Summary
Changes in phospholipid composition and consequent loss of membrane integrity are correlated with loss of seed viability. Furthermore, phospholipid compositional changes affect the composition of the triacylglycerols, i.e. the storage lipids. Phospholipase D (PLD) catalyzes the hydrolysis of phospholipids to phosphatidic acid, and PLDα is an abundant PLD isoform. Although wild type seeds stored for 33 months were non-viable, 30 to 50% of PLDα-knockdown (PLD-KD) soybean seeds stored for 33 months germinated. Wild type and PLD-KD seeds increased in lysophospholipid levels and in triacylglycerol fatty acid unsaturation during aging, but the levels of lysophospholipids increased more in wild type than in PLD-KD seeds. The loss of viability of wild type seeds was correlated with alterations in ultrastructure, including detachment of the plasma membrane from the cell wall complex and disorganization of oil bodies. The data demonstrate that, during natural aging, PLDα affects the soybean phospholipid profile and the triacylglycerol profile. Suppression of PLD activity in soybean seed has potential for improving seed quality during long-term storage.
Keywords: genetic transformation, phospholipase D, seed viability, phospholipid, triacylglycerol, seed oil
Introduction
The preservation of seed viability and quality in storage is an important trait both for food usage and for seed use (Bentsink et al., 2000). Generally, viability and quality of seeds gradually deteriorate after harvest (Coolbear, 1995; McDonald, 1999), but the deterioration in long-term storage depends on environment, biochemical, biological, and genetic factors. Changes characterized during the aging process in seeds include alterations in membrane protein composition (Nowakowska & Rakowski, 2002), disruption of the nuclear envelope (Haithcock et al., 2005), protein degradation (Kumar et al., 1999), decreases in lipid content (Lin & Pearce, 1990), oxidative stress, and decreases in mRNA translation (Gidrol et al., 1990) and DNA replication capabilities (de Castro et al., 1995). Reduced levels of antioxidant enzymes such as superoxide dismutase, catalase, and ascorbate peroxidase can also lead to oxidative damage (Bailly et al., 1996; 1998).
Degradation of phospholipids and structural deterioration of biological membranes have been suggested to be particularly significant issues during aging in soybean seeds (Parrish & Leopold, 1978). When Ca2+ is added to young bean cotyledons, the action of phospholipase D (PLD) and phosphatidic acid phosphatase can induce lipid breakdown in smooth microsomal membranes (Duxbury et al., 1991). Duxbury et al. suggested that degradation of the phospholipid bilayer during natural aging of soybean cotyledons, facilitated by lipolytic enzymes, stimulates loss of protein function and proteolysis. In natural aging of seeds, the loss of seed vigor and germination ability is also coincident with increased electrolyte leakage (Leinonen, 1998; Rakowski et al., 1998). Moreover, changes in lipid composition, including oxidation and peroxidation can lead to loss of the economic and nutritional value of soybean seed through modifications in color, taste, odor, and viability during shipment and storage (Nakayama et al., 1981; Samama & Pearce, 1993).
Lipid degrading enzymes may play important roles in the natural aging process in seeds (Devaiah et al., 2007). PLD can catalyze degradation of membrane phospholipids, including phosphatidylcholine (PC), phosphatidylethanolamine (PE), and phosphatidylglycerol (PG), producing phosphatidic acid (PA) and a free head group. Accumulation of PA in the cell may lead to formation of hexagonal II-phase lipid particles with loss of cell membrane integrity (Kooijman et al., 2003). PA produced by phospholipase D is also known to regulate other cellular activities (Wang et al., 2006). It has been suggested that degradation of membrane lipids by PLDs also can lead to senescence and cell death (Fan et al., 1997).
Recently, we investigated alterations in phospholipid and triacylglycerol profiles caused by suppression of PLD in transgenic soybean (Lee et al., 2011). Knockdown of PLD induced increased unsaturation of phospholipids, mainly PC and PE, and decreased unsaturation of the triacylglycerol (TAG) fraction in fresh soybean seeds. These changes may have reflected a slowdown in formation of TAG from PC in the PLD-KD seeds compared to wild type seeds. Suppressing and ablating PLDα1 in Arabidopsis seeds enhanced seed viability and lipid stability resulting from reduced loss of oil fatty acids during aging (Devaiah et al., 2007). However, the effects of PLDα suppression on storage lipid profiles and particularly phospholipid membrane properties during seed aging are still not fully understood. Here, we investigate how knockdown of PLDα affects membrane phospholipids and storage lipids during aging and how these effects relate to stability of soybean seeds.
Results
Changes in the level of PLDα protein and enzyme activity in seeds of T2, null-transgenic, and the background soybean cultivar
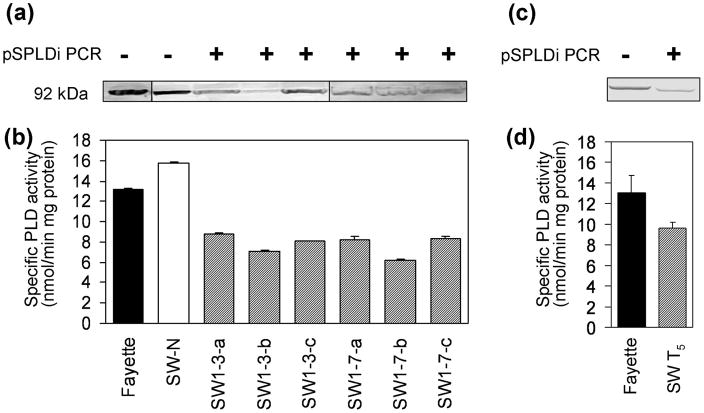
PLDα-suppressed (PLD-knockdown or PLD-KD) soybean lines were generated by transferring pSPLDi and pHG1 plasmids via particle inflow gun bombardment, resulting in knockdown of PLDα activity by RNA interference (RNAi) in the lines transformed with pSPLDi. PLD gene expression, physiological cost, and biochemical changes in PLD-KD transgenic soybean seeds were characterized in a previous report (Lee et al., 2011). To confirm the suppression of PLDα in pSPLDi transgenic soybean seed, pSPLDi PCR, western blotting for PLDα, and a PLDα enzyme assay were performed using immature cotyledons of T2 and T5 SW lines. The presence of the transgene in T2 SW1 and SW-derived T5 line seeds was confirmed by PCR, specifically amplifying the pSPLDi construct (Figure 1a and c). Monitoring soybean PLD protein in the soybean extract using an Arabidopsis thaliana PLDα1 (AtPLDα1) antibody showed that there was decreased PLDα in the immature cotyledons of transgenic SW1-3 and SW1-7 lines compared to wild type (Fayette) and the null-transgenic segregating seed, SW-N (Figure 1a). The size of the PLDα protein, 92 kDa in denaturing gels, was consistent with previous data (Ryu et al., 1996). When the endogenous PLD gene expression was suppressed by the RNAi machinery, the immature cotyledons of T2 and T5 transgenic lines showed a 40 to 50 percent reduction in PLDα activity compared to wild type, whereas null-transgenic segregating seed did not show a PLDα activity reduction (Figure 1b and d).
Figure 1.
pSPLDi PCR, western blotting for PLDα, and a PLDα enzyme activity in immature cotyledons from, (a) and (b), heterozygous T2 SW1 and, (c) and (d), a T5 SW line. (a) and (c) The presence of the pSPLDi transgene was confirmed with GOI PCR. + indicates PCR-positive for pSPLDi, and − indicates PCR-negative. Immunoblot analysis of PLDα was performed with Arabidopsis PLDα1 antibody, and PLDα proteins were detected at the 92 kDa position, based on protein markers. (b) and (d) PLDα enzyme assay of T2 SW1 and T5 SW seeds. PLDα enzyme activity was expressed as nmol of PC hydrolyzed min−1 mg−1total protein. Error bars indicate ± SE from three independent measurements .
Changes in PLDα-suppressed soybean seeds response to natural aging
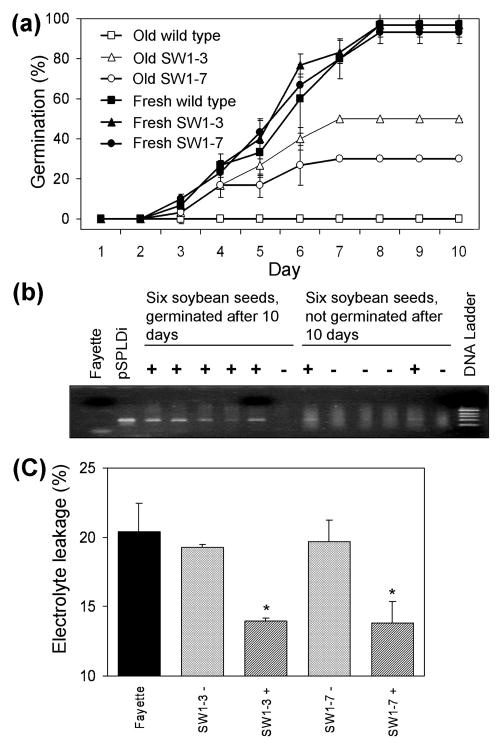
To determine whether PLDα activity affects soybean seed viability, seed germination and electrolyte leakage of fresh and naturally aged PLD-KD and wild type seeds were evaluated. The germination percentage of fresh soybean seeds from each genotype was approximately 95 percent. After 33 months storage at room temperature, no aged wild type seeds had germinated after 10 days of imbibition, whereas the germination of aged PLD-KD seeds of the SW1-3 and SW1-7 lines was observed to be 50 and 30%, respectively (Figure 2a). Because T2 seeds might be null, heterozygous, or homozygous for the transgene, we performed PCR analysis to determine the relationship between the presence of the transgene and seed viability in the SW1-3 progeny line (Figure 2b). PCR analysis showed that five out of 6 seedlings that germinated were transgenic, and only two out of 6 seeds that didn’t germinate were transgenic. These results suggest that the viability of 33 month-old seeds was associated with suppression of PLDα.
Figure 2.
Effect of PLDα suppression on seed germination of soybean seeds stored for 33 months. T2 SW1-3 and SW1-7 seeds were tested for germination, the presence of the pSPLDi transgene, and electrolyte leakage. (a) Fresh and aged soybean seeds were placed on a wet paper roll for 10 days, and soybean sprouts were counted to calculate germination (%, number of germinated seeds/total number of seeds × 100). Values are mean ± SD (n = 30 in three replications). (b) PCR analysis for the presence of pSPLDi transgene in soybean sprout from 33 month-old soybean seeds. (c) Effect of PLDα suppression on electrolyte leakage of 33 month-old soybean seeds. Wild type (Fayette), SW1-3 transgene PCR negative (−), SW1-3 transgene PCR positive (+), SW1-7 transgene PCR negative (−), and SW1-7 transgene PCR positive (+) seeds were tested for ion leakage by conductivity measurements. * indicates a significant difference at the p < 0.05 level in transgenic (PCR positive) or null-transgenic (PCR negative) compared to the wild type genotype (n = 3).
Leakage of endogenous electrolyte is indicative of poor membrane integrity. Electrolyte leakage from aged null-transgenic and wild type seeds was significantly greater than from aged PLD-suppressed lines (Figure 2c). Ion leakage from SW1-3 and SW1-7 was around 14%, whereas leakage from wild type seeds and null-transgenic soybean seeds was about 20%. Thus, the improved viability of PLD-KD seeds compared to the wild type seeds under natural aging was correlated with increased membrane stability.
Changes in ultrastructure of cotyledon tissues in response to natural aging
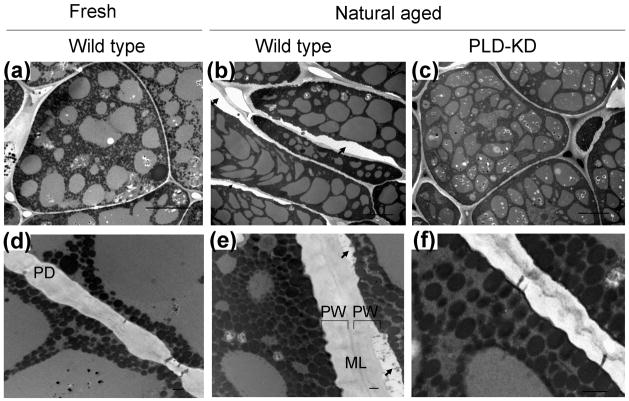
Transmission electron microscopy was used to investigate ultrastructural changes in plasma membrane and the cell wall complex of fresh wild type, aged wild type, and aged PLD-KD cotyledons (Figure 3). Obvious changes in cell structure were detected by light microscopy with 100× magnification (Figure S1). The plasma membrane in aged wild type cotyledon tissue was partially plasmolyzed and detached from the cell wall complex (Figure 3b). Plasma membrane detachment was observed in both aged PLD-KD and wild type cotyledon tissues; however, the plasma membrane in wild type cotyledons was irregularly piled, resulting from cell dehydration (Figure 3b and e). The presence of voids between the plasma membrane and secondary cell wall in aged wild type cotyledons indicated that a structural alteration of the plasma membrane occurred during the aging process. The greater structural changes that occurred in wild type seeds, in comparison to PLD-KD seeds, were coincident with the greater loss of seed viability and higher electrolyte leakage.
Figure 3.
Ultrastructure of cotyledon tissues andcell wall complex from fresh wild type [(a) and (d)], aged wild type [(b) and (e)], and aged PLD-KD [(c) and (f)] cotyledon tissues. In (b) and (e), black arrows indicate signs of partial plasmolysis. Bars in a, b, and e = 10 μm. Bars in d, e, and f= 0.5 μm. ML, middle lamella; PW, primary walls; PD, plasmodesmata.
Changes in phospholipid and triacylglycerol profiles after natural aging of wild type and PLD-KD soybean seeds
To determine the metabolic basis for the changes in membrane structure and electrolyte leakage during aging and to identify differences in lipid metabolism between wild type and PLD-KD seeds, the lipid compositions of the soybean seeds were analyzed by ESI-MS/MS. Total lysophosphatidylcholine (lysoPC), lysophosphatidylethanolamine (lysoPE), and lysophosphatidylglycerol (lysoPG) were dramatically increased in both wild type and PLD-KD seeds by natural aging, but increases were higher for lysoPC and lysoPG in wild type seeds compared to PLD-KD seeds (Table 1). PA was not significantly increased in aged seeds, compared to fresh seeds.
Table 1.
Total amount of lipid in each head group class in fresh and aged seeds of wild type and PLD-KD soybean. All PL classes and TAG were represented as mean ± SD (n = 5), and the same lower case letter indicates no significant difference at LSD (α < 0.05) level among fresh wild type seed, aged wild type seed, fresh PLD-KD, and aged PLD-KD seed.
| Polar lipid | Wild type | PLD-KD | ||
|---|---|---|---|---|
|
| ||||
| Fresh seed | Aged seed | Fresh seed | Aged seed | |
| Lipid per dry weight (nmol/mg)
| ||||
| PC | 15.90 ± 3.09a | 16.59 ± 2.00a | 14.92 ± 1.79a | 18.43 ± 6.15a |
| PE | 7.75 ± 1.27a | 5.56 ± 0.62a | 7.67 ± 1.18a | 7.47 ± 2.82a |
| PG | 0.52 ± 0.08a | 0.83 ± 0.15a | 0.52 ± 0.09a | 0.75 ± 0.35a |
| PI | 8.38 ± 1.52a | 8.75 ± 1.27a | 7.54 ± 0.80a | 9.63 ± 2.58a |
| PS | 0.26 ± 0.04a | 0.22 ± 0.05a | 0.23 ± 0.03a | 0.24 ± 0.07a |
| PA | 1.35 ± 0.24a | 1.53 ± 0.24a | 1.11 ± 0.20a | 1.48 ± 0.67a |
| LysoPC | 0.13 ± 0.02c | 0.96 ± 0.16a | 0.14 ± 0.02c | 0.56 ± 0.16b |
| LysoPE | 0.07 ± 0.02b | 0.27 ± 0.03a | 0.08 ± 0.01b | 0.23 ± 0.05a |
| LysoPG | 0.01 ± 0.00b | 0.05 ± 0.01a | 0.01 ± 0.00b | 0.02 ± 0.01b |
| MGDG | 0.04 ± 0.01a | 0.03 ± 0.02a | 0.04 ± 0.01a | 0.03 ± 0.01a |
| DGDG | 0.34 ± 0.11ab | 0.47 ± 0.13a | 0.23 ± 0.06b | 0.45 ± 0.12a |
| Total polar lipids | 34.75 ± 6.40a | 35.27 ± 4.11a | 32.49 ± 4.10a | 39.29 ± 11.75a |
|
| ||||
| Triacylglycerol | Wild type | PLD-KD | ||
|
| ||||
| Fresh seed | Aged seed | Fresh seed | Aged seed | |
|
| ||||
| Lipid per dry weight (normalized mass spectral signal/mg)
| ||||
| 16:0 containing | 14.75 ± 4.46a | 11.95 ± 1.72a | 9.73 ± 1.63a | 13.88 ± 3.83a |
| 18:0 containing | 4.99 ± 1.25a | 4.17 ± 0.90a | 3.60 ± 0.33a | 4.90 ± 1.64a |
| 18:1 containing | 38.72 ± 11.57a | 19.80 ± 2.65b | 40.03 ± 5.58a | 24.54 ± 4.97b |
| 18:2 containing | 31.02 ± 1.52ab | 32.96 ± 5.64ab | 20.89 ± 3.98b | 38.65 ± 10.14a |
| 18:3 containing | 5.41 ± 0.36a | 4.97 ± 1.00a | 4.65 ± 1.04a | 6.20 ± 2.11a |
| Total TAG signals | 94.89 ± 19.16a | 73.96 ± 11.92a | 78.90 ± 12.56a | 88.16 ± 22.68a |
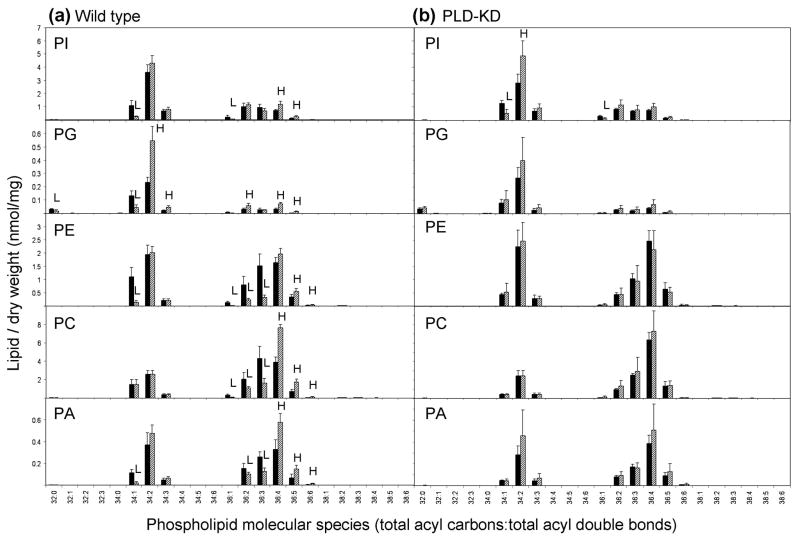
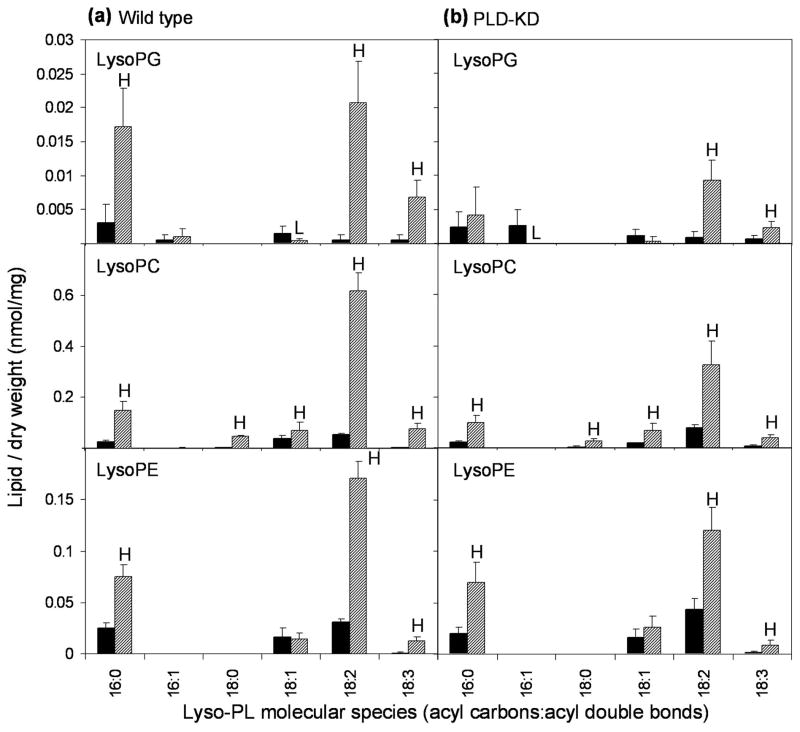
Analysis of lipids of wild type seeds at the level of molecular species showed that the phospholipids of WT seeds exhibited more significant changes during aging than did the phospholipids of PLD-KD seeds (Figure 4). The phospholipids of wild type seeds increased in unsaturation during natural aging, while the molecular species of PLD-KD phospholipids did not change as much during aging. In particular, diacyl 34-C phospholipid species with two or more double bonds and diacyl 36-C phospholipid species with four or more double bonds in the two acyl chains tended to increase during aging of wild type seeds, while those with fewer double bonds tended to decrease. For example, the lipid profiling data showed that 34:2-PG and 36:4-PC were dramatically increased in wild type seeds after natural aging, while 36:2- and 36:3- species of PE, PC, and PA were decreased.
Figure 4.
Lipid molecular species of wild type and PLD-KD fresh and aged seeds. (a) The black bars and the hatched bars represent wild type fresh and aged seeds, respectively. (b) The black bars and the hatched bars represent PLD-KD fresh and aged seeds, respectively. The values are the mean ± SD (n = 5). An H or L in the hatched bars indicates that the value is significantly higher or lower than that of fresh seeds at p < 0.05, respectively.
Table 2 delineates the individual fatty acyl composition of PCs, PEs, and PIs, with diacyl species designated 36:4, 36:3, and 36:2. Phospholipids with 36:4 species are primarily di18:2 combinations, and this combination becomes more predominant during aging in wild type seeds, while species containing 18:1 with 18:3 decline during aging. Phospholipids with 36:3, which tend to decrease during aging of wild type seeds, are primarily 18:1 with 18:2. PC and PE with 36:2, which decline during aging of wild type seeds, are a mixture of di18:1 and 18:0 with 18:2; during aging, di18:1 species exhibit a large decline in both genotypes. Overall, phospholipid data, shown in Figure 4 and Table 2, and summarized in Supplementary Figure S2, show that phospholipids containing 18:1 decline during aging in wild type seeds.
Table 2.
Individual fatty acyl groups of the major molecular species of PC, PE and PI of soybean seeds. Data were obtained by ESI-MS/MS in negative product ion scanning mode. Each PL species was determined to be a combination of two 18:n fatty acyl chains. PL species levels are indicated as mean mole % of that diacyl combination ± SD (n = 5) and differentiated at p < 0.05 among fresh wild type seed, aged wild type seed, fresh PLD-KD, and aged PLD-KD seed.
| 36:4-PC | 36:3-PC | 36:2-PC | ||||
|---|---|---|---|---|---|---|
|
| ||||||
| Di18:2 | 18:1+18:3 | 18:0+18:3 | 18:1+18:2 | Di18:1 | 18:0+18:2 | |
| Fresh wild type | 92.8±1.9b | 7.2±1.9a | 0.3±0.2b | 99.7±0.2a | 77.1±9.2a | 22.9±9.2c |
| Aged wild type | 97.6±1.4a | 2.4±1.4b | 2.1±1.7a | 97.9±1.7b | 17.4±6.5c | 82.6±6.5a |
| Fresh PLD-KD | 96.5±1.1a | 3.5±1.1b | 0.3±0.2b | 99.7±0.2a | 49.9±11.4b | 50.1±11.4b |
| Aged PLD-KD | 98.1±1.0a | 1.9±1.0b | 0.6±0.7b | 99.4±0.7a | 37.4±13.0b | 62.6±13.0b |
|
| ||||||
| 36:4-PE | 36:3-PE | 36:2-PE | ||||
|
| ||||||
| Di18:2 | 18:1+18:3 | 18:0+18:3 | 18:1+18:2 | Di18:1 | 18:0+18:2 | |
|
| ||||||
| Fresh wild type | 94.1±1.5b | 5.9±1.5a | 0.5±0.3a | 99.5±0.3a | 64.0±14.0a | 36.0±14.0c |
| Aged wild type | 98.3±0.8a | 1.7±0.8b | 1.5±2.1a | 98.5±2.1a | 7.2±3.3c | 92.8±3.3a |
| Fresh PLD-KD | 95.5±0.8b | 4.5±0.8a | 1.0±1.0a | 99.0±1.0a | 47.5±16.8ab | 52.5±16.8bc |
| Aged PLD-KD | 97.8±0.8a | 2.2±0.8b | 0.8±0.7a | 99.2±0.7a | 29.6±18.1b | 70.4±18.1b |
|
| ||||||
| 36:4-PI | 36:3-PI | 36:2-PI | ||||
|
| ||||||
| Di18:2 | 18:1+18:3 | 18:0+18:3 | 18:1+18:2 | Di18:1 | 18:0+18:2 | |
|
| ||||||
| Fresh wild type | 89.1±4.9b | 10.9±4.9a | 8.6±3.4ab | 91.4±3.4ab | 27.4±6.5a | 72.6±6.5b |
| Aged wild type | 95.8±3.4a | 4.2±3.4b | 14.4±7.4ab | 85.6±7.4ab | 1.6±0.5b | 98.4±0.5a |
| Fresh PLD-KD | 94.6±4.6ab | 5.4±4.6ab | 7.8±6.3b | 92.2±6.3a | 27.7±7.6a | 72.3±7.6b |
| Aged PLD-KD | 95.4±4.6a | 4.6±4.6b | 17.9±9.7a | 82.1±9.7b | 5.0±6.8b | 95.0±6.8a |
Figure 5 shows the molecular species of lysolipids formed during aging. As seen here and in Table 2, PLD-KD seeds produced fewer of these lipolytic products during natural aging than wild type seeds. Nearly every molecular species of lysoPC, lysoPE, and lysoPG levels increased during seed aging. The significantly greater gains during aging in lysoPC and lysoPG formation in wild-type compared to PLD-KD seeds (Table 1) were due mainly to increases in 18:2-lysoPLs.
Figure 5.
Changes in molecular species of lysoPC, lysoPE and lysoPG caused by natural aging of (a) wild type and (b) PLD-KD seeds, as revealed by ESI-MS/MS. The black bars represent fresh seeds and the hatched bars represent old seeds. The values are the mean ± SD (n = 5). An H or L indicates that the value is significantly higher or lower than that of fresh seeds at p < 0.05.
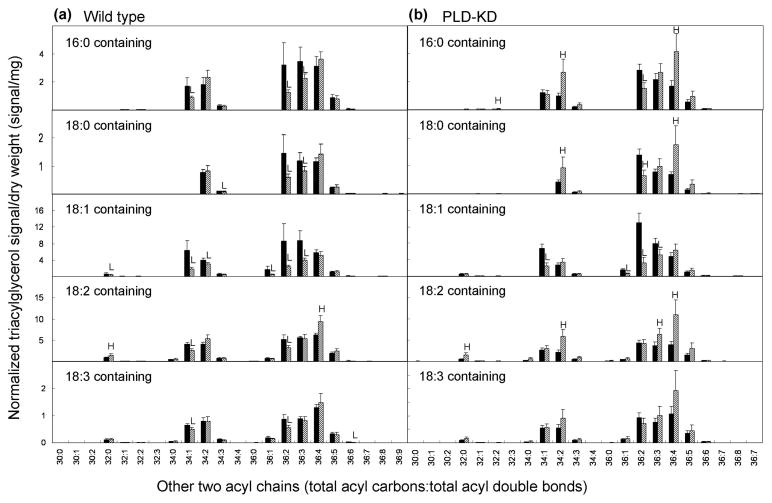
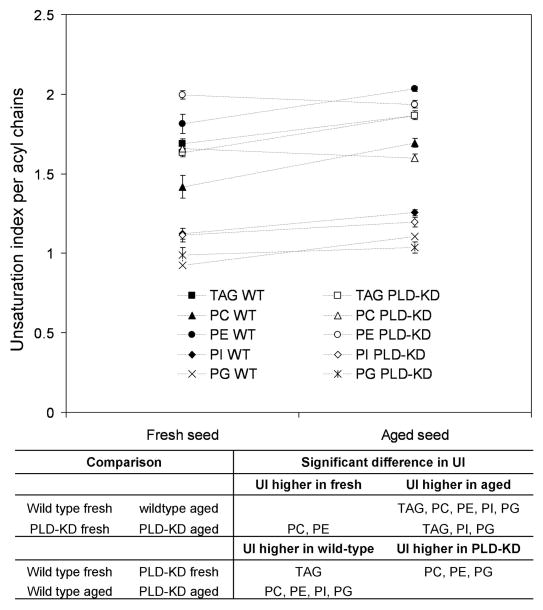
Triacylglycerols containing particular fatty acyl species were measured by ESI-MS/MS (as normalized mass spectral signal; Table 1 and Figure 6). This technique reveals overlapping subsets of TAG molecular species with one acyl chain specified and the other two determined together (Lee et al., 2011). During aging, the total amount of triacylglycerol signal in wild type seeds tended to decrease, albeit not quite significantly (p = 0.069) (Table 1). In addition TAG species in both genotypes became more unsaturated (Figures 6 and 7). Unsatuation index (UI), the average number of fatty acyl double bonds per fatty acid present, provides a simple measure of overall unsaturation (Figures 7 and S3).
Figure 6.
Changes in triacylglycerol molecular species resulted by natural aging in (a) wild type and (b) PLD-KD seeds measured by ESI-MS/MS with standards. The black bars and the hatched bars represent fresh and aged respectively. The value are the mean ± SD (n = 5). An H or L in the aged seed indicates that the value is significantly higher or lower than that of fresh seed of each cultivar at p < 0.05.
Figure 7.
Indexes of PL and TAG species in wild type and PLD-KD soybean seeds during natural aging. Unsaturation index of PL and TAG from fresh wild type (WT) seeds, fresh PLD-KD seeds, aged wild type seeds, and aged PLD-KD seeds. All values are mean ± SD of five biological replicates, and the significance data is shown belowat p <0.05 level.
There were significant decreases in 18:1-containing TAG molecular species in both genotypes (Table 1 and Figure 6), and in most TAG molecular species containing 36:2- and 36:3-diacyl components (Figure 6). 36:3 and 36:2 molecular species were demonstrated to contain 18:1 fatty acyl chains in PC (Table 2); because TAG diacyl groups are derived from PC (Bates et al., 2009), it is likely that 36:3 and 36:2 TAG molecular species also contain 18:1. Thus, these data, which are summarized in Supplementary Figure S3, are consistent with a tendency toward an overall decrease in 18:1 in TAG species in seeds; this trend was also detected directly by fatty acid compositional analysis (Supplementary Table 1).
Discussion
Post-harvest degradation of phospholipids reduces seed viability and oil quality during long-term storage of soybean seed (Wilson & McDonald, 1986; Danielle et al., 1993; Bailly et al., 1996). PLDs can be involved in membrane phospholipid degradation and loss of seed viability by natural and accelerated aging (Devaiah et al., 2007). We demonstrated that suppression of PLDα preserves soybean seed viability and quality during long-term storage. Immature seeds of PLD-suppressed T2 and T5 transgenic lines showed lower levels of PLDα protein and PLDα activity than null-transgenic and wild type cultivars (Figure 1). Agricultural traits of PLD-suppressed soybean were described in our previous report (Lee et al., 2011).
During storage, temperature, moisture, and time are critical factors affecting physical, physiological and biochemical changes that impact seed properties. These changes include loss of viability, grain color changes, alterations in moisture content, decomposition of lipids, degradation of protein, and damage to plasma and organelle membranes (Bailly et al., 1996; 1998; Kumar et al., 1999). After storage for 33 months, wild type seeds failed to germinate, whereas aged PLD-KD transgenic seeds maintained 30 to 50% germination (Figure 2a). The greater viability of PLD-KD seeds compared to wild type after aging was associated with (1) decreased electrolyte leakage (Figure 2) (2) fewer ultrastructural changes (Figure 3), and (3) fewer lipid compositional changes. In particular, PLD-KD seeds produced lower levels of lysoPLs than wild type seeds, and TAG levels in PLD-KD seeds were maintained during aging, while TAG levels in wild type seeds tended to decrease during aging.
Loss of viability has been shown to be positively correlated with an increase in electrolyte leakage in sunflower seeds during natural aging (Corbineau et al., 2002). Electrolyte leakage could be caused by membrane phospholipid deterioration. Sphingolipids, ubiquitous and particularly abundant integral components of the plasma membrane in eukaryotic cells, are involved in various cellular functions (Hait et al, 2006). Phospholipase D and phosphatidic acid have been shown to affect activity of plant sphingolipid-metabolizing enzymes and sphingolipid profiles (Guo et al., 2011). Recently, altered sphingolipid profiles have been shown to change the leaf ionome in Arabidopsis (Chao et al., 2011). Thus, we cannot rule out the notion that differences in membrane integrity between non-transgenic and transgenic seeds are caused by differences in sphingolipid metabolism, secondary to the phospholipase D differences, during natural aging. Additionally, the storage lipids, mainly TAG, in soybean seeds are coated with a phospholipid monolayer to form a lipid body in the cytosol. Lipolysis of phospholipids at the oil body surface may destabilize the oil bodies, contributing to their degradation, eventually causing deterioration of seed quality. PLDα is located in the cytosol and can act on lipids in the plasma membrane or in intracellular (microsomal) membranes (Xu et al., 1996).
It has been suggested that high levels of PLDα1 activity cause formation of PA, resulting in membrane and storage lipid degradation in seeds (List et al., 1992). In this study, PA levels in seeds at 33 months were not significantly increased in either genotype (Table 1 and Figure 4). Devaiah et al. (2007) reported that levels of PA increased 30% after accelerated aging treatment in wild type Arabidopsis. Despite the lack of a measurable change, PLDα may be acting locally to create PA involved in signaling responses (Wang et al., 2006). Such responses may lead to local alterations in enzyme activities that result in compromised membrane integrity. For example, the lower level of lysophospholipids observed in PLD-KD seed, compared to wild type seed, may reflect lower levels of lipolytic activity by phopholipase As during natural aging. It has been reported that endogenously accumulated PA formed by PLD action can enhance PLA2 activity in mammalian cells (Sato et al., 1993; Kinkald et al., 1998), and it’s possible that PLD or its PA may also stimulate phospholipase A activity in soybean seeds. LysoPLs may increase membrane permeability during natural aging, and the somewhat higher levels of these lipolytic products in the wild type seeds compared to the PLD-KD seeds may contribute to the higher electrolyte leakage observed in the wild type seeds.
Significant differences in the unsaturation of the PC of wild type and PLD-KD seeds were observed (Figures 4 and 7; Lee et al., 2011). Our previous report suggested that knockdown of PLD may retard the conversion of PC to DAG to TAG in developing soybean seed, causing fatty acids retained in PC, which is the substrate for desaturases, to become more unsaturated in fresh seeds (Lee et al., 2011). Here, we show that the overall levels of TAG unsaturation in aged wild type and PLD-KD seeds are similar, but aged seeds of both wild type and PLD-KD lines contain more unsaturated TAG species than fresh seeds. The data suggest the possibility that lipid metabolism, including acyl desaturation, occurs during aging of mature seeds. Further work is needed to test this notion. Stable isotope labeling would be an attractive approach, but labeling intact seeds and analyzing over an extended time is logistically challenging.
In the current study, RNAi silencing with a partial PLDα sequence and seed specific β-conglycinin α′ subunit promoter showed around 40% reduction in PLDα activity compared to wild type seed. Devaiah et al. (2007) showed that, in Arabidopsis, seed vigor and germination of PLDα-knockout Arabidopsis was better than that of the PLDα-ablated pldα1-1 after natural aging, and they proposed that PLDα activity is needed for optimal germination and seedling growth. PLDα-attenuation did affect some agronomic traits of soybean, such as seed quality, plant height, and seed weight (Lee et al., 2011). Considering the data described herein, we propose that suppression of PLDα in soybean seeds improves seed oil storability and long-term viability of seeds after harvest.
Experimental Procedures
Plant material, seed storage, germination test, and electrolyte leakage
The soybean cultivar, Fayette (background cultivar or “wild type” in this study) (Bernard et al., 1988), was co-transformed with pSPLDi harboring RNAi structure (800 bp sense and 1300 bp antisense soybean PLD fragments) and pHG1 (CaMV35S:HPH) via particle inflow gun bombardment and regenerated (Lee et al., 2011). T1 soybean lines SW1-3, SW1-7, SW1-5-N (null transgenic line) and wild type were grown in a greenhouse under 25 ± 2 °C day and 20 ± 2 °C night temperatures and a 16 h light/8 h dark photoperiod. Immature pods were analyzed at approximately 26 days after fertilization (DAF) by PCR, enzyme assay, and western blotting for PLDα. T2 seeds were harvested from each T1 plant, and dried to 13% moisture. T2 transgenic and wild type seeds were stored in paper envelopes at 25 °C and 17% relative humidity for 33 months, and T5 seeds derived from SW1-3 and SW1-7 lines were harvested and stored under the same conditions for two months. The viability of transgenic, SW1-3 and SW1-7, and wild type seeds stored for 2 and 33 months was evaluated using a standard germination test. Three replicates of ten seeds from each genotype were wrapped in a 25 cm-long roll of paper and drenched in water. One end of the paper was submerged. The water level was maintained 5 cm below the seeds in order to supply the same amount of water to each seed. The number of soybean sprouts was counted after 10 days.
Electrolyte leakage from seeds stored 2 and 33 months was measured by a modification of the method of Fan et al. (1997). Briefly, three soybean seeds were agitated in 3 ml of 0.4 M mannose for 6 h to induce ion leakage. Conductivity of the solution was measured with a YSI model 32 conductance meter (Yellow Springs Instrument Co., Inc.) after vortexing the samples for 30 s. The total electrolyte concentration was determined after boiling samples for 15 min and cooling to 25 °C. Electrolyte leakage was expressed as a percentage of the initial electrolytic concentration divided by total the electrolyte concentration in the solution. Three replicates with three seeds per replication were evaluated for each soybean line.
PCR analyses
Genomic DNA was extracted from young soybean leaves and immature seeds using a simplified CTAB (cetyl trimethylammonium bromide) DNA extraction method by Saghai-Maroof et al. (1984). Two hundred ng of genomic DNA were used as a template in a 50 μl PCR reaction containing 20 pmol each of forward and reverse primer, 0.2 mM of deoxynucleotide triphosphate (dNTPs), 1.25 U Taq DNA polymerase, and 1.5 mM MgCl2 in PCR buffer (New England BioLabs, MA, USA). Specific primer sequences of the transgene left arm (forward: 5′-TATTTCAACACCCGTCA-3′ and reverse: 5′-CCACCATGACATGCGGAAACC-3′), the transgene right arm (forward: 5′-TTTCCGCATGTCATTGTGGTA-3′ and reverse: TAGCCCGATACTTTCCT-3′), and hygromycin phosphotransferase (forward: 5′-GCACAATCCCACTATCCTTCGCAA-3′ and reverse: 5′-CTTCTACACAGCCATCGGTCCAG-3′), designed by primer3 software (http://frodo.wi.mit.edu), were used for transgene PCR using a Hybaid PCR Exress Thermocycler (Hybaid, UK) and a Multigene Thermocycler (Labnet International Inc., NJ, USA). The left arm and right arm of the pSPLDi construct were amplified using one cycle at 94 °C for 5 min; 30 cycles at 94 °C for 30 sec, 52 °C for 60 sec, and 72 °C for 90 sec; and a final cycle at 72 °C for 7 min with forward βCONGF (5′-taa ttc aac acc cgt ca-3′) and reverse SQSP1 (5′-cca caa tga cat gcg gaa acc-3′). PCR products were run on 0.8% agarose gel with 1X TAE buffer at 50 V. The gel was stained in ethidium bromide solution for 10 min, and the stained gel image was acquired with Kodak 1D™ image analysis software (Eastman Kodak Company, New Haven, CT, USA) on a UV transilluminator.
Western blot analysis and PLD enzyme assay
Total protein from a T2 immature cotyledon was extracted using a chilled pestle in a 1.5 mL microcentrifuge tube with liquid N2 and a homogenization buffer (50 mM Tris-HCl, pH 7.5, 10 mM KCl, 1 mM EDTA, 0.5 mM phenyl methylsulfonyl fluoride, and 2 mM DTT) at 4 °C (Fan et al.,1997). The concentration of total native protein was measured with a Nanodrop ND1000 spectrophotometer (Nanodrop, DE, USA). For western blot analysis, 20 μg of total native proteins were incubated at 95 °C for 15 min with SDS-PAGE loading buffer (100 μl of 50 mM Tris-HCL, pH 6.8, 10 mM DTT, 2% SDS, 0.01% bromophenol blue, and 10% glycerol). Denatured protein was separated in 10% SDS-PAGE gel at 100 V until the loading dye reached the bottom of the gel. Gels were transferred to a nitrocellulose membrane (Millipore, Bedford, MA, USA) using electrophoretic transfer with a Bio-Rad criterion blotter (Bio-Rad, Hercules, CA, USA) according to the manufacturer’s instructions. The transferred membrane was blotted with Arabidopsis PLDα antibodies as described by Zhang et al. (2003). Briefly, non-fat milk was used for membrane blocking for 1 h with agitation at 4° C, and membranes were soaked in AtPLDα antibody (1:1000 in TBST containing 10 mM Tris, 140 mM NaCl and Tween 20, pH 7.4) overnight. The blots were washed 3 times in 1X TBST for 5 min per wash. The blots were incubated in a secondary antibody (1:1000 diluted Goat anti-rabbit IgG (H+L) horseradish peroxide conjugate (Bio-Rad, CA, USA) in TBST buffer) for 1 h and then washed two times in 1X TBST followed by one wash in TBS (10 mM Tris, 140 mM NaCl, pH 7.5). The protein bands binding to antibodies were visualized with a HRP color development reagent (Bio-Rad, CA, USA).
Aliquots of 20 μg of native protein were used for the PLD enzyme assay as described previously (Fan et al., 1997). Briefly, the reaction mixture contained 100 mM MES, pH 6.5, 25 mM CaCl2, 0.5 mM SDS, 1% (v/v) ethanol, and 2 mM PC (egg yolk) containing dipalmitoyl glycerol-3-phosphate-(methyl-3H)-choline in a final volume of 200 μl. Substrate preparation, reaction conditions, and product separation were based on a previously described procedure (Wang et al., 1993). The release of 3H-labeled choline into the aqueous phase was quantified by liquid scintillation counting (Beckman-Coulter, CA, USA).
Transmission electron microscopy (TEM)
Soybean tissues were prepared for TEM and the image captured using a protocol modified from that of Boyle and Takemoto (1998). Briefly, soybean cotyledon tissues were immersed and fixed in 2% paraformaldehyde (Ladd, Burlington, VT, USA), 2% glutaraldehyde, and 10% sucrose in 0.1 M sodium cacodylate buffer pH 6.8 for 16 h at room temperature with constant rotation. Samples were post-fixed with 2% osmium tetroxide and 10% sucrose in 0.1 M sodium cacodylate buffer pH 6.8. Fixed tissues were dehydrated in an ascending series of acetone with 50% – 100% at room temperature, embedded in Spurr’s embedding media in gelatin capsules, and polymerized at 65 °C for 24 h. Embedded soybean tissues were cut with glass knives on a Reichert Ultracut S microtome (Leica, Austria) and held on 100 nickel slotted grids. All images were captured using an Advanced Microscopy Techniques digital image capturing system (ATM, Chazy, NY, USA).
Polar lipid, triacylglycerol, and major fatty acid analyses
Lipids were extracted from soybean seed and analyzed by ESI-MS/MS as described by Lee et al. (2011). Fatty acid composition was determined by gas-liquid chromatography on a Supelco 30-m Omegawax 250 capillary column (Sigma-Aldrich, St. Louis, MO, USA) after derivatization of the fatty acids to methyl esters in 1.5 M methanolic HCl, followed by addition of water, and extraction with pentane.
Unsaturation index and percentage of MUFA and PUFA
The unsaturation index (UI) was calculated as the average number of double bonds per fatty acid present (the sum of the percentage of each lipid × number of double bonds)/(number of fatty acid species per lipid, i.e. 2 for PLs and 3 for TAGs) with molecular species composition of TAG and PLs determined by ESI-MS/MS. UI data are presented as mean ± SD and Tukey’s studentized range test with five determinations. Percentage of monounsaturated fatty acid (% MUFA) and percentage of polyunsaturated fatty acid (% PUFA) were calculated from molecular species composition of each PL class and total TAG, and then % unsaturated fatty acid was calculated from each molecular species composition (sum of MUFA and PUFA divided by total molecular composition × 100). The molecular species compositions used for the calculations (for phospholipids and TAGs) were: 36:6, di18:3; 36:5, 18:3/18:2; 36:4, 36:3, and 36:2, as indicated in Table 2; 34:3, 16:0/18:3; 34:2, 16:0/18:2; 34:1, 16:0/18:1; 32:0, di16:0.
Statistical analysis
Data collected from the fatty acid composition, electrolyte leakage analysis, PLD enzyme assays, and UI were examined by analysis of variance using SAS (SAS Institute Inc., NC, USA) PROC general linear model (GLM) procedure. Means were separated using the LSD or Tukey test at the p < 0.05 probability level.
Supplementary Material
Acknowledgments
This work was supported by a Kansas Soybean Commission grant. The lipid analyses described in this work were performed at the Kansas Lipidomics Research Center Analytical Laboratory with funding from KSU’s Targeted Excellence program. Equipment acquisition and method development at the Kansas Lipidomics Research Center were funded by National Science Foundation (EPS 0236913, MCB 0455318 and 0920663, DBI 0521587), Kansas Technology Enterprise Corporation, K-IDeA Networks of Biomedical Research Excellence (INBRE) of National Institute of Health (P20RR16475), and Kansas State University. We thank Juliane S. Essig and Marcy L. Main for their technical support with soybean genetic transformations, Dr. Richard Jeannotte for lipid analysis, Suqin Zheng of Dr. Xuemin Wang’s laboratory for donating soybean PLD cDNA, Dr. Shivakumar Devaiah of Dr. Wang’s lab for PLD enzyme analysis, Dr. Daniel Boyle of Kansas State University for TEM imaging, Dr. Yoon-Sup So of North Carolina State University for statistical advice. This article is contribution no. 11-170-J from the Kansas Agricultural Experimental Station, Kansas State University, Manhattan, KS.
Footnotes
Additional supporting information may be found in the online version of this article.
References
- Bailly C, Benamar A, Corbineau F, Come D. Changes in malondialdehyde content and in superoxide dismutase, catalase and glutathione reductase activities in sunflower seeds as related to deterioration during accelerated aging. Physiol Plant. 1996;97:104–110. [Google Scholar]
- Bailly C, Benamar A, Corbineau F, Come D. Free radical scavenging as affected by accelerated ageing and subsequent priming in sunflower seeds. Physiol Plant. 1998;104:646–652. [Google Scholar]
- Bates PD, Durrett TP, Ohlrogge JB, Pollard M. Analysis of acyl fluxes through multiple pathways of triacylglycerol synthesis in developing soybean embryos. Plant Physiol. 2009;150:55–72. doi: 10.1104/pp.109.137737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentsink L, Alonso-Blanco C, Vreugdenhil D, Tesnier K, Groot SPC, Koornneef M. Genetic analysis of seed-soluble oligosaccharides in relation to seed storability of Arabidopsis. Plant Physiol. 2000;124:1595–1604. doi: 10.1104/pp.124.4.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard RL, Noel GR, Anand SC, Shannon JG. Registration of Fayette soybean. Crop Sci. 1988;28:1028–1029. [Google Scholar]
- Boyle DL, Takemoto LJ. Finger-like projections of plasma membrane in the most senescent fiber cells of human lenses. Current Eye Res. 1998;17:1118–1123. doi: 10.1076/ceyr.17.12.1118.5130. [DOI] [PubMed] [Google Scholar]
- Chao DY, Gable K, Chen M, Baxter I, Dietrich CR, Cahoon EB, Guerinot ML, Lahner B, Lü S, Markham JE, Morrissey J, Han G, Gupta SD, Harmon JM, Jaworski JG, Dunn TM, Salt DE. Sphingolipids in the root play an important role in regulating the leaf ionome in Arabidopsis thaliana. Plant Cell. 2011;23:1061–1081. doi: 10.1105/tpc.110.079095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coolbear P. Mechanism of seed deterioration. In: Basra AS, editor. Seed Quality: Basic Mechanisms and Agricultural Implications. New York: Food Product Press; 1995. pp. 223–277. [Google Scholar]
- Corbineau F, Gay-Mathieu C, Vinel D, Côme D. Decrease in sunflower (Helianthus annuus) seed viability caused by high temperature as related to energy metabolism, membrane damage and lipid composition. Physiol Plant. 2002;116:489–496. [Google Scholar]
- de Castro RD, Zheng X, Bergervoet JHW, De Vos CHR, Bino RJ. β-tubulin accumulation and DNA replication in imbibing tomato seeds. Plant Physiol. 1995;109:499–504. doi: 10.1104/pp.109.2.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaiah SP, Pan X, Hong Y, Roth M, Welti R, Wang X. Enhancing seed quality and viability by suppressing phospholipase D in Arabidopsis. Plant J. 2007;50:950–957. doi: 10.1111/j.1365-313X.2007.03103.x. [DOI] [PubMed] [Google Scholar]
- Duxbury CL, Legge RL, Thompson JE. Lipid breakdown in smooth microsomal membranes from bean cotyledons alters membrane proteins and induces proteolysis. J Exp Bio. 1991;42:103–112. [Google Scholar]
- Fan L, Zheng S, Wang X. Antisense suppression of phospholipase Dα retards abscisic acid- and ethylene-promoted senescence of postharvest Arabidopsis leaves. Plant Cell. 1997;9:2183–2196. doi: 10.1105/tpc.9.12.2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gidrol S, Noubhani A, Pradet A. Biochemical changes induced by accelerated aging in sunflower seeds. II RNA populations and protein synthesis. Physiol Plant. 1990;80:598–604. [Google Scholar]
- Guo L, Mishra G, Taylor K, Wang X. Phosphatidic acid binds and stimulates Arabidopsis sphingosine kinases. J Biol Chem. 2011;286:13336–13345. doi: 10.1074/jbc.M110.190892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hait NC, Oskeritzian CA, Paugh SW, Milstien S, Spiegel S. Sphingosine kinases, sphingosine 1-phosphate, apoptosis and disease. Biochim Biophys Acta. 2006;1758:2016–2026. doi: 10.1016/j.bbamem.2006.08.007. [DOI] [PubMed] [Google Scholar]
- Haithcock E, Dayani Y, Neufeld E, Zahand AJ, Feinstein N, Mattout A, Gruenbaum Y, Liu J. Age-related changes of nuclear architecture in Caenorhabditis elegans. Proc Natl Acad Sci USA. 2005;102:16690–16695. doi: 10.1073/pnas.0506955102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinkald AR, Othman R, Voysey J, Wilton DC. Phospholipase D and phosphatidic acid enhance the hydrolysis of phospholipids in vesicles and in cell membranes by human secreted phospholipase A2. Biochim Biophys Acta. 1998;1390:173–185. doi: 10.1016/s0005-2760(97)00181-1. [DOI] [PubMed] [Google Scholar]
- Kooijman EE, Chupin V, de Kruijff B, Burger KN. Modulation of membrane curvature by phosphatidic acid and lysophosphatidic acid. Traffic. 2003;4:162–174. doi: 10.1034/j.1600-0854.2003.00086.x. [DOI] [PubMed] [Google Scholar]
- Kumar GNM, Houtz LR, Knowles NR. Age-induced protein modifications and increased proteolysis in potato seed-tubers. Plant Physiol. 1999;119:89–100. doi: 10.1104/pp.119.1.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Welti R, Schapaugh WT, Trick HN. Phospholipid and triacylglycerol profiles modified by PLD suppression in soybean seed. Plant Biotech J. 2011;9:359–372. doi: 10.1111/j.1467-7652.2010.00562.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leinonen K. Effect of storage conditions on dormancy and vigor of Picea abies seeds. New Forest. 1998;16:231–249. [Google Scholar]
- Lin SS, Pearce RS. Changes in lipids of bean seed (Phaseolus vulgaris) and corn caryopses (Zea mays) aged in contrasting environments. Ann Botany. 1990;65:451–456. [Google Scholar]
- List GR, Mounts TL, Lanser AC. Factors promoting the formation of nonhydratable soybean phosphatides. JAOCS. 1992;69:443–446. [Google Scholar]
- McDonald MB. Seed deterioration: physiology, repair and assessment. Seed Sci Technol. 1999;27:177–237. [Google Scholar]
- Nakayama Y, Sayo K, Kito M. Decomposition of phospholipids in soybean during storage. Cereal Chem. 1981;58:260–264. [Google Scholar]
- Nowakowska J, Rakowski K. Accelerated and natural ageing processes change the properties of plasma membrane in Norway spruce (Picea abies L. Karst) seeds during storage. Dendrobiology. 2002;47:79–82. [Google Scholar]
- Parrish DJ, Leopold AC. On the mechanism of aging in soybean seeds. Plant Physiol. 1978;61:365–368. doi: 10.1104/pp.61.3.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakowski K, Behzadipour M, Ratajczak R, Kluge M. Effect of seeds freezing and storage conditions on plasma membrane properties of Norway spruce (Picea abies Karst) seedlings. Botanica Acta. 1998;111:236–240. [Google Scholar]
- Ryu SB, Zheng L, Wang X. Changes in phospholipase D expression in soybeans during seed development and germination. JAOCS. 1996;73:1171–1176. [Google Scholar]
- Saghai-Maroof MA, Soliman KM, Jorgensen RA, Allard RW. Ribosomal DNA spacer-length polymorphisms in barley: Mendelian inheritance, chromosomal location, and population dynamics. Proc Natl Acad Sci USA. 1984;81:8014–8018. doi: 10.1073/pnas.81.24.8014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samama AM, Pearce RS. Ageing of cucumber and onion seeds-phospholipase-D, lipoxygenase activity and changes in phospholipid content. J Exp Bot. 1993;44:1253–1265. [Google Scholar]
- Sang Y, Cui D, Wang X. Phospholipase D and phosphatidic acid-mediated generation of superoxide in Arabidopsis. Plant Physiol. 2001;126:1449–1458. doi: 10.1104/pp.126.4.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, Ishimoto T, Akiba S, Fujii T. Enhancement of phospholipase A2 activation by phosphatidic acid endogenously formed through phospholipase D action in rat peritoneal mast cell. FEBS Lett. 1993;24:23–26. doi: 10.1016/0014-5793(93)81440-b. [DOI] [PubMed] [Google Scholar]
- Wang X, Dyer JH, Zheng L. Purification and immunological analysis of phospholipase D from Cater Bean Endosperm. Arch Biochem Biophys. 1993;306:486–494. doi: 10.1006/abbi.1993.1541. [DOI] [PubMed] [Google Scholar]
- Wang X, Devaiah SP, Zhang W, Welti R. Signaling functions of phosphatidic acid. Biochim Biophys Acta. 2006;45:250–278. doi: 10.1016/j.plipres.2006.01.005. [DOI] [PubMed] [Google Scholar]
- Wilson DO, Jr, McDonald MB., Jr The lipid peroxidation model of seed ageing. Seed Science and Technology Zürich. 1986;14:269–300. [Google Scholar]
- Xu L, Paulsen AQ, Ryu SB, Wang X. Intracellular localization of phospholipase D in leaves and seedling tissues of castor bean. Plant Physiol. 1996;111:101–107. doi: 10.1104/pp.111.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Wang C, Qin C, Wood T, Olafsdottir G, Welti R, Wang X. The oleate-stimulated phospholipase D, PLDδ and phosphatidic acid decrease H2O2-induced cell death in Arabidopsis. Plant Cell. 2003;15:2285–2295. doi: 10.1105/tpc.013961. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.