Abstract
Streptococcus pneumoniae (the pneumococcus) is a common commensal inhabitant of the nasopharynx and a frequent etiologic agent in serious diseases such as pneumonia, otitis media, bacteremia, and meningitis. Multiple pneumococcal strains can colonize the nasopharynx, which is also home to many other bacterial species. Intraspecies and interspecies interactions influence pneumococcal carriage in important ways. Co-colonization by two or more pneumococcal strains has implications for vaccine serotype replacement, carriage detection, and pneumonia diagnostics. Interactions between the pneumococcus and other bacterial species alter carriage prevalence, modulate virulence, and affect biofilm formation. By examining these interactions, this review highlights how the bacterial ecosystem of the nasopharynx changes the nature and course of pneumococcal carriage.
Keywords: Streptococcus pneumoniae, the pneumococcus, co-colonization, carriage, nasopharynx, interaction
Streptococcus pneumoniae: a commensal and a pathogen
The Gram-positive bacterium Streptococcus pneumoniae (the pneumococcus) is a common asymptomatic resident of the human nasopharynx [1] and is also a primary etiologic agent in pneumonia, otitis media, meningitis, and sepsis. Pneumococcal pneumonia is the leading cause of childhood death in developing countries [2] and imposes a significant burden of morbidity, mortality, and healthcare costs in developed countries such as the USA [3]. Colonization of the nasopharynx is a necessary – but not sufficient – step along the path to pneumococcal disease [4,5]. On entering the nasopharynx, and during its residence there, the pneumococcus shares this anatomical and physiological niche with an array of other viral and bacterial inhabitants. Although there have been recent reviews of host–pneumococcal [6,7] and viral–pneumococcal interactions [8], there has been less written on the direct bacterial–bacterial interactions that influence pneumococcal carriage. This review highlights recent advances in our understanding of the bacterial interactions that alter the nature of pneumococcal carriage and its potential to progress to pneumococcal disease.
The nasopharynx lies at a vital crossroads connecting the nose, ears, mouth, and lower respiratory tract (Figure 1). Directly posterior to the nasal cavity and superior to the oropharynx, the nasopharynx is also close to the paranasal sinuses and directly connected to the middle ear by the Eustachian tubes. A dynamic environment with a constant flow of air, blood, and mucus, the nasopharynx modulates incoming air quality and temperature for the host, while also providing a home to millions of microbes. Colonization by the pneumococcus may occur at any time during a person’s life, but is most common during infancy, with acquisition even documented in the first day of life [9]. The prevalence of pneumococcal carriage increases in the first few years of life, peaking at approximately 50% to >70% in hosts 2–3 years of age [4,10] and decreasing thereafter until stabilizing at 5–10% in hosts over 10 years of age in developed countries [4,11,12] but as high as 25–60% in some developing countries [13,14]. The median duration of carriage is estimated to be 31 days in adults and 60.5 days in children, with length of carriage dependent on capsular serotype and previous immunologic exposure, as well as the host’s age and immunocompetence [15,16]. However, a recent study using intensive sampling of African children found a shorter duration of pediatric carriage: an overall mean of 31.3 days, with serotype-specific means ranging from 6.7 to 50 days [17]. Similar studies in adults may also reveal that the duration of carriage is shorter than previously deduced from studies using monthly assessments.
Figure 1.
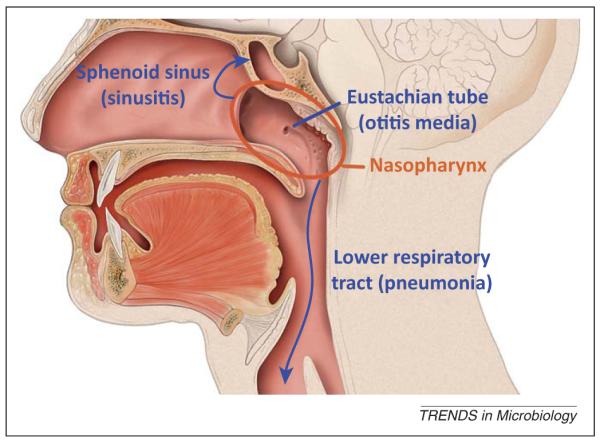
Diagram of the nasopharynx and pathways to pneumococcal diseases. The nasopharynx lies directly posterior to the nasal cavity and superior to the oropharynx. The pneumococcus can spread from its home in the nasopharynx through the Eustachian tube to cause otitis media, to the sinus cavities to cause sinusitis, through the larynx to the lower respiratory tract to cause pneumonia, and then in some cases, bacteremia. Image adapted from an illustration by Patrick J. Lynch distributed under Creative Commons Attribution 2.5 License.
Once carriage is established in the nasopharynx, the pneumococcus can migrate through the Eustachian tubes to cause otitis media [18], descend the respiratory tract to cause pneumonia [19], or invade the bloodstream through the respiratory epithelium to cause bacteremia or meningitis (Figure 1) [20]. The success of the pneumococcus as both a commensal and a pathogen can be attributed to a repertoire of cellular components that are useful during both carriage and disease [21]. Ultimately, regulation of these dual-use cellular components and changing conditions in the nasopharynx mediate if and when the pneumococcus progresses from a commensal resident to a pathogen. In this review, we argue that the abilities of the pneumococcus to persist and harm are influenced by within-species and cross-species population dynamics.
Effects of co-colonization on serotype distribution, genetic exchange, and density
Pneumococcal strains interact directly when they co-colonize a single host, an occurrence more prevalent than previously believed. Co-colonization is clinically and epidemiologically important because pneumococcal strains interfere with or aid in subsequent colonization by additional strains [22–24], and the pneumococcus is highly prone to homologous recombination and transformation of exogenous DNA [24–28]. In fact, it has recently been demonstrated that strains with poor colonization efficiency are better at colonizing a mouse model when a second strain with high colonization efficiency is already present [24]. Furthermore, this phenomenon appears to occur without genetic exchange, possibly through biofilm formation [24].
Recent work has begun to explore specific molecular mechanisms of interaction, including bacteriocin production and competence-induced fratricide in inter-strain dynamics. The blp locus in the pneumococcal genome encodes an antimicrobial bacteriocin peptide and corresponding immunity peptide. A strain harboring the blp locus has a competitive advantage in a mouse colonization model [22], and the importance of this locus in the wild is supported by a comparative genomic study that demonstrated extensive variation at the blp locus, indicative of diversifying selection [23]. Autolysis is another important mediator of inter-strain dynamics; long recognized for its ability to rapidly release the toxin pneumolysin, autolysis may actually be a form of competence-induced fratricide regulated by the Com quorum-sensing (QS) system [29]. In addition to upregulating factors necessary for uptake and transformation of DNA, the Com system activates the two-peptide bacteriocin CibAB and its immunity factor CibC; these bacteriocins target non-competent pneumococci that do not produce CibC, causing LytA-, LytC-, and CbpD-mediated lysis and DNA release from sister cells [30,31]. The role of bacteriocins and competence-induced fratricide in vivo is not yet clear, but it has become increasingly important to understand the mechanisms of strain interaction because it is now apparent that co-colonization is far more common than previously believed.
The original study of co-colonization by Gundel and Okura used a technique involving serial intraperitoneal saliva injections into mice to detect multiple serotypes [32]. The cost of this animal work led to a standard assay with streaking out of enriched nasopharyngeal specimens and serotyping of a handful of colonies. However, this approach yielded low rates of co-colonization; only 1.3–1.9% of samples from pediatric populations in South Africa were found to have more than one serotype using this laborious and costly method [33]. Testing of enriched cultures directly with the capsular reaction test using pooled and type-specific serum proved more sensitive, detecting co-colonization in 9.9% of pediatric Danish carriers [34]. However, a more cost-effective approach using PCR amplification of a noncoding region adjacent to the ply gene, followed by restriction fragment length polymorphism (RFLP) analysis yielded similar rates of co-colonization, 7.9% and 9.5% of pediatric carriers in two studies [35,36]. More recently, studies have demonstrated the utility of sweep techniques in which nasopharyngeal specimens are plated onto blood agar plates and incubated before all growth is swept up and assayed by multiplex PCR or latex agglutination [37]. A study of indigenous children in Venezuela using the sweep approach followed by multiplex PCR revealed a co-colonization rate of 20% [38]. In a careful comparison of the sweep technique and a microarray-based approach against traditional WHO culture methods, sweep–agglutination and microarray resulted in co-colonization rates of 43.2% and 48.8%, respectively, compared to 11.2% by the WHO-recommended culture [39]. Clearly, multiplex PCR, sweep, and microarray techniques demonstrate that co-colonization is more common than previously believed, and our ability to accurately detect co-colonization will only improve with the refinement of new techniques such as metagenomic sequencing and sequence-based serotype identification [40].
New studies are beginning to examine the theoretical and practical aspects of co-colonization on serotype-specific interactions and the implications for pneumococcal vaccine development and deployment. Current formulations of the pneumococcal conjugate vaccine (PCV) protect against 7, 10, or 13 capsular vaccine types (VTs), but have no direct effect on the more than 80 non-vaccine types (NVTs). Introduction of PCV unmasks the presence of NVTs that may have been at lower density or under less scrutiny during pre-PCV epidemiological studies [41]. In addition, NVTs are likely to gain a competitive advantage, both within hosts and within populations, when strains capable of competition are removed by vaccination [42,43]. PCV introduction has also precipitated a shift towards more NVT-caused invasive pneumococcal disease (IPD) [43]. In general, even with the increase in NVT-caused IPD, the rates of pneumococcal disease are still far below the pre-PCV era [44]. However, in certain populations, namely in Native Alaskan children and adults, IPD has actually increased [45], and in the USA, NVT 19A emerged prior to the introduction of PCV13 as a more virulent and drug-resistant cause of IPD [46,47]. This increase in IPD due to serotype 19A is likely to be because of its increased carriage prevalence in a population immunized with PCV7.
Co-colonization is also important because the pneumococcus has a significant propensity for genetic exchange through transformation and homologous recombination. A longitudinal study of chronic otitis media demonstrated extensive genomic rearrangements in vivo [26], and animal models of pneumococcal disease have demonstrated that even minor genetic differences can significantly alter virulence [23]. A multi-locus sequence typing (MLST) study of pediatric strains in Gambia found extensive recombination and serotype switching among carriage strains; the authors suggested this finding may be due to high rates of co-colonization in this population [48]. Furthermore, a study of 240 full genome sequences from the PMEN1 lineage revealed more than 700 recombination events and 10 capsule-switching events [28], suggesting extensive diversity within a relatively contemporary highly related clone and thus a high frequency of recombination events. More studies using whole-genome sequencing of isolates obtained sequentially from carriers are needed to pinpoint the frequency of these recombination and capsule-switching events, especially in relation to the changing flora of the nasopharynx post-vaccination.
It has recently been suggested that co-colonization with multiple strains of S. pneumoniae increases the density of nasopharyngeal carriage [36]. Brugger and colleagues found that subjects co-colonized with two or more strains had higher carriage density than subjects colonized with one strain [36]. Lower density carriage is less likely to be detected by culture methods than by real-time quantitative PCR (qPCR) [49,50], so the findings of previous, culturebased studies may be distorted by the relative frequency or rarity of co-colonization. Nasopharyngeal pneumococcal density is also showing promise as a diagnostic tool for pneumonia [51,52]. If co-colonization is correlated with nasopharyngeal pneumococcal density, then this factor will need to be controlled for when exploring density-based pneumonia diagnostics.
Interactions between nasopharyngeal bacterial species
The nasopharyngeal microbial community is established in the first year after birth and varies throughout a person’s lifetime [53]. The five most common bacterial families in the nasopharynx are Moraxellaceae, Streptococcaceae, Corynebacteriaceae, Pasteurellaceae (including the genus Haemophilus), and Staphylococcaceae (Table 1) [10,54–56]. The first microbiome studies to analyze 16S rRNA sequences have revealed high inter-individual variability in nasopharyngeal communities, as well as possible correlations with season [55,56]. In addition to bacteria, viruses and the host immune system are also major determinants of nasopharyngeal dynamics. The synergy between viral co-infections and bacterial pneumonia has been an ongoing area of research [8,57] and a recent review by Lijek and Weiser examines the evidence that mucosal immunity mediates interactions between S. pneumoniae, Haemophilus influenzae, and Staphylococcus aureus [7]. Here we consider how direct bacterial–bacterial interactions may impact clinically important nasopharyngeal dynamics.
Table 1.
Average frequency of 16S rRNA sequences present in nasal swabs of children
| Operational taxonomic unit (OTU)a | Frequency (%)b |
|---|---|
| Unclassified Moraxellaceae | 19.0 |
| Streptococcus | 17.9 |
| Corynebacterium | 7.0 |
| Moraxella | 6.5 |
| Haemophilus | 4.7 |
| Unclassified Pasteurellaceae | 4.1 |
| Staphylococcus | 3.8 |
| Acinetobacter | 3.4 |
| Dolosigranulum | 3.2 |
| Propionibacterium | 3.1 |
| Unclassified Proteobacteria | 2.6 |
| Lactococcus | 2.6 |
Adapted with permission from Laufer et al. [55].
Average frequency, among 108 children, of 16S rRNA sequences in nasal swab samples. Only OTUs with an average frequency >2% are listed.
With the widespread deployment of PCV and a seemingly concurrent rise in community-associated methicillin-resistant Staphylococcus aureus (MRSA) infections, there has been speculation as to whether these two trends may be related [54,58,59]. Epidemiological studies have indeed revealed an inverse correlation between carriage of S. pneumoniae and carriage of S. aureus [50,54,60,61]. The presence of the pneumococcus in the nasopharynx appears to inhibit S. aureus colonization. In vitro studies demonstrated that hydrogen peroxide (H2O2) produced by the enzyme pyruvate oxidase (SpxB) of S. pneumoniae kills S. aureus strains. In addition, targeted mutagenesis of staphylococcal catalase results in decreased survival of S. aureus in the presence of the pneumococcus [62]. The mechanistic explanation is that without catalase to degrade hydrogen peroxide, the staphylococcal SOS response is triggered, leading to bacteriophage-induced lysis [63,64]. However, the in vivo relevance of hydrogen peroxide-mediated killing is in question because a neonatal rat study using an SpxB-deficient S. pneumoniae strain or a catalase-deficient S. aureus strain demonstrated no significant difference in co-colonization densities [65]. The finding that carriage of S. aureus was less likely in individuals carrying piliated strains of S. pneumoniae suggests that the pneumococcal pilus may somehow reduce the likelihood of staphylococcal colonization or sustained carriage [66].
Regardless of mechanism, for PCV to contribute to the rise in MRSA there would need to be an inverse correlation specifically between VT the pneumococcus and MRSA. Cross-sectional studies have demonstrated an inverse association between carriage of S. aureus and vaccine-type S. pneumoniae [54,60]. However, interventional studies that assessed bacterial carriage before and after PCV introduction are divided. Some studies found that carriage prevalence of S. aureus remained steady [67–69] whereas others found increased prevalence of S. aureus following PCV [61,70,71]. Importantly, although some have searched for it [58], no study has demonstrated an association between pneumococcal carriage and MRSA carriage. Given that the inverse correlation between S. aureus and S. pneumoniae is absent in immunodeficient HIV-positive populations [61,72], an immune-mediated mechanism may be responsible for this relationship. Thus, although there does appear to be a link between carriage of S. aureus and carriage of VT S. pneumoniae, there is not sufficient evidence to assert that the increase in MRSA is a result of PCV rollout and no increase in community-acquired MRSA has been seen in the years following PCV rollout in Europe [73].
Pneumococcal carriage in the nasopharynx has also been associated with carriage of two other major nasophar-ynx-based pathogens, H. influenzae and Moraxella catarrhalis. Although there are exceptions, most studies find a positive association in humans between the presence in the nasopharynx of S. pneumoniae and H. influenzae and between S. pneumoniae and M. catarrhalis [74–76]. The epidemiologic association between S. pneumoniae and H. influenzae was recently extended by the observation that their carriage densities are also positively correlated [50]. Animal studies concur with the human epidemiology, with a neonatal rat study demonstrating that resident S. pneumoniae facilitated colonization of H. influenzae [77]. In contrast to epidemiologic data showing a positive correlation between the pneumococcus and H. influenzae, in vitro studies by Pericone and colleagues demonstrated that S. pneumoniae inhibits the growth of H. influenzae, M. catarrhalis, and Neisseria meningitidis via hydrogen peroxide production [78]. This apparent paradox suggests that cultivation of the two pathogens together in vitro does not accurately reflect their physiological interactions in vivo. In a longitudinal study of PCV introduction in the Netherlands, researchers found that whereas overall pneumococcal prevalence remained steady (with VTs decreasing and NVTs increasing), H. influenzae prevalence increased [71], suggesting that if there is an association between colonization with S. pneumoniae and H. influenzae, the relationship may be serotype-specific. Ongoing and future human studies of pneumococcal colonization [79] have the potential to help us better understand the relationship between the pneumococcal colonization event and the nasopharyngeal carriage of other bacterial species.
Although the mechanisms behind inter-species interactions are still being investigated, it is likely that bacteriocins play a role. It has been shown that S. pneumoniae bacteriocins inhibit the growth of other streptococcal species [80], and bacteriocins produced by Streptococcus mitis and Streptococcus salivarius inhibit the growth of the pneumococcus [81]. In addition to growth inhibition via secreted signals, there is also a growing body of evidence that cross-species pheromones alter bacterial gene expression. Recent studies have shown that cross-species signals can modify the expression of pneumococcal virulence genes. In contrast to the findings of Pericone et al. [78], Cope and colleagues demonstrated that co-culture of clinical strains of S. pneumoniae with H. influenzae does not change planktonic growth rates [82]. However, co-culture of these two strains did upregulate expression of SpxB and downregulate expression of ply, the gene that encodes pneumolysin, and pavA, the gene that encodes pneumococcal adherence and virulence factor A [82]. These results suggest that bacteria sharing the nasopharynx have the potential to alter one another’s gene expression. The mechanisms of interspecies gene regulation are just beginning to be explored, but the well-studied QS system LuxS/autoinducer-2 (AI-2) may be partly responsible for interspecies communication. AI-2, synthesized by the enzyme LuxS, is a molecule secreted and recognized by many bacterial species and has the potential to alter growth and virulence [83]. As reviewed in the next section, pneumococcal LuxS/AI-2 plays an important role in biofilm formation.
Pneumococcal biofilms in the nasopharynx
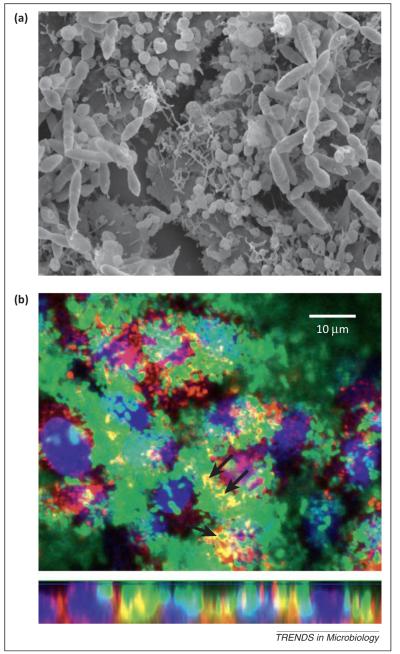
There is mounting evidence that the nasopharynx hosts biofilms created by many bacterial species, including the pneumococcus [84,85], S. aureus, Staphylococcus epidermidis [86], N. meningitidis [87], and H. influenzae [88]. Pneumococcal biofilms form in stages, starting with attachment to the substratum mediated by pneumococcal adhesion factors such as CbpA, PcpA, and PspA [85,89], and followed by bacterial aggregation and matrix maturation [90,91]. A mature matrix is mainly composed of extracellular DNA (eDNA), proteins, and polysaccharides [91,92] linking pneumococcal cells and attaching them to host cells in a tight mesh (Figure 2). Recent studies suggest that biofilm formation, cellular autolysis, and genetic competence are all intertwined and regulated by the Com and LuxS/AI-2 QS systems.
Figure 2.
Streptococcus pneumoniae forms multicellular structures in vitro on top of human lung cells. (a) Scanning electron micrograph of pneumococcal cells in 8-h biofilms grown on top of human lung epithelial cells show bacterial cells connected by an extracellular matrix. (b) Confocal images of GFP-expressing S. pneumoniae biofilms demonstrate cells co-localizing with sialic acid residues (stained red) present on human lung epithelial cells (nuclei stained blue). The top image is an optical xy section and the bottom is an xz section.
The Com system is well known for its ability to control the genetic competence of the pneumococcus and mediate killing of non-competent sister cells [30]. Competence-stimulating peptide (CSP), the signal secreted by the Com QS system, is strain-specific and regulates autolysis and genetic competence, which allows for release and uptake of DNA from other streptococci [93]. Recent studies have shown that gene transference among streptococci is more efficient in early biofilm structures [24,94], and LytC, a lysozyme implicated in ComC-mediated lysis of non-competent neighbors, is a common component of DNA–protein complexes in pneumococcal biofilms [92]. Two recent studies have also demonstrated that strains deficient in LuxS are unable to form biofilms at early time points and have decreased ability for eDNA release and competence [90,95]. How these two QS systems work together in biofilm formation is unknown, but perhaps LuxS/AI-2 is necessary for attachment of early biofilms and pneumococcal biofilm maturation can only be completed with activation of the Com system and release of the eDNA required for matrix development.
The pneumococcus is also capable of participating in multi-species biofilms. Co-incubation of H. influenzae and S. pneumoniae increases the biomass of pneumococcal biofilms [96]. In addition, these interspecies biofilms are able to confer protection against antibiotics to strains of susceptible S. pneumoniae through passive transference of β-lactamase produced by resistant H. influenzae [97]. Bio-film formation in S. pneumoniae and H. influenzae is in part regulated by the LuxS/AI-2 QS mechanism [90,95,98]. Accordingly, animal studies also demonstrate that the LuxS/AI-2 system regulates persistence of the pneumococcus in the mouse nasopharynx [99] and persistence of H. influenzae biofilms in the ear epithelium of chinchillas [98].
AI-2 is produced by most bacterial pathogens and normal flora strains, so it is quite possible that the pneumococcus secretes AI-2 in the nasopharynx to modify the behaviors of other bacterial species or that the pneumococcus responds to AI-2 secreted by other colonizers. In support of this theory, exogenous addition of purified AI-2 significantly increased the biofilm biomass of S. pneumoniae strain D39 [90]. Furthermore, M. catarrhalis, which does not possess a luxS homolog, produced more robust biofilms in response to AI-2 secreted by H. influenzae strains [100]. A number of other streptococcal species that colonize the pharynx, such as S. intermedius, S. oralis, S. gordonii, and S. mutans, utilize LuxS/AI-2 to control biofilms [83]. Whether AI-2 is used for competition or cooperation among species in the pharynx requires further investigation.
Concluding remarks
Our understanding of how microbes and humans live together in health and disease is rapidly evolving. The pneumococcus is a clinically important and biologically fascinating organism that we can better understand by observing its interactions with the human host and with other microbes. Additional pneumococcal strains and other bacterial species play key roles in pneumococcal colonization. Metagenomic sequencing of all DNA in the nasopharynx will not only allow us to take a better census of the microbes and viruses, but will also help us to understand the full genetic and transcriptional activity of these populations [101]. Furthermore, experimental human studies of pneumococcal colonization of the nasopharynx have the potential to distinguish cause and effect when a correlation or change in bacterial composition is observed [79,102]. Ultimately, the challenge will be to identify biological and clinical relevance when examining large complex networks of intraspecies and interspecies interactions inside the human host.
Acknowledgments
We thank Bruce Levin, Bill Shafer, and Tim Read for comments on an earlier draft of this manuscript and Hong Yi, Elizabeth Wright, and Jeannette Taylor for assistance at the Robert P. Apkarian Integrated Electron Microscopy Core. J.R.S. gratefully acknowledges the financial support of the Molecules to Mankind Program at the Laney Graduate School, Emory University, and the Medical Scientist Training Program at Emory University School of Medicine. J.E.V. was supported in part by PHS Grant UL1 RR025008 from the Clinical and Translational Science Award program, NIH, National Center for Research Resources.
References
- 1.van der Poll T, Opal SM. Pathogenesis, treatment, and prevention of pneumococcal pneumonia. Lancet. 2009;374:1543–1556. doi: 10.1016/S0140-6736(09)61114-4. [DOI] [PubMed] [Google Scholar]
- 2.O’Brien KL, et al. Burden of disease caused by Streptococcus pneumoniae in children younger than 5 years: global estimates. Lancet. 2009;374:893–902. doi: 10.1016/S0140-6736(09)61204-6. [DOI] [PubMed] [Google Scholar]
- 3.Huang SS, et al. Healthcare utilization and cost of pneumococcal disease in the United States. Vaccine. 2011;29:3398–3412. doi: 10.1016/j.vaccine.2011.02.088. [DOI] [PubMed] [Google Scholar]
- 4.Bogaert D, et al. Streptococcus pneumoniae colonisation: the key to pneumococcal disease. Lancet Infect. Dis. 2004;4:144–154. doi: 10.1016/S1473-3099(04)00938-7. [DOI] [PubMed] [Google Scholar]
- 5.Simell B, et al. The fundamental link between pneumococcal carriage and disease. Expert Rev. Vaccines. 2012;11:841–855. doi: 10.1586/erv.12.53. [DOI] [PubMed] [Google Scholar]
- 6.Koppe U, et al. Recognition of Streptococcus pneumoniae by the innate immune system. Cell. Microbiol. 2012;14:460–466. doi: 10.1111/j.1462-5822.2011.01746.x. [DOI] [PubMed] [Google Scholar]
- 7.Lijek RS, Weiser JN. Co-infection subverts mucosal immunity in the upper respiratory tract. Curr. Opin. Immunol. 2012;24:417–423. doi: 10.1016/j.coi.2012.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Klugman KP, et al. Pneumococcal pneumonia and influenza: a deadly combination. Vaccine. 2009;27(Suppl. 3):C9–C14. doi: 10.1016/j.vaccine.2009.06.007. [DOI] [PubMed] [Google Scholar]
- 9.Tigoi CC, et al. Rates of acquisition of pneumococcal colonization and transmission probabilities, by serotype, among newborn infants in Kilifi district, Kenya. Clin. Infect. Dis. 2012;55:180–188. doi: 10.1093/cid/cis371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Watson K, et al. Upper respiratory tract bacterial carriage in Aboriginal and non-Aboriginal children in a semi-arid area of Western Australia. Pediatr. Infect. Dis. J. 2006;25:782–790. doi: 10.1097/01.inf.0000232705.49634.68. [DOI] [PubMed] [Google Scholar]
- 11.Hussain M, et al. A longitudinal household study of Streptococcus pneumoniae nasopharyngeal carriage in a UK setting. Epidemiol. Infect. 2005;133:891–898. doi: 10.1017/S0950268805004012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jounio U, et al. Pneumococcal carriage is more common in asthmatic than in non-asthmatic young men. Clin. Respir. J. 2010;4:222–229. doi: 10.1111/j.1752-699X.2009.00179.x. [DOI] [PubMed] [Google Scholar]
- 13.Roca A, et al. Effects of community-wide vaccination with PCV-7 on pneumococcal nasopharyngeal carriage in the Gambia: a cluster-randomized trial. PLoS Med. 2011;8:e1001107. doi: 10.1371/journal.pmed.1001107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Adetifa IM, et al. Pre-vaccination nasopharyngeal pneumococcal carriage in a Nigerian population: epidemiology and population biology. PLoS ONE. 2012;7:e30548. doi: 10.1371/journal.pone.0030548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Turner P, et al. A longitudinal study of Streptococcus pneumoniae carriage in a cohort of infants and their mothers on the Thailand–Myanmar border. PLoS ONE. 2012;7:e38271. doi: 10.1371/journal.pone.0038271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hill PC, et al. Transmission of Streptococcus pneumoniae in rural Gambian villages: a longitudinal study. Clin. Infect. Dis. 2010;50:1468–1476. doi: 10.1086/652443. [DOI] [PubMed] [Google Scholar]
- 17.Abdullahi O, et al. Rates of acquisition and clearance of pneumococcal serotypes in the nasopharynges of children in Kilifi district, Kenya. J. Infect. Dis. 2012;206:1020–1029. doi: 10.1093/infdis/jis447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bluestone CD. Pathogenesis of otitis media: role of Eustachian tube. Pediatr. Infect. Dis. J. 1996;15:281–291. doi: 10.1097/00006454-199604000-00002. [DOI] [PubMed] [Google Scholar]
- 19.Murphy TF, et al. Microbial interactions in the respiratory tract. Pediatr. Infect. Dis. J. 2009;28:S121–S126. doi: 10.1097/INF.0b013e3181b6d7ec. [DOI] [PubMed] [Google Scholar]
- 20.Mook-Kanamori BB, et al. Pathogenesis and pathophysiology of pneumococcal meningitis. Clin. Microbiol. Rev. 2011;24:557–591. doi: 10.1128/CMR.00008-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weiser JN. The pneumococcus: why a commensal misbehaves. J. Mol. Med. 2010;88:97–102. doi: 10.1007/s00109-009-0557-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dawid S, et al. The blp bacteriocins of Streptococcus pneumoniae mediate intraspecies competition both in vitro and in vivo. Infect. Immun. 2007;75:443–451. doi: 10.1128/IAI.01775-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hiller NL, et al. Differences in genotype and virulence among four multidrug-resistant Streptococcus pneumoniae isolates belonging to the PMEN1 clone. PLoS ONE. 2011;6:e28850. doi: 10.1371/journal.pone.0028850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marks LR, et al. High levels of genetic recombination during nasopharyngeal carriage and biofilm formation in Streptococcus pneumoniae. MBio. 2012;3:e00200–e00212. doi: 10.1128/mBio.00200-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Donati C, et al. Structure and dynamics of the pan-genome of Streptococcus pneumoniae and closely related species. Genome Biol. 2010;11:R107. doi: 10.1186/gb-2010-11-10-r107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hiller NL, et al. Generation of genic diversity among Streptococcus pneumoniae strains via horizontal gene transfer during a chronic polyclonal pediatric infection. PLoS Pathog. 2010;6:e1001108. doi: 10.1371/journal.ppat.1001108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Muzzi A, Donati C. Population genetics and evolution of the pan-genome of Streptococcus pneumoniae. Int. J. Med. Microbiol. 2011;301:619–622. doi: 10.1016/j.ijmm.2011.09.008. [DOI] [PubMed] [Google Scholar]
- 28.Croucher NJ, et al. Rapid pneumococcal evolution in response to clinical interventions. Science. 2011;331:430–434. doi: 10.1126/science.1198545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Claverys J-P, Håvarstein LS. Cannibalism and fratricide: mechanisms and raisons d’être. Nat. Rev. Microbiol. 2007;5:219–229. doi: 10.1038/nrmicro1613. [DOI] [PubMed] [Google Scholar]
- 30.Guiral S, et al. Competence-programmed predation of noncompetent cells in the human pathogen Streptococcus pneumoniae: genetic requirements. Proc. Natl. Acad. Sci. U.S.A. 2005;102:8710–8715. doi: 10.1073/pnas.0500879102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eldholm V, et al. Fratricide in Streptococcus pneumoniae: contributions and role of the cell wall hydrolases CbpD, LytA and LytC. Microbiology. 2009;155:2223–2234. doi: 10.1099/mic.0.026328-0. [DOI] [PubMed] [Google Scholar]
- 32.Gundel M. Bakteriologische und epidemiologische Untersuchungen uber die Besiedlung der oberen Atmungswege Gesunder mit Pneumokokken. Zeitschr. Hyg. Infektkrankh. 1933;114:659–677. [Google Scholar]
- 33.Huebner RE, et al. Lack of utility of serotyping multiple colonies for detection of simultaneous nasopharyngeal carriage of different pneumococcal serotypes. Pediatr. Infect. Dis. J. 2000;19:1017–1020. doi: 10.1097/00006454-200010000-00019. [DOI] [PubMed] [Google Scholar]
- 34.Kaltoft MS, et al. An easy method for detection of nasopharyngeal carriage of multiple Streptococcus pneumoniae serotypes. J. Microbiol. Methods. 2008;75:540–544. doi: 10.1016/j.mimet.2008.08.010. [DOI] [PubMed] [Google Scholar]
- 35.Brugger SD, et al. Detection of Streptococcus pneumoniae strain cocolonization in the nasopharynx. J. Clin. Microbiol. 2009;47:1750–1756. doi: 10.1128/JCM.01877-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brugger SD, et al. Multiple colonization with S. pneumoniae before and after introduction of the seven-valent conjugated pneumococcal polysaccharide vaccine. PLoS ONE. 2010;5:e11638. doi: 10.1371/journal.pone.0011638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hill PC, et al. Nasopharyngeal carriage of Streptococcus pneumoniae in Gambian infants: a longitudinal study. Clin. Infect. Dis. 2008;46:807–814. doi: 10.1086/528688. [DOI] [PubMed] [Google Scholar]
- 38.Rivera-Olivero IA, et al. Multiplex PCR reveals a high rate of nasopharyngeal pneumococcal 7-valent conjugate vaccine serotypes co-colonizing indigenous Warao children in Venezuela. J. Med. Microbiol. 2009;58:584–587. doi: 10.1099/jmm.0.006726-0. [DOI] [PubMed] [Google Scholar]
- 39.Turner P, et al. Improved detection of nasopharyngeal cocolonization by multiple pneumococcal serotypes by use of latex agglutination or molecular serotyping by microarray. J. Clin. Microbiol. 2011;49:1784–1789. doi: 10.1128/JCM.00157-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leung MH, et al. Sequetyping: serotyping Streptococcus pneumoniae by a single PCR sequencing strategy. J. Clin. Microbiol. 2012;50:2419–2427. doi: 10.1128/JCM.06384-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lipsitch M. Interpreting results from trials of pneumococcal conjugate vaccines: a statistical test for detecting vaccine-induced increases in carriage of nonvaccine serotypes. Am. J. Epidemiol. 2001;154:85–92. doi: 10.1093/aje/154.1.85. [DOI] [PubMed] [Google Scholar]
- 42.Lipsitch M, et al. Competition among Streptococcus pneumoniae for intranasal colonization in a mouse model. Vaccine. 2000;18:2895–2901. doi: 10.1016/s0264-410x(00)00046-3. [DOI] [PubMed] [Google Scholar]
- 43.Weinberger DM, et al. Serotype replacement in disease after pneumococcal vaccination. Lancet. 2011;378:1962–1973. doi: 10.1016/S0140-6736(10)62225-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Klugman KP. Contribution of vaccines to our understanding of pneumococcal disease. Philos. Trans. R. Soc. Lond. Ser. B: Biol. Sci. 2011;366:2790–2798. doi: 10.1098/rstb.2011.0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Singleton RJ, et al. Invasive pneumococcal disease caused by nonvaccine serotypes among Alaska native children with high levels of 7-valent pneumococcal conjugate vaccine coverage. J. Am. Med. Assoc. 2007;297:1784–1792. doi: 10.1001/jama.297.16.1784. [DOI] [PubMed] [Google Scholar]
- 46.Pelton SI, et al. Emergence of 19A as virulent and multidrug resistant pneumococcus in Massachusetts following universal immunization of infants with pneumococcal conjugate vaccine. Pediatr. Infect. Dis. J. 2007;26:468–472. doi: 10.1097/INF.0b013e31803df9ca. [DOI] [PubMed] [Google Scholar]
- 47.Tyrrell GJ. The changing epidemiology of Streptococcus pneumoniae serotype 19A clonal complexes. J. Infect. Dis. 2011;203:1345–1347. doi: 10.1093/infdis/jir056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Donkor ES, et al. High levels of recombination among Streptococcus pneumoniae isolates from the Gambia. MBio. 2011;2:e00222–11. doi: 10.1128/mBio.00040-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.da Gloria Carvalho M, et al. Revisiting pneumococcal carriage by use of broth enrichment and PCR techniques for enhanced detection of carriage and serotypes. J. Clin. Microbiol. 2010;48:1611–1618. doi: 10.1128/JCM.02243-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chien YW, et al. Density interactions between Streptococcus pneumoniae, Haemophilus influenzae and Staphylococcus aureus in the nasopharynx of young Peruvian children. Pediatr. Infect. Dis. J. 2013;32:72–77. doi: 10.1097/INF.0b013e318270d850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vu HT, et al. Association between nasopharyngeal load of Streptococcus pneumoniae, viral coinfection, and radiologically confirmed pneumonia in Vietnamese children. Pediatr. Infect. Dis. J. 2011;30:11–18. doi: 10.1097/INF.0b013e3181f111a2. [DOI] [PubMed] [Google Scholar]
- 52.Albrich WC, et al. Use of a rapid test of pneumococcal colonization density to diagnose pneumococcal pneumonia. Clin. Infect. Dis. 2012;54:601–609. doi: 10.1093/cid/cir859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Faden H, et al. Relationship between nasopharyngeal colonization and the development of otitis media in children. J. Infect. Dis. 1997;175:1440–1445. doi: 10.1086/516477. [DOI] [PubMed] [Google Scholar]
- 54.Regev-Yochay G, et al. Association between carriage of Streptococcus pneumoniae and Staphylococcus aureus in children. J. Am. Med. Assoc. 2004;292:716–720. doi: 10.1001/jama.292.6.716. [DOI] [PubMed] [Google Scholar]
- 55.Laufer AS, et al. Microbial communities of the upper respiratory tract and otitis media in children. MBio. 2011;2:e00245–10. doi: 10.1128/mBio.00245-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bogaert D, et al. Variability and diversity of nasopharyngeal microbiota in children: a metagenomic analysis. PLoS ONE. 2011;6:e17035. doi: 10.1371/journal.pone.0017035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nair H, et al. Global burden of respiratory infections due to seasonal influenza in young children: a systematic review and meta-analysis. Lancet. 2011;378:1917–1930. doi: 10.1016/S0140-6736(11)61051-9. [DOI] [PubMed] [Google Scholar]
- 58.Lee GM, et al. Epidemiology and risk factors for Staphylococcus aureus colonization in children in the post-PCV7 era. BMC Infect. Dis. 2009;9:110. doi: 10.1186/1471-2334-9-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.David MZ, Daum RS. Community-associated methicillin-resistant Staphylococcus aureus: epidemiology and clinical consequences of an emerging epidemic. Clin. Microbiol. Rev. 2010;23:616–687. doi: 10.1128/CMR.00081-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bogaert D, et al. Colonisation by Streptococcus pneumoniae and Staphylococcus aureus in healthy children. Lancet. 2004;363:1871–1872. doi: 10.1016/S0140-6736(04)16357-5. [DOI] [PubMed] [Google Scholar]
- 61.Madhi SA, et al. Long-term effect of pneumococcal conjugate vaccine on nasopharyngeal colonization by Streptococcus pneumoniae – and associated interactions with Staphylococcus aureus and Haemophilus influenzae colonization – in HIV-infected and HIV-uninfected children. J. Infect. Dis. 2007;196:1662–1666. doi: 10.1086/522164. [DOI] [PubMed] [Google Scholar]
- 62.Park B, et al. Role of Staphylococcus aureus catalase in niche competition against Streptococcus pneumoniae. J. Bacteriol. 2008;190:2275–2278. doi: 10.1128/JB.00006-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Regev-Yochay G, et al. Interference between Streptococcus pneumoniae and Staphylococcus aureus: in vitro hydrogen peroxide-mediated killing by Streptococcus pneumoniae. J. Bacteriol. 2006;188:4996–5001. doi: 10.1128/JB.00317-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Selva L, et al. Killing niche competitors by remote-control bacteriophage induction. Proc. Natl. Acad. Sci. U.S.A. 2009;106:1234–1238. doi: 10.1073/pnas.0809600106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Margolis E. Hydrogen peroxide-mediated interference competition by Streptococcus pneumoniae has no significant effect on Staphylococcus aureus nasal colonization of neonatal rats. J. Bacteriol. 2009;191:571–575. doi: 10.1128/JB.00950-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Regev-Yochay G, et al. The pneumococcal pilus predicts the absence of Staphylococcus aureus co-colonization in pneumococcal carriers. Clin. Infect. Dis. 2009;48:760–763. doi: 10.1086/597040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vestrheim DF, et al. Impact of a pneumococcal conjugate vaccination program on carriage among children in Norway. Clin. Vaccine Immunol. 2010;17:325–334. doi: 10.1128/CVI.00435-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kuo CY, et al. Nasopharyngeal carriage of Streptococcus pneumoniae in Taiwan before and after the introduction of a conjugate vaccine. Vaccine. 2011;29:5171–5177. doi: 10.1016/j.vaccine.2011.05.034. [DOI] [PubMed] [Google Scholar]
- 69.Dunne EM, et al. Effect of pneumococcal vaccination on nasopharyngeal carriage of Streptococcus pneumoniae, Haemophilus influenzae, Moraxella catarrhalis, and Staphylococcus aureus in Fijian children. J. Clin. Microbiol. 2012;50:1034–1038. doi: 10.1128/JCM.06589-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.van Gils EJ, et al. Effect of seven-valent pneumococcal conjugate vaccine on Staphylococcus aureus colonisation in a randomised controlled trial. PLoS ONE. 2011;6:e20229. doi: 10.1371/journal.pone.0020229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Spijkerman J, et al. Long-term effects of pneumococcal conjugate vaccine on nasopharyngeal carriage of S. pneumoniae, S. aureus, H. influenzae and M. catarrhalis. PLoS ONE. 2012;7:e39730. doi: 10.1371/journal.pone.0039730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.McNally LM, et al. Lack of association between the nasopharyngeal carriage of Streptococcus pneumoniae and Staphylococcus aureus in HIV-1-infected South African children. J. Infect. Dis. 2006;194:385–390. doi: 10.1086/505076. [DOI] [PubMed] [Google Scholar]
- 73.Mediavilla JR, et al. Global epidemiology of community-associated methicillin resistant Staphylococcus aureus (CA-MRSA) Curr. Opin. Microbiol. 2012;15:588–595. doi: 10.1016/j.mib.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 74.Pettigrew MM, et al. Microbial interactions during upper respiratory tract infections. Emerg. Infect. Dis. 2008;14:1584–1591. doi: 10.3201/eid1410.080119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jacoby P, et al. Modelling the co-occurrence of Streptococcus pneumoniae with other bacterial and viral pathogens in the upper respiratory tract. Vaccine. 2007;25:2458–2464. doi: 10.1016/j.vaccine.2006.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mackenzie GA, et al. Epidemiology of nasopharyngeal carriage of respiratory bacterial pathogens in children and adults: cross-sectional surveys in a population with high rates of pneumococcal disease. BMC Infect. Dis. 2010;10:304. doi: 10.1186/1471-2334-10-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Margolis E, et al. The ecology of nasal colonization of Streptococcus pneumoniae, Haemophilus influenzae and Staphylococcus aureus: the role of competition and interactions with host’s immune response. BMC Microbiol. 2010;10:59. doi: 10.1186/1471-2180-10-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pericone CD, et al. Inhibitory and bactericidal effects of hydrogen peroxide production by Streptococcus pneumoniae on other inhabitants of the upper respiratory tract. Infect. Immun. 2000;68:3990–3997. doi: 10.1128/iai.68.7.3990-3997.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ferreira DM, et al. Experimental human pneumococcal carriage models for vaccine research. Trends Microbiol. 2011;19:464–470. doi: 10.1016/j.tim.2011.06.003. [DOI] [PubMed] [Google Scholar]
- 80.Lux T, et al. Diversity of bacteriocins and activity spectrum in Streptococcus pneumoniae. J. Bacteriol. 2007;189:7741–7751. doi: 10.1128/JB.00474-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Santagati M, et al. Bacteriocin-producing oral streptococci and inhibition of respiratory pathogens. FEMS Immunol. Med. Microbiol. 2012;65:23–31. doi: 10.1111/j.1574-695X.2012.00928.x. [DOI] [PubMed] [Google Scholar]
- 82.Cope EK, et al. Regulation of virulence gene expression resulting from Streptococcus pneumoniae and nontypeable Haemophilus influenzae interactions in chronic disease. PLoS ONE. 2011;6:e28523. doi: 10.1371/journal.pone.0028523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pereira CS, et al. AI-2-mediated signalling in bacteria. FEMS Microbiol. Rev. 2013;37:156–181. doi: 10.1111/j.1574-6976.2012.00345.x. [DOI] [PubMed] [Google Scholar]
- 84.Sanchez CJ, et al. The pneumococcal serine-rich repeat protein is an intra-species bacterial adhesin that promotes bacterial aggregation in vivo and in biofilms. PLoS Pathog. 2010;6:e1001044. doi: 10.1371/journal.ppat.1001044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Muñoz-Elías EJ, et al. Isolation of Streptococcus pneumoniae biofilm mutants and their characterization during nasopharyngeal colonization. Infect. Immun. 2008;76:5049–5061. doi: 10.1128/IAI.00425-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Iwase T, et al. Staphylococcus epidermidis Esp inhibits Staphylococcus aureus biofilm formation and nasal colonization. Nature. 2010;465:346–349. doi: 10.1038/nature09074. [DOI] [PubMed] [Google Scholar]
- 87.Neil RB, et al. Biofilm formation on human airway epithelia by encapsulated Neisseria meningitidis serogroup B. Microbes Infect. 2009;11:281–287. doi: 10.1016/j.micinf.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 88.Jurcisek JA, et al. The PilA protein of non-typeable Haemophilus influenzae plays a role in biofilm formation, adherence to epithelial cells and colonization of the mammalian upper respiratory tract. Mol. Microbiol. 2007;65:1288–1299. doi: 10.1111/j.1365-2958.2007.05864.x. [DOI] [PubMed] [Google Scholar]
- 89.Moscoso M, et al. Biofilm formation by Streptococcus pneumoniae: role of choline, extracellular DNA, and capsular polysaccharide in microbial accretion. J. Bacteriol. 2006;188:7785–7795. doi: 10.1128/JB.00673-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Vidal JE, et al. The LuxS-dependent quorum-sensing system regulates early biofilm formation by Streptococcus pneumoniae strain D39. Infect. Immun. 2011;79:4050–4060. doi: 10.1128/IAI.05186-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Allegrucci M, et al. Phenotypic characterization of Streptococcus pneumoniae biofilm development. J. Bacteriol. 2006;188:2325–2335. doi: 10.1128/JB.188.7.2325-2335.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Domenech M, et al. Insight into the composition of the intercellular matrix of Streptococcus pneumoniae biofilms. Environ. Microbiol. 2013;15:502–516. doi: 10.1111/j.1462-2920.2012.02853.x. [DOI] [PubMed] [Google Scholar]
- 93.Johnsborg O, Håvarstein LS. Regulation of natural genetic transformation and acquisition of transforming DNA in Streptococcus pneumoniae. FEMS Microbiol. Rev. 2009;33:627–642. doi: 10.1111/j.1574-6976.2009.00167.x. [DOI] [PubMed] [Google Scholar]
- 94.Wei H, Havarstein LS. Fratricide is essential for efficient gene transfer between pneumococci in biofilms. Appl. Environ. Microbiol. 2012;78:5897–5905. doi: 10.1128/AEM.01343-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Trappetti C, et al. LuxS mediates iron-dependent biofilm formation, competence, and fratricide in Streptococcus pneumoniae. Infect. Immun. 2011;79:4550–4558. doi: 10.1128/IAI.05644-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Weimer KED, et al. Coinfection with Haemophilus influenzae promotes pneumococcal biofilm formation during experimental otitis media and impedes the progression of pneumococcal disease. J. Infect. Dis. 2010;202:1068–1075. doi: 10.1086/656046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Weimer KE, et al. Divergent mechanisms for passive pneumococcal resistance to beta-lactam antibiotics in the presence of Haemophilus influenzae. J. Infect. Dis. 2011;203:549–555. doi: 10.1093/infdis/jiq087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Armbruster CE, et al. LuxS promotes biofilm maturation and persistence of nontypeable haemophilus influenzae in vivo via modulation of lipooligosaccharides on the bacterial surface. Infect. Immun. 2009;77:4081–4091. doi: 10.1128/IAI.00320-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Joyce EA, et al. LuxS is required for persistent pneumococcal carriage and expression of virulence and biosynthesis genes. Infect. Immun. 2004;72:2964–2975. doi: 10.1128/IAI.72.5.2964-2975.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Armbruster CE, et al. Indirect pathogenicity of Haemophilus influenzae and Moraxella catarrhalis in polymicrobial otitis media occurs via interspecies quorum signaling. MBio. 2010;1:e00102–e00110. doi: 10.1128/mBio.00102-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Abubucker S, et al. Metabolic reconstruction for metagenomic data and its application to the human microbiome. PLoS Comput. Biol. 2012;8:e1002358. doi: 10.1371/journal.pcbi.1002358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.McCool TL, et al. The immune response to pneumococcal proteins during experimental human carriage. J. Exp. Med. 2002;195:359–365. doi: 10.1084/jem.20011576. [DOI] [PMC free article] [PubMed] [Google Scholar]