0.64 | 0.64 | 0 | 0.00 | 0.00 | |
| Po | 3 | 1.73 | 0.93 | 0 | 0.26 | 0.26 | |
| Pf + Pv | 5 | 4.85 | 0.00 | 5 | 4.53 | 0.05 | |
| Pf + Pm | 1 | 0.46 | 0.63 | 0 | 0.00 | 0.00 | |
| Pf + Po | 1 | 1.26 | 0.05 | 1 | 1.22 | 0.04 | |
| Pv + Pm | 1 | 0.37 | 1.07 | 0 | 0.00 | 0.00 | |
| Pv + Po | 0 | 1.01 | 1.01 | 0 | 0.09 | 0.09 | |
| Pm + Po | 0 | 0.10 | 0.10 | 0 | 0.00 | 0.00 | |
| Pf + Pv + Pm | 0 | 0.27 | 0.27 | 0 | 0.00 | 0.00 | |
| Pf + Pv + Po | 1 | 0.73 | 0.10 | 1 | 0.43 | 0.75 | |
| Pf + Pm + Po | 0 | 0.07 | 0.07 | 0 | 0.00 | 0.00 | |
| Pv + Pm + Po | 0 | 0.06 | 0.06 | 0 | 0.0 | 0.00 | |
| Pf + Pv + Pm + Po | 0 | 0.04 | 0.04 | 0 | 0.00 | 0.00 | |
| Not infected | 11 | 11.43 | 0.02 | 4 | 2.70 | 0.63 | |
| Total | 38 | 23 | |||||
| chi-square (15 df) | 5.03 | 2.82 | |||||
| P value | 0.99 | >0.995 | |||||
| 5–9 yr | Pf | 15 | 13.26 | 0.23 | 16 | 13.27 | 0.56 |
| Pv | 12 | 10.81 | 0.13 | 9 | 6.71 | 0.78 | |
| Pm | 2 | 0.84 | 1.59 | 1 | 0.61 | 0.25 | |
| Po | 1 | 0.35 | 1.24 | 0 | 0.39 | 0.39 | |
| Pf + Pv | 26 | 25.94 | 0.00 | 16 | 18.70 | 0.39 | |
| Pf + Pm | 1 | 2.02 | 0.52 | 2 | 1.69 | 0.06 | |
| Pf + Po | 1 | 0.83 | 0.04 | 1 | 1.08 | 0.01 | |
| Pv + Pm | 1 | 1.65 | 0.26 | 1 | 0.86 | 0.02 | |
| Pv + Po | 1 | 0.68 | 0.16 | 1 | 0.55 | 0.37 | |
| Pm + Po | 0 | 0.05 | 0.05 | 0 | 0.05 | 0.05 | |
| Pf + Pv + Pm | 4 | 3.96 | 0.00 | 2 | 2.39 | 0.06 | |
| Pf + Pv + Po | 0 | 1.62 | 1.62 | 2 | 1.53 | 0.15 | |
| Pf + Pm + Po | 0 | 0.13 | 0.13 | 0 | 0.14 | 0.14 | |
| Pv + Pm + Po | 0 | 0.10 | 0.10 | 0 | 0.07 | 0.07 | |
| Pf + Pv + Pm + Po | 1 | 0.25 | 2.29 | 0 | 0.19 | 0.19 | |
| Not infected | 3 | 5.52 | 1.15 | 2 | 4.76 | 1.60 | |
| Total | 68 | 53 | |||||
| chi-square (15 df) | 9.43 | 5.08 | |||||
| P value | 0.85 | 0.99 | |||||
| ≥ 10 yr | Pf | 40 | 36.61 | 0.31 | 90 | 87.27 | 0.09 |
| Pv | 48 | 41.44 | 1.04 | 27 | 24.89 | 0.18 | |
| Pm | 17 | 14.09 | 0.60 | 7 | 8.97 | 0.43 | |
| Po | 0 | 1.91 | 1.91 | 2 | 3.41 | 0.58 | |
| Pf + Pv | 38 | 40.71 | 0.18 | 25 | 28.94 | 0.54 | |
| Pf + Pm | 13 | 13.84 | 0.05 | 10 | 10.43 | 0.02 | |
| Pf + Po | 2 | 1.87 | 0.01 | 5 | 3.97 | 0.27 | |
| Pv + Pm | 13 | 15.67 | 0.45 | 5 | 2.97 | 1.38 | |
| Pv + Po | 1 | 2.12 | 0.59 | 0 | 1.13 | 1.13 | |
| Pm + Po | 3 | 0.72 | 7.21 | 0 | 0.41 | 0.41 | |
| Pf + Pv + Pm | 14 | 15.39 | 0.13 | 3 | 3.46 | 0.06 | |
| Pf + Pv + Po | 3 | 2.08 | 0.40 | 2 | 1.32 | 0.36 | |
| Pf + Pm + Po | 0 | 0.71 | 0.71 | 1 | 0.47 | 0.58 | |
| Pv + Pm + Po | 0 | 0.80 | 0.80 | 1 | 0.14 | 5.53 | |
| Pf + Pv + Pm + Po | 2 | 0.79 | 1.87 | 0 | 0.16 | 0.16 | |
| Not infected | 32 | 37.26 | 0.74 | 75 | 75.08 | 0.08 | |
| Total | 226 | 253 | |||||
| chi-square (15 df) | 17.01 | 11.48 | |||||
| P value | 0.32 | 0.72 | |||||
Pf = Plasmodium falciparum; Pv = P. vivax; Pm = P. malariae; Po = P. ovale.
DISCUSSION
In this malaria epidemiology study, we were interested in determining if Plasmodium species assemblages followed the random distribution pattern previously observed5 in another malaria holoendemic region of PNG. We examined this issue by studying the distribution of the 4 human malaria parasite species in 2 additional holoendemic communities, including children <5 years old, at different time points. The probability that we would observe by chance the same random outcome as observed previously was assumed to be remote.
From a technical perspective, we developed the PCR/SSOPH assay applied in this study owing to advantages over nested PCR/gel electrophoresis15 following strategies previously described.25–27 This approach assesses presence of each species in individual study subjects through positive probe identification based on species-specific sequence recognition. Previous methods have relied on accurate interpretation of the presence or absence of amplicons observed after agarose gel electrophoresis without interrogating the species specificity of the intervening sequence between PCR primers.15 The PCR/SSOPH diagnostic approach is consistent with efficient large-scale evaluation of samples from malaria epidemiologic surveys and offers improved specificity in the interpretation of Plasmodium species diagnosis.
Our overall epidemiologic findings in this expanded study have reached the same conclusion we reported previously5: The prevalence of Plasmodium species is randomly distributed in holoendemic regions of PNG. We reached this conclusion regardless of whether we analyzed data generated by blood smear or the more sensitive PCR/SSOPH diagnostic approach.
Because previous studies have suggested that immunity acquired through exposure to Plasmodium influences the nature of mixed-species infections,11–13 it was important to determine whether malaria exposure influenced acquisition of immunity and whether we might observe its impact in epidemiologic survey data. Acquired immunity against malaria in endemic individuals is marked by 2 generally acknowledged phenomena as childhood progresses: antiparasite and antidisease immunity.28–30 Our overall observations after PCR/SSOPH and blood smear analyses for each study area suggest that prevalence of Plasmodium species infections increased among children through 5 years of age, then decreased in individuals >5 years old (Table 3). Consistent with these observations, we found that parasite density, assessed through blood smear examination, decreased with age in each of the study populations. Similar overall findings have been reported in previous malaria epidemiologic surveys in PNG.18,31,32 Despite observations consistent with exposure-based acquisition of immunity in the holoendemic regions surveyed in this study, we were not able to find support for the hypothesis that heterologous immunity acts to suppress the occurrence of mixed malaria species infections of any assemblage. It is difficult to conclude that development of immunity against 1 human malaria parasite species reduces susceptibility to any of the other 3 species. Consistent with these observations, it would be unlikely for a vaccine against 1 malaria parasite species to offer protection against cross-species infection.
From our studies, a review of similar malaria epidemiology surveys,8 and the literature on epidemiologic measures of risk of malaria, we find that the calculation of expected prevalence of infection assemblages is similar to the straightforward calculation of prevalence of malaria. Both calculations require that the denominator be the total number of individuals surveyed, not just the portion of the population that is infected. This is not to be confused with approaches for calculating the “parasite formula”33 that compares relative rates of infection by endemic parasite species. For example, in a survey of 100 individuals in which there were 45 P. falciparum, 10 P. vivax, and 2 P. malariae infections, the prevalence of infection for each species would be (45/100) · 100 = 45% for P. falciparum, (10/100) · 100 = 10% for P. vivax, and (2/100) · 100 = 2% for P. malariae. The parasite rates, or relative prevalence of each species, would be calculated using a denominator comprised only of individuals in the survey who were positive for parasitemia (45 + 10 + 2 = 57). The parasite rates would then be (45/57) · 100 = 78.9% for P. falciparum, (10/57) · 100 = 17.5% for P. vivax, and (2/57) · 100 = 3.5% for P. malariae. Infection prevalence determined without including “not infected” individuals is not classic prevalence per se but rather the relative proportion of malaria infection resulting from a given species. Exclusion of uninfected individuals in the calculation of expected prevalence of infection assemblages results in artificially concentrating the study population and may lead to overestimating expected mixed-species infection frequencies, underestimating expected single-species infection frequencies, and ultimately reporting an erroneous suppression of observed mixed-species infection in malaria epidemiology surveys. The specification of the measure of risk used needs to be identified clearly and applicable to the specific question of interest within the context of the study.
Because clinical data were not collected during this study, we were not able to assess the impact of mixed-species infections on clinical manifestations of malaria. Previous field studies conducted in the malaria holoendemic Wosera region by Smith et al34 suggested that P. vivax and P. malariae infections were correlated with heterologous protection against clinical P. falciparum disease. In Thailand, in regions experiencing low transmission of P. falciparum, P. vivax, and P. malariae, Luxemburger et al35 reported that severe P. falciparum malaria was 4.2 times less common in patients with P. vivax coinfection than in patients infected with P. falciparum alone. These studies suggest that P. vivax coinfection is observed to be associated with reduced severity of P. falciparum malaria in settings characterized by different extremes of malaria endemicity (the Wosera—holoendemic; Thailand—hypoendemic). If species interactions in mixed-species infections influence clinical outcomes associated with malaria infections, it would be important for vaccine and drug treatment studies to include safeguards to ensure that the potential clinical benefits of mixed infections would not be thrown into imbalance by malaria control efforts and increase the severity of malaria illness in the community.
Finally, many studies have suggested that human genetic polymorphisms may influence not only disease severity, but also susceptibility to infection by Plasmodium species parasites.29,36–38 Our malaria epidemiology studies in PNG have reported on many of these polymorphisms affecting erythrocyte membrane proteins and hemoglobin.39,40 Because of the intimate relationship between these human polymorphisms and malaria endemicity, it is important for future studies to determine if these erythrocyte polymorphisms influence the distribution of mixed Plasmodium species assemblages or the clinical manifestations of disease, or both.
Acknowledgments
We thank Dr. Charles H. King, Dr. Chandy John, David McNamara, and Gabriel Mattera for review of the manuscript, and Will Kastens and Dr. Christopher King for organization and assistance with sample collection. We also thank all the study volunteers for their willing participation.
Financial support: This study was supported by grants from the National Institutes of Health (AI-36478, JWK; AI-46919, PAZ) and the Fogarty International Center (support for RKM).
References
- 1.Molineaux L, Storey J, Cohen JE, Thomas A. A longitudinal study of human malaria in the West African Savanna in the absence of control measures: relationships between different Plasmodium species, in particular P. falciparum and P. malariae. Am J Trop Med Hyg. 1980;29:725–737. doi: 10.4269/ajtmh.1980.29.725. [DOI] [PubMed] [Google Scholar]
- 2.Snounou G, Viriyakosol S, Jarra W, Thaithong S, Brown KN. Identification of the four human malaria parasite species in field samples by the polymerase chain reaction and detection of a high prevalence of mixed infections. Mol Biochem Parasitol. 1993;58:283–292. doi: 10.1016/0166-6851(93)90050-8. [DOI] [PubMed] [Google Scholar]
- 3.Snounou G, Pinheiro L, Goncalves A, Fonseca L, Dias F, Brown KN, do Rosario VE. The importance of sensitive detection of malaria parasites in the human and insect hosts in epidemiological studies, as shown by the analysis of field samples from Guinea Bissau. Trans R Soc Trop Med Hyg. 1993;87:649–653. doi: 10.1016/0035-9203(93)90274-t. [DOI] [PubMed] [Google Scholar]
- 4.May J, Mockenhaupt FP, Ademowo OG, Falusi AG, Olumese PE, Bienzle U, Meyer CG. High rate of mixed and sub-patent malarial infections in southwest Nigeria. Am J Trop Med Hyg. 1999;61:339–343. doi: 10.4269/ajtmh.1999.61.339. [DOI] [PubMed] [Google Scholar]
- 5.Mehlotra RK, Lorry K, Kastens W, Miller SM, Alpers MP, Bockarie M, Kazura JW, Zimmerman PA. Random distribution of mixed species malaria infections in Papua New Guinea. Am J Trop Med Hyg. 2000;62:225–231. doi: 10.4269/ajtmh.2000.62.225. [DOI] [PubMed] [Google Scholar]
- 6.Knowles R, White RS. Studies in the parasitology of malaria. Indian Medical Research Memoirs. 1930;18:436. [Google Scholar]
- 7.Rosenberg R, Andre RG, Ngampatom S, Hatz C, Burge R. A stable, oligosymptomatic malaria focus in Thailand. Trans R Soc Trop Med Hyg. 1990;84:14–21. doi: 10.1016/0035-9203(90)90366-m. [DOI] [PubMed] [Google Scholar]
- 8.McKenzie FE, Bossert WH. Mixed-species Plasmodium infections of humans. J Parasitol. 1997;83:593–600. [PMC free article] [PubMed] [Google Scholar]
- 9.McKenzie FE, Bossert WH. Multispecies Plasmodium infections of humans. J Parasitol. 1999;85:12–18. [PMC free article] [PubMed] [Google Scholar]
- 10.Bruce MC, Donnelly CA, Alpers MP, Galinski MR, Barnwell JW, Walliker D, Day KP. Cross-species interactions between malaria parasites in humans. Science. 2000;287:845–848. doi: 10.1126/science.287.5454.845. [DOI] [PubMed] [Google Scholar]
- 11.Cohen JE. Heterologous immunity in human malaria. Q Rev Biol. 1973;48:467–489. doi: 10.1086/407705. [DOI] [PubMed] [Google Scholar]
- 12.Richie TL. Interactions between malaria parasites infecting the same vertebrate host. Parasitology. 1988;96:607–639. doi: 10.1017/s0031182000080227. [DOI] [PubMed] [Google Scholar]
- 13.Maitland K, Williams TN, Bennett S, Newbold CI, Peto TE, Viji J, Timothy R, Clegg JB, Weatherall DJ, Bowden DK. The interaction between Plasmodium falciparum and P. vivax in children on Espiritu Santo island, Vanuatu. Trans R Soc Trop Med Hyg. 1996;90:614–620. doi: 10.1016/s0035-9203(96)90406-x. [DOI] [PubMed] [Google Scholar]
- 14.Bockarie MJ, Alexander ND, Hyun P, Dimber Z, Bockarie F, Ibam E, Alpers MP, Kazura JW. Randomised community-based trial of annual single-dose diethylcarbamazine with or without ivermectin against Wuchereria bancrofti infection in human beings and mosquitoes. Lancet. 1998;351:162–168. doi: 10.1016/S0140-6736(97)07081-5. [DOI] [PubMed] [Google Scholar]
- 15.Snounou G, Viriyakosol S, Zhu XP, Jarra W, Pinheiro L, do Rosario VE, Thaithong S, Brown KN. High sensitivity of detection of human malaria parasites by the use of nested polymerase chain reaction. Mol Biochem Parasitol. 1993;61:315–320. doi: 10.1016/0166-6851(93)90077-b. [DOI] [PubMed] [Google Scholar]
- 16.Janovy J, Jr, Clopton RE, Clopton DA, Snyder SD, Efting A, Krebs L. Species density distributions as null models for ecologically significant interactions of parasite species in an assemblage. Ecol Model. 1995;77:189–196. [Google Scholar]
- 17.Bockarie M, Kazura J, Alexander N, Dagoro H, Bockarie F, Perry R, Alpers M. Transmission dynamics of Wuchereria bancrofti in East Sepik Province, Papua New Guinea. Am J Trop Med Hyg. 1996;54:577–581. doi: 10.4269/ajtmh.1996.54.577. [DOI] [PubMed] [Google Scholar]
- 18.Genton B, al-Yaman F, Beck HP, Hii J, Mellor S, Narara A, Gibson N, Smith T, Alpers MP. The epidemiology of malaria in the Wosera area, East Sepik Province, Papua New Guinea, in preparation for vaccine trials: I. malariometric indices and immunity. Ann Trop Med Parasitol. 1995;89:359–376. doi: 10.1080/00034983.1995.11812965. [DOI] [PubMed] [Google Scholar]
- 19.Cattani JA, Tulloch JL, Vrbova H, Jolley D, Gibson FD, Moir JS, Heywood PF, Alpers MP, Stevenson A, Clancy R. The epidemiology of malaria in a population surrounding Madang, Papua New Guinea. Am J Trop Med Hyg. 1986;35:3–15. doi: 10.4269/ajtmh.1986.35.3. [DOI] [PubMed] [Google Scholar]
- 20.Bockarie MJ, Alexander N, Bockarie F, Ibam E, Barnish G, Alpers M. The late biting habit of parous Anopheles mosquitoes and pre-bedtime exposure of humans to infective female mosquitoes. Trans R Soc Trop Med Hyg. 1996;90:23–25. doi: 10.1016/s0035-9203(96)90465-4. [DOI] [PubMed] [Google Scholar]
- 21.Hii JL, Smith T, Mai A, Ibam E, Alpers MP. Comparison between anopheline mosquitoes (Diptera: Culicidae) caught using different methods in a malaria endemic area of Papua New Guinea. Bull Entomol Res. 2000;90:211–219. doi: 10.1017/s000748530000033x. [DOI] [PubMed] [Google Scholar]
- 22.Hii JL, Smith T, Vounatsou P, Alexander N, Mai A, Ibam E, Alpers MP. Area effects of bednet use in a malaria-endemic area in Papua New Guinea. Trans R Soc Trop Med Hyg. 2001;95:7–13. doi: 10.1016/s0035-9203(01)90315-3. [DOI] [PubMed] [Google Scholar]
- 23.Burkot TR, Graves PM, Paru R, Wirtz RA, Heywood PF. Human malaria transmission studies in the Anopheles punctulatus complex in Papua New Guinea: sporozoite rates, inoculation rates, and sporozoite densities. Am J Trop Med Hyg. 1988;39:135–144. doi: 10.4269/ajtmh.1988.39.135. [DOI] [PubMed] [Google Scholar]
- 24.Mehlotra RK, Fujioka H, Roepe PD, Janneh O, Ursos LM, Jacobs-Lorena V, McNamara DT, Bockarie MJ, Kazura JW, Kyle DE, Fidock DA, Zimmerman PA. Evolution of a unique Plasmodium falciparum chloroquine-resistance phenotype in association with pfcrt polymorphism in Papua New Guinea and South America. Proc Natl Acad Sci U S A. 2001;98:12689–12694. doi: 10.1073/pnas.221440898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li J, Wirtz RA, McConkey GA, Sattabongkot J, McCutchan TF. Transition of Plasmodium vivax ribosome types corresponds to sporozoite differentiation in the mosquito. Mol Biochem Parasitol. 1994;65:283–289. doi: 10.1016/0166-6851(94)90079-5. [DOI] [PubMed] [Google Scholar]
- 26.Perandin F, Manca N, Galati L, Piccolo G, Calderaro A, Viani I, Ricci L, Dettori G, Chezzi C, Turano A. Usefulness of genus-specific PCR and Southern blot species-specific hybridization for the detection of imported malaria cases in Italy. New Microbiol. 2001;24:69–76. [PubMed] [Google Scholar]
- 27.Kimura M, Kaneko O, Liu Q, Zhou M, Kawamoto F, Wataya Y, Otani S, Yamaguchi Y, Tanabe K. Identification of the four species of human malaria parasites by nested PCR that targets variant sequences in the small subunit rRNA gene. Parasitol Int. 1997;46:91–95. [Google Scholar]
- 28.Bruce-Chwatt LJ. A longitudinal survey of natural malaria infection in a group of West African adults. West Afr Med J. 1963;12:199–200. [PubMed] [Google Scholar]
- 29.Marsh K. Malaria—a neglected disease? Parasitology. 1992;104:S53–S69. doi: 10.1017/s0031182000075247. [DOI] [PubMed] [Google Scholar]
- 30.Mohan K, Stevenson MM. Acquired immunity to asexual blood stages. In: Sherman IW, editor. Malaria: Parasite Biology, Pathogenesis, and Protection. Washington, DC: American Society of Microbiology Press; 1998. pp. 467–993. [Google Scholar]
- 31.Graves PM, Burkot TR, Carter R, Cattani JA, Lagog M, Parker J, Brabin BJ, Gibson FD, Bradley DJ, Alpers MP. Measurement of malarial infectivity of human populations to mosquitoes in the Madang area, Papua New Guinea. Parasitology. 1988;96:251–263. doi: 10.1017/s003118200005825x. [DOI] [PubMed] [Google Scholar]
- 32.Cox MJ, Kum DE, Tavul L, Narara A, Raiko A, Baisor M, Alpers MP, Medley GF, Day KP. Dynamics of malaria parasitaemia associated with febrile illness in children from a rural area of Madang, Papua New Guinea. Trans R Soc Trop Med Hyg. 1994;88:191–197. doi: 10.1016/0035-9203(94)90292-5. [DOI] [PubMed] [Google Scholar]
- 33.Bruce-Chwatt LJ. Essential Malariology. London: William Heinemann Medical Books; 1985. [Google Scholar]
- 34.Smith T, Genton B, Baea K, Gibson N, Narara A, Alpers MP. Prospective risk of morbidity in relation to malaria infection in an area of high endemicity of multiple species of Plasmodium. Am J Trop Med Hyg. 2001;64:262–267. doi: 10.4269/ajtmh.2001.64.262. [DOI] [PubMed] [Google Scholar]
- 35.Luxemburger C, Ricci F, Nosten F, Raimond D, Bathet S, White NJ. The epidemiology of severe malaria in an area of low transmission in Thailand. Trans R Soc Trop Med Hyg. 1997;91:256–262. doi: 10.1016/s0035-9203(97)90066-3. [DOI] [PubMed] [Google Scholar]
- 36.Haldane JBS. The rate of mutation of human genes. Hereditas. 1949;35:267–273. [Google Scholar]
- 37.Hill AV. The genomics and genetics of human infectious disease susceptibility. Annu Rev Genomics Hum Genet. 2001;2:373–400. doi: 10.1146/annurev.genom.2.1.373. [DOI] [PubMed] [Google Scholar]
- 38.Rihet P, Abel L, Traore Y, Traore-Leroux T, Aucan C, Fumoux F. Human malaria: segregation analysis of blood infection levels in a suburban area and a rural area in Burkina Faso. Genet Epidemiol. 1998;15:435–450. doi: 10.1002/(SICI)1098-2272(1998)15:5<435::AID-GEPI1>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 39.Zimmerman PA, Woolley I, Masinde GL, Miller SM, McNamara DT, Hazlett F, Mgone CS, Alpers MP, Genton B, Boatin BA, Kazura JW. Emergence of FY*A(null) in a Plasmodium vivax–endemic region of Papua New Guinea. Proc Natl Acad Sci U S A. 1999;96:13973–13977. doi: 10.1073/pnas.96.24.13973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Patel SS, Mehlotra RK, Kastens W, Mgone CS, Kazura JW, Zimmerman PA. The association of the glycophorin C exon 3 deletion with ovalocytosis and malaria susceptibility in the Wosera, Papua New Guinea. Blood. 2001;98:3489–3491. doi: 10.1182/blood.v98.12.3489. [DOI] [PubMed] [Google Scholar]