Abstract
The marRAB operon is conserved in seven genera of enteric bacteria (Escherichia, Shigella, Klebsiella, Enterobacter, Salmonella, Cronobacter, and Citrobacter). MarA is a transcriptional regulator affecting many genes involved in resistance to stresses, and MarR is an autorepressor of the operon, but a role for the marB gene has been unclear. A recent work reported that deletion of marB causes resistance to certain stresses and increases the amount of marA transcript. We show here that the small (216 bp) marB gene encodes a protein, not an sRNA, since two different stop codons within the predicted open reading frame of marB prevented plasmid-borne marB from complementing ΔmarB::Kan. The ΔmarB::Kan mutation did not increase the stability of the marA transcript, suggesting that MarB does not destabilize the marA transcript but rather reduces its rate of transcription. Placing the putative signal sequence of MarB upstream of signal-sequence-less alkaline phosphatase guided the phosphatase to its normal periplasmic location. We conclude that MarB is a small periplasmic protein that represses the marRAB promoter by an indirect mechanism, possibly involving a signal to one of the cytoplasmic regulators of that promoter.
Keywords: transcript stability, small protein, regulation
Introduction
Regulation of the expression of the marRAB (multiple antibiotic resistance) operon in Escherichia coli includes both autorepression by MarR and autoactivation by MarA (Martin et al., 1995; Seone & Levy 1995; Alekshum & Levy, 1997). This operon is expressed from a promoter upstream of marR that contains the marO operator targeted by MarR. MarR is a member of a more widely–recognized regulatory “MarR” family (Sulavik et al., 1995) and is inactivated by binding to salicylate, menadione, plumbagin, and 2,4-dinitrophenol (Alekshun & Levy, 1999). MarA is known to directly regulate about forty genes, mostly as an activator (Martin & Rosner, 2002). It mediates multidrug resistance in E. coli by up-regulating expression of the AcrAB-TolC multidrug efflux pump (Li & Nikaido, 2004) and of MicF (Cohen et al., 1988), a small inhibitory RNA that down-regulates translation of the outer membrane porin OmpF. The genes of the MarA regulon are involved in a wide spectrum of activities including oxidative stress, resistances to antibiotics, solvents, bile salts, and to other hazardous substances, as well as functions in energy metabolism and nutrient transport (Alekshun & Levy, 1997).
Although the marRAB operon is conserved in seven genera of enteric bacteria (Escherichia, Shigella, Klebsiella, Enterobacter, Salmonella, Cronobacter, and Citrobacter), the role of the small marB gene has been unclear. Nichols et al. (2011) found that deletion of marB in E. coli increases the amount of marA transcript and increases the resistance to a number of drugs and stresses in a manner correlating with the effects of a deletion of marR. Lee & Mitchell (2012) reported that a marB deletion increases expression of inaA, known to be directly upregulated by the MarA protein. To better understand the role of the marB gene, we investigated whether the gene product MarB acts as a protein or an sRNA, how MarB decreases the level of marA transcript, and in which compartment of the cell MarB is located.
Materials and Methods
Strains and plasmids
The bacterial strains and plasmids used in this study are described in Table 1. The ΔmarB deletion parental strain LV22 was obtained from strain JW1525 by removal of the kanamycin resistance gene, using plasmid pCP20 as previously described (Datsenko & Wanner, 2000). The complementation plasmid pMB102-marB was constructed by replacing marA of pMB102 (between HindIII/BamHI sites) with marB with its native Shine-Dalgarno motif. We used primers MarB-pMB102-F (5′-TTACCCAAGCTTAACAGCTAGTTGAAAACGTGAC-3′ with HindIII restriction site underlined) and MarB-pMB102-R (5′-AATCGCGGATCCGATGTCGGGGCCAGAACA-3′ with BamHI restriction site underlined) to create the 303 bp marB amplicon used in this construction. The DNA sequence of all constructs was confirmed (Tufts University Core Facility).
Table 1.
Strains and plasmids used in this study
| Strain/Plasmid | Genotype or relevant characteristic | Reference/source |
|---|---|---|
| E. coli K-12 strains | ||
| BW25113 | Wild type; F− λ− Δ(araD-araB)567 ΔlacZ4787(::rrnB-3) rph-1 Δ(rhaD-rhaB)568 hsdR514 | CGSC (Keio) (Baba et al., 2006) |
| JW1525 | BW25113 ΔmarB::Kan | CGSC (Keio) (Baba et al., 2006) |
| LV22 | BW25113 ΔmarB | This study |
| plasmids | ||
| pJPBH (control) | ori colE1 lacI, Ampr | (Barbosa & Levy, 2002) |
| pMB102 | ori colE1 lacI lacZp::marA; Ampr | (Pomposiello et al., 2001) |
| pMB102-marB | ori colE1 lacI lacZp::marB; Ampr | This study |
| pMB102-marB-42-STOP | ori colE1 lacI lacZp::marB-42-stop codon; Ampr | This study |
| pMB102-marB-23-STOP | ori colE1 lacI lacZp::marB-23-stop codon; Ampr | This study |
| pCH58 | ori pMB1, phoA fusion vector, Tetr | (Hoffman & Wright, 1985) |
| pCH58-marB | ori pMB1, marB-phoA fusion, Tetr | This study |
| pCH58-marB-SS | ori pMB1, marB signal sequence-phoA fusion, Tetr | This study |
| pCH58-SS | ori pMB1, phoA signal sequence-phoA fusion, Tetr | This study |
| pCP20 | oriR101, plasmid for excision of kan and cat markers by FLP-mediated site-specific recombination; Ampr Chlr | (Datsenko & Wanner, 2000) |
Gene expression and transcript half-life
Reverse transcription followed by real-time quantitative PCR (RT-qPCR) was used to determine the expression levels of marA in the wild type (BW25113), ΔmarB::Kan (JW1525), and ΔmarB (LV22) strains, and of marR in the wild type and ΔmarB::Kan strains. gapA, which encodes glyceraldehyde-3-phosphate dehydrogenase, was used as an endogenous reference gene according to the method of Viveiros et al. (2007). The specific primers for RT-qPCR of marA were: marA–F: CATAGCATTTTGGACTGGAT and marA–R: TACTTTCCTTCAGCTTTTGC (Viveiros et al., 2007), yielding a 187 bp fragment from the middle of marA. The primers for gapA and marR are described elsewhere (Viveros et al., 2007). These primers were also employed to generate the PCR DNA products used to perform qPCR standard plots for each gene.
Cultures of the different strains were inoculated from −80°C stocks into LB broth, grown at 37°C overnight, diluted 1:1000 into fresh LB, and grown for 2 to 3 h to an optical density at 600 nm (OD600) of about 0.3 (Beckman DU530 spectrophotometer). Then, total RNA in the cultures was stabilized using RNAprotect bacterial reagent (Qiagen), bacteria were digested with lysozyme-proteinase K, and total RNA was isolated using an RNeasy minikit (Qiagen) and On-Column DNase I digestion (Qiagen) according to the manufacturer’s specifications. RNAs were then treated with RQ1 RNase-free DNase (Promega) to remove any remaining DNA, repurified with the RNeasy minikit, and RNA purity and concentration were determined using a NanoDrop ND-1000 spectrophotometer. Reverse transcription was performed using the SuperScript III first-strand synthesis system (Invitrogen) with the specific primers aforementioned and 200 ng of RNA (“RT-plus” reactions). Reactions without reverse transcriptase (“RT-minus” reactions) were used as controls to confirm the lack of contaminating DNA in the RNA samples. The cDNAs obtained from the RT-plus and RT-minus reactions were quantified after 45 cycles using a Mx3000P detection system (Stratagene) with 25 μl reaction mixtures of primers (300 nM each), cDNA (4 μl of a 1:10 dilution of the RT-plus or RT-minus reaction mixture), and 2X QuantiTect SYBR Green qPCR master mix from Qiagen (12.5 μl). Absolute transcript numbers were calculated using standard plots obtained by qPCR of serial dilutions of gene-specific, gel-purified PCR products of known concentrations.
For the marA mRNA half-life experiments in the parental strain and ΔmarB::Kan mutant, overnight cultures of each strain were diluted and grown as before. RNAs were then extracted before (0 min) and 0.5, 1, 2, 3, 4, 6, and 8 min after addition of rifampin (500 μg ml−1 final concentration) to stop RNA synthesis. The number of marA transcripts was determined by RT-qPCR in three independent experiments for each strain as described above and the values were averaged. The slope (−k) of a linear regression fit of the curve for a plot of ln[average marA] vs time in rifampin was determined using Microsoft Excel software and the half-life (= ln2/k) was calculated.
Stop codons in MarB
Two different stop codons were placed singly in marB by a single base pair change at codons 23 and 42 of the 72-codon MarB open reading frame in the pMB102-marB complementation plasmid. To make these mutations we used the megaprimer method (Colosimo et al., 1999) with pMB102-marB as template; a first round of PCR was done to create the first stop codon, named 42-STOP, with an external forward primer MarB-pMB102-F (mentioned above) and a mutated reverse primer MarB-mut42-R (5′-CAAACGGTGGGTGTTCCTACGATGGGGGAACAACC-3′ with the mutation A), generating a 194 bp mutant megaprimer. In the case of the second stop codon, named 23-STOP, we used the same MarB-pMB102-F forward primer mentioned above and a different mutated reverse primer MarB-mut23-R (5′-CAACTGGCTGCGTGGTTTATTCCGCAACGCCCTGCGC-3′ with the mutation A), generating a 137 bp mutant megaprimer. In each case, a second PCR was carried out using each mutant megaprimer plus forward (MarB-pMB102-F) and reverse (MarB-pMB102-R) external primers, both described above. The 303 bp marB amplicon thus generated was cloned between the HindIII and BamHI sites of pMB102. The mutations were verified by sequencing and the mutated plasmids were introduced into wild type BW25113 and its ΔmarB::kan derivative by electroporation.
Fusion of marB to the gene for alkaline phosphatase (phoA)
PhoA localizes to the periplasm because of its native amino-terminal signal sequence. The following three regions were cloned at the single PstI site of plasmid pCH58 (Hoffman & Wright, 1985; Hoffman, 1986) upstream of phoA lacking its native signal sequence: 1) the entire marB gene; 2) the isolated putative signal sequence of marB; and 3) the known native signal sequence of phoA (as a control). In pCH58, upstream of the PstI site is the bla (β-lactamase) promoter followed by the first 4 codons of Bla (insufficient for localization), while downstream of the PstI site is a modified phoA gene missing both its entire signal sequence and the following 13 codons. The phoA sequences of pCH58 had been derived from a PstI-BamHI fragment from pCH48 (Hoffman, 1986), while the bla/Bla sequences came from the PstI-BamHI small fragment of pKT218 (Talmadge et al., 1981).
Cloning details are as follows. In each primer, the PstI site is underlined. The entire marB gene lacking its stop codon was cloned between (and in frame with) Bla and PhoA using a wild type chromosomal template amplified by forward primer marB-PstI-F (5′-AAAGCTGCAGCTATGAAACCACTTTCATCCGCAA-3′) and reverse primer marB-PstI-R-new (5′-AAAGCTGCAGACATAGCGTGTTGATTATAATAG-3′). The resulting plasmid was named pCH58-marB. The marB signal sequence alone was inserted using the marB-PstI-F primer described above and reverse primer marB-SS-PstI-R (5′-AAAGCTGCAGCTTCCGCAACGCCCTGCGCGGAA-3′), creating plasmid pCH58-marB-SS. In a similar fashion, the native phoA signal sequence was inserted using the following primer pair: PhoA-PstI-F (5′-AAAGCTGCAGCTGTGAAACAAAGCACTATTGC-3′) and PhoA-PstI-R-new (5′-AAAGCTGCAGCCCGGTTTTCCAGAACAGG-3′), creating plasmid pCH58-SS, which was used a positive control. Transformants were selected on LB plates supplemented with 20 μg tetracycline ml−1. Plasmids containing the sequences cloned in the forward orientation were found/verified using DNA sequencing.
Alkaline phosphatase activity was measured on cells grown in LB broth by the production of p-nitrophenol from p-nitrophenyl phosphate as described by Manoil et al. (2000). Units of alkaline phosphatase activity were defined as 1000 X [change in A420 per OD600 Unit per minute]. The number of OD600 Units was found by multiplying the OD600 by the volume of culture assayed (ml).
Statistical analysis
At least three determinations were made for each experiment; we report the mean and the standard error of the mean (SEM). The statistical significance of differences between two means was determined by unpaired Student’s t test (two independent samples with equal variance, with two-tailed distribution), using Microsoft Excel software.
Results and Discussion
MarB reduces the expression of marA and marR
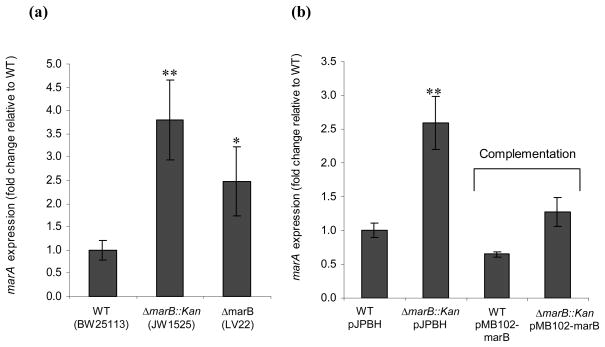
A recent work reported that deletion of the marB gene of E. coli enhances transcription of marA (Nichols et al., 2011). To confirm this unexpected finding, we studied the expression level of marA by RT-qPCR in wild type, ΔmarB::Kan, and ΔmarB strains (BW25113, JW1525, and LV22 respectively). We found that the ΔmarB::Kan strain had 3.8 fold more marA transcript than did the wild type strain; while the ΔmarB strain had 2.5 fold more (Fig. 1a); the difference between ΔmarB::Kan and ΔmarB was statistically insignificant (P = 0.32). These results confirmed that a marB deletion increased the level of marA.
Fig. 1.
marA gene expression (a) in wild type and ΔmarB mutant strains and (b) when complemented by wild type marB. Shown are relative amounts of marA gene transcript in wild type (WT) and ΔmarB::kan mutant hosts carrying no plasmid (a) or carrying either complementing plasmid pMB102-marB or control vector pJPBH (b). Fold 1 means no change in marA expression in a mutant compared to the level for the parental strain. The results in each chart are normalized to the first sample and are presented as the average +/− the standard error of the mean (Fig. 1a, n=5; Fig. 1b, n=3). Statistically significant differences for a mutant compared to the level for the parental strain are shown as * (P < 0.05) or ** (P <0.01).
Complementation of the ΔmarB::Kan mutation with marB on a plasmid (pMB102-marB) reduced transcription of marA to wild type levels (Fig. 1b), showing that the effect of ΔmarB::Kan was due to the loss of marB and not to the insertion of Kan per se. In the wild type strain, the number of marR transcripts per ng total RNA (5790+/−566) was very similar to that of marA (5541+/−1152), consistent with cotranscription of both genes in the marRAB operon regulated by the known mar promoter. The number of marR transcripts was enhanced 2.2-fold by the ΔmarB::Kan mutation (up to 12700 +/− 1520). The difference between the marR result and the 3.8-fold increase seen for the marA transcript was of borderline statistical significance (P = 0.056), suggesting that both marR and marA were affected similarly by ΔmarB::Kan.
ΔmarB::Kan did not increase the stability of the marA transcript
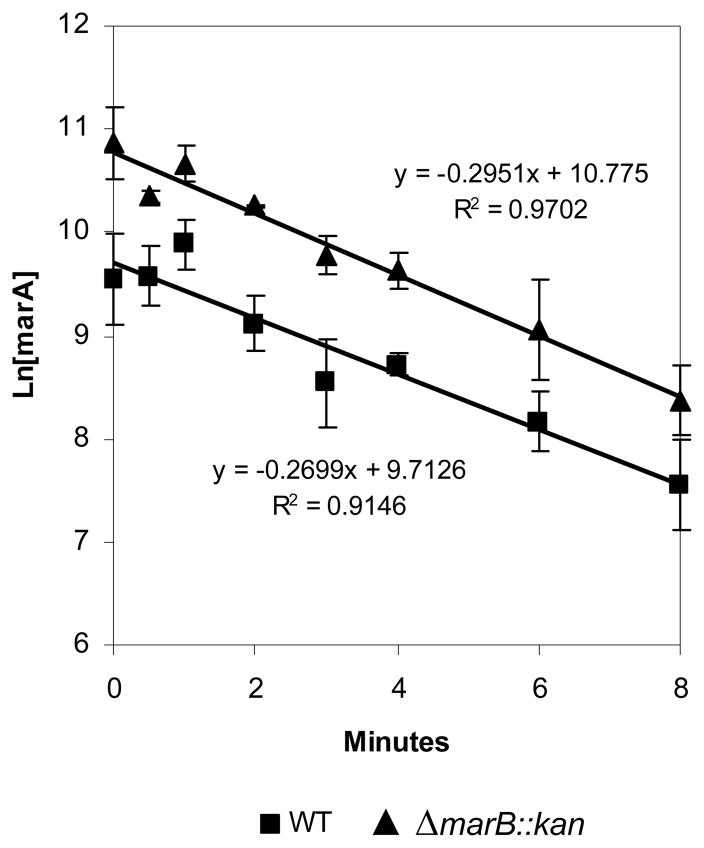
There are two ways in which ΔmarB::Kan might enhance the amount of marA and marR transcripts: 1) increase transcript stability and 2) increase the rate of transcription. We compared the stabilities of the marA mRNA transcripts of the parental strain and the ΔmarB::kan mutant. We found no differences between their half-lives (2.6 min vs 2.3 min; Fig. 2). Therefore, ΔmarB::Kan presumably caused an increase in the rate of marA transcription, probably from the marRAB promoter.
Fig. 2.
Deletion of marB had no effect on stability of marA mRNA. The half-life of marA mRNA was compared in wild type (WT) and ΔmarB::kan strains using rifampin as detailed in Materials and Methods. Error bars show SEM. No difference was found: the marA half-life was 2.6 min for the wild type strain and 2.3 min for its ΔmarB::kan derivative. The correlation coefficient (R2) describes how well the data fit the linear equations shown and can range from 0 to 1, the latter being a perfect fit. A correlation coefficient greater than 0.8 is generally described as strong. The absolute numbers of marA transcripts per ng of total RNA before addition of rifampin (time zero) were 17000 for the wild type strain BW25113 (■), and 64500 for the ΔmarB::kan mutant (▲).
MarB is a protein
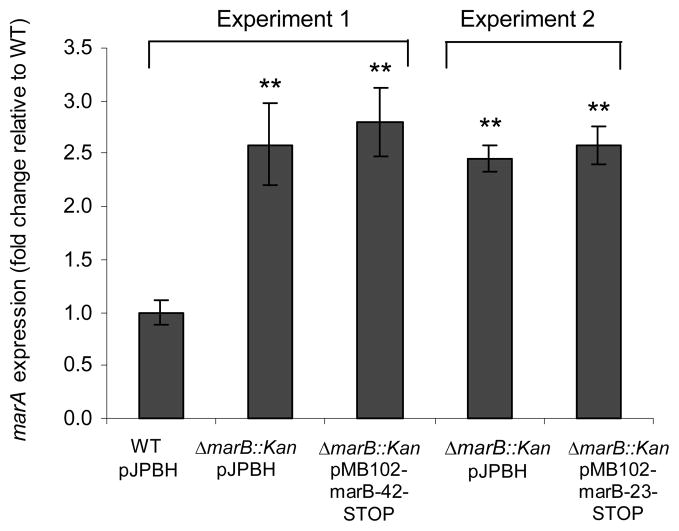
Since the marB gene has a consensus ribosomal binding site (Cohen et al., 1993) and the open reading frame has a clear periplasmic signal sequence, it has been assumed that the small marB gene (216 bp) encodes a protein rather than a small RNA (Misra et al., 2005). We investigated whether this assumption was valid. If MarB acted as a protein, it would be inactivated by a single bp change that created a stop codon within the open reading frame. We therefore converted the CAG codon for residue Glu42 of MarB in pMB102-marB to TAG (42-STOP). A second stop codon was also made in a second marB plasmid: CAA for residue Glu23 was converted to TAA (23-STOP). Neither of these minute changes was predicted by MFOLD (MFOLD, 2003) to influence the folding of marB mRNA. As before, we measured the level of marA transcript in the wild type harboring vector pJPBH and in the ΔmarB::Kan strain harboring pMB102-marB-42-STOP, pMB102-marB-23-STOP, or the vector control pJPBH. We found that each stop codon prevented complementation by marB: the number of marA transcripts did not decrease (Fig. 3), though they had decreased with wild type marB (Fig. 1b). These results proved that MarB indeed acts as a protein.
Fig. 3.
Lack of complementation by MarB mutants containing an internal stop codon. Shown are relative amounts of marA gene transcript in the WT strain bearing control vector pJPBH (set to 1.0) and in the ΔmarB::kan strain carrying the control vector or carrying pMB102-marB-(STOP) with stop codons at residue 42 or residue 23 within MarB. Two separate experiments are shown. Complementation by the wild type marB can be seen in Fig. 1b, where the same WT/pJPBH control was used. Results (n=3 for each condition) are presented as in Fig. 1. The absolute number of marA transcripts per ng of total RNA for WT/pJPBH was 12600.
Localization of MarB to the periplasm
If MarB protein was in the periplasm, as predicted (Misra et al., 2005), this would put constraints on how it might function. For example, a previous suggestion that MarB might interact with and inhibit the cytoplasmic MarA/RNA polymerase complex (Nichols et al., 2011) would not be possible. It was important therefore to test for the location of MarB. Efforts to localize MarB using a MarB-6H fusion protein constructed via the expression vector pET21b (Novagen) failed: no polyhistidine fusion protein was detectable at any IPTG level in any cellular fraction by Western blotting using anti-6H.
Therefore, we took an approach involving phoA, the gene for the periplasmic enzyme alkaline phosphatase (PhoA). PhoA is active only when it has been brought to the periplasm by an amino terminal signal sequence, and this fact has been used to identify signal sequences (Hoffman & Wright, 1985; Varga & Kaplan 1989, Okamoto et al., 1991). We fused the entire marB gene, including its predicted signal sequence, to a downstream modified phoA gene missing its own native signal sequence, to find whether PhoA activity would result in cells. The vector was plasmid pCH58, in which transcription occurs from the bla (β-lactamase) promoter; the 4 amino-terminal codons of the Bla signal sequence were fused in frame to modified PhoA (see Materials and Methods). The marB gene, cloned at the PstI site, was in frame both with the 4 Bla codons (upstream) and the modified PhoA (downstream), generating a Bla-MarB-PhoA fusion protein in pCH58-marB. We also cloned just the putative 21-codon MarB signal sequence itself, without the rest of MarB, creating pCH58-marB-SS. Finally, as a control, the native 21-codon signal sequence of PhoA was similarly placed, creating pCH58-SS.
The results in Table 2 show that wild type strain BW25113 carrying the plasmid with the entire marB sequence produced a much higher PhoA activity (390 units, pCH58-marB) than did the strain itself (4.3 units) or the strain with vector pCH58 (22 units). As anticipated, the 4 residues of the signal of periplasmic Bla protein in vector pCH58 were not enough to locate PhoA to the periplasm. The construct with the MarB signal sequence alone produced the same level (410 units, pCH58-marB-SS) as did the construct with the entire marB sequence, showing that the rest of the MarB sequence had no effect. Therefore the MarB signal sequence alone was sufficient to export PhoA to the periplasm, showing that MarB is indeed a periplasmic protein.
Table 2.
PhoA enzymatic activities in wild type strain BW25113 carrying plasmids containing the indicated phoA gene fusions
| Plasmid | Promoter | Signal sequence | PhoA activity units‡ (SEM) |
|---|---|---|---|
| — | — | — | 4.3 (1.3) |
| pCH58 | bla | — | 22 (6.7) |
| pCH58-marB* | bla | marB | 390 (40) |
| pCH58-marB-SS# | bla | marB | 410 (19) |
| pCH58-SS | bla | phoA | 1600 (250) |
pCH58-marB contains full marB (except stop codon) cloned upstream of phoA.
pCH58-marB-SS contains only marB signal sequence cloned upstream of phoA.
These values are the averages of at least 6 independent experiments, with SEM in parentheses.
The 4-fold higher activity seen for the control plasmid containing the native PhoA signal sequence (1600 units, pCH58-SS) when compared to the MarB signal could reflect a more efficient export of PhoA by the native PhoA signal sequence or a greater number of native PhoA molecules in the cell.
Possible mode of action of MarB
We found that negative regulation of marA expression by MarB appears to be at the level of transcript formation rather than transcript stability. Nichols et al. (2011) reported, although without presentation of data, that MarB does not act via MarR and does not affect the degradation of MarA via Lon protease. Taken together these results suggest that MarB may decrease the function of the canonical marRAB promoter by altering the activity of one of its non-MarR direct regulators (MarA, SoxS, Rob, Fis, Crp, or FruR (Martin & Rosner, 1997; Martin et al., 1999; Shimada et al., 2011; Zheng et al., 2004) or of an indirect regulator such as PAP I (Ruiz & Levy, 2010). This possibility awaits further study.
As a periplasmic protein, MarB obviously cannot directly contact any of these cytoplasmic proteins that regulate the known promoter of marRAB. Instead, MarB presumably exerts its control indirectly, perhaps via a chemical signal which alters (either directly or indirectly) the activity of one of the cytoplasmic regulators of the operon. The small MarB protein (predicted to contain 51 amino acids once processed) may function as a homomultimer or by interacting with another protein or component of the periplasm or the membrane (see Hobbs et al., 2011) to respond to a stress and/or to create a signal.
MarB has no homology with any protein of known function. Among the seven bacterial genera having a marRAB operon, the open reading frame for MarB is more poorly conserved (41–55% identity) than are those of MarR (82–98% identity) and MarA (90–98% identity). Nevertheless, a periplasmic signal is predicted for all of the diverse MarBs using the SignalP 4.1 server (Petersen et al., 2011), and a conserved multi-charged sequence motif (G)SDKSD (starting about 16 residues before the end of the protein) would be present in all of the mature MarBs (Punta et al., 2012). A β strand of 5–6 residues starting at the 14th residue is predicted for mature MarB proteins of all genera using Jpred 3 (Cole et al., 2008). The predicted isoelectric points of the mature proteins range from 4.2 to 5.4, indicating that all MarBs are acidic. These observations suggest that MarB, like the other two more highly conserved proteins in the operon, likely has a similar function in all seven genera.
Acknowledgments
We thank Andrew Wright for kindly providing plasmid pCH58 and giving advice. Laura Vinué was supported by a postdoctoral fellowship (PDOC-2010) from the Gobierno de La Rioja of Spain. This work was supported by United States Public Health Service grant AI56012 from the National Institutes of Health.
References
- Alekshun MN, Levy SB. Regulation of chromosomally mediated multiple antibiotic resistance: the mar regulon. Antimicrob Agents Chemother. 1997;41:2067–2075. doi: 10.1128/aac.41.10.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alekshun MN, Levy SB. Alteration of the repressor activity of MarR, the negative regulator of the Escherichia coli marRAB locus, by multiple chemicals in vitro. J Bacteriol. 1999;181:4669–4672. doi: 10.1128/jb.181.15.4669-4672.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba T, Ara T, Hasegawa M, et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2:2006.0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa TM, Levy SB. Activation of the Escherichia coli nfnB gene by MarA through a highly divergent marbox in a class II promoter. Mol Microbiol. 2002;45:191–202. doi: 10.1046/j.1365-2958.2002.03006.x. [DOI] [PubMed] [Google Scholar]
- Cohen SP, McMurry LM, Levy SB. marA locus causes decreased expression of OmpF porin in multiple-antibiotic-resistant (Mar) mutants of Escherichia coli. J Bacteriol. 1988;170:5416–5422. doi: 10.1128/jb.170.12.5416-5422.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen SP, Hächler H, Levy SB. Genetic and functional analysis of the multiple antibiotic resistance (mar) locus in Escherichia coli. J Bacteriol. 1993;175:1484–1492. doi: 10.1128/jb.175.5.1484-1492.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole C, Barber JD, Barton GJ. The Jpred 3 secondary structure prediction server. Nucl Acids Res. 2008;36(Web Server issue):W197–W201. doi: 10.1093/nar/gkn238. http://www.compbio.dundee.ac.uk/www-jpred/index.html. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colosimo A, Xu Z, Novelli G, Dallapiccola B, Gruenert DC. Simple version of “megaprimer” PCR for site-directed mutagenesis. Biotechniques. 1999;26:870–873. doi: 10.2144/99265bm15. [DOI] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbs EC, Fontaine F, Yin X, Storz G. An expanding universe of small proteins. Curr Opin Microbiol. 2011;14:167–173. doi: 10.1016/j.mib.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman CS, Wright A. Fusions of secreted proteins to alkaline phosphatase: an approach for studying protein secretion. Proc Natl Acad Sci USA. 1985;82:5107–5111. doi: 10.1073/pnas.82.15.5107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman CS. PhD thesis. Tufts University; Boston: 1986. Studies on protein secretion to the Escherichia coli K-12 outer membrane using phoA gene fusions. [Google Scholar]
- Lee S, Mitchell RJ. Detection of toxic lignin hydrolysate-related compounds using an inaA::luxCDABE fusion strain. J Biotechnol. 2012;157:598–604. doi: 10.1016/j.jbiotec.2011.06.018. [DOI] [PubMed] [Google Scholar]
- Li XZ, Nikaido H. Efflux-mediated drug resistance in bacteria: an update. Drugs. 2004;69:1555–1623. doi: 10.2165/11317030-000000000-00000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manoil C. Tagging exported proteins using Escherichia coli alkaline phosphatase gene fusions. Methods in Enzymology. 2000;326:35–47. doi: 10.1016/s0076-6879(00)26045-x. [DOI] [PubMed] [Google Scholar]
- Martin RG, Nyantakyi PS, Rosner JL. Regulation of the multiple antibiotic resistance (mar) regulon by marORA sequences in Escherichia coli. J Bacteriol. 1995;177:4176–4178. doi: 10.1128/jb.177.14.4176-4178.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin RG, Rosner JL. Fis, an accessorial factor for transcriptional activation of the mar (multiple antibiotic resistance) promoter of Escherichia coli in the presence of the activator MarA, SoxS, or Rob. J Bacteriol. 1997;179:7410–7419. doi: 10.1128/jb.179.23.7410-7419.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin RG, Gillette WK, Rhee S, Rosner JL. Structural requirements for marbox function in transcriptional activation of mar/sox/rob regulon promoters in Escherichia coli: sequence, orientation and spatial relationship to the core promoter. Mol Microbiol. 1999;34:431–441. doi: 10.1046/j.1365-2958.1999.01599.x. [DOI] [PubMed] [Google Scholar]
- Martin RG, Rosner JL. Genomics of the marA/soxS/rob regulon of Escherichia coli: identification of directly activated promoters by application of molecular genetics and informatics to microarray data. Mol Microbiol. 2002;44:1611–1624. doi: 10.1046/j.1365-2958.2002.02985.x. [DOI] [PubMed] [Google Scholar]
- MFOLD. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. http://mfold.rna.albany.edu/?q=mfold/RNA-Folding-Form. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra RV, Horler RS, Reindl W, Goryanin II, Thomas GH. EchoBASE: an integrated post-genomic database for Escherichia coli. Nucleic Acids Res. 2005;33(Database issue):D329–333. doi: 10.1093/nar/gki028. http://www.york.ac.uk/res/thomas/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols RJ, Sen S, Choo YJ, et al. Phenotypic landscape of a bacterial cell. Cell. 2011;144:143–156. doi: 10.1016/j.cell.2010.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto S, Toyoda-Yamamoto A, Ito K, Takebe I, Machida Y. Localization and orientation of the VirD4 protein of Agrobacterium tumefaciens in the cell membrane. Mol Gen Genet. 1991;228:24–32. doi: 10.1007/BF00282443. [DOI] [PubMed] [Google Scholar]
- Petersen TN, Brunak S, Von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8:785–786. doi: 10.1038/nmeth.1701. http://www.cbs.dtu.dk/services/SignalP. [DOI] [PubMed] [Google Scholar]
- Pomposiello PJ, Bennik MH, Demple B. Genome-wide transcriptional profiling of the Escherichia coli responses to superoxide stress and sodium salicylate. J Bacteriol. 2001;183:3890–3902. doi: 10.1128/JB.183.13.3890-3902.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punta M, Coggill PC, Eberhardt RY, et al. The Pfam protein families database. Nucleic Acids Research. 2012;(Database Issue 40):D290–D301. doi: 10.1093/nar/gkr1065. http://pfam.sanger.ac.uk/family/PF13999. [DOI] [PMC free article] [PubMed]
- Ruiz C, Levy SB. Many chromosomal genes modulate MarA-mediated multidrug resistance in Escherichia coli. Antimicrob Agents Chemother. 2010;54:2125–2134. doi: 10.1128/AAC.01420-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seoane AS, Levy SB. Characterization of MarR, the repressor of the multiple antibiotic resistane (mar) operon in Escherichia coli. J Bacteriol. 1995;177:3414–3419. doi: 10.1128/jb.177.12.3414-3419.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada T, Yamamoto K, Ishihama A. Novel members of the Cra regulon involved in carbon metabolism in Escherichia coli. J Bacteriol. 2011;193:649–659. doi: 10.1128/JB.01214-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulavik MC, Gambino LF, Miller PF. The MarR repressor of the multiple antibiotic resistance (mar) operon in Escherichia coli: prototypic member of a family of bacterial regulatory proteins involved in sensing phenolic compounds. Mol Med. 1995;1:436–446. [PMC free article] [PubMed] [Google Scholar]
- Talmadge K, Brosius J, Gilbert W. An ‘internal’ signal sequence directs secretion and processing or proinsulin in bacteria. Nature. 1981;294:176–178. doi: 10.1038/294176a0. [DOI] [PubMed] [Google Scholar]
- Varga AR, Kaplan S. Construction, expression, and localization of a CycA::PhoA fusion protein in Rhodobacter sphaeroides and Escherichia coli. J Bacteriol. 1989;171:5830–5839. doi: 10.1128/jb.171.11.5830-5839.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viveiros M, Dupont M, Rodrigues L, Couto I, Davin-Regli A, Martins M, Pagès JM, Amaral L. Antibiotic stress, genetic response and altered permeability of E. coli. PLoS One. 2007;2:e365. doi: 10.1371/journal.pone.0000365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng D, Constantinidou C, Hobman JL, Minchin SD. Identification of the CRP regulon using in vitro and in vivo transcriptional profiling. Nucleic Acids Res. 2004;32:5874–5893. doi: 10.1093/nar/gkh908. [DOI] [PMC free article] [PubMed] [Google Scholar]