Abstract
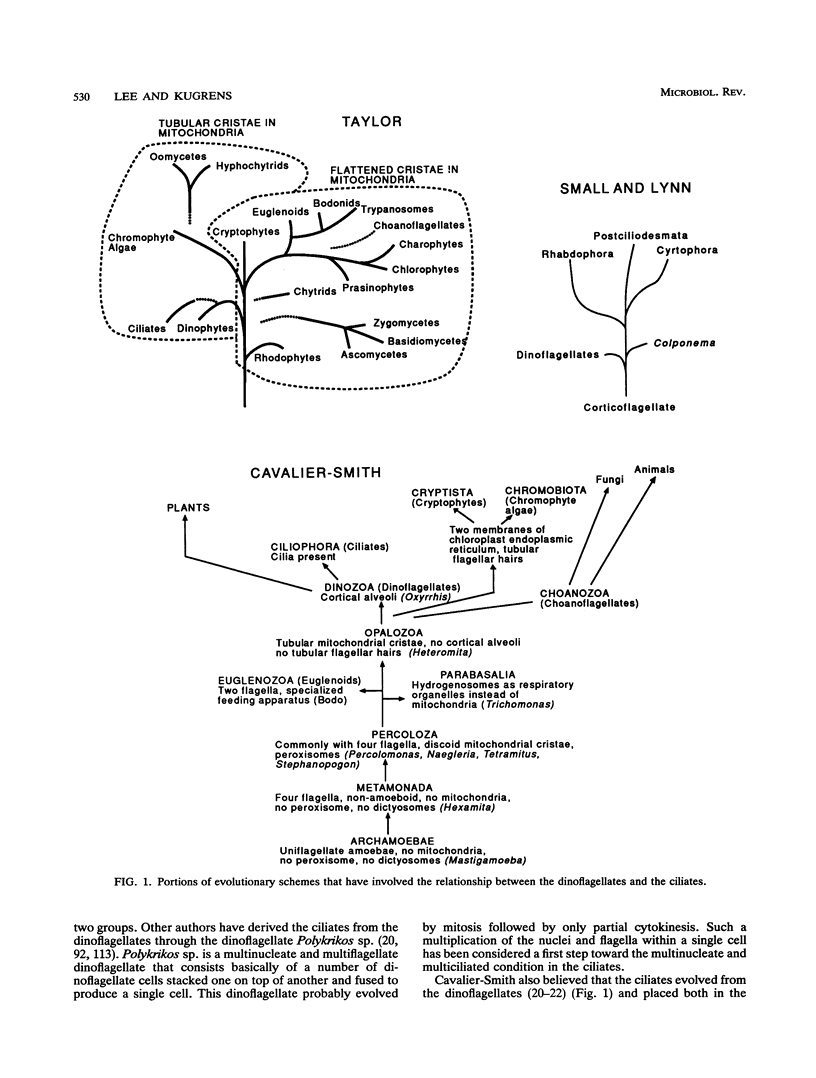
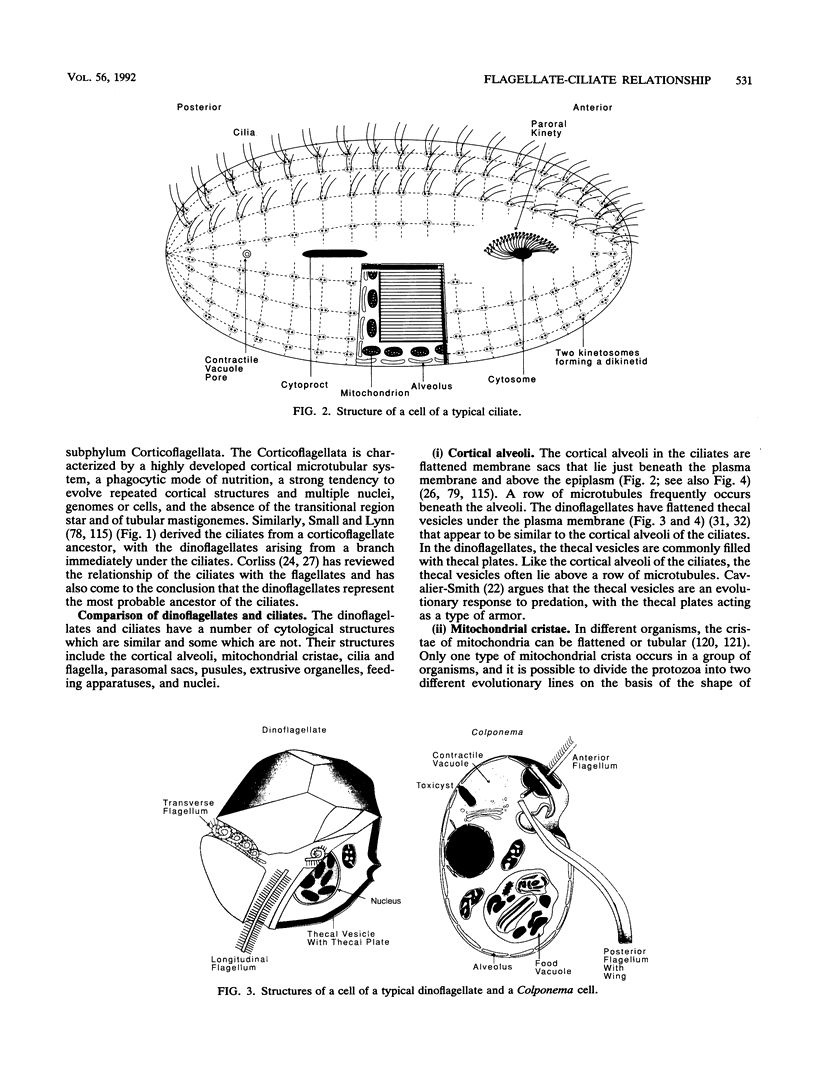
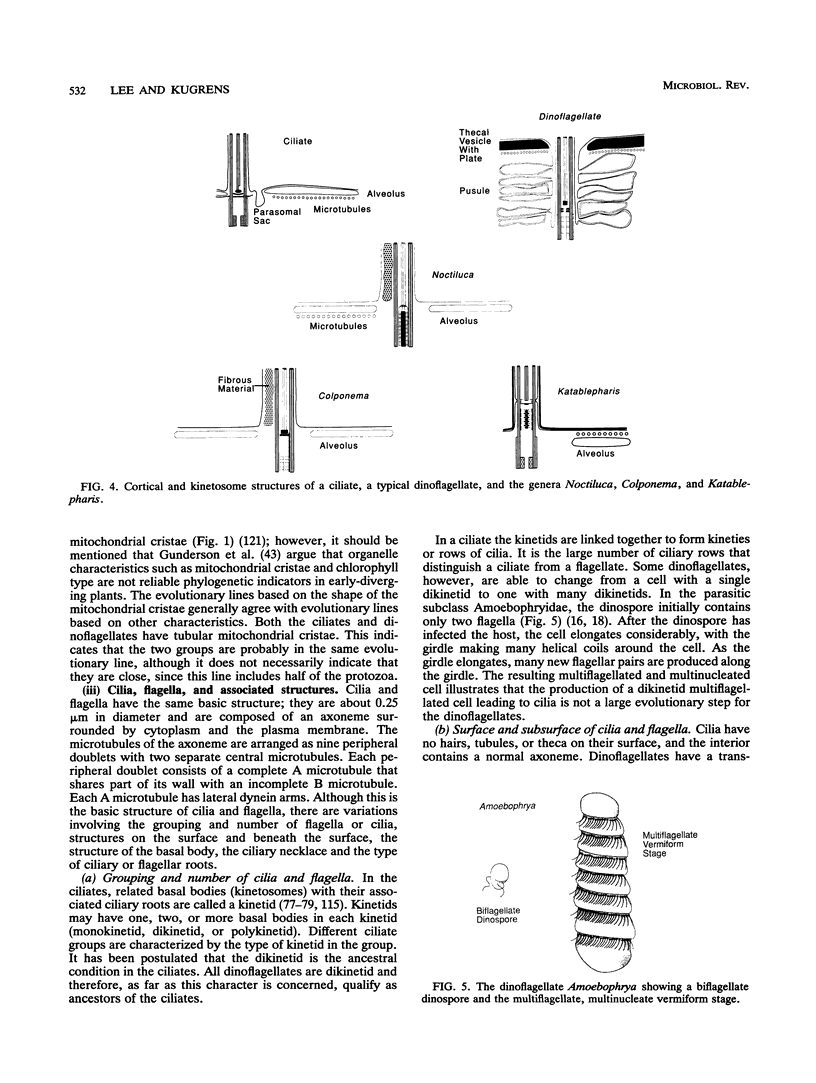
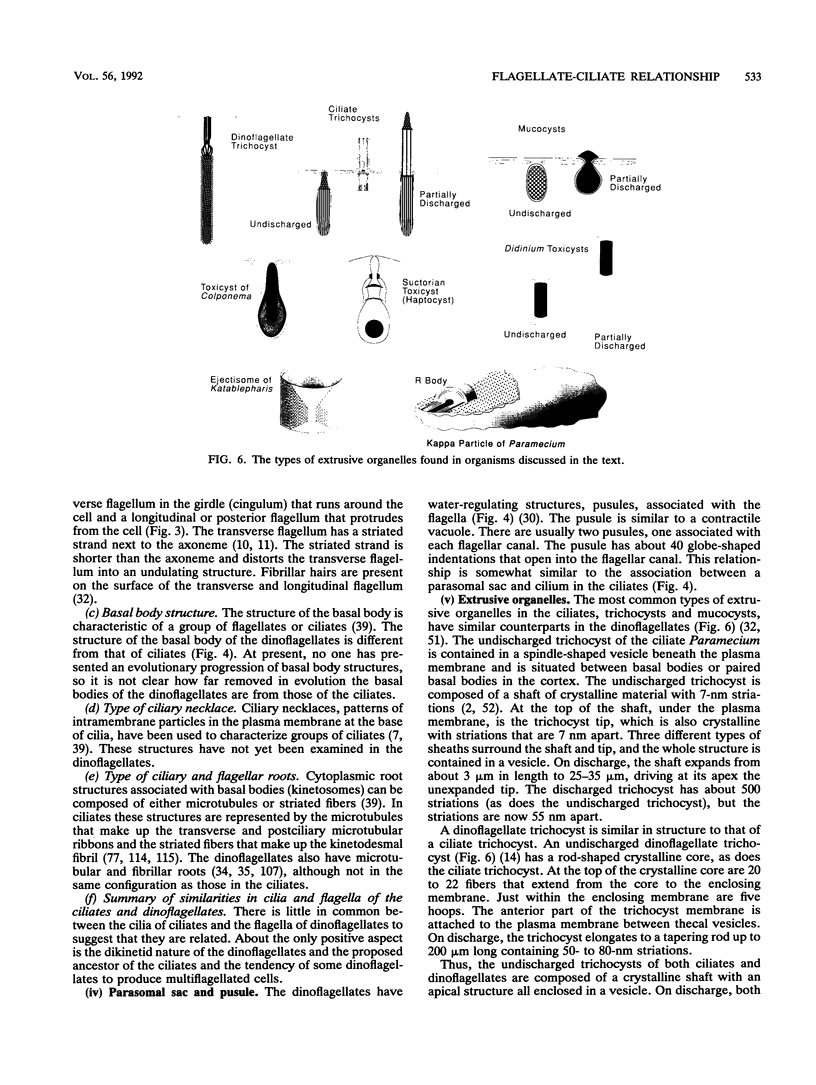
The flagellates and the ciliates have long been considered to be closely related because of their unicellular nature and the similarity in the structures of the axoneme of the flagella and cilia in both groups. Most protozoologists believe that the ciliates arose from a flagellate. The flagellates that are most similar in structure to the ciliates are the dinoflagellates and two genera of uncertain taxonomic position, Colponema and Katablepharis. Structurally, dinoflagellates have a number of similarities with ciliates. These include the similarity of the cortical alveoli in the ciliates to the thecal vesicles in the dinoflagellates, the possession of tubular cristae, the similarity of the parasomal sac of the ciliates to the pusule of the dinoflagellates, the possession of similar trichocysts and mucocysts, and some similarity in the feeding apparatus. Colponema spp. are probably related to the dinoflagellates and have many of the same similarities with the ciliates. Katablepharis spp. are very similar in structure to the swarmer (embryo) of the suctorian ciliates. Indeed, reduction in the number of cilia to two in the suctorian swarmer and elimination of the macronucleus would result in a cell that is very similar to the Katablepharis cell. The feeding apparatus of Katablepharis spp. and the rest of the ciliates consists of two concentric microtubular arrays associated with vesicles. Information available from nucleotide sequencing of rRNA places the dinoflagellates in an ancestral position to the ciliates. The rRNA of Colponema and Katablepharis spp. has not yet been investigated. The use of stop codons in mRNA is discussed in relation to phylogeny.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bannister L. H. The structure of trichocysts in Paramecium caudatum. J Cell Sci. 1972 Nov;11(3):899–929. doi: 10.1242/jcs.11.3.899. [DOI] [PubMed] [Google Scholar]
- Barahona I., Soares H., Cyrne L., Penque D., Denoulet P., Rodrigues-Pousada C. Sequence of one alpha- and two beta-tubulin genes of Tetrahymena pyriformis. Structural and functional relationships with other eukaryotic tubulin genes. J Mol Biol. 1988 Aug 5;202(3):365–382. doi: 10.1016/0022-2836(88)90271-9. [DOI] [PubMed] [Google Scholar]
- Bardele C. F. A microtubule model for ingestion and transport in the suctorian tentacle. Z Zellforsch Mikrosk Anat. 1972;126(1):116–134. doi: 10.1007/BF00306784. [DOI] [PubMed] [Google Scholar]
- Bardele C. F. Transport of materials in the suctorian tentacle. Symp Soc Exp Biol. 1974;(28):191–208. [PubMed] [Google Scholar]
- Baroin A., Perasso R., Qu L. H., Brugerolle G., Bachellerie J. P., Adoutte A. Partial phylogeny of the unicellular eukaryotes based on rapid sequencing of a portion of 28S ribosomal RNA. Proc Natl Acad Sci U S A. 1988 May;85(10):3474–3478. doi: 10.1073/pnas.85.10.3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodansky S., Mintz L. B., Holmes D. S. The mesokaryote Gyrodinium cohnii lacks nucleosomes. Biochem Biophys Res Commun. 1979 Jun 27;88(4):1329–1336. doi: 10.1016/0006-291x(79)91126-4. [DOI] [PubMed] [Google Scholar]
- Bouck G. B., Sweeney B. M. The fine structure and ontogeny of trichocysts in marine dinoflagellates. Protoplasma. 1966;61(1):205–223. doi: 10.1007/BF01247920. [DOI] [PubMed] [Google Scholar]
- Brunk C. F., Kahn R. W., Sadler L. A. Phylogenetic relationships among Tetrahymena species determined using the polymerase chain reaction. J Mol Evol. 1990 Mar;30(3):290–297. doi: 10.1007/BF02099999. [DOI] [PubMed] [Google Scholar]
- Caron F., Meyer E. Does Paramecium primaurelia use a different genetic code in its macronucleus? Nature. 1985 Mar 14;314(6007):185–188. doi: 10.1038/314185a0. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. The evolutionary origin and phylogeny of microtubules, mitotic spindles and eukaryote flagella. Biosystems. 1978 Apr;10(1-2):93–114. doi: 10.1016/0303-2647(78)90033-3. [DOI] [PubMed] [Google Scholar]
- Corliss J. O. Nuclear characteristics and phylogeny in the protistan phylum Ciliophora. Biosystems. 1975 Nov;7(3-4):338–349. doi: 10.1016/0303-2647(75)90012-x. [DOI] [PubMed] [Google Scholar]
- Corliss J. O. The quest for the ancestor of the Ciliophora: a brief review of the continuing problem. Biosystems. 1988;21(3-4):323–331. doi: 10.1016/0303-2647(88)90029-9. [DOI] [PubMed] [Google Scholar]
- Eschbach S., Wolters J., Sitte P. Primary and secondary structure of the nuclear small subunit ribosomal RNA of the cryptomonad Pyrenomonas salina as inferred from the gene sequence: evolutionary implications. J Mol Evol. 1991 Mar;32(3):247–252. doi: 10.1007/BF02342747. [DOI] [PubMed] [Google Scholar]
- Fox T. D. Natural variation in the genetic code. Annu Rev Genet. 1987;21:67–91. doi: 10.1146/annurev.ge.21.120187.000435. [DOI] [PubMed] [Google Scholar]
- Gao X. P., Li J. Y. Nuclear division in the marine dinoflagellate Oxyrrhis marina. J Cell Sci. 1986 Sep;85:161–175. doi: 10.1242/jcs.85.1.161. [DOI] [PubMed] [Google Scholar]
- Greenwood S. J., Schlegel M., Sogin M. L., Lynn D. H. Phylogenetic relationships of Blepharisma americanum and Colpoda inflata within the phylum ciliophora inferred from complete small subunit rRNA gene sequences. J Protozool. 1991 Jan-Feb;38(1):1–6. doi: 10.1111/j.1550-7408.1991.tb04783.x. [DOI] [PubMed] [Google Scholar]
- Greenwood S. J., Sogin M. L., Lynn D. H. Phylogenetic relationships within the class Oligohymenophorea, phylum Ciliophora, inferred from the complete small subunit rRNA gene sequences of Colpidium campylum, Glaucoma chattoni, and Opisthonecta henneguyi. J Mol Evol. 1991 Aug;33(2):163–174. doi: 10.1007/BF02193631. [DOI] [PubMed] [Google Scholar]
- Gunderson J. H., Elwood H., Ingold A., Kindle K., Sogin M. L. Phylogenetic relationships between chlorophytes, chrysophytes, and oomycetes. Proc Natl Acad Sci U S A. 1987 Aug;84(16):5823–5827. doi: 10.1073/pnas.84.16.5823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haapala O. K., Soyer M. O. Absence of longitudinal differentiation of dinoflagellate (Prorocentrum micans) chromosomes. Hereditas. 1974;78(1):141–145. doi: 10.1111/j.1601-5223.1974.tb01436.x. [DOI] [PubMed] [Google Scholar]
- Hanyu N., Kuchino Y., Nishimura S., Beier H. Dramatic events in ciliate evolution: alteration of UAA and UAG termination codons to glutamine codons due to anticodon mutations in two Tetrahymena tRNAs. EMBO J. 1986 Jun;5(6):1307–1311. doi: 10.1002/j.1460-2075.1986.tb04360.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper D. S., Jahn C. L. Differential use of termination codons in ciliated protozoa. Proc Natl Acad Sci U S A. 1989 May;86(9):3252–3256. doi: 10.1073/pnas.86.9.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassouna N., Michot B., Bachellerie J. P. The complete nucleotide sequence of mouse 28S rRNA gene. Implications for the process of size increase of the large subunit rRNA in higher eukaryotes. Nucleic Acids Res. 1984 Apr 25;12(8):3563–3583. doi: 10.1093/nar/12.8.3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser M. Elektronemikroskopische Unteruschung an dem Suktor Paracineta limbata Maupas. Z Zellforsch Mikrosk Anat. 1970;106(4):584–614. [PubMed] [Google Scholar]
- Hauser M., Van Eys H. Microtubules and associated microfilaments in the tentacles of the suctorian Heliophrya erhardi Matthes. J Cell Sci. 1976 May;20(3):589–617. doi: 10.1242/jcs.20.3.589. [DOI] [PubMed] [Google Scholar]
- Hausmann K. Extrusive organelles in protists. Int Rev Cytol. 1978;52:197–276. doi: 10.1016/s0074-7696(08)60757-3. [DOI] [PubMed] [Google Scholar]
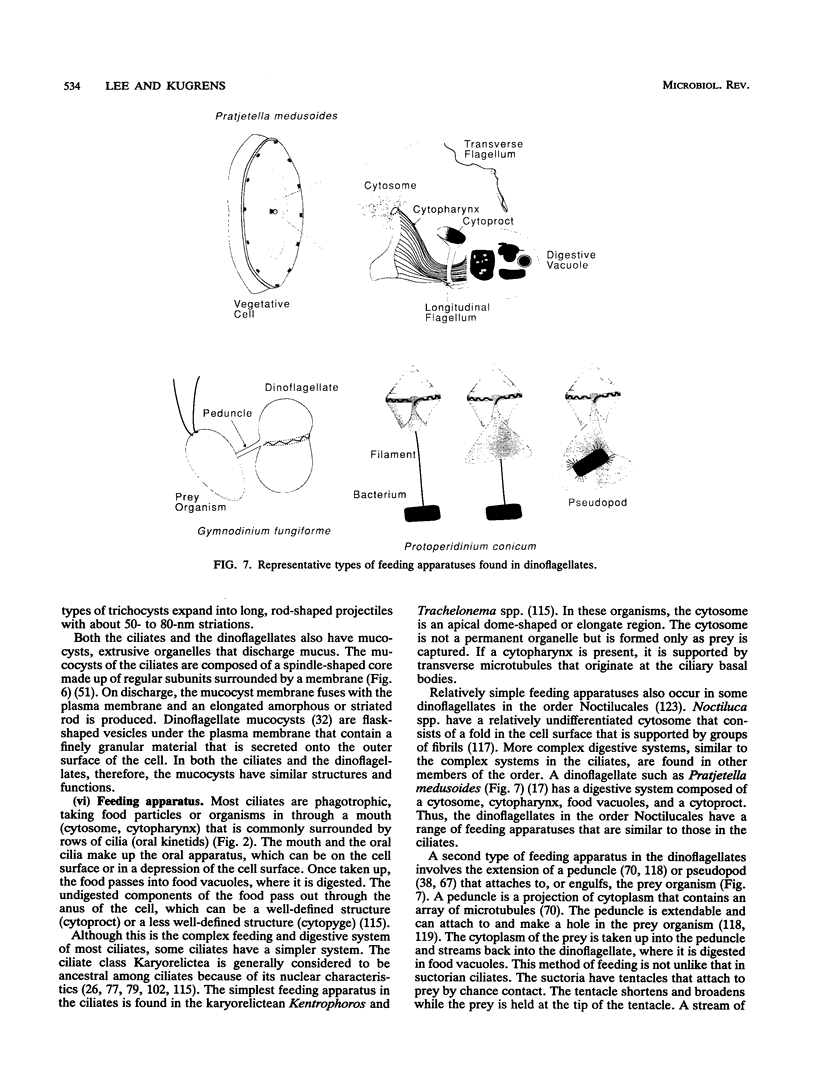
- Helftenbein E., Müller E. Both alpha-tubulin genes are transcriptionally active in Stylonychia lemnae. Curr Genet. 1988 May;13(5):425–432. doi: 10.1007/BF00365664. [DOI] [PubMed] [Google Scholar]
- Helftenbein E. Nucleotide sequence of a macronuclear DNA molecule coding for alpha-tubulin from the ciliate Stylonychia lemnae. Special codon usage: TAA is not a translation termination codon. Nucleic Acids Res. 1985 Jan 25;13(2):415–433. doi: 10.1093/nar/13.2.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendriks L., De Baere R., Van de Peer Y., Neefs J., Goris A., De Wachter R. The evolutionary position of the rhodophyte Porphyra umbilicalis and the basidiomycete Leucosporidium scottii among other eukaryotes as deduced from complete sequences of small ribosomal subunit RNA. J Mol Evol. 1991 Feb;32(2):167–177. doi: 10.1007/BF02515389. [DOI] [PubMed] [Google Scholar]
- Hendriks L., Van Broeckhoven C., Vandenberghe A., Van de Peer Y., De Wachter R. Primary and secondary structure of the 18S ribosomal RNA of the bird spider Eurypelma californica and evolutionary relationships among eukaryotic phyla. Eur J Biochem. 1988 Oct 15;177(1):15–20. doi: 10.1111/j.1432-1033.1988.tb14339.x. [DOI] [PubMed] [Google Scholar]
- Herrick G., Hunter D., Williams K., Kotter K. Alternative processing during development of a macronuclear chromosome family in Oxytricha fallax. Genes Dev. 1987 Dec;1(10):1047–1058. doi: 10.1101/gad.1.10.1047. [DOI] [PubMed] [Google Scholar]
- Herzog M., Soyer M. O. Distinctive features of dinoflagellate chromatin. Absence of nucleosomes in a primitive species Prorocentrum micans E. Eur J Cell Biol. 1981 Feb;23(2):295–302. [PubMed] [Google Scholar]
- Hitchen E. T., Butler R. D. The ultrastructure and function of the tentacle in Rhyncheta cyclopum Zenker (Ciliatea, Suctorida). J Ultrastruct Res. 1974 Feb;46(2):279–295. doi: 10.1016/s0022-5320(74)80062-6. [DOI] [PubMed] [Google Scholar]
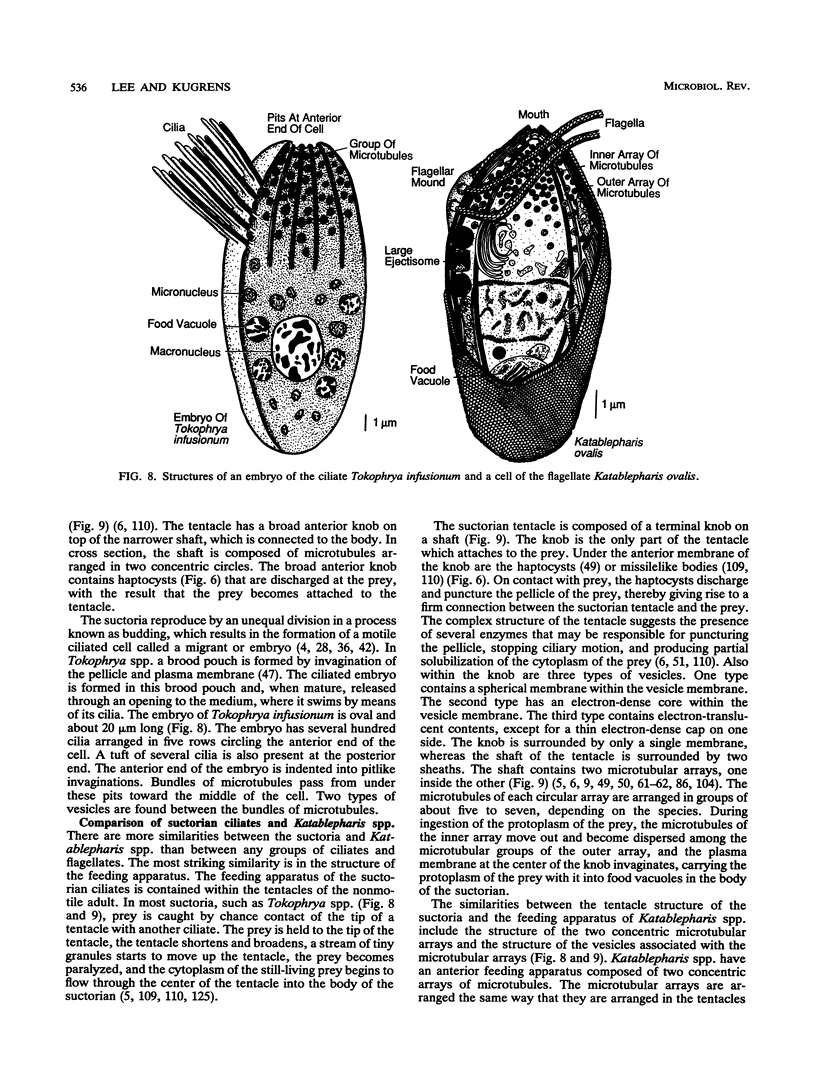
- Hitchen E. T., Butler R. D. Ultrastructural studies of the commensal suctorian, Choanophrya infundibulifera Hartog. I. Tentacle structure, movement and feeding. Z Zellforsch Mikrosk Anat. 1973 Oct 30;144(1):37–57. doi: 10.1007/BF00306685. [DOI] [PubMed] [Google Scholar]
- Hitchen E. T., Butler R. D. Ultrastructural studies of the commensal suctorian, Choanophrya infundibulifera Hartog. II. Tentacle morphogenesis. Z Zellforsch Mikrosk Anat. 1973 Oct 30;144(1):59–73. doi: 10.1007/BF00306686. [DOI] [PubMed] [Google Scholar]
- Hori H., Osawa S. Evolutionary change in 5S rRNA secondary structure and a phylogenic tree of 352 5S rRNA species. Biosystems. 1986;19(3):163–172. doi: 10.1016/0303-2647(86)90037-7. [DOI] [PubMed] [Google Scholar]
- Horowitz S., Gorovsky M. A. An unusual genetic code in nuclear genes of Tetrahymena. Proc Natl Acad Sci U S A. 1985 Apr;82(8):2452–2455. doi: 10.1073/pnas.82.8.2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchino Y., Hanyu N., Tashiro F., Nishimura S. Tetrahymena thermophila glutamine tRNA and its gene that corresponds to UAA termination codon. Proc Natl Acad Sci U S A. 1985 Jul;82(14):4758–4762. doi: 10.1073/pnas.82.14.4758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenaers G., Maroteaux L., Michot B., Herzog M. Dinoflagellates in evolution. A molecular phylogenetic analysis of large subunit ribosomal RNA. J Mol Evol. 1989 Jul;29(1):40–51. doi: 10.1007/BF02106180. [DOI] [PubMed] [Google Scholar]
- Lenaers G., Scholin C., Bhaud Y., Saint-Hilaire D., Herzog M. A molecular phylogeny of dinoflagellate protists (pyrrhophyta) inferred from the sequence of 24S rRNA divergent domains D1 and D8. J Mol Evol. 1991 Jan;32(1):53–63. doi: 10.1007/BF02099929. [DOI] [PubMed] [Google Scholar]
- Loeblich A. R., 3rd Dinoflagellate evolution: speculation and evidence. J Protozool. 1976 Feb;23(1):13–28. doi: 10.1111/j.1550-7408.1976.tb05241.x. [DOI] [PubMed] [Google Scholar]
- Lynn D. H., Small E. B. An update on the systematics of the phylum Ciliophora doflein, 1901: the implications of kinetid diversity. Biosystems. 1988;21(3-4):317–322. doi: 10.1016/0303-2647(88)90028-7. [DOI] [PubMed] [Google Scholar]
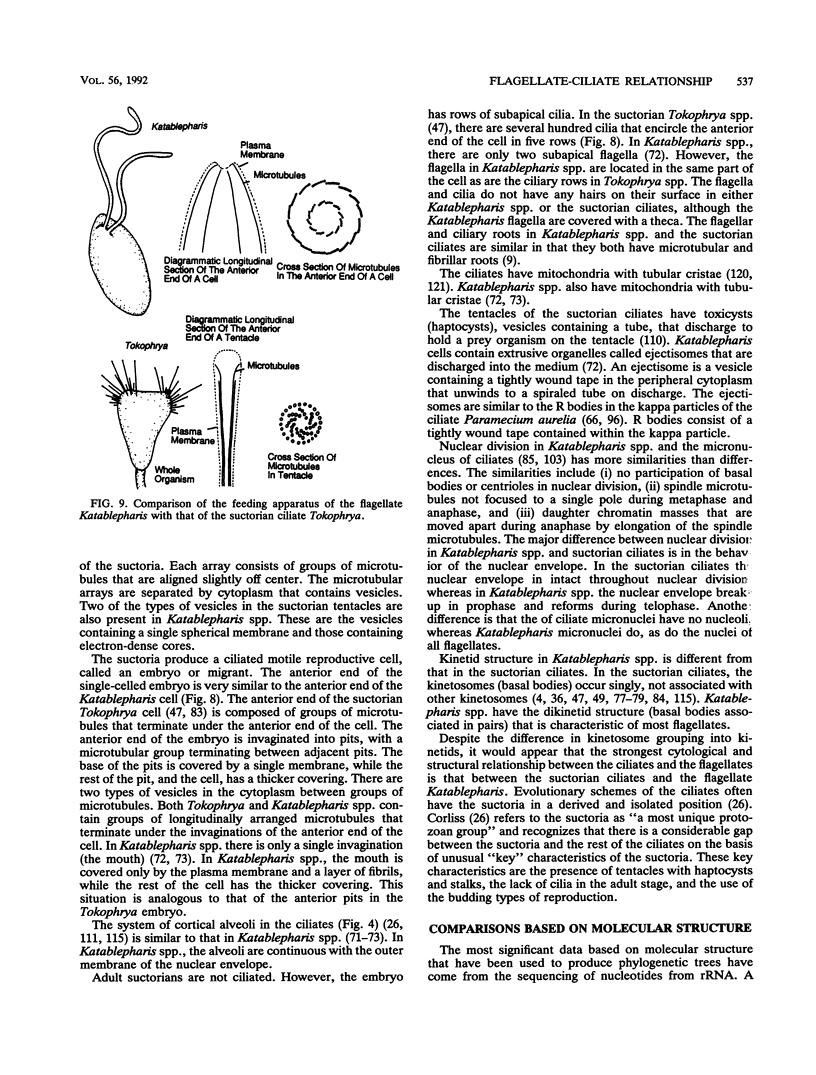
- Lynn D. H., Sogin M. L. Assessment of phylogenetic relationships among ciliated protists using partial ribosomal RNA sequences derived from reverse transcripts. Biosystems. 1988;21(3-4):249–254. doi: 10.1016/0303-2647(88)90020-2. [DOI] [PubMed] [Google Scholar]
- Michot B., Hassouna N., Bachellerie J. P. Secondary structure of mouse 28S rRNA and general model for the folding of the large rRNA in eukaryotes. Nucleic Acids Res. 1984 May 25;12(10):4259–4279. doi: 10.1093/nar/12.10.4259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millecchia L. L., Rudzinska M. A. The ultrastructure of nuclear division in a suctorian, Tokophrya infusionum. Z Zellforsch Mikrosk Anat. 1971;115(2):149–164. doi: 10.1007/BF00391122. [DOI] [PubMed] [Google Scholar]
- Nanney D. L., Meyer E. B., Simon E. M., Preparata R. M. Comparison of ribosomal and isozymic phylogenies of tetrahymenine ciliates. J Protozool. 1989 Jan-Feb;36(1):1–8. doi: 10.1111/j.1550-7408.1989.tb02661.x. [DOI] [PubMed] [Google Scholar]
- Nanney D. L., Mobley D. O., Preparata R. M., Meyer E. B., Simon E. M. Eukaryotic origins: string analysis of 5S ribosomal RNA sequences from some relevant organisms. J Mol Evol. 1991 Apr;32(4):316–327. doi: 10.1007/BF02102190. [DOI] [PubMed] [Google Scholar]
- Nanney D. L., Preparata R. M., Preparata F. P., Meyer E. B., Simon E. M. Shifting ditypic site analysis: heuristics for expanding the phylogenetic range of nucleotide sequences in Sankoff analyses. J Mol Evol. 1989 May;28(5):451–459. doi: 10.1007/BF02603080. [DOI] [PubMed] [Google Scholar]
- Nomoto M., Imai N., Saiga H., Matsui T., Mita T. Characterization of two types of histone H2B genes from macronuclei of Tetrahymena thermophila. Nucleic Acids Res. 1987 Jul 24;15(14):5681–5697. doi: 10.1093/nar/15.14.5681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen G. J. Earliest phylogenetic branchings: comparing rRNA-based evolutionary trees inferred with various techniques. Cold Spring Harb Symp Quant Biol. 1987;52:825–837. doi: 10.1101/sqb.1987.052.01.090. [DOI] [PubMed] [Google Scholar]
- Orias E. Derivation of ciliate architecture from a simple flagellate: an evolutionary model. Trans Am Microsc Soc. 1976 Jul;95(3):415–429. [PubMed] [Google Scholar]
- Osawa S., Jukes T. H., Watanabe K., Muto A. Recent evidence for evolution of the genetic code. Microbiol Rev. 1992 Mar;56(1):229–264. doi: 10.1128/mr.56.1.229-264.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perasso R., Baroin A., Qu L. H., Bachellerie J. P., Adoutte A. Origin of the algae. Nature. 1989 May 11;339(6220):142–144. doi: 10.1038/339142a0. [DOI] [PubMed] [Google Scholar]
- Prat A., Katinka M., Caron F., Meyer E. Nucleotide sequence of the Paramecium primaurelia G surface protein. A huge protein with a highly periodic structure. J Mol Biol. 1986 May 5;189(1):47–60. doi: 10.1016/0022-2836(86)90380-3. [DOI] [PubMed] [Google Scholar]
- Preer J. R., Jr, Hufnagel L. A., Preer L. B. Structure and behavior of R bodies from killer paramecia. J Ultrastruct Res. 1966 Apr;15(1):131–143. doi: 10.1016/s0022-5320(66)80100-4. [DOI] [PubMed] [Google Scholar]
- Preer J. R., Jr, Preer L. B., Rudman B. M., Barnett A. J. Deviation from the universal code shown by the gene for surface protein 51A in Paramecium. Nature. 1985 Mar 14;314(6007):188–190. doi: 10.1038/314188a0. [DOI] [PubMed] [Google Scholar]
- Preparata R. M., Beam C. A., Himes M., Nanney D. L., Meyer E. B., Simon E. M. Crypthecodinium and Tetrahymena: an exercise in comparative evolution. J Mol Evol. 1992 Mar;34(3):209–218. doi: 10.1007/BF00162970. [DOI] [PubMed] [Google Scholar]
- Qu L. H., Nicoloso M., Bachellerie J. P. Phylogenetic calibration of the 5' terminal domain of large rRNA achieved by determining twenty eucaryotic sequences. J Mol Evol. 1988 Dec;28(1-2):113–124. doi: 10.1007/BF02143502. [DOI] [PubMed] [Google Scholar]
- Rae P. M. Hydroxymethyluracil in eukaryote DNA: a natural feature of the pyrrophyta (dinoflagellates). Science. 1976 Dec 3;194(4269):1062–1064. doi: 10.1126/science.988637. [DOI] [PubMed] [Google Scholar]
- Rizzo P. J. Comparative aspects of basic chromatin proteins in dinoflagellates. Biosystems. 1981;14(3-4):433–443. doi: 10.1016/0303-2647(81)90048-4. [DOI] [PubMed] [Google Scholar]
- Rothschild L. J., Ragan M. A., Coleman A. W., Heywood P., Gerbi S. A. Are rRNA sequence comparisons the Rosetta stone of phylogenetics? Cell. 1986 Dec 5;47(5):640–640. doi: 10.1016/0092-8674(86)90505-2. [DOI] [PubMed] [Google Scholar]
- Rudzinska M. A. The fine structure and function of the tentacle in Tokophrya infusionum. J Cell Biol. 1965 Jun;25(3):459–477. doi: 10.1083/jcb.25.3.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satir B. H., Wissig S. L. Alveolar sacs of Tetrahymena: ultrastructural characteristics and similarities to subsurface cisterns of muscle and nerve. J Cell Sci. 1982 Jun;55:13–33. doi: 10.1242/jcs.55.1.13. [DOI] [PubMed] [Google Scholar]
- Schneider S. U., Leible M. B., Yang X. P. Strong homology between the small subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase of two species of Acetabularia and the occurrence of unusual codon usage. Mol Gen Genet. 1989 Sep;218(3):445–452. doi: 10.1007/BF00332408. [DOI] [PubMed] [Google Scholar]
- Small E. B., Lynn D. H. A new macrosystem for the phylum Ciliophora doflein, 1901. Biosystems. 1981;14(3-4):387–401. doi: 10.1016/0303-2647(81)90045-9. [DOI] [PubMed] [Google Scholar]
- Sogin M. L., Elwood H. J., Gunderson J. H. Evolutionary diversity of eukaryotic small-subunit rRNA genes. Proc Natl Acad Sci U S A. 1986 Mar;83(5):1383–1387. doi: 10.1073/pnas.83.5.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soyer M. O. Les ultrastructures liées aux fonctions de relation chez Noctiluca miliaris S. (Dinoflagellata) Z Zellforsch Mikrosk Anat. 1970;104(1):29–55. [PubMed] [Google Scholar]
- Taylor F. J. On dinoflagellate evolution. Biosystems. 1980;13(1-2):65–108. doi: 10.1016/0303-2647(80)90006-4. [DOI] [PubMed] [Google Scholar]
- Taylor F. J. Problems in the development of an explicit hypothetical phylogeny of the lower eukaryotes. Biosystems. 1978 Apr;10(1-2):67–89. doi: 10.1016/0303-2647(78)90031-x. [DOI] [PubMed] [Google Scholar]
- Tucker J. B. Microtubule arms and cytoplasmic streaming and microtubule bending and stretching of intertubule links in the feeding tentacle of the suctorian ciliate Tokophrya. J Cell Biol. 1974 Aug;62(2):424–437. doi: 10.1083/jcb.62.2.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzzell T., Corbin K. W. Fitting discrete probability distributions to evolutionary events. Science. 1971 Jun 11;172(3988):1089–1096. doi: 10.1126/science.172.3988.1089. [DOI] [PubMed] [Google Scholar]






