Retroviral capsid proteins are conserved structurally but assemble into different morphologies1. The mature human immunodeficiency virus-1 (HIV-1) capsid is best described by a ‘fullerene cone’model2,3, in which hexamers of the capsid protein are linked to form a hexagonal surface lattice that is closed by incorporating 12 capsid-protein pentamers. HIV-1 capsid protein contains an amino-terminal domain (NTD) comprising seven α-helices and a β-hairpin4,5, a carboxy-terminal domain (CTD) comprising four α-helices6,7, and a flexible linker with a 310-helix connecting the two structural domains8. Structures of the capsid-protein assembly units have been determined by X-ray crystallography9,10; however, structural information regarding the assembled capsid and the contacts between the assembly units is incomplete. Here we report the cryo-electron microscopy structure of a tubular HIV-1 capsid-protein assembly at 8Å resolution and the three-dimensional structure of a native HIV-1 core by cryo-electron tomography. The structure of the tubular assembly shows, at the three-fold interface11, a three-helix bundle with critical hydrophobic interactions. Mutagenesis studies confirm that hydrophobic residues in the centre of the three-helix bundle are crucial for capsid assembly and stability, and for viral infectivity. The cryo-electron-microscopy structures enable modelling by large-scale molecular dynamics simulation, resulting in all-atom models for the hexamer-of-hexamer and pentamer-of-hexamer elements as well as for the entire capsid. Incorporation of pentamers results in closer trimer contacts and induces acute surface curvature. The complete atomic HIV-1 capsid model provides a platform for further studies of capsid function and for targeted pharmacological intervention.
HIV-1 capsid protein (CA) spontaneously assembles in vitro into helical tubes and cones that resemble authentic viral capsids3, allowing direct structural investigation of the CA assembly using cryo-electron microscopy (cryo-EM). Previous studies of helical tubes provided low-resolution views of the curved hexagonal lattice, which is stabilized by important inter-hexamer CTD–CTD contacts across local two- and three-fold axes3,11. However, accurate and reliable atomic modelling of the HIV-1 capsid assembly has not been possible, owing to limited resolution (up to 16Å). In fact, high-resolution structural determination of in vitro assembled samples is very challenging: HIV-1 capsid assembly in vitro is inefficient, requires high salt (1 M), and exhibits great variability in tube diameters and helical symmetries (Supplementary Fig. 1a)3,11. We devised a rapid back-dilution method to improve cryo-EM sample quality and recorded more than 1,500 cryo-EM images on film, many of which showed diffraction layer lines to 10Å resolution. Using 27 high-resolution tubes in a single helical symmetry (−12, 11), along with a real-space, single-particle, helical reconstruction method12,13, we determined the structure of the CA tubular assembly at 8.6Å resolution (Fig. 1a–c and Supplementary Fig. 1b–d).
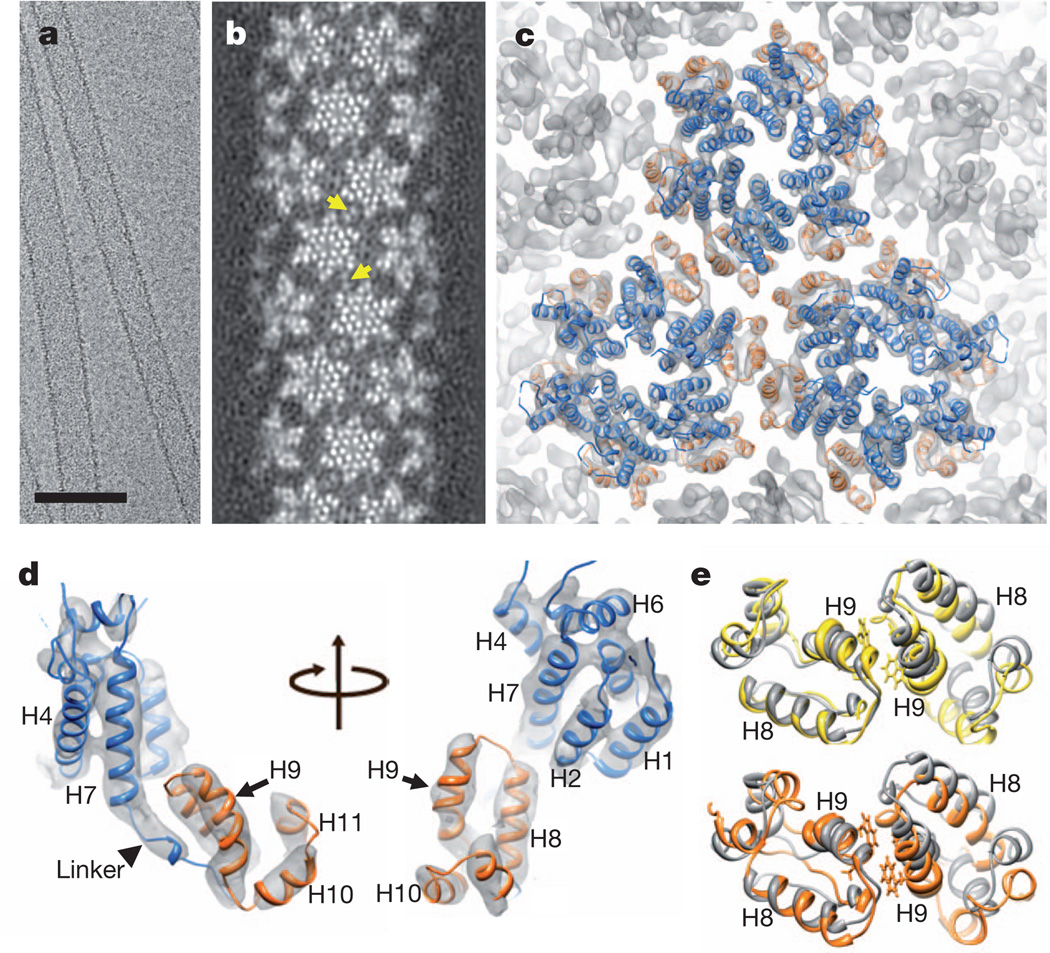
Figure 1. Cryo-EM reconstruction of HIV-1 CA tubular assembly at 8Å resolution andMDFF.
a, A cryo-EM image of recombinant A92E CA tubular assembly. Scale bar, 100 nm. b, Electron density map of the A92E CA tube with (−12, 11) helical symmetry. Yellow arrows indicate pairs of helix H9, located between adjacent hexamers. c, MDFF model of the HIV-1 capsid assembly, superimposed with the electron density map contoured at 4.0σ. Three CA hexamers, with NTDs (blue) and CTDs (orange), are shown. d,MDFF model of a CA monomer viewed from two angles. e, Two CTD dimer structures along -1 (orange) and 11 (yellow) helical directions, superimposed onto the NMR solution dimer structure (grey, 2KOD).
The density map clearly delineates all the α-helical motifs in the assembled structure (Fig. 1b–d), including the CTD dimerization helix, H9, with a distinct kink in the middle (Fig. 1d). The linker region (residues 146–150), followed by a 310-helix, shows strong electron density (Fig. 1d), suggesting that it adopts a relatively ordered and stable conformation. This linker region is a critical determinant for proper capsid assembly and stability8. The map also shows detailed intermolecular interactions at all four interfaces critical for assembly: the inter-hexamer CTD interfaces at the pseudo-two-fold and pseudo-three-fold axes, and the hexamer forming NTD–NTD and NTD–CTD interfaces (Fig. 1c and Supplementary Fig. 1c)9,11,14.
The 8Å resolution density map allowed reliable atomic modelling for the entire assembly, using molecular dynamics flexible fitting (MDFF)15. We docked atomic structures (3H47 for CA NTD9 and 2KOD for CA CTD11) into the electron density, built the linker and missing loops using homology modelling, and then applied MDFF (Supplementary Video 1). The resulting atomic model for the tubular assembly comprises 71 CA hexamers and 13 million atoms, including water molecules and ions. The final model fits the experimental map remarkably well, as indicated by the excellent overlap between α-helices and their corresponding rod-like densities (Fig. 1c, d), with an overall cross-correlation function value of 0.96. All the densities are accounted for in the model, including those of the linker region (Fig. 1d) and β-hairpin region. An equivalent pseudo-atomic model was obtained independently, using a normal mode flexible-fitting method (Supplementary Fig. 2a).
The MDFF model of the full CA assembly contains an almost invariant hexameric NTD structure with an average root mean squared deviation of 1.5 ± 1.0Å for backbone atoms between all 71 hexamers. The hexameric arrangement of the NTD is very similar to that observed in the crystal structure of cross-linked hexameric CA protein9 (Supplementary Fig. 2b). Further, the intermolecular interactions in a single hexameric unit are the same as those observed in the crystal structure (Supplementary Fig. 2c, d). Considering that retroviral CA proteins share a common NTD fold and all form hexamers, hexameric CA NTD appears to constitute a building block in the final assembly.
In contrast, the CTD dimers, which connect adjacent assembly units on the inner, more curved surface, are more variable than the NTD interfaces, with a root mean squared deviation of 3.0Å ± 0.9Å for all 71 dimers. Three distinct CTD dimer conformations are seen in the assembled helical tube, with dimerization helix H9 crossing angles of 36°, 44° or 54°. Several conformers for the CTD dimer and full-length dimeric CA were previously observed in solution16, demonstrating the plastic nature of the dimer interface. Nonetheless, key contacts are formed by the hydrophobic residues, W184 and M185, albeit with small variations in their local environment (Fig. 1e). The CTD girdle of the CA hexamer, helices H9 and H10 in particular, seems more variable, compared with the CTD seen in the crystal structure (Supplementary Fig. 2b, e)9. Thus, the curvature needed for the helical tube assembly is accommodated by variable orientations in the CTD, mediated by the intrinsic flexibility of helix H9 in the monomer.
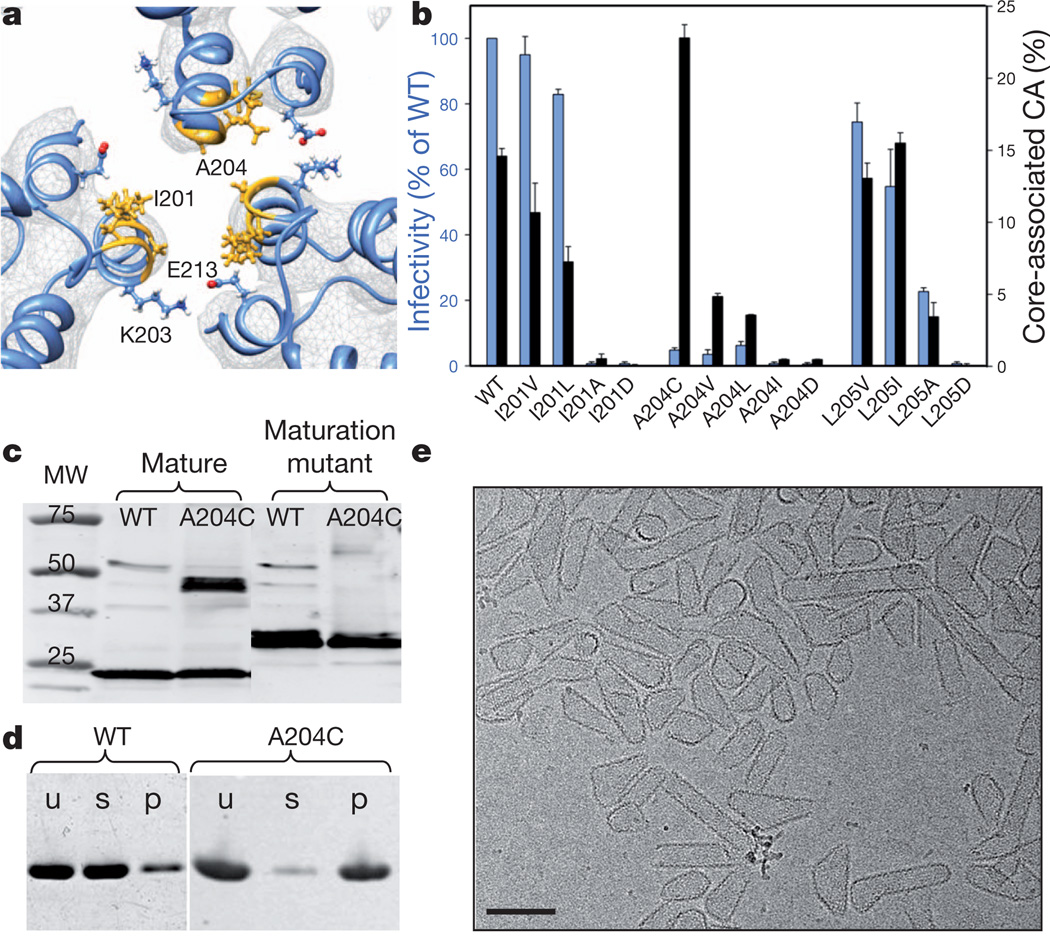
Important new structural detail is seen at the CTD trimer interface. The MDFF model of the tubular assembly shows that this interface involves a patch of hydrophobic residues, specifically I201, L202, A204 and L205, all situated on one face of the amphipathic CTD helix H10 (Supplementary Fig. 3a, b). At the centre of the hydrophobic core reside residues I201 and A204 (Fig. 2a and Supplementary Fig. 3b). These residues contribute a large portion of the total buried surface area (8,758Å2) at the pseudo-three-fold axis. The central hydrophobic core is surrounded by polar residues of opposite charges, for example K203 and E213 (Figs 2a and 3c), further stabilizing the interface. Residues K203 and E213 are highly conserved and were previously shown to be critical for optimal HIV-1 capsid stability and for viral infectivity11,17. To test further the involvement of the hydrophobic interactions in HIV-1 capsid function, we performed mutagenesis and chemical crosslinking studies. Remarkably, hydrophobic replacements (I201V/L, A204V/L and L205V/I) largely retained the infectivity and capsid stability (Fig. 2b)18, but mutant virions with polar residue substitutions (I201D, A204D and L205D) were non-infectious (Fig. 2b) and contained unstable or abnormal cores (Supplementary Fig. 3c–g). Interestingly, these mutations had no apparent effect on the morphology of immature virions (Supplementary Fig. 3h–j). Purified CA proteins with the same polar substitutions failed to assemble in vitro (Supplementary Fig. 3m, n). The A204C CA mutant, which exhibits a Cys-Cys backbone distance of 5.3Å between adjacent molecules at the pseudo-three-fold axis, exhibited disulphide crosslinking of CA into dimers, in vitro and in mature virions (Fig. 2c). This was not the case for maturation-defective virions in which cleavage of nucleocapsid protein was prohibited (Fig. 2c), implying that the hydrophobic CTD trimer interface is specific to the mature capsid19. In addition, the A204C CA protein exhibited higher assembly efficiency (>90%) compared with wild-type CA (10–15%) (Fig. 2d) and assembled under physiological salt concentrations. More intriguingly, the A204C protein (and A204L) assembled into closed cones and short tubes that closely resembled authentic lentiviral cores (Fig. 2e and Supplementary Fig. 3l, o), rather than the long tubes typically formed by wild-type CA (Supplementary Fig. 3k). This suggests that CA pentamers are readily incorporated in A204C assemblies. Consistent with the in vitro results, virions carrying the A204C substitution showed enhanced capsid stability and reduced infectivity (Fig. 2b). These results indicate that hydrophobic interactions at the CTD trimer interface play a critical role in mature capsid assembly and stability. This role is further supported by semi-three-dimensional Monte Carlo simulations of 36 CA dimers, on the basis of the assembly phase diagram established previously (Supplementary Fig. 4)20.
Figure 2. Mutational analysis of the hydrophobic trimer interface.
a, Detailed viewof the trimer interface, with the structural model superimposed on the density map (grey mesh, contoured at 3.5σ). Selected residues are depicted in stick-and-ball representation. b, Virus infectivity (blue) and capsid stability (black), as percentage of total CA,± standard deviation (n=53), of wild-type (WT) and CA trimer interface mutants. c, Spontaneous disulphide crosslinking of A204C mature and maturation-defective virions analysed by immunoblotting for CA. d, In vitro assembly of recombinant wild-type and A204C CA proteins, assayed with high-speed sedimentation and polyacrylamide gel electrophoresis analysis. Letters u, s and p denote the unassembled reaction mixture (u) and the supernatant (s) and pellet (p) after assembly. e, Cryo-EM image of A204C in vitro assembly. Scale bar, 100 nm.
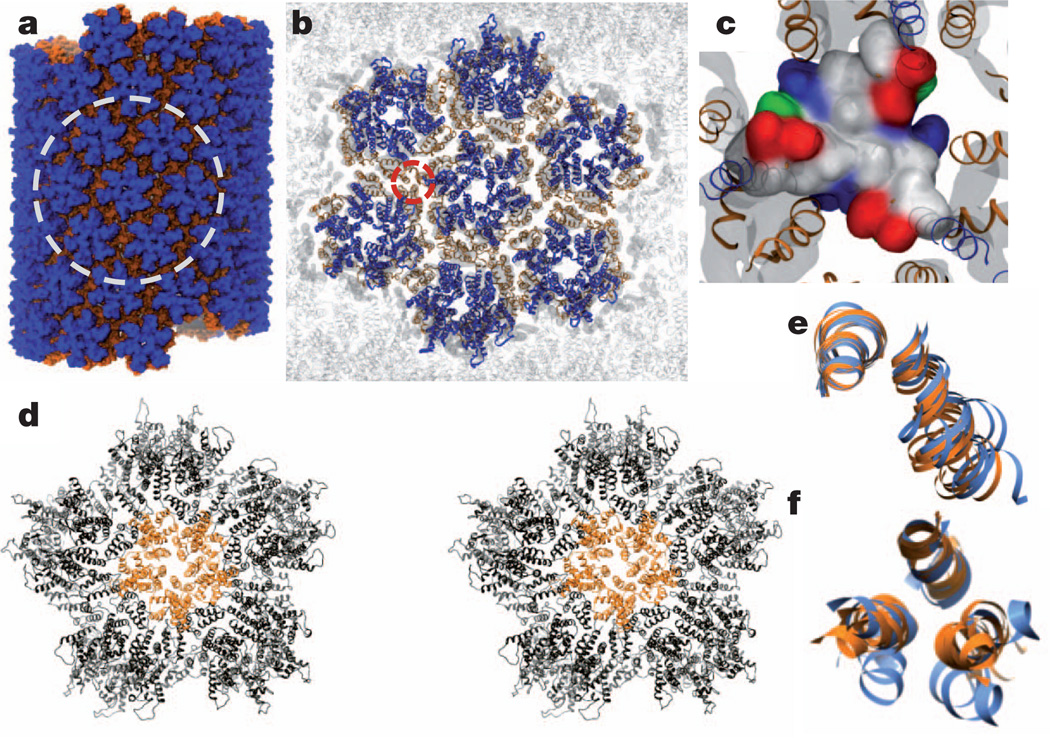
A ‘fullerene cone’ model of the HIV-1 capsid requires insertion of 12 CA pentamers into the hexagonal surface lattice to close the ovoid. Although the atomic structures of pentameric and hexameric HIV-1 CA mutants have been determined9,10, and their structural similarity suggests that they are quasi-equivalent10, detailed contacts between pentamers and hexamers for building a closed fullerene cone are not known. On the basis of the hexamer-of-hexamers (HOH) motif (six hexamers surrounding a central hexamer) extracted from the helical tube assembly (Fig. 3a, b), we generated a model for a pentamer of hexamers (POH; five hexamers encompassing a central pentamer), by replacing the central hexamer with a pentamer. The resulting model contains gaps between the surrounding hexamers because of the deletion of one hexamer, and poor dimer and trimer interactions (Supplementary Video 2). However, during a molecular dynamics simulation (Supplementary Table 1), these gaps closed quickly (Supplementary Video 2), converting the relatively flat starting model into a highly curved dome-like structure (Fig. 3d, Supplementary Fig. 5a, b and Supplementary Video 3). More specifically, sharp bite angles between adjacent subunits were formed, namely 137° and 147° for pentamer–hexamer and hexamer–hexamer edges, respectively, compared with 167° and 147° in the case of HOH(Supplementary Fig. 5b). The sharp bite angle closely matches the 135° value seen in the fullerene cone geometric model21. All pre-existing intra-hexamer (or pentamer) and CTD dimer intermolecular interfaces were preserved in the simulation, whereas new dimer and trimer interactions between hexamers and between pentamer and hexamers were formed in 300 ns (Fig. 3d and Supplementary Video 2), validating the previously suggested quasi-equivalence in the capsid lattice22,23. Although the CTD dimer interface remained slightly variable, as in the HOH model (Fig. 3e), the three-helix bundle at the CTD trimer interface was conserved but more closely packed in the POH(Fig. 3f and Supplementary Fig. 5c, orange) compared with the HOH (Fig. 3f, blue). This is intriguing and suggests that enhancing the interaction at the three-fold axis, for example by A204C crosslinking, facilitates incorporation of pentamers into the assembly (Fig. 2e). Electrostatic destabilization at the six-fold axis in CA NTD10,22, counterbalanced by the stabilizing hydrophobic interactions at the three-fold axis, probably controls pentamer incorporation. Given that the trimer contact is absent in the planar two-dimensional crystal of the CA assembly14, but clearly present in the helical tubular assembly, and even tighter in the highly curved POH model, we suggest that the three-fold interface is essential for the curved asymmetric assembly of the conical capsid.
Figure 3. All-atom molecular dynamics simulation of CA tubular assembly.
a, All-atom tubular assembly model comprising 71 CA hexamers (blue NTD and orange CTD) equilibrated for 125 ns. b, Ribbon representation of the tubular assembly, highlighting an HOH, circled area in a, superimposed on the density map (grey). c, Surface representation of the trimer interface (circled area in b) in a 425 ns equilibrated HOH model (hydrophobic, polar, negative and positive residues are grey, green, red and blue, respectively). d, Stereo view of a POH model after 1.5 µs equilibration. A pentamer (orange) is surrounded by five hexamers (black). e, f, Superposition of CTD H9 dimer (e) or H10 trimer (f) interfaces from HOH (blue) and POH (orange).
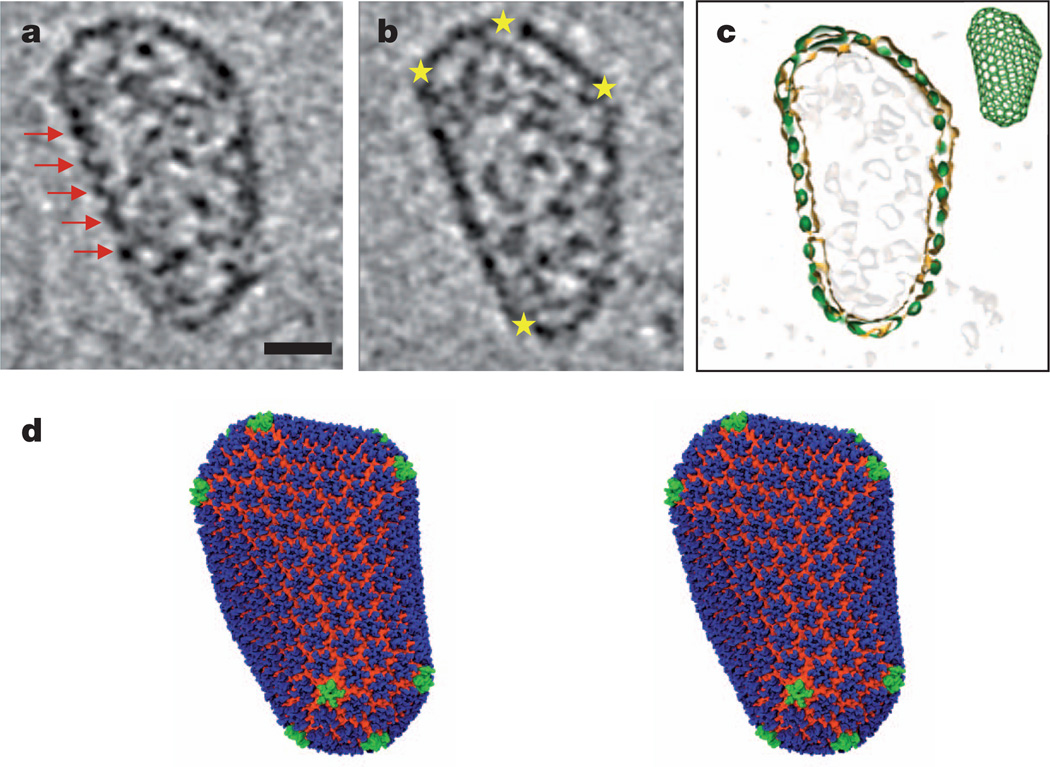
The HOH and POH assembly structures allow construction of an improved geometric HIV-1 capsid model10. Building a realistic atomic model of the HIV-1 capsid, however, requires an experimentally determined native HIV-1 core structure. Because of the non-uniform, asymmetric nature of the native cores, we performed structural analysis by cryo-electron tomography (cryo-ET) of HIV-1 cores isolated from virions (Supplementary Fig. 6 and Supplementary Video 4). The tomographic slices of a best-quality HIV-1 core, with a cone angle of 23° and representing the most common core24 (Supplementary Fig. 6b), clearly exhibit arrays of CA hexamers (Fig. 4a, red arrows) and declinations made by CA pentamers (Fig. 4b, yellow stars) at the outer capsid density layer. The native core also includes internal densities corresponding to the viral RNA genome and proteins enclosed within the capsid (Fig. 4a, b). At the current tomography resolution, individual hexamers or pentamers in the capsid cannot be discerned. Thus, guided by the shape, size and structural features of the capsid layer, extensive fullerene models, including classes containing 252, 216, 186 or 166 hexamers, were evaluated and top candidates from each class (119 of a total of 724) were tested by cross-correlation between the model and the capsid density (Supplementary Fig. 7). Two fullerene models, I and II, comprising 12 pentamers, as well as 216 and 186 hexamers, respectively, were selected to generate all-atom molecular dynamics models of the entire HIV-1 capsid, comprising 1,356 (model I, Fig. 4c, d) or 1,176 (model II, Supplementary Fig. 7b) CA subunits. These models were confirmed and analysed through a fully solvated, unconstrained 100 ns, 64 million atom, molecular dynamics simulation as described in Supplementary Material (Supplementary Table 1).
Figure 4. All-atom HIV-1 capsid model.
a, b, Cryo-ET analysis of an isolated, native HIV-1 core, shown as two representative slices through the three-dimensional volume. Red arrows indicate arrays of CA hexamers; yellow stars indicate locations of sharp curvature change. Scale bar, 20 nm. c, A fullerene cone model (216H+12P, green inset) matches the shape and size of the capsid, shown by the overlay of densities from the segmented capsid (orange contour) and the fullerene model (green). HIV-1 core internal densities are shown in light grey. d, Stereo view of the final molecular dynamics equilibrated all-atom capsid model (model I, see text) comprising 216 CA hexamers (blue, NTD; orange, CTD) and 12 CA pentamers (green).
The resulting capsid model I contains a variable CTD structure, with an average root mean squared deviation of 3.6 ± 0.5Å for backbone atoms between all 216 hexamers, compared with 2.3 ± 0.4Å for the NTD. Analysis of the bite angle between the neighbouring hexamers (or pentamers) shows a bimodal distribution (Supplementary Fig. 8a), with a minor population of sharp bite angles (mean = 139°) located around pentamers (Supplementary Fig. 8b), in addition to most continuously varying bite angles (144–180°). Compared with the recent cryo-EM structure of an immature intermediate of a different retrovirus25, our mature capsid structure shows a very different set of inter-subunit interactions, suggesting large conformational changes accompany retrovirus maturation. Our atomic models of the complete HIV-1 capsid further highlight the three-fold CA CTD trimer interface as an attractive therapeutic target and provide a platform for future studies of capsid function and for targeted pharmacological intervention.
METHODS SUMMARY
CA tubes were assembled at 2 mg ml−1 in a buffer containing 1 M NaCl and 50 mM Tris-HCl, pH 8.0. Cryo-EM micrographs were collected on Kodak SO-163 films, on an FEI Polara microscope operated at 200 kV, and digitized at a pixel size of 1.09 Å. Iterative Helical Real Space Reconstruction13 was used to estimate the helical parameters. The final refinement was performed using a previously established method12. Cryo-ET tilt series of native HIV-1 cores were collected on a Gatan 4k × 4k CCD (charge-coupled device) camera by tilting the specimen from −70° to 66°, with a total dose of approximately 120 e−/Å2 and a defocus value around 8 µm. Three-dimensional tomograms were reconstructed using IMOD and TOMO3D software26,27. MDFF15 was applied for 10 ns using NAMD2.9 (ref. 28) at 2 fs time-steps and with helical symmetry restraints. The MDFF-derived HOH structure was further equilibrated for 425 ns. On the basis of the equilibrated HOH model and a published pentamer structure 3P05 (ref. 10), a POH model was built and equilibrated for 1.5 µs.A total of 71 MDFF-derived hexamers were docked into the cryo-EM map and equilibrated for 125 ns. Simulations of the complete HIV-1 capsid (64 million atoms, 100 ns) were performed with NAMD. Virus particles were derived by transfection of the full-length HIV-1 proviral construct R9 and mutant derivatives into 293T cells, and infectivity was quantified by titration on HeLa-P4 cells. Capsid stability was determined by measuring the amount of CA protein present after purification of HIV-1 cores, and the rate of uncoating was analysed by quantifying CA release from purified cores at 37 °C. For crosslinking analysis, particles were pelleted from the supernatants of transfected 293T cells, analysed by non-reducing SDS–PAGE, and CA was detected by immunoblotting.
Supplementary Material
Acknowledgements
We thank P. Schwerdtfeger for access to his software for creating fullerene models, T. Brosenitsch for reading the manuscript, and M. DeLucia and J. Mehrens for technical assistance. This work was supported by the National Institutes of Health (GM082251, GM085043 and GM104601) and the National Science Foundation (PHY0822613, MCB0744057). Large-scale molecular dynamics simulations were performed on the Blue Waters Computer, financed by the National Science Foundation (OCI 07-25070).
Footnotes
Supplementary Information is available in the online version of the paper.
Author Contributions G.Z., J.R.P., E.L.Y., A.M.G., K.S., C.A. and P.Z. designed the research. J.N. and J.A. prepared samples for electron microscopy. G.Z. collected cryo-EM data. G.Z., X.M. and P.Z. analysed cryo-EM and cryo-ET data. E.L.Y. and C.A. performed biochemical and functional analysis. K.S. developed large-scale modelling methodology; J.R.P. performed molecular dynamics simulations and B.C. performed CG-MC simulations. G.Z., J.R.P., K.S. and P.Z. analysed atomic models. G.Z., J.R.P., A.M.G., K.S., C.A. and P.Z wrote the paper with support from all the authors.
Author Information Cryo-EM structural data have been deposited in the EMDataBank under accession codes EMD-5582 and EMD-5639, and the MDFF atomic model of the CA HOH and models of HIV-1 capsid have been deposited in the Protein Data Bank under accession numbers 3J34, 3J3Q, 3J3Y.
The authors declare no competing financial interests.
References
- 1.Sundquist WI, Krausslich HG. HIV-1 assembly, budding, and maturation. Cold Spring Harb. Perspect. Med. 2012;2:a006924. doi: 10.1101/cshperspect.a006924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ganser BK, Li S, Klishko VY, Finch JT, Sundquist WI. Assembly and analysis of conical models for the HIV-1 core. Science. 1999;283:80–83. doi: 10.1126/science.283.5398.80. [DOI] [PubMed] [Google Scholar]
- 3.Li S, Hill CP, Sundquist WI, Finch JT. Image reconstructions of helical assemblies of the HIV-1 CA protein. Nature. 2000;407:409–413. doi: 10.1038/35030177. [DOI] [PubMed] [Google Scholar]
- 4.Gitti RK, et al. Structure of the amino-terminal core domain of the HIV-1 capsid protein. Science. 1996;273:231–235. doi: 10.1126/science.273.5272.231. [DOI] [PubMed] [Google Scholar]
- 5.Momany C, et al. Crystal structure of dimeric HIV-1 capsid protein. Nature Struct. Biol. 1996;3:763–770. doi: 10.1038/nsb0996-763. [DOI] [PubMed] [Google Scholar]
- 6.Du S, et al. Structure of the HIV-1 full-length capsid protein in a conformationally trapped unassembled state induced by small-molecule binding. J. Mol. Biol. 2011;406:371–386. doi: 10.1016/j.jmb.2010.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gamble TR, et al. Structure of the carboxyl-terminal dimerization domain of the HIV-1 capsid protein. Science. 1997;278:849–853. doi: 10.1126/science.278.5339.849. [DOI] [PubMed] [Google Scholar]
- 8.Jiang J, et al. The interdomain linker region of HIV-1 capsid protein is a critical determinant of proper core assembly and stability. Virology. 2011;421:253–265. doi: 10.1016/j.virol.2011.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pornillos O, et al. X-ray structures of the hexameric building block of the HIV capsid. Cell. 2009;137:1282–1292. doi: 10.1016/j.cell.2009.04.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pornillos O, Ganser-Pornillos BK, Yeager M. Atomic-level modelling of the HIV capsid. Nature. 2011;469:424–427. doi: 10.1038/nature09640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Byeon IJ, et al. Structural convergence between cryo-EM and NMR reveals intersubunit interactions critical for HIV-1 capsid function. Cell. 2009;139:780–790. doi: 10.1016/j.cell.2009.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meng X, Zhao G, Zhang P. Structure of HIV-1 capsid assemblies by cryo-electron microscopy and iterative helical real-space reconstruction. J. Vis. Exp. 2011;54:e3041. doi: 10.3791/3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Egelman EH. The iterative helical real space reconstruction method: surmounting the problems posed by real polymers. J. Struct. Biol. 2007;157:83–94. doi: 10.1016/j.jsb.2006.05.015. [DOI] [PubMed] [Google Scholar]
- 14.Ganser-Pornillos BK, Cheng A, Yeager M. Structure of full-length HIV-1 CA: a model for the mature capsid lattice. Cell. 2007;131:70–79. doi: 10.1016/j.cell.2007.08.018. [DOI] [PubMed] [Google Scholar]
- 15.Trabuco LG, Villa E, Schreiner E, Harrison CB, Schulten K. Molecular dynamics flexible fitting: a practical guide to combine cryo-electron microscopy and X-ray crystallography. Methods. 2009;49:174–180. doi: 10.1016/j.ymeth.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Byeon IJ, et al. Motions on the millisecond timescale and multiple conformations of HIV-1 capsid protein: implications for structural polymorphism of CA assemblies. J. Am. Chem. Soc. 2012;134:6455–6466. doi: 10.1021/ja300937v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Forshey BM, von Schwedler U, Sundquist WI, Aiken C. Formation of a human immunodeficiency virus type 1 core of optimal stability is crucial for viral replication. J. Virol. 2002;76:5667–5677. doi: 10.1128/JVI.76.11.5667-5677.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Joshi A, Nagashima K, Freed EO. Mutation of dileucine-like motifs in the human immunodeficiency virus type 1 capsid disrupts virus assembly, gag-gag interactions, gag-membrane binding, and virion maturation. J. Virol. 2006;80:7939–7951. doi: 10.1128/JVI.00355-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meng X, et al. Protease cleavage leads to formation of mature trimer interface in HIV-1 capsid. PLoS Pathog. 2012;8:e1002886. doi: 10.1371/journal.ppat.1002886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen B, Tycko R. Simulated self-assembly of the HIV-1 capsid: protein shape and native contacts are sufficient for two-dimensional lattice formation. Biophys. J. 2011;100:3035–3044. doi: 10.1016/j.bpj.2011.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yeager M. Design of in vitro symmetric complexes and analysis by hybrid methods reveal mechanisms of HIV capsid assembly. J. Mol. Biol. 2011;410:534–552. doi: 10.1016/j.jmb.2011.04.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cardone G, Purdy JG, Cheng N, Craven RC, Steven AC. Visualization of a missing link in retrovirus capsid assembly. Nature. 2009;457:694–698. doi: 10.1038/nature07724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Caspar DL, Klug A. Physical principles in the construction of regular viruses. Cold Spring Harb. Symp. Quant. Biol. 1962;27:1–24. doi: 10.1101/sqb.1962.027.001.005. [DOI] [PubMed] [Google Scholar]
- 24.Briggs JA, Wilk T, Welker R, Krausslich HG, Fuller SD. Structural organization of authentic, mature HIV-1 virions and cores. EMBO J. 2003;22:1707–1715. doi: 10.1093/emboj/cdg143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bharat TA, et al. Structure of the immature retroviral capsid at 8 A resolution by cryo-electron microscopy. Nature. 2012;487:385–389. doi: 10.1038/nature11169. [DOI] [PubMed] [Google Scholar]
- 26.Mastronarde DN. Correction for non-perpendicularity of beam and tilt axis in tomographic reconstructions with the IMOD package. J. Microsc. 2008;230:212–217. doi: 10.1111/j.1365-2818.2008.01977.x. [DOI] [PubMed] [Google Scholar]
- 27.Agulleiro JI, Fernandez JJ. Fast tomographic reconstruction on multicore computers. Bioinformatics. 2011;27:582–583. doi: 10.1093/bioinformatics/btq692. [DOI] [PubMed] [Google Scholar]
- 28.Phillips JC, et al. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.