Abstract
Drosophila Hrp38, a homolog of human hnRNP A1, has been shown to regulate splicing, but its function can be modified by poly(ADP-ribosyl)ation. Notwithstanding such findings, our understanding of the roles of poly(ADP-ribosyl)ated Hrp38 on development is limited. Here, we have demonstrated that Hrp38 is essential for fly eye development based on a rough-eye phenotype with disorganized ommatidia observed in adult escapers of the hrp38 mutant. We also observed that Poly(ADP-ribose) Glycohydrolase (Parg) loss-of-function, which caused increased Hrp38 poly(ADP-ribosyl)ation, also resulted in the rough-eye phenotype with disrupted ommatidial lattice and reduced number of photoreceptor cells. In addition, ectopic expression of DE-cadherin, which is required for retinal morphogenesis, fully rescued the rough-eye phenotype of the hrp38 mutant. Similarly, Parg mutant eye clones had decreased expression level of DE-cadherin with orientation defects, which is reminiscent of DE-cadherin mutant eye phenotype. Therefore, our results suggest that Hrp38 poly(ADP-ribosyl)ation controls eye pattern formation via regulation of DE-cadherin expression, a finding which has implications for understanding the pathogenic mechanisms of Hrp38-related Fragile X syndrome and PARP1-related retinal degeneration diseases.
Keywords: Drosophila, Hrp38, poly(ADP-ribosyl)ation, pattern formation, eye development
1. Introduction
Drosophila Hrb98DE/Hrp38 gene, a member of the hnRNP family of proteins, has two RNA-binding motifs and one glycine-rich region, having highest identity with human hnRNP A1 (Haynes et al., 1990). The hrp38 gene has been shown to regulate alternative splicing, both in vivo and in vitro, using the S2 cell line (Shen et al., 1995; Blanchette et al., 2009; Borah et al., 2009). Alternative splicing is used extensively to produce the different mRNA isoforms of a gene to increase the complexity of the transcriptome in higher eukaryotic genomes (Nilsen and Graveley, 2010). It is generally believed that two groups of RNA-binding proteins, hnRNPs and serine-arginine-rich splicing factor (SR protein), regulate alternative splicing by binding with exonic and intronic splicing silencers (ESEs and ISEs) and enhancers (ESSs and ISSs), respectively (Matlin et al., 2005). However, emerging evidence suggests that hnRNPs can also bind with ESSs and ISSs to enhance splicing (Kiesler et al., 2005; Blanchette et al., 2009; Borah et al., 2009). In addition, the genome-wide analysis of the mRNA bound by hnRNP and SR proteins revealed that Drosophila hnRNPs regulate quite different sets of genes compared with SR proteins in Drosophila cell lines, challenging the current model that hnRNP and SR proteins have an antagonistic effect on splicing regulation (Blanchette et al., 2005).
As an alternative to changing the concentration of splicing factors in different tissues or developmental stages (Matlin et al., 2005), regulating the splicing activities of hnRNPs and SR proteins may be accomplished by post-translational modification via poly(ADP-ribosyl)ation (Gagne et al., 2003; Malanga et al., 2008; Ji and Tulin, 2009; Ji and Tulin, 2010). Specifically, Hrp38 is modified at the post-translational level by poly(ADP-ribosyl)ation through the activity of Poly(ADP-ribose) Polymerase 1 (PARP1) in Drosophila (Ji and Tulin, 2009). In addition, protein poly(ADP-ribosyl)ation can be reversed by Poly(ADP-ribose) Glycohydrolase (PARG), which degrades poly(ADP-ribose) polymer (Hanai et al., 2004; Tulin et al., 2006). Consequently, Hrp38 is highly poly(ADP-ribosyl)ated in the Parg mutant (Ji and Tulin, 2009). Furthermore, it appears that poly(ADP-ribosyl)ation inhibits the RNA-binding ability of hnRNPs and can modulate the alternative splicing pathways (Ji and Tulin, 2009). Our recent study suggested that poly(ADP-ribosyl)ation regulates Hrp38-dependent translation of DE-cadherin by the inhibition of Hrp38 binding to 5′UTR of DE-cadherin mRNA (Ji and Tulin, 2012). Based on this evidence, it could be reasonably concluded that post-translational modification of hnRNPs by poly(ADP-ribose) is a novel mechanism that regulates such hnRNP-dependent pathways as splicing and translation.
Therefore, we have been further assessing whether hnRNP poly(ADP-ribosyl)ation regulates gene expression during development. In our previous study, we have demonstrated that Hrp38 poly(ADP-ribosyl)ation controls germline stem cell (GSC) self-renewal and oocyte localization during Drosophila oogenesis by regulating DE-cadherin translation (Ji and Tulin 2012). Importantly, we note that DE-cadherin-mediated adherens junctions are required for retinal morphogenesis by organizing photoreceptor cell patterns and regulating ommatidial rotation (Tepass and Harris, 2007). Accordingly, in the present study, we further found that both Hrp38 loss-of-function and its poly(ADP-ribosy)lation cause a rough-eye phenotype displaying disorganized ommatidia. As expected, the rough-eye phenotype in the hrp38 mutant was rescued by overexpression of DE-cadherin in the eye, while the Parg mutant eye clones showed decreased expression of DE-cadherin. These results suggest that Hrp38 poly(ADP-ribosyl)ation plays a role during eye pattern formation by regulating DE-cadherin expression.
2. Material and Methods
2.1 Drosophila strains
Flies were cultured on standard cornmeal-molasses-agar media at 22°C. Hrb98DE/hrp38 GFP trap line (ZCL588) (Morin et al., 2001; Ji and Tulin, 2009), P{w[+mC]=GAL4-ninaE.GMR}12 (stock number: 1104) and P{w[+mC]=longGMR-GAL4}2 (stock number: 8605) were from the Bloomington Drosophila Stock Center. A P-element insertion of the hrp38 gene (w*,P[XP]d05172/TM6B, Tb1), a hrp38 region deficiency line (w1118; Df(3R)Exel6209, P{XP-U}Exel6209/TM6B, Tb1) and the UAS-Hrp38:RFP transgenic line were previously described (Ji and Tulin, 2012). The UAS-DE-cadherin:GFP (UAS-DEFL) transgenic line is a gift from the laboratory of Dr. Yamashita (Inaba et al., 2010). Tubulin-DE-cadherin transgenic line is a gift from Dr. Mark Van Doren (Mathews et al., 2006). Two Hrp38 RNAi lines (w1118; P{GD14939}v29523 and w1118; P{GD14939}v29524/CyO) were from the Vienna Drosophila RNAi Centre.
2.2 FRT/FLP Clonal Analysis
The Parg female heterozygotes (Parg27.1/FM7a,wa) (Hanai et al., 2004; Tulin et al., 2006) were crossed with w1118, sn3, P{neoFRT}19A/Y (Xu and Rubin, 1993) to generate the FRT-bearing Parg mutations (Parg27.1, P{neoFRT}19A/FM7a,wa) by genetic recombination. To induce the adult Parg mutant and wild-type eye clones, Parg27.1, P{neoFRT}19A/FM7a,wa or w1118, sn3, P{neoFRT}19A/Y was crossed with P{GMR-hid}SS1, y1 w* P{neoFRT}19A; P{GAL4-ey.H}SS5, P{UAS-FLP1.D}JD2 using the ey-Gal4/UAS-FLP/GMR-hid method (Stowers and Schwarz, 1999). To induce the Parg mutant eye disc clones in the third-instar larvae stage, Parg27.1, P{FRT(whs)101/FM7a,wa (Ji and Tulin, 2012) was crossed with Ubi-GFP, P{FRT(whshs)101/Y; P{GAL4-ey.H}SS5, P{UAS-FLP1.D}JD2 to select the GFP mosaic eye imaginal disc.
2.3 Western Blotting
Total protein (50 ug) from the wild-type fly, mutants (non-GFP homozygotes) at the wandering third-instar larvae, and the head and body of the Parg mutant mosaic adult was isolated and measured as described previously (Ji and Tulin, 2009). The proteins were then resolved in SDS-PAGE and transferred to nitrocellulose membrane (0.45 μm, Bio-Rad). The blot was incubated with rabbit anti-pADPr antibody (Calbiochem) at 1:1000 dilution. The signals were detected with horseradish peroxidase-conjugated secondary antiserum and ECL™ reagents (GE Healthcare). The blots were stripped and detected with mouse anti α-tubulin antibody (DM1A, Sigma) at 1:1000 dilution.
2.4 Immunohistochemistry
The eye imaginal discs of the third-instar larvae were dissected in Grace’s insect medium and fixed in 4% paraformaldehyde + 0.1% Triton X-100 in PBS for 20 min. Afterwards, the discs were incubated with mouse anti-Elav (1:10; DSHB) and Alexa Fluor 488 goat anti-mouse antibody (1:400; Invitrogen), respectively. The nuclear DNA was stained with DRAQ5 dye (Biostatus). The pupal retinae at 44 hours after puparation were dissected and stained with Alexa Fluor 633 phalloidin (1:40; Invitrogen) for 30 minutes. The adult eyes were dissected and stained with anti-rhodopsin (4C5) antibody (1:10, DSHB) based on the published protocol (Williamson and Hiesinger, 2010). All the images were visualized using the Leica TCS-NT confocal microscope.
2.5 Electron Microscopy
For scanning EM, the dissected heads were fixed as for TEM, postfixed in 1% OsO4 for 3 hours, dehydrated in ethanol and critical point dried as described (Anderson 1951). The samples were viewed on an Autoscan scanning electron microscope (ETEC, Hayward, CA). For ultrastructural analysis by transmission EM, the heads were dissected, fixed with 2% formaldehyde/2% glutaraldehyde in 0.1 M cacodylate buffer (pH 7.2) in 0.1% Triton X-100 overnight, postfixed for 1 hour with osmium tetroxide, dehydrated in ethanol and propylenoxide, and embedded in EMbed-812 (EMS, Fort Washington, PA) in flat molds. After polymerization for 60 hours at 65°C, 70 nm sections were cut on a Leica UC6 microtome (Leica, Austria), placed on formvar/carbon-coated grids, and stained with 2% uranyl acetate/lead citrate. Sections were viewed on a Tecnai 12 transmission electron microscope (FEI, Hillsboro, OR).
3. Results
3.1 Loss-of-function of the hrp38 gene causes the rough-eye phenotypes
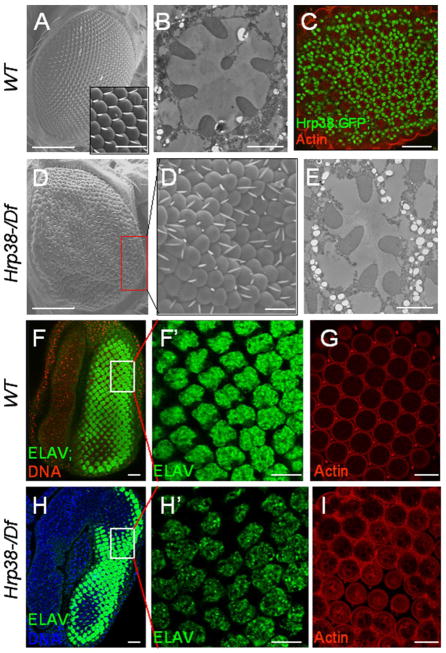
Our previous study suggested that the hrp38 gene is important for fly development because a full 75% of the hrp38 hemizygotes (hrp38d05172/Df) died before the pupa stage (Ji and Tulin 2012). Interestingly, we observed that the adult escapers of the hrp38 hemizygotes showed rough-eye phenotype with disorganized ommatidia and bristles (Figure 1D and 1D′), while the wild-type fly had no defects (Figure 1A). Occasionally, loss of one photoreceptor cell was observed in the ommatidia (Figure 1E) compared to the wild type (Figure 1B). In addition, Hrp38 expression was observed in the photoreceptor cells in the ommatidia in a Hrp38:GFP protein trap line (Figure 1C). We further examined the larval eye disc stained with anti-Elav (a neuron marker) antibody in the wild-type and hrp38 mutant. It appears that both hrp38 mutant cells (Figure 1H) and wild type (Figure 1F) had normal specification of the photoreceptor fate. However, the pattern of photoreceptor cells in the hrp38 mutant cells was irregular (Figure 1H′) compared to the wild-type cells (Figure 1F′). Moreover, the hrp38 mutant pupal eye showed a disrupted ommatidial lattice (Figure 1I) compared with the wild-type eye (Figure 1G). Based on this evidence, we concluded that the rough-eye phenotype caused by Hrp38 loss-of-function results from disrupted pattern formation shown as early as at the larval stage.
Figure 1. The hrp38 gene is required for normal pattern formation of the fly eye.
(A) Scanning electron microscopy (SEM) image of wild-type eye. Insert shows magnified view of the regular organization of ommatidia and bristles. (B) Tangential sections of wild-type adult retinae showing seven photoreceptor cells in ommatidia. (C) Confocal section of the pupal eye (44h AP) of the Hrp38:GFP strain. F-actin is enriched in rhabodomere. (D,D′) SEM image of the hrp38 hemizygous eye (hrp38d05172/Df) showing the disrupted pattern of ommatidia and bristles. (D′) shows magnified view of boxed area in (D). (E) Tangential sections of the hrp38 hemizygous retinae showing that one photoreceptor cell is missing in an ommatidium. (F) and (F′) The eye imaginal disc of the wild-type fly stained with Elav showing the regular array of the specified photoreceptor cells. The nuclear DNA was stained with Draq5 (red) in (F). (F′) shows magnified view of boxed area in (F). (G) Confocal section of the pupal eye (44h AP) of the wild-type strain stained for F-actin. (H,H′) The eye imaginal disc of the hrp38 hemizygous fly stained with Elav showing the organization of the specified photoreceptor cells. The nuclear DNA in (H) was stained with Draq5 (blue). (H′) shows a disrupted pattern of the photoreceptor cells magnified in the boxed area in (H). (I) Confocal section of the pupal eye (44h AP) of the hrp38 hemizygous fly stained for F-actin, showing the disrupted ommatidial lattice. Scale bars: 100 μm (A, D); 10μm (A insert, C, D′, F, F′,G, H, H′ and I); 2 μm (B, E).
3.2 Expression of Hrp38:RFP transgene in the eye rescued the rough-eye phenotype of the Hrp38 mutant
In addition, we used RNAi to knock down hrp38 expression in the eye using two eye-specific GAL4 drivers (ninaE.GMR-Gal4 and longGMR-Gal4). While ninaGMR-Gal4 heterozygotes, as the control, had the wild-type eye phenotype (Figure 2A and 2A′), hrp38 RNAi induced by ninaE.GMR-Gal4 driver also caused the rough-eye phenotype with disrupted ommatidia and bristle organizations (Figure 2B and 2B′). However, hrp38 knockdown by longGMR-Gal4 driver did not show any mutated phenotype. This observation may reflect the different Gal4 expression level of two GMR-Gal4 drivers because the promoter of ninaE-GMR transgene, which is composed of a pentamerized 29bp glass binding site named as “short GMR”, has a stronger ability of inducing GAL expression than the longGMR-Gal4 driver whose promoter has a pentamerized 38 bp glass binding site termed as “long GMR” (Freeman, 1996; Wernet, et al. 2003). Indeed, the expression of a UAS-hrp38-RFP transgene induced by the longGMR-Gal4 driver rescued the observed rough-eye phenotypes of the hemizygotes (hrp38d05172/Df) (Figure 2C and 2C′), which further demonstrates that loss-of-function of the hrp38 gene in the eye results in the observed phenotypes in the hrp38 hemizygotes. In contrast, overexpression of the UAS-hrp38-RFP transgene induced by the ninaE.GMR-Gal4 (short-GMR) driver could not rescue the rough-eye phenotype of the hrp38 mutant (Figure 2D and 2D′), suggesting that the expression level of the hrp38 gene in the eye is critical for normal eye development. Therefore, Hrp38 protein regulates eye pattern formation.
Figure 2. The rescue of the rough-eye phenotype of the Hrp38 mutant by the expression of Hrp38:RFP transgene.
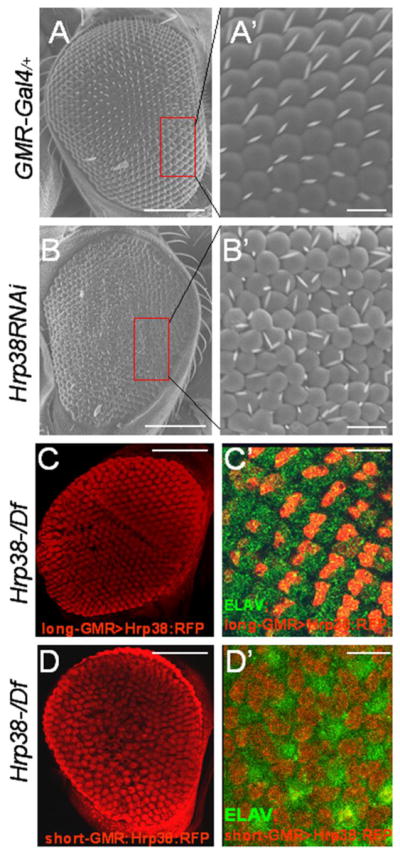
(A,A′) SEM eye image of RNAi control line (ninaE.GMR-Gal4/+), showing the regular array of ommatidia and bristles. (A′) shows magnified view of boxed area in (A). (B,B′) SEM image of the Hrp38 knockdown eye by RNAi (ninaE.GMR-Gal4/UAS-Hrp38RNAi), showing the disrupted pattern of ommatidia and bristles. (C,C′) Confocal image of the adult eye structure (C) and the third-instar eye imaginal disc (C′) of the hrp38 mutant expressing Hrp38:RFP transgene driven by longGMR:Gal4 line (UAS>Hrp38:RFP/Long-GMR:Gal4;hrp38d05172/Df). (D,D′) Confocal image of the adult eye structure (D) and the third-instar eye imaginal disc (D′) of the hrp38 mutant expressing Hrp38:RFP transgene driven by the ninaE.GMR:Gal4 line (UAS>Hrp38:RFP/ninE-GMR:Gal4;hrp38d05172/Df). In (C′,D′), the photoreceptor cells are labeled with anti-ELAV antibody (green). Scale bars: 100 μm (A, B, C, D); 10μm (A′, B′, C′, D′).
3.3 Hrp38 Poly(ADP-ribosyl)ation causes rough-eye phenotypes
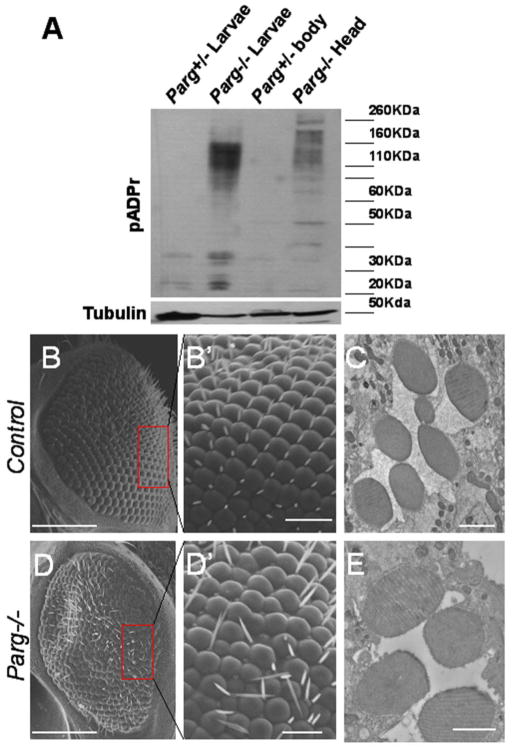
We have shown that Hrp38 is highly poly(ADP-ribosyl)ated in the Parg mutant which affects hnRNP-dependent pathways, such as splicing and translation, by changing the RNA-binding ability of hnRNP proteins (Ji and Tulin, 2009; Ji and Tulin, 2012). In order to determine if Hrp38 poly(ADP-ribosyl)ation also plays a role during eye development, we examined eye development in the Parg mutant. Since Parg mutants are completely lethal and died at the pharate stages (Hanai et al. 2004), we used the ey-Gal4/UAS-FLP/GMR-hid method (Stowers and Schwarz, 1999) to generate the Parg mutant eye. Western blotting also showed that the Parg mutant eye does indeed accumulate a large amount of poly(ADP-ribose) (pADPr) (Figure 3A). The homozygous FRT19A eyes generated by Flippase induction showed normal eye structure (Figure 3B, 3B′ and 3C). However, knocking out the Parg gene in the eye resulted in the rough-eye phenotype with the disrupted pattern of ommatidia and bristles (Figure 3D and D′). Furthermore, we found that around half of the ommatidia in the Parg mutant eye were missing two or three photoreceptor cells (Figure 3E), which suggests that the Parg gene is required for the growth and differentiation of photoreceptor cells. Taken together, these results suggest that Parg genes are also required for eye development. The phenotype similarity between the hrp38 and Parg mutations implies that the Parg gene may regulate the same pathway as Hrp38 through Hrp38 poly(ADP-ribosyl)ation during eye development.
Figure 3. The Parg gene is required for normal pattern formation of the fly eye.
(A) Western blotting analysis of pADPr in the wild-type and Parg mutant eye. Accumulation of pADPr was shown in the adult head, including the Parg mutant eye (Parg−/− head), but not in the separated body (Parg+/− body) dissected from the adult fly (Parg 27.1, FRT19A/FRT19A,GMR-hid; ey-Gal4,UAS-FLP). Parg−/− and Parg+/− larvae were used as the positive and negative controls, respectively. (B,B′) SEM image of the control eye clones (FRT19A/FRT19A,GMR-hid; ey-Gal4,UAS-FLP), showing the regular array of ommatidia and bristles. (B′) shows magnified view of boxed area in (B). (C) Tangential sections of the adult retinae of the control fly (FRT19A/FRT19A,GMR-hid; ey-Gal4,UAS-FLP), showing seven photoreceptor cells in an ommatidium. (D,D′) SEM image of the Parg−/− eye clones (Parg27.1, FRT19A/FRT19A,GMR-hid; ey-Gal4,UAS-FLP), showing the disrupted organization of ommatidia and bristles. (D′) shows magnified view of boxed area in (D). (E) Tangential sections of the adult retinae of the Parg−/− eye clones, showing missing photoreceptor cells in an ommatidium. Scale bars; 100 μm (B, D); 10μm (B′, D′); 2 μm (C, E).
3.4 Hrp38 Poly(ADP-ribosyl)ation regulates DE-cadherin expression in the eye imaginal disc
Our previous study has demonstrated that both hrp38 and Parg mutants had a lower expression level of DE-cadherin compared with the wild-type fly (Ji and Tulin, 2012), suggesting that both hrp38 and Parg genes are required for normal DE-cadherin expression during fly development. Since DE-cadherin-mediated adherens junctions are very important for retinal morphogenesis (Grzeschik and Knust, 2005; Mirkovic and Mlodzik, 2006; Tepass and Harris, 2007), we also examined DE-cadherin expression in the Parg mutant eye imaginal disc clones generated by the eye-FLP in the 3rd instar larvae stage. As shown in Figure 4A, 4A′ and 4A″, DE-cadherin was enriched between the members of the photoreceptor precursor cells behind the morphogenic furrow in the wild-type eye imaginal disc, as reported before (Grzeschik and Knust, 2005). However, it appeared that DE-cadherin expression was lost in the Parg mutant photoreceptor precursor cell clones (Figure 4B,4B′,4B″). These Parg eye imaginal disc clones also showed rotation defects (Figure 4B‴), which is reminiscent of the phenotype shown in DE-cadherin mutant eye clones (Mirkovic and Mlodzik, 2006).
Figure 4. Hrp38 poly(ADP-ribosyl)ation controls DE-cadherin expression in the eye.
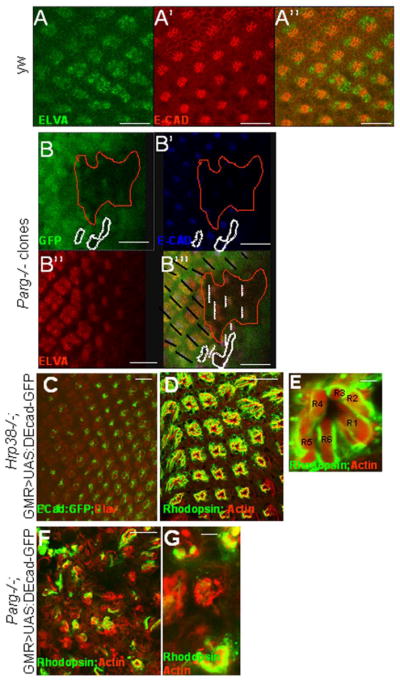
(A,A′,A″) DE-cadherin expression pattern in the third-instar eye discs of the wild-type fly (y,w). (B,B′,B″,B‴) Decreased DE-cadherin expression in the Parg mutant clones in the third-instar eye imaginal discs {Parg27.1, P{FRT(whs)101/Ubi-GFP,P{FRT(whshs)101; P{GAL4-ey.H}SS5, P{UAS-FLP1.D}JD2/+}. The Parg mutant clones are GFP-negative and circled in (B,B″,B‴). The black lines indicate the orientation of the wild-type ommatidia in (B‴). The dashed white lines indicate the orientation of the mutant ommatidia in (B‴). The photoreceptor precursor cells were labeled with anti-ELAV antibody in (A) and (B″). (C,D,E) Rescue of the rough-eye phenotype by expression of DE-cadherin:GFP transgene in the Hrp38 mutant background (UAS:DEF/longGMR:Gal4; hrp38do5172/Df). (C) The regular array of the photoreceptor cells labeled with anti-ELAV antibody with DE-cadherin:GFP expression induced by longGMR:Gal4 driver in the third-instar eye disc. (D) The regular ommatidia structure in the adult eye of the rescued fly visualized by confocal imaging. (E) Confocal image of an ommatidium from (D) with the normal seven photoreceptor cells. (F,G) Failed rescue of the Parg mutant phenotype with ubiquitous expression of DE-cadherin (Parg27.1, FRT19A/FRT19A,GMR-hid; Tub-DE-cadherin/eye-Gal4,UAS-FLP). (F) The irregular ommatidia structure in the Parg mutant eye. (G) Reduced photoreceptor cell numbers in the Parg mutant eye. Adult photoreceptor cells in (D,E,F,G) were labeled with anti-Rhodopsin antibody and F-actin. Scale bars: 10 μm (A, A′, A″, B, B′,B″, B‴, C); 100 μm (D, F); 5 μm (E, G).
Because Hrp38 has been shown to be required for DE-cadherin translation by binding to 5′ UTR of DE-cadherin (Ji and Tulin, 2012), decreased DE-cadherin expression may also be responsible for the rough-eye phenotype in the hrp38 mutant. Indeed, expression of a UAS-DE-cadherin:GFP transgene (Oda et al., 1994) induced by GMR-Gal4 driver fully rescued the rough-eye phenotypes in the hrp38 mutants in both the third-instar larvae stage (Figure 4C) and adult stage (Figure 4D and 4E). However, even expression of DE-cadherin transgene with ubiquitous tubulin promoter (Tub-DE-cad) (Mathews et al. 2006) could not restore normal eye pattern in the adult Parg mutant eye (Figure 4F and 4G), suggesting that decreased DE-cadherin expression is not the sole reason for eye development defects in the Parg mutant.
4. Discussion
4. 1 The overlapping functions between Hrp38 and Hrp36
It has been reported that hrp36 deficiency also results in the rough-eye phenotype (Sengupta and Lakhotia, 2006). Therefore, it appears that both hrp36 and hrp38 genes are required during eye development. In addition, the genome-wide analysis of the mRNA bound by Hrp36 and Hrp38 proteins revealed that hrp36 and hrp38 regulate overlapping sets of genes in Drosophila cell lines (Blanchette et al., 2005; Blanchette et al., 2009). Hrp38 and Hrp36 are highly conserved with 76% identity in the RNA-binding motifs and 67% identity in the glycine-rich region (Haynes et al., 1991). A recent study also suggested that Hrp36 is required for normal development, especially under stress conditions (Singh and Lakhotia, 2012). Therefore, this accumulated evidence suggests that the hrp38 gene has overlapping, but not redundant, functions with the hrp36 gene, despite high similarity between the two genes.
4. 2 Decreased DE-cadherin in a Drosophila model of Fragile X syndrome may provide clues for understanding its pathogenesis in human
Our data showed that decreased DE-cadherin expression is an underlying cause of the rough-eye phenotype in the hrp38 mutant. Interestingly, the rough-eye phenotype caused by hrp38 loss-of-function was also shown in a Drosophila model of Fragile X syndrome as a result of the titration of RNA-binding proteins, including Hrp38, by binding to the CGG repeats (Sofola et al., 2007). It was also shown that Hrp38 overexpression can suppress the rCGG-mediated rough-eye phenotype (Sofola et al., 2007). If decreased DE-cadherin does result in the rCGG-mediated rough-eye phenotype in a Drosophila model of Fragile X syndrome, it would provide additional clues for understanding the pathogenesis of Fragile X syndrome.
4.3 PARG activity regulates cellular pADPr level
PARG is responsible for the cleavage of pADPr into the single ADP-ribose unit by hydrolyzing the ribose-ribose bonds within the polymer chain (Miwa and Sugimura 1971; Lin et al., 1997). Therefore, Parg knockout in model animals, such as Drosophila and mice, resulted in the accumulation of pADPr in the whole animals (Koh et al., 2004; Hanai et al., 2004; Tulin et al., 2006), as well as in the specific tissues shown in this study (Fig. 3A), suggesting that PARG activity controls the level of cellular pADPr. However, a recent study showed that knockdown of PARG expression by siRNA reduced PARP1 expression at the transcriptional level, thereby decreasing pADPr level in treated HeLa S3 cells (Uchiumi et al., 2013). The observed difference in pADPr level between the whole animals and the cultured cells most likely results from the fact that Drosophila PARP1 gene has strong maternal expression (Hanai et al., 1998; Tulin et al., 2002). Thus, even if PARG knockout had such effect on PARP1 expression in the fly, maternal PARP1 activity could continue to generate pADPr in the PARG mutants. Furthermore, PARG loss-of-function on PARP1 activity could also result in the failure of PARP1 automodification. This could also inhibit PARP1 activity in the PARG knockout animals because PARP automodification disables its enzymatic ability to synthesize new pADPr (Satoh and Lindahl, 1992). However, two recent studies on the enzymes which cleave the terminal ADP-ribose unit from auto(ADP-ribosyl)ated PARP1 (Sharifi et al., 2013; Jankevicius et al., 2013) have shed light on this long-standing puzzle. It is well known that PARG cannot hydrolyze the bond between terminal ADP-ribose and glutamate residues of automodifed PARP1 (Slade et al., 2011). It has been reported that two macrodomain proteins, including human MacroD2 and TARG1/C6orf130, have the ability to remove the terminal modification of PARP1 (Sharifi et al., 2013; Jankevicius et al., 2013). It is therefore possible that these two enzymes could directly cleave the whole pADPr chain from the terminal bonds of weakly automodified PARP, thus partially restoring PARP1 activity in the Parg mutant background. Consequently, the residual activities of MacroD2 and TARG1 may contribute to the accumulation of pADPr in the Parg knockout animals.
4.4 The new insight into the pathogenesis of PARP1-related retinal degeneration diseases
Previous studies have found that excessive pADPr from PARP1 activation contributes to degeneration of photoreceptor cells in the retinal degeneration 1 (rd1) mouse model of retinitis pigmentosa (RP) (Paquet-Durand et al., 2007). Our present study further confirms this phenomenon by demonstrating that accumulation of pADPr exclusively in the fly eye by conditional Parg knockout also causes eye developmental defects with the rough-eye phenotype and reduced photoreceptor cell numbers. Our finding also implies that decreased Parg expression or activation may also play a role in the rd1 mouse model or human RP disease (Sahaboglu et al., 2010; McLaughlin et al., 1995). In addition, since most human RP shows progressive severity from night blindness in the early stage to total vision loss with aging (Hartong et al., 2006), it is likely that cell death is not the only causal factor underlying retina developmental defects, even in RP patients. Therefore, based on the findings of the present study, other pathways involving PARP1 and PARG genes should be investigated in the future. We found that decreased expression of DE-cadherin in the Parg mutant is associated with the rough-eye phenotypes, and this discovery gives additional clues for understanding RP. However, because overexpression of DE-cadherin cannot rescue the developmental defects of Parg mutant eye, it is very likely that poly(ADP-risosyl)ation affects other Hrp38-dependent pathways during eye pattern formation (Figure 5).
Figure 5. Model explaining the regulatory role of hnRNP poly(ADP-ribosyl)ation in eye pattern development.
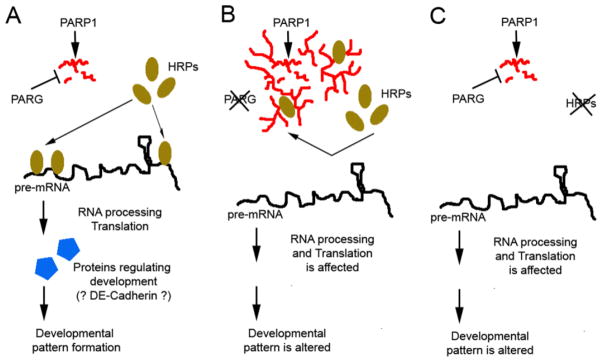
(A) In a temporal- or spatial-specific way, protein poly(ADP-ribosyl)ation is reversed by PARG activity. Therefore, HRP proteins are not poly(ADP-ribosyl)ated and will bind to their target pre-mRNAs for RNA processing or translational control, which further regulates developmental patterns. (B) Once PARG activity is downregulated in different tissues or developmental stages, HRP proteins are poly(ADP-ribosyl)ated, which inhibits Hrps from binding to pre-mRNA. Therefore, the pre-mRNA bound by Hrps are not processed or translated, and developmental patterns are changed. (C) Mutating HRP proteins blocks pre-mRNA processing or translation.
4.5 Conclusion
In summary, this study has further demonstrated that post-translational modification of hnRNPs, such as poly(ADP-ribosyl)ation, is an important mechanism in the regulation of gene expression during development, such as eye pattern formation.
Highlights.
Hrp38 knockout caused a rough-eye phenotype with disorganized ommatidia.
Parg knockout fly also exhibited a rough-eye phenotype with reduced photoreceptor cells.
Ectopic expression of DE-cadherin rescued the rough-eye phenotype of the hrp38 mutant.
Parg mutant eye clones showed decreased DE-cadherin expression with orientation defects.
Hrp38 poly(ADP-ribosyl)ation controls eye pattern formation via regulation of DE-cadherin expression.
Acknowledgments
We thank Drs. Van Doren and Yamashita for providing materials. Drs. M. Robinson, A. O’Reilly, D. Martin and A. Liubimova provided comments on the manuscript. The research was supported by grants from the National Institutes of Health (R01 GM077452) to A.V.T.
ABBREVIATIONS for GENE38629
- PARG
Poly(ADP-ribose) Glycohydrolase
- PARP1
Poly(ADP-ribose) Polymerase 1
- pADPr
poly(ADP-ribose)
- hnRNP/Hrp
Heterogeneous nuclear ribonucleoproteins
- SR protein
serine-arginine-rich protein
- ESSs
exonic splicing silencers
- ISSs
intronic splicing silencers
- ESEs
exonic splicing enhancers
- ISEs
intronic splicing enhancers
- GSC
germline stem cell
- 5′UTR
5′ untranslated region
- DE-cadherin
Drosophila Epithelial cadherin
- Df
deficiency
- UAS
upstream activation sequence
- rd1
retinal degeneration 1
- RP
retinitis pigmentosa
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anderson TF. Technique for preservation of three-dimensional structure in preparing specimens for the electron microscope. Trans N Y Acad Sci. 1951;13:130–134. doi: 10.1111/j.2164-0947.1954.tb00377.x. [DOI] [PubMed] [Google Scholar]
- Blanchette M, Green RE, Brenner SE, Rio DC. Global analysis of positive and negative pre-mRNA splicing regulators in Drosophila. Genes & Development. 2005;19:1306–1314. doi: 10.1101/gad.1314205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchette M, Green RE, Macarthur S, Brooks AN, Brenner SE, et al. Genome-wide analysis of alternative pre-mRNA splicing and RNA-binding specificities of the Drosophila hnRNP A/B family members. Molecular Cell. 2009;33:438–449. doi: 10.1016/j.molcel.2009.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borah S, Wong AG, Steitz JA. Drosophila hnRNP A1 homologs Hrp36/Hrp38 enhance U2-type versus U12-type splicing to regulate alternative splicing of the prospero twintron. PNAS. 2009;106:2577–2582. doi: 10.1073/pnas.0812826106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman M. Reiterative use of the EGF receptor triggers differentiation of all cell types in the Drosophila eye. Cell. 1996;87:651–660. doi: 10.1016/s0092-8674(00)81385-9. [DOI] [PubMed] [Google Scholar]
- Gagne JP, Hunter JM, Labrecque B, Chabot B, Poirier GG. A proteomic approach to the identification of heterogeneous nuclear ribonucleoproteins as a new family of poly(ADP-ribose)-binding proteins. Biochem J. 2003;371:331–340. doi: 10.1042/BJ20021675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grzeschik NA, Knust E. IrreC/rst-mediated cell sorting during Drosophila pupal eye development depends on proper localisation of DE-cadherin. Development. 2005;132:2035–2045. doi: 10.1242/dev.01800. [DOI] [PubMed] [Google Scholar]
- Hanai M, Uchida M, Kobayashi S, Miwa M, Uchida K. Genomic organization of Drosophila poly(ADP-ribose) polymerase and distribution of its mRNA during development. J Biol Chem. 1998;273:11881–11886. doi: 10.1074/jbc.273.19.11881. [DOI] [PubMed] [Google Scholar]
- Hanai S, Kanai M, Ohashi S, Okamoto K, Yamada M, et al. Loss of poly(ADP- ribose) glycohydrolase causes progressive neurodegeneration in Drosophila melanogaster. PNAS. 2004;10:82–86. doi: 10.1073/pnas.2237114100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartong DT, Berson EL, Dryja TP. Retinitis pigmentosa. The Lancet. 2006;368:1795–1809. doi: 10.1016/S0140-6736(06)69740-7. [DOI] [PubMed] [Google Scholar]
- Haynes S, Raychaudhuri G, Beyer AL. The Drosophila Hrb98DE locus encodes four protein isoforms homologous to the A1 protein of mammalian heterogeneous nuclear ribonucleoprotein complexes. Mol Cell Biol. 1990;10:316–323. doi: 10.1128/mcb.10.1.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes SR, Johnson D, Raychaudhuri G, Beyer AL. The Drosophila Hrb87F gene encodes a new member of the A and B hnRNP protein group. Nucleic Acids Research. 1991;19:25–31. doi: 10.1093/nar/19.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inaba M, Yuan H, Salzmann V, Fuller MT, Yamashita YM. E-Cadherin Is Required for Centrosome and Spindle Orientation in Drosophila Male Germline Stem Cells. PLoS ONE. 2010;5:e12473. doi: 10.1371/journal.pone.0012473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankevicius G, Jankevicius G, Hassler M, Golia B, Rybin V, et al. A family of macrodomain proteins reverses cellular mono-ADP-ribosylation. Nat Struct Mol Biol. 2013;20:508–514. doi: 10.1038/nsmb.2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh DW, Lawler AM, Poitras MF, Sasaki M, Wattler S, et al. Failure to degrade poly(ADP-ribose) causes increased sensitivity to cytotoxicity and early embryonic lethality. Proc Natl Acad Sci USA. 2004;101:17699–17704. doi: 10.1073/pnas.0406182101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Y, Tulin AV. Poly(ADP-ribosyl)ation of heterogeneous nuclear ribonucleoproteins modulates splicing. Nucleic Acids Research. 2009;37:3501–3513. doi: 10.1093/nar/gkp218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Y, Tulin AV. The roles of PARP1 in gene control and cell differentiation. Current Opinion in Genetics & Development. 2010;20:512–518. doi: 10.1016/j.gde.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Y, Tulin AV. Poly(ADP-ribose) controls DE-cadherin-dependent stem cell maintenanceand oocyte localization. Nature Communications. 2012;3:760. doi: 10.1038/ncomms1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiesler E, Hase ME, Brodin D, Visa N. Hrp59, an hnRNP M protein in Chironomus and Drosophila, binds to exonic splicing enhancers and is required for expression of a subset of mRNAs. J Cell Biol. 2005;168:1013–1025. doi: 10.1083/jcb.200407173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W, Ame JC, Aboul-Ela N, Jacobson EL, Jacobson MK. Isolation and characterization of the cDNA encoding bovine poly(ADP-ribose) glycohydrolase. J Biol Chem. 1997;272:11895–11901. doi: 10.1074/jbc.272.18.11895. [DOI] [PubMed] [Google Scholar]
- Matlin AJ, Clark F, Smith CW. Understanding alternative splicing: towards a cellular code. Nat Rev Mol Cell Biol. 2005;6:386–398. doi: 10.1038/nrm1645. [DOI] [PubMed] [Google Scholar]
- Malanga M, Czubaty A, Girstun A, Staron K, Althaus FR. Poly(ADP-ribose) binds to the splicing factor ASF/SF2 and regulates its phosphorylation by DNA topoisomerase I. J Biol Chem. 2008;283:19991–19998. doi: 10.1074/jbc.M709495200. [DOI] [PubMed] [Google Scholar]
- Mathews WR, Ong D, Milutinovich AB, Van Doren M. Zinc transport activity of Fear of Intimacy is essential for proper gonad morphogenesis and DE-cadherin expression. Development. 2006;133:1143–1153. doi: 10.1242/dev.02256. [DOI] [PubMed] [Google Scholar]
- McLaughlin ME, Ehrhart TL, Berson EL, Dryja TP. Mutation spectrum of the gene encoding the beta subunit of rod phosphodiesterase among patients with autosomal recessive retinitis pigmentosa. PNAS. 1995;92:3249–3253. doi: 10.1073/pnas.92.8.3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miwa M, Sugimura T. Splitting of the riboseribose linkage of poly(adenosine diphosphate-robose) by a calf thymus extract. J Biol Chem. 1971;246:6362–6364. [PubMed] [Google Scholar]
- Mirkovic I, Mlodzik M. Cooperative activities of Drosophila DE-Cadherin and DN-Cadherin regulate the cell motility process of ommatidial rotation. Development. 2006;133:3283–3293. doi: 10.1242/dev.02468. [DOI] [PubMed] [Google Scholar]
- Morin X, Daneman R, Zavortink M, Chia W. A protein trap strategy to detect GFP-tagged proteins expressed from their endogenous loci in Drosophila. PNAS. 2001;98:15050–15055. doi: 10.1073/pnas.261408198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsen TW, Graveley BR. Expansion of the eukaryotic proteome by alternative splicing. Nature. 2010;463:457–463. doi: 10.1038/nature08909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda H, Uemura T, Harada Y, Iwai Y, Takeichi M. A Drosophila Homolog of Cadherin Associated with Armadillo and Essential for Embryonic Cell-Cell Adhesion. Developmental Biology. 1994;165:716–726. doi: 10.1006/dbio.1994.1287. [DOI] [PubMed] [Google Scholar]
- Paquet-Durand F, Silva J, Talukdar T, Johnson LE, Azadi S, et al. Excessive activation of poly(ADP-ribose) polymerase contributes to inherited photoreceptor degeneration in the retinal degeneration 1 mouse. The Journal of Neuroscience. 2007;27:10311–10319. doi: 10.1523/JNEUROSCI.1514-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahaboglu A, Tanimoto N, Kaur J, Sancho-Pelluz J, Huber G, et al. PARP1 Gene Knock-Out Increases Resistance to Retinal Degeneration without Affecting Retinal Function. PLoS ONE. 2010;5:e15495. doi: 10.1371/journal.pone.0015495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh Masahiko S, Lindahl T. Role of poly (ADP-ribose) formation in DNA repair. Nature. 1992;356:356–358. doi: 10.1038/356356a0. [DOI] [PubMed] [Google Scholar]
- Sharifi R, Morra R, Appel DC, Tallis M, Chioza B, et al. Deficiency of terminal ADP-ribose protein glycohydrolase TARG1/C6orf130 in neurodegenerative disease. EMBO J. 2013 doi: 10.1038/emboj.2013.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J, Zu K, Cass CL, Beyer AL. Exon skipping by overexpression of a Drosophila heterogeneous nuclear ribonucleoprotein in vivo. PNAS. 1995;92:1822–1825. doi: 10.1073/pnas.92.6.1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slade D, Dunstan MS, Barkauskaite E, Weston R, Lafite P, et al. The structure and catalytic mechanism of a poly(ADP-ribose) glycohydrolase. Nature. 2011;477:616–620. doi: 10.1038/nature10404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stowers RS, Schwarz TL. A Genetic Method for Generating Drosophila Eyes Composed Exclusively of Mitotic Clones of a Single Genotype. Genetics. 1999;152:1631–1639. doi: 10.1093/genetics/152.4.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sofola OA, Jin P, Qin Y, Duan R, Liu H, et al. RNA-Binding Proteins hnRNP A2/B1 and CUGBP1 Suppress Fragile X CGG Premutation Repeat-Induced Neurodegeneration in a Drosophila Model of FXTAS. Neuron. 2007;55:565–571. doi: 10.1016/j.neuron.2007.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta S, Lakhotia SC. Altered expressions of the noncoding hsromega gene enhances poly-Q-induced neurotoxicity in Drosophila. RNA Biology. 2006;3:e1–e8. doi: 10.4161/rna.3.1.2559. [DOI] [PubMed] [Google Scholar]
- Singh A, Lakhotia SC. The hnRNP A1 homolog Hrp36 is essential for normal development, female fecundity, omega speckle formation and stress tolerance in Drosophila melanogaster. Journal of Biosciences. 2012;37:659–678. doi: 10.1007/s12038-012-9239-x. [DOI] [PubMed] [Google Scholar]
- Tepass U, Harris KP. Adherens junctions in Drosophila retinal morphogenesis. Trends in Cell Biology. 2007;17:26–35. doi: 10.1016/j.tcb.2006.11.006. [DOI] [PubMed] [Google Scholar]
- Tulin A, Stewart D, Spradling AC. The Drosophila heterochromatic gene encoding poly(ADP-ribose) polymerase (PARP) is required to modulate chromatin structure during development. Genes Dev 2002. 2002;16:2108–2119. doi: 10.1101/gad.1003902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulin AV, Naumova NM, Menon AK, Spradling AC. Drosophila Poly(ADP-Ribose) Glycohydrolase Mediates Chromatin Structure and SIR2-Dependent Silencing. Genetics. 2006;172:363–371. doi: 10.1534/genetics.105.049239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchiumi F, Watanabe T, Ohta R, Abe H, Tanuma S. PARP1 gene expression is downregulated by knockdown of PARG gene. Oncology Reports. 2013;29:1683–1688. doi: 10.3892/or.2013.2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernet MF, Labhart T, Baumann F, Mazzoni EO, Pichaud F, et al. Homothorax switches function of Drosophila photoreceptors from color to polarized light sensors. Cell. 2003;115:267–279. doi: 10.1016/s0092-8674(03)00848-1. [DOI] [PubMed] [Google Scholar]
- Williamson WR, Hiesinger PR. Preparation of Developing and Adult Drosophila Brains and Retinae for Live Imaging. J Vis Exp. 2010;37:e1936. doi: 10.3791/1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T, Rubin GM. Analysis of genetic mosaics in developing and adult Drosophila tissues. Development. 1993;117:1223–1237. doi: 10.1242/dev.117.4.1223. [DOI] [PubMed] [Google Scholar]