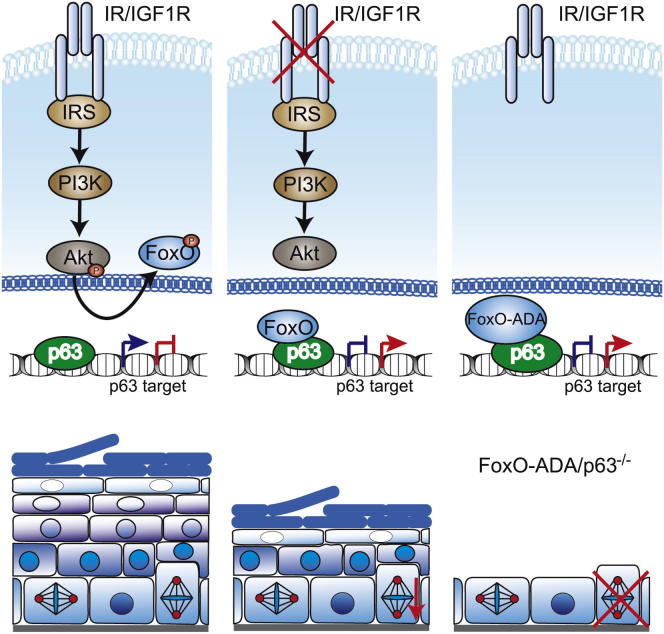
Summary
The multilayered epidermis is established through a stratification program, which is accompanied by a shift from symmetric toward asymmetric divisions (ACD), a process under tight control of the transcription factor p63. However, the physiological signals regulating p63 activity in epidermal morphogenesis remain ill defined. Here, we reveal a role for insulin/IGF-1 signaling (IIS) in the regulation of p63 activity. Loss of epidermal IIS leads to a biased loss of ACD, resulting in impaired stratification. Upon loss of IIS, FoxO transcription factors are retained in the nucleus, where they bind and inhibit p63-regulated transcription. This is reversed by small interfering RNA-mediated knockdown of FoxOs. Accordingly, transgenic expression of a constitutive nuclear FoxO variant in the epidermis abrogates ACD and inhibits p63-regulated transcription and stratification. Collectively, the present study reveals a critical role for IIS-dependent control of p63 activity in coordination of ACD and stratification during epithelial morphogenesis.
Graphical Abstract
Highlights
-
•
Epidermal insulin/IGF-1 signaling (IIS) regulates asymmetric cell division and mitosis
-
•
IIS-controlled FoxOs bind p63 and negatively regulate p63 transcriptional activity
-
•
Constitutive nuclear FoxO disturbs epidermal stratification
-
•
The IIS/FoxO/p63 pathway is a major mechanism controlling epidermal stratification
Insulin/IGF signaling (IIS) is required for epidermal morphogenesis. Günschmann et al. show that IIS-mediated control of FoxOs regulates p63. Loss of epidermal IIS activates FoxOs that inhibit p63 transcriptional activity independent of FoxO DNA binding. The IIS/P63 pathway is critical to coordinate asymmetric division and stratification.
Introduction
The mammalian epidermis is a constantly self-renewing protective barrier against external challenges and dehydration that is formed and maintained by basal progenitor cells with high proliferative potential. The epidermis arises from a single ectodermal layer with a stratification program initiated at approximately embryonic day 12.5 (E12.5). In the single layered developing epidermis the majority of divisions are symmetric (SCD), producing two basal cells, but at the onset of stratification the balance shifts toward asymmetric divisions (ACD), resulting in a basal and a more differentiated suprabasal cell (Lechler and Fuchs, 2005; Poulson and Lechler, 2010; Williams et al., 2011). The epidermal master regulator p63 controls epidermal stratification and differentiation, proliferative potential, and division orientation (Ferone et al., 2013; Koster and Roop, 2007; Lechler and Fuchs, 2005; Lefkimmiatis et al., 2009; Senoo et al., 2007; Truong and Khavari, 2007). However, how p63 is regulated to execute these different functions and if extracellular signals are required to couple division orientation to epidermal stratification is largely unclear. We previously identified insulin- and IGF-1 receptor (IR/IGF-1R) signaling in keratinocytes as key regulators of epidermal morphogenesis and proliferative potential as epidermal loss of IR, IGF-1R, or both increasingly impaired stratification (Stachelscheid et al., 2008). Insulin/IGF-1 signaling-mediated inhibition of the FoxO transcription family of Forkhead proteins is important for the regulation of longevity, metabolism, and stem cell behavior (Calnan and Brunet, 2008; Tothova and Gilliland, 2007). FoxOs are phosphorylated by IIS-activated PKB/Akt, and this results in their export from the nucleus (Kloet and Burgering, 2011). Here, we show that IIS-mediated regulation of the forkhead transcription factor (FoxO) controls p63-dependent transcription via a FoxO DNA-binding-independent interaction with p63 to couple cell cycle progression to spindle orientation and thereby drive stratification.
Results
Epidermal IIS Promotes ACD
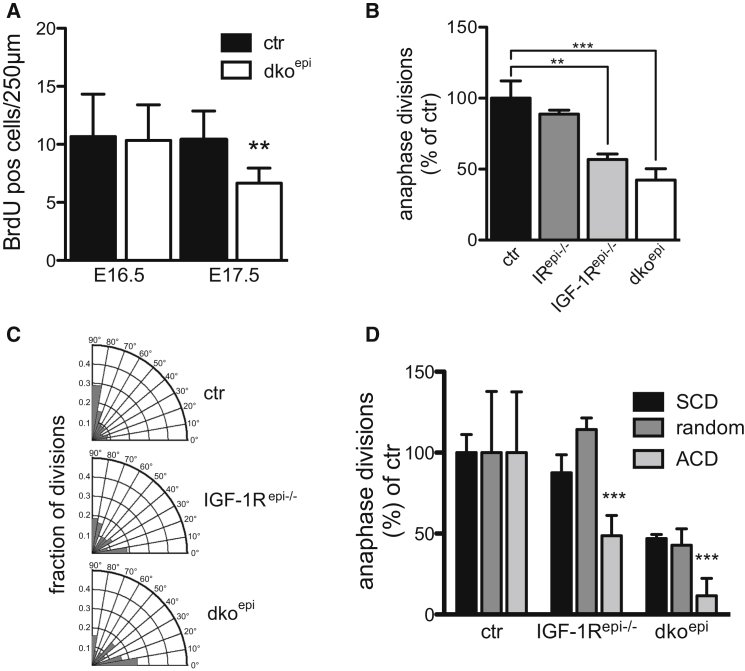
To determine how IIS regulates epidermal stratification, we initially focused on E16.5 mouse embryos carrying a K14-Cre-mediated (Hafner et al., 2004) epidermal IR and/or IGF-1R deletion (IRepi−/−, IGF-1Repi−/−, or dkoepi), the time point when the microscopic phenotype first becomes obvious. In control mice, the transition from E15.5 to E16.5 coincides with an expansion of the number of suprabasal layers. This expansion is reduced in the IRepi−/−, more reduced in the IGF-1Repi−/− with dkoepi mice being most affected. In the IGF-1Repi−/− and dkoepi mice this is accompanied by less polarized basal cells (Stachelscheid et al., 2008; Figure S1A available online). Surprisingly, short term in vivo labeling of BrdU did not reveal a difference in the E16.5 epidermis whereas at E17.5 incorporation was significantly reduced in dkoepi mice (Figures 1A and S1B), similar to what was observed using Ki67 (Stachelscheid et al., 2008). As BrdU is incorporated into the DNA during S-phase, a late block in the cell cycle provides a possible explanation why the E16.5 morphological phenotype induced by loss of IGF-1R/IR is not accompanied by changes in proliferation markers. In agreement, the number of anaphase spindles was significantly reduced in E16.5 epidermis of IGF-1Repi−/− and dkoepi mice whereas the IRepi−/− mice, which only have a mild defect in stratification, showed a slight, albeit nonsignificant, reduction (Figure 1B).
Figure 1.
IIS Controls Epidermal Stratification through Promoting Asymmetric Cell Division
(A) Quantification of short term BrdU-labeled cells in the basal layer of E16.5 and E17.5 ctr and dkoepi epidermis (n = 3 embryos/genotype, mean ± SD).
(B) Reduction of anaphase keratinocytes in IRepi−/−, IGF-1Repi−/−, and dkoepi epidermis at E16.5 (n = 5 embryos/genotype, mean ± SEM). Number of cells in anaphase in control was set at 100%.
(C and D) Biased loss of asymmetric cell divisions (ACD) in the basal epidermal layer of IGF-1Repi−/− and dkoepi mice at E16.5. (C) Radial histogram quantification of division angles. (n = 3 E16.5 embryos per genotype). (D) Relative comparison of different division orientation shows a significant biased loss of ACD in the IGF-1Repi−/− (n = 49 divisions) and dkoepi (n = 25) compared to control (n = 71). Each of the number of symmetric divisions (SCD), undefined divisions or ACDs of the control were set to 100%. Significance was tested separately for each type of division (SCD, random, and ACDs) using one-way ANOVA and indicated by ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001 (n = 3 E16.5 embryos/genotype, mean ± SEM).
See also Figure S1.
As the expansion of suprabasal layers at E16.5 is at least in part driven by ACD (Poulson and Lechler, 2010; Williams et al., 2011), we asked whether epidermal loss of IR/IGF-1R affected SCD and/or ACD. Ablation of IR, IGF-1R, or both resulted in biased loss of ACDs most prominent in the epidermis of IGF-1Repi−/− and dkoepi mice (Figures 1C, 1D, S1C, and S1D), in line with the severity of the stratification phenotype observed in these mice. Thus, by preferentially promoting ACD, IIS couples the regulation of proliferative potential to the expansion of suprabasal differentiated layers.
Epidermal IIS Drives Mitosis
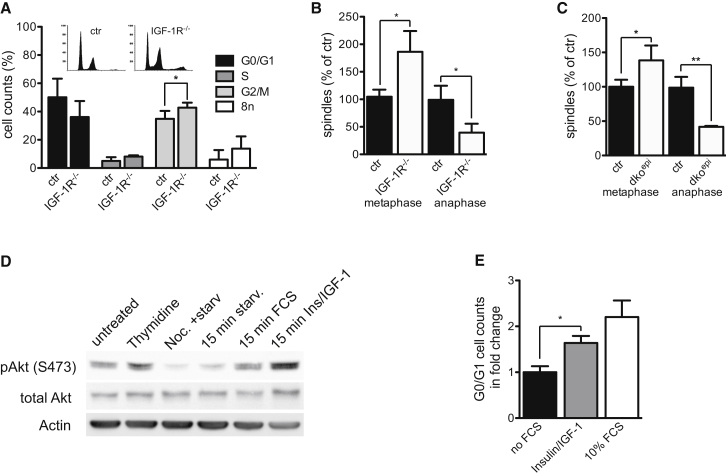
To determine at which cell cycle stage loss of IIS-induced arrest, we performed cell cycle analysis. Significantly more cells were in G2/M in IGF-1R−/− keratinocytes compared to control (Figure 2A). To determine if this was related to an arrest in mitosis, we counted the number of metaphase and anaphase spindles, as the mitotic checkpoint is activated between these two mitotic phases. This revealed an increase in number of metaphase spindles accompanied by a decrease in number of anaphase spindles either in IGF-1R−/− keratinocytes (Figure 2B) or in vivo in E16.5 dko epidermis (Figure 2C). Thus, loss of epidermal insulin/IGF-1 signaling is associated with a mitotic checkpoint arrest.
Figure 2.
Insulin/IGF-1 Is Required for Progression of Mitosis
(A) FACS cell cycle analysis on primary cultured keratinocytes shows an increased number of IGF-1R−/− cells in G2/M (n = 5/genotype, mean ± SD). Insert shows representative propidium iodide cell cycle histograms.
(B and C) Increase in metaphase and reduction in anaphase cells in IGF-1R−/− keratinocytes in vitro (B) and in E16.5 dkoepi embryos (C) (n = 3/genotype, mean ± SD).
(D) Western blot analysis showing increased Akt phosphorylation in serum starved HaCat keratinocytes upon release of mitotic arrest after addition of FCS or Insulin/IGF-1.
(E) Cell cycle analysis showing that insulin/IGF-1 is sufficient to release mitotic arrest of HaCat keratinocytes, as indicated by an increase of cells in G0/G1. Serum-starved mitotically arrested keratinocytes were set to 1 (n = 5, mean ± SD).
Significance was tested using Student’s t test and indicated by ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001.
To ask if IIS is required during mitosis, we synchronized human keratinocytes in S-phase and subsequently serum-starved these cells during late G2 and arrested cells in mitosis. Release of the mitotic block combined with addition of either Insulin/IGF-1 or fetal calf serum (FCS)-induced phosphorylation of Akt (Figure 2D), indicating that IIS is activated during mitosis, whereas no activation was observed in the absence of growth factors. More importantly, addition of Insulin/IGF-1 or the positive-control FCS was sufficient to release mitotic arrest and drive significantly more cells into G1 upon release of the block compared to serum-starved cells (Figure 2E). Together, these results indicate that IIS activity is directly required during mitosis.
IIS Signaling Regulates p63 Activity
The transcription factor p63 controls proliferative potential, epidermal stratification, ACD, and cell cycle progression of keratinocytes (Koster and Roop, 2007; Lechler and Fuchs, 2005; Lefkimmiatis et al., 2009; Senoo et al., 2007; Truong and Khavari, 2007). These processes are also affected by loss of epidermal IIS (Figure 1) (Stachelscheid et al., 2008). Regulation of p63 might thus provide a mechanism for how IR/IGF-1R control epidermal morphogenesis. We therefore asked whether epidermal loss of IIS altered the transcriptional activity of p63. A significant increase in luciferase activity was observed in IGF-1R−/− keratinocytes compared to control upon transfection of a reporter that is repressed by p63 (pG13-Luc) (Hermeking et al., 1997; Yang et al., 1998) (Figure 3A), whereas the activity of the p63-transactivated reporter (BDS-2 (3x)) (Hermeking et al., 1997; King et al., 2003) was repressed in IGF-1R−/− keratinocytes (Figure 3B). Thus, the loss of IGF-1R reduced both p63 transactivation and repressive activities. Importantly, overexpression of p63 in IGF-1R−/− keratinocytes reversed inhibition of the p63-transactivated reporter (Figure 3B).
Figure 3.
IIS Regulates p63 Transcriptional Activity
(A) Luciferase reporter assay showing an increased activity of the p63-repressed pG13 reporter in IGF-1R−/− keratinocytes compared to control, which was set to 1 (n = 4/genotype, mean ± SEM).
(B) Luciferase reporter assay showing a reduced activity of the p63-transactivated BDS-2 (3x) luciferase gene reporter in IGF-1R−/− keratinocytes and restoration of luciferase activity upon transient overexpression of p63. Control activity was set to 1 (n = 4 independent experiments, mean ± SEM).
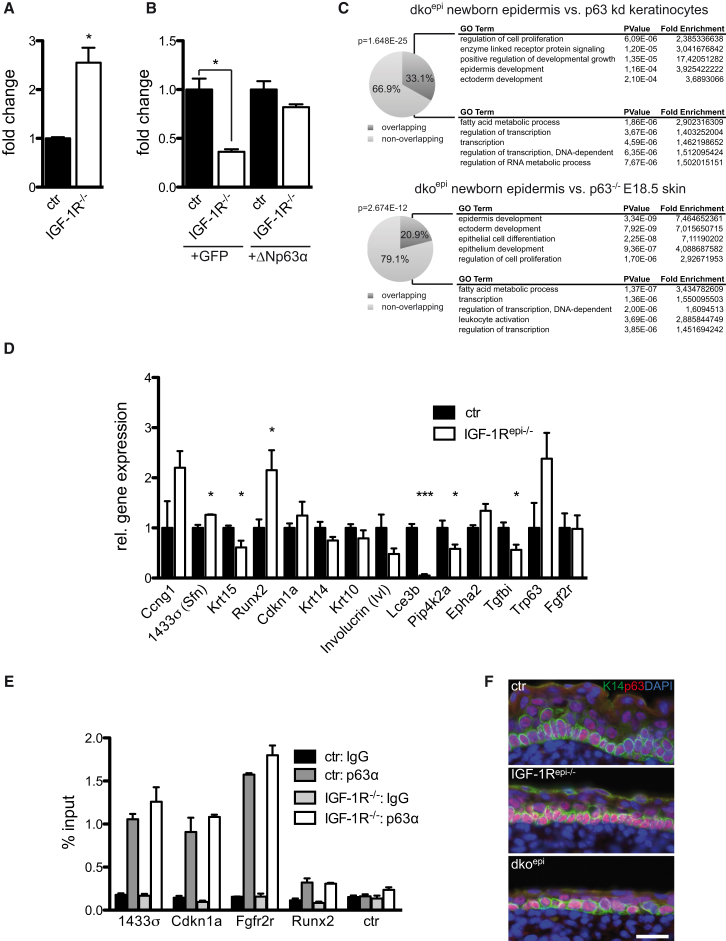
(C) Gene expression microarray analysis of newborn dkoepi epidermis and comparison with arrays of p63 kd keratinocytes (upper panel) or p63−/− E18.5 skin show overlap in gene sets (Pie charts). The overlap of the arrays is highly significant and was calculated using hypergeometric distribution algorithm (indicated by p values). Gene ontology (GO) terms for overlapping genes reveal significant enrichment for epidermal and ectodermal development compared to nonoverlapping GO terms.
(D) Quantitative real-time PCR analysis showing relative gene expression of p63 regulated genes in control (ctr, set as 1) and IGF-1Repi−/− E16.5 epidermis (n = 3 E16.5 embryos/genotype, mean ± SEM). K15, 14-3-3σ, and lce3B were only weakly expressed at E16.5.
(E) Chromatin immunoprecipitation (ChIP)-qPCR assays in primary keratinocytes showing that the binding of p63 to p63-binding regions in different endogenous target promoters is not affected by loss of IGF-1R (mean of three technical replicates ± SD). Shown is a representative example of three independent experiments.
(F) Immunofluorescence analysis for p63 (red) and keratin14 (green) in the epidermis of E16.5 embryos showing that p63 localization is not affected by the loss of IGF-1/IR.
Statistical significance was tested using Student’s t test. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001.
See also Figure S2.
We next asked if these alterations in p63 transcriptional activity have functional consequences for p63 target gene expression. To obtain an unbiased overview, we performed gene expression analysis on RNA isolated from newborn epidermis of control and dkoepi mice and compared this to the gene expression data derived from either mouse keratinocytes in which p63 was knocked down (Della Gatta et al., 2008) or p63−/− E18.5 whole skin (Koster et al., 2006). This revealed a statistically highly significant overlap of the dkoepi gene expression set with both p63-regulated sets (Figure 3C), the percentage of which was in a similar range as the overlap of the two p63 gene expression sets (Figure S2A). More importantly, a statistically significant enrichment for gene ontology terms related to epidermal development was only observed in the overlapping gene sets of dko with either the p63 KD keratinocytes (∼4-fold) or the p63−/− E18.5 skin (∼7-fold) but not in the nonoverlapping sets (Figures 3C and S2B).
We next analyzed the expression of several genes that are regulated by both IIS and p63, most of which are also direct targets of p63 (Ferone et al., 2012; Kouwenhoven et al., 2010; Westfall et al., 2003; Yang et al., 2006). We mostly focused on genes that have been implicated in epidermal differentiation and/or cell cycle regulation. Several p63 repressed targets (e.g., 14-3-3σ or Runx2) were upregulated, whereas other targets (e.g., K15 or Tgfbi) are downregulated in the epidermis of both E16.5 IGF-1Repi−/− and newborn dkoepi mice (Figures 3D and S2C). For 14-3-3σ, this increase was further confirmed at the protein level (Figure S2F). Surprisingly, the global gene expression analysis revealed that several members of the late envelope (Lce) protein families and small proline rich (Sprr) protein, which are part of the epidermal differentiation complex (EDC), were highly upregulated not only in the newborn dkoepi but also in the different p63 knockout or knockdown gene expression sets (Della Gatta et al., 2008; Koster et al., 2006). This is consistent with the complex function of p63 in balancing the promotion of epidermal stratification while inhibiting a subset of terminal differentiation genes or counteracting Notch-induced differentiation (Koster and Roop, 2007; Nguyen et al., 2006). Real-time PCR analysis confirmed the strong increase in Lce3B, Sprr2i, and Sprr2f in the dkoepi (Figure S2D). In contrast, Lce3B was downregulated in IGF-1Repi−/− E16.5 (Figure 3D), albeit that control expression levels were very low and Sprr2i and Sprr2f mRNA levels were below detection at this developmental stage. Other putative p63 targets, such as Fgfr2 (Ferone et al., 2012) were not affected by loss of IGF-1R or IR/IGF-1R either in vivo (Figures 3D and S2C) or in vitro (Figure S2E). Thus, epidermal IIS signaling and p63 regulate the expression of an overlapping set of genes.
If a decrease in p63 transcriptional activity is responsible for the altered expression of a range of p63 target genes in the epidermis of IGF-1Repi−/− or dkoepi mice, then one would predict less p63 binding to p63 consensus elements in the endogenous promoters of these altered genes. Based on previously identified p63-binding sites in promoters in human keratinocytes and cells (Kouwenhoven et al., 2010; Westfall et al., 2003; Yang et al., 2006) we identified conserved p63 binding sites in promoters/enhancers of murine Runx2, Stratifin (Sfn, encoding 14-3-3σ), and Cdkn1a and used the already known site in Fgfr2 (Ferone et al., 2012) as a control as its expression is not altered upon IR/IGF-1R loss. We next asked whether p63 binding to these sites was changed upon loss of IGF-1R. Surprisingly, p63 binding to the p63 consensus site in these targets was similar in control and IGF-1R−/− keratinocytes in chromatin immunoprecipitation quantitative PCR (ChIP-qPCR) (Figure 3E), even though RNA expression of these targets is altered. In agreement, loss of IIS signaling did not induce significant changes in p63 expression in either E16.5 IGF-1Repi−/− (Figure 3F) or in keratinocytes (Figure S2G). Taken together, these findings suggest that insulin/IGF-1 regulate p63 transcriptional activity independent of p63 protein expression and promoter binding.
IIS Regulates Epidermal FoxO Activity
The FoxO family of forkhead transcription factors is one of the key targets through which IIS exerts its biological effects. IIS negatively controls the transcriptional activity of FoxOs through Akt phosphorylation-dependent nuclear exclusion of FoxOs (Calnan and Brunet, 2008). FoxOs play central roles in longevity, stem cell regulation, metabolism, and tumor suppression (Accili and Arden, 2004; Partridge and Brüning, 2008). More importantly, FoxOs were shown to regulate transcriptional activity of other proteins independent of binding to their consensus sites in promoters (Jensen et al., 2011; Nemoto et al., 2004). Thus, FoxO transcription factors are potential downstream candidates to mediate IIS regulation of genes also regulated by p63. However, little is known on the expression of FoxO members in the epidermis.
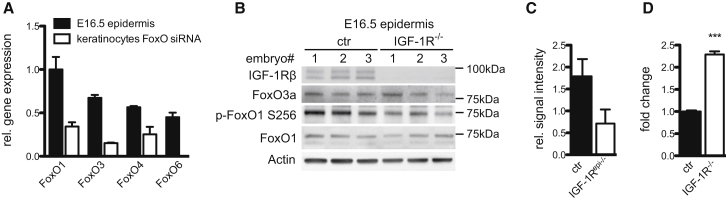
Real-time PCR analysis showed that all four members, FoxO1, FoxO3, FoxO4, and FoxO6, were expressed in E16.5 epidermis (Figure 4A), the time point when the phenotype induced by loss of IIS signaling becomes obvious. As IIS does not regulate nuclear shuttling of FoxO6 (Jacobs et al., 2003), we did not further analyze this member. Western blot analysis for FoxO1 and FoxO3 also showed protein expression at E16.5 (Figure 4B) and in newborn (not shown) as well as in primary keratinocytes (Figure S3A), whereas only a specific band could be detected for FoxO4 in primary keratinocytes but not E16.5 epidermis, despite RNA expression (Figure S3A). Thus, FoxO1 and FoxO3 proteins are expressed in E16.5 epidermis.
Figure 4.
Epidermal FoxO Transcription Factors Are Regulated by IIS
(A) Quantitative real-time PCR analysis of the FoxO family in E16.5 epidermis and in keratinocytes treated with a combination of siRNAs to FoxO1/3/4 as a negative control (mean of n = 3, embryos or n = 3 knockdown cell lines ± SEM). Expression of FoxO1 was set to 1.
(B) Western blot analysis showing expression of FoxO1 and FoxO3 in E16.5 epidermis. Loss of IGF-1R does not affect overall expression levels but does reduce phosphorylation of FoxO1.
(C) Quantification of Phospho-FoxO1 intensities in (B) normalized to total FoxO1 with control set to 1 (mean of n = 3 E16.5 epidermis/genotype ± SD).
(D) Luciferase reporter assay showing an increase in FoxO reporter (6xDBE) activity in IGF-1R−/− keratinocytes compared to control, which is set to 1 (n = 4 independent experiments, mean ± SEM).
Statistical significance was tested using Student’s t test. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001.
See also Figure S3.
Loss of IGF-1R did not alter FoxO1 and FoxO3 expression levels in E16.5 embryos (Figures 4B and S3B). Importantly, using western blot analysis, we detected less phospho-FoxO1 in E16.5 IGF-1R−/− epidermis compared to control (Figures 4B and 4C), indicating a loss of IIS-stimulated Akt phosphorylation activity. This was associated with an increase in nuclear FoxO1 protein (Figure S3C). Most importantly, FoxO-specific reporter assays showed increased FoxO transcriptional activity in IGF-1R−/− keratinocytes (Figures 4D and S5A). Together, these data suggest that loss of epidermal IIS activates FoxOs.
IIS-Regulated FoxO Inhibits p63 Independent of Its DNA Binding Properties
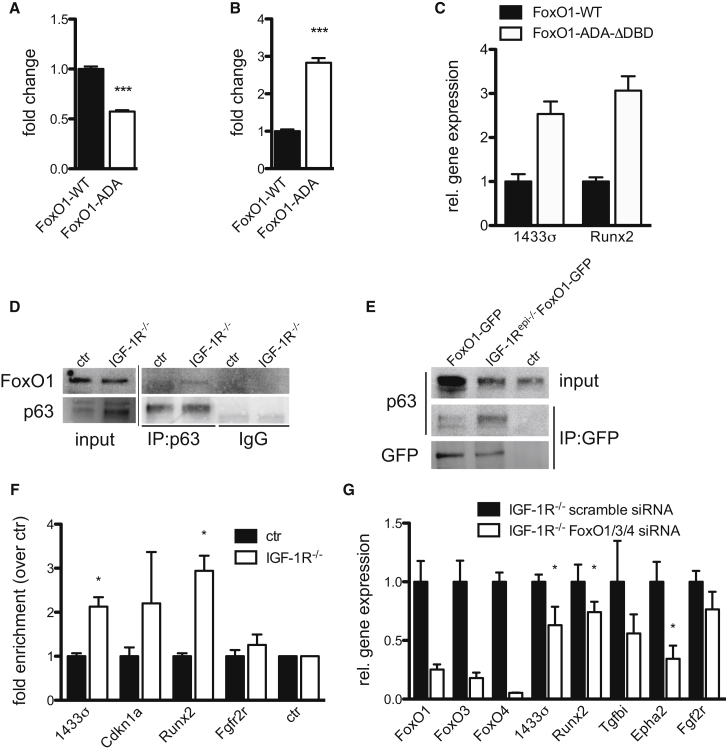
To examine whether FoxO can regulate p63, we transfected primary keratinocytes with either the p63-transactivated reporter or the p63-repressed reporter in the presence of either GFP, FoxO1-WT, or of a FoxO1 with mutated Akt phosphorylation sites (FoxO1-ADA) resulting in a constitutively nuclear FoxO that thus mimics loss of IR/IGF-1R (Nakae et al., 2000). FoxO1-WT was used as a control as this is excluded from the nucleus due to the presence of Insulin/IGF-1 in serum. FoxO1-ADA inhibited the p63-transactivated reporter (Figure 5A), whereas the activity of the p63-repressed reporter was upregulated by FoxO1-ADA (Figure 5B). Thus, nuclear FoxO counteracts p63-transcriptional activity. Mutation of the FoxO DNA binding domain (FoxO1-ADA-ΔDBD) (Kitamura et al., 2007) did not affect the ability of FoxO1-ADA to repress the p63-transactivated reporter in primary keratinocytes (Figure S4A). To rule out that FoxO-ADA can directly bind p63-binding sites, we transfected CHO cells as these cells do not express p63. FoxO-ADA-mediated repression required cotransfection with p63 (Figure S4B).
Figure 5.
Insulin/IGF-1-Regulated FoxO Interacts with p63 at p63 Binding Sites
(A and B) Luciferase reporter assays showing that transfection of the constitutive nuclear FoxO1-ADA mutant in primary mouse keratinocytes (A) repressed the activity of the p63-transactivated reporter BDS-2 (3x) and (B) increased the activity of the p63-repressed reporter pG13 p63 reporter. Luciferase activity was compared to primary mouse keratinocytes transfected with WT-FoxO1 in (A) and (B) and set to 1 for WT-FoxO1 (mean of n = 4 independent experiments ± SEM).
(C) Real-time PCR analysis showing that a DNA binding deficient mutant of FoxO1 (FoxO1-ADA-ΔDBD) enhances expression of the p63-repressed targets 14-3-3σ and Runx2. Primary keratinocytes were transiently transfected with either WT-FoxO1 or FoxO1-ADA-ΔDBD (mean of n = 2 independent experiments ± SD).
(D) Coimmunoprecipitation of endogenous FoxO1 with p63 from primary IGF-1R−/− but not control keratinocytes.
(E) Coimmunoprecipitation of p63 with FoxO1-GFP from the epidermis of newborn control and IGF-1Repi−/− mice that also express a FoxO1-GFP in the epidermis using GFP antibodies.
(F) Chromatin immunoprecipitation (ChIP)-qPCR analysis in primary mouse control and IGF-1R−/− keratinocytes using FoxO1 antibodies showing an increased interaction of FoxO1 with p63-binding regions in promoters of p63 targets. Results were normalized to a negative binding region and are shown as enrichment over control keratinocyte IP, which was set to 1 (mean of n = 3 independent experiments ± SD; n = 2 for Fgfr2r).
(G) Real-time PCR analysis on primary mouse IGF-1R−/− keratinocytes transiently transfected with either scramble or combined knockdown of FoxO1/3/4 using smart pool siRNAs to each of these FoxOs. FoxO1/3/4 knockdown, but not scrambled, siRNAs reduced expression of different p63 targets that are upregulated upon loss of IGF-1R−/− (mean of n = 4 independent experiments ± SEM).
Values in (A)–(C) and (G) are means ± SEM. Values in (F) are means ± SD. Statistical significance was tested by Student’s t test and indicated as ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001.
See also Figure S4.
To examine whether FoxO directly interacts with p63, we transfected CHO cells with p63 and FoxO1-ADA and found that FoxO1-ADA precipitated p63 and vice versa, p63 precipitated FoxO1-ADA (Figure S4C), thus providing evidence that FoxO1-ADA interacts with p63. More importantly, more endogenous FoxO1 was immunoprecipitated by p63 from IGF-1R−/− than from control keratinocytes (Figure 5D). Using K14-Cre mice carrying a flox-stop-flox-FoxO-GFP cassette under the control of the Rosa26 promoter (Fukuda et al., 2008), we expressed FoxO1-GFP in the epidermis either in a control or IGF-1Repi−/− background. GFP immunoprecipitations showed an increased interaction of p63 with FoxO1-GFP in the absence of IGF-1R compared to control newborn epidermis (Figure 5E).
Thus, loss of IIS results in nuclear translocation of FoxOs, where they interact with p63 to counteract p63 transcriptional activity. This would predict that loss of IGF-1R would promote an association of FoxOs with p63 sitting on p63 binding sites present in endogenous promoters. As already shown, p63 binding to p63 consensus site containing promoters was not altered by IIS loss (Figure 3E). FoxO1 ChIP-qPCR experiments in control and IGF-1R−/− keratinocytes showed that upon loss of IGF-1R FoxO1 was enriched 2- to 3-fold on p63 consensus sites in the promoters of targets that showed altered expression upon loss of IGF-1 (e.g., Sfn or Runx2) but not on p63 sites located in targets with unaffected expression, such as e.g., Fgfr2r (Figure 5F). In agreement, transfection of FoxO1-ADA-ΔDBD DNA binding mutant in control primary keratinocytes increased expression of these same targets, Runx2 and 14-3-3α (Figure 5C), similar to in vivo loss of IIS (Figure 3D). These results indicate that FoxO interacts with p63 on p63-DNA consensus sites to regulate expression of these p63 targets in a FoxO-DNA-binding-independent manner. Most importantly, siRNA mediated knockdown of FoxO1, -3, and -4 but not a scrambled siRNA reversed the expression of e.g., 14-3-3σ and Runx2 in IGF-1R−/− primary keratinocytes (Figure 5G), whereas these siRNAs showed no effect in control (Figure S4D), indicating that the increased expression of these targets upon loss of IGF-1R is a direct consequence of increased FoxO activity. Together, these findings identify a DNA binding-independent function for FoxOs in the regulation of p63 activity.
Strongly Impaired Stratification in Mice Expressing Epidermal FoxO1-ADA
If FoxO functionally counteracts p63 in vivo in a DNA binding-independent manner, then expression of the constitutive nuclear FoxO1-ADA, but not a FoxO1-DNA binding-domain-only mutant (FoxO-DN) (Nakae et al., 2000) in the epidermis, should result in a skin phenotype that is similar to that caused by loss of p63. In addition, FoxO-DN should not be able to rescue the impaired stratification in IGF-1Repi−/−. We thus crossed mice that carry a flox-stop-flox cassette followed by either a FoxO1-ADA-IRES-GFP or FoxO1-DN-IRES-GFP cassette in the Rosa26 locus (Belgardt et al., 2008; Stöhr et al., 2013) to the Keratin14-Cre transgene (Hafner et al., 2004) and in case of the FoxO-DN also to IGF-1RFl/Fl mice, to induce epidermal expression of the ADA mutants (FoxO1-DNepi or FoxO1-ADAepi).
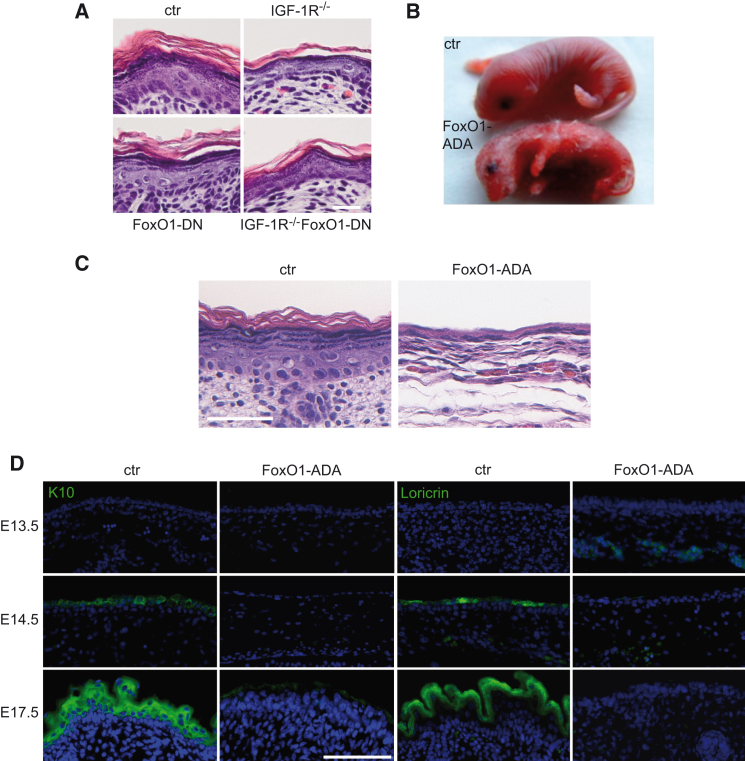
FoxO1-DN expression reversed FoxO reporter activity induced by the loss of IGF-1R in primary keratinocytes isolated from these mice (Figure S5A), showing that the FoxO DNA binding mutant was indeed a dominant negative toward FoxO DNA-binding-dependent transcriptional activity. Expression of FoxO1-DN did not affect epidermal morphogenesis (Figure 6A), as predicted based on our observations that the FoxO1-ADA DNA binding mutant was still able to alter p63 reporter activity and gene expression. More importantly, FoxO1-DN was unable to rescue stratification in IGF-1Repi−/− mice (Figure 6A).
Figure 6.
Epidermal Expression of Constitutive Nuclear FoxO1 Strongly Impairs Epidermal Differentiation
(A) H&E staining showing that epidermal expression of dominant negative FoxO1 (FoxO1-DN) did not affect epidermal morphogenesis and did not rescue the impaired stratification of IGF-1Repi−/− mice. Scale bar represents 25 μm.
(B and C) Expression of FoxO1-ADA in the epidermis interferes with stratification. Macroscopic appearance of control and FoxO-1ADAepi newborn mice (B) and H&E stainings of paraffin sections of newborn mice (C). Scale bar represents 25 μm.
(D) Immunofluorescence analysis for the epidermal differentiation markers K10 (green, left) and loricrin (green) in E13.5, E14.5, and E17.5 control and FoxO1-ADAepi embryos revealed a strong reduction in differentiation in the FoxO1-ADAepi mice. Nuclei are counterstained by DAPI. Scale bar represents 50 μm.
See also Figure S5.
FoxO1-ADA was expressed in the epidermis already at E13.5 (Figures S5B and S5C) and resulted in fragile, translucent skin and perinatal death (Figure 6B). Histochemical analysis of newborn mice revealed a striking hypoplastic epidermis (Figure 6C), which strongly resembled that of p63 knockout mice (Mills et al., 1999; Yang et al., 1999). This phenotype became first apparent at E14.5 (Figure S5D) accompanied by a failure to properly induce the suprabasal differentiation markers keratin10 and loricrin (Figure 6D), indicating that FoxO1-ADA interferes with proper initiation of stratification.
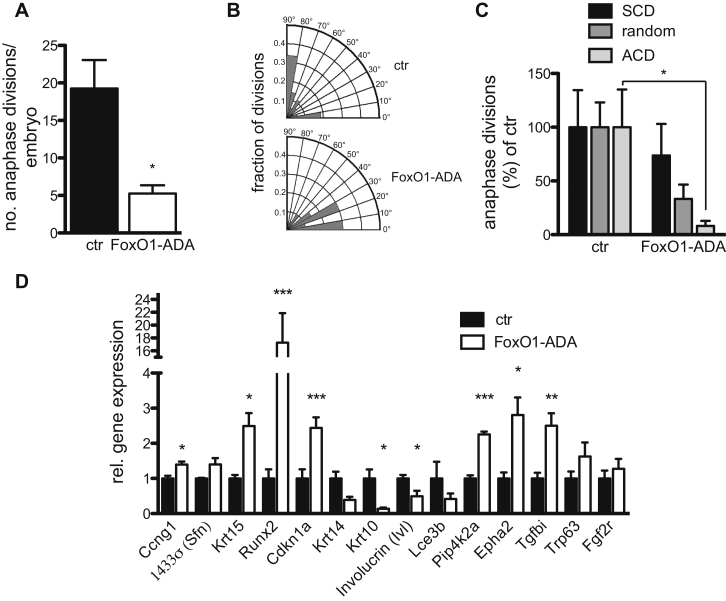
Biased Loss of ACD and Altered p63 Target Gene Expression
TUNEL staining revealed no increase in apoptosis in the developing epidermis of K14-Cre-FoxO1-ADA mice (Figure S6A), similar to what was observed in dko epidermis (Stachelscheid et al., 2008) and thus likely is not causative of the phenotype. In contrast, E16.5 FoxO1-ADA mice showed a strong reduction in the number of anaphase divisions in the epidermis (Figure 7A), further confirming that FoxO is downstream of IIS in the regulation of epidermal morphogenesis. This was even more pronounced than in dko epidermis (Figure 1B), thus reflecting the more severe phenotype. This was due to a biased loss of ACDs (Figures 7B and 7C), as was also observed in IRepi−/−, IGF-1Repi−/−, dkoepi (Figures 1C, 1D, and S1D), and p63 knockout mice (Lechler and Fuchs, 2005).
Figure 7.
Constitutive Nuclear FoxO1 Results in Biased Loss of ACD and Altered p63 Target Gene Expression
(A) Reduction of number of basal keratinocytes in anaphase in E16.5 FoxO1-ADA mice compared to control (n = 3 embryos each genotype, mean of divisions ± SD).
(B and C) Biased loss of ACD in FoxO1-ADA E16.5 epidermis. (B) Radial histograms quantification of division angles (n = 3 E16.5 embryos/genotype). (C) Relative comparison of different division orientations shows a significant biased loss of ACD in FoxO1-ADA E16.5 epidermis (n = 22 divisions) compared to control (n = 70). Each of the number of symmetric divisions (SCD), undefined divisions, or ACDs of the control was set to 100%. Significance was tested separately for each division type (SCD, undefined and ACDs) using Student’s t test (n = 3 E16.5 embryos/genotype).
(D) Real-time qPCR analysis of control (set to 1) and E16.5 FoxO1-ADA epidermis showing altered expression of several p63 target genes compared to control, which was set to 1 (mean of n = 4 embryos/genotype ± SEM). K15, Lce3B, and 14-3-3σ are only weakly expressed at E16.5.
Statistical significance was tested using Student’s t test and indicated as ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001.
See also Figure S6.
Most importantly, expression analysis revealed that FoxO1-ADA expression regulated a similar subset of P63 target genes in E16.5 epidermis (Figure 7D), most of which were also altered in E16.5 IGF-1Repi−/− epidermis (Figure 3D), whereas targets not altered by loss of IIS (e.g., FGFR2) were again not changed. As expected, basal p63 localization was not obviously affected by epidermal FoxO1-ADA expression (Figure S6B). Thus, insulin/IGF-1-controlled FoxOs are crucial regulators of p63, ACD, and epidermal morphogenesis.
Discussion
Our data demonstrate a role for IIS signaling in FoxO-dependent control of p63, a key determinant of epidermal cell fate, to regulate ACD and progression of mitosis during epidermal morphogenesis. This allows IGF-1, and to a much lesser extent insulin, to couple the maintenance of progenitors with high proliferative potential to suprabasal differentiation. We identify p63 as an interacting partner of FoxOs and show that FoxO is a negative regulator of p63 transcriptional activity on a subset of targets important for epidermal stratification. Surprisingly, this is independent of FoxO binding to DNA. This is based on the following observations: a constitutive nuclear FoxO, FoxO1-ADA, inhibits transcription of a luciferase reporter that only contains p63 sites and only in the presence of p63. More importantly, a point mutation that prevents DNA binding, as shown by FoxO reporter assays FoxO1-ADA-ΔDBD, still inhibits p63-dependent reporters. Last, epidermal expression of a mutant FoxO1 that consists only of its DNA binding domain did not obviously affect epidermal morphogenesis and, more importantly, was not able to rescue the phenotype caused by inactivation of IGF-1 (results not shown). This same FoxO-1 mutant was shown to rescue loss of IIS in the hypothalamus (Belgardt et al., 2008). In agreement with our findings, FoxOs can also regulate transcriptional activity of Myc and p53 independent of binding to the consensus FoxO recognition element (Jensen et al., 2011; Nemoto et al., 2004).
The p63 transcription factor is crucial for the formation of stratifying epithelia such as epidermis and mutations in human p63 result in ectodermal dysplasia disorders (Koster and Roop, 2007; Vanbokhoven et al., 2011). Except for BMP signaling in zebrafish (Bakkers et al., 2002), the dermal signals that regulate p63 have so far remained largely elusive. Here, we identify p63 as a target of IIS, which is necessary during epidermal morphogenesis to inhibit FoxO activity, likely through nuclear export and thereby prevent FoxO to negatively regulate p63 function. We find that loss of IIS resulted in a decrease in Akt-dependent FoxO1 phosphorylation accompanied by an increase in nuclear FoxO1 protein and, more importantly, increased FoxO transcriptional activity as judged by reporter assays. We were unable to detect nuclear FoxO by immunohistochemistry. As the FoxO1-ADA is readily detected in the nucleus (Figure S5C) and expressed ∼5-fold more as endogenous FoxO (Figure S5B), endogenous FoxOs are likely expressed at levels too low to be detected by immunostaining. Thus far, only a few publications show nuclear translocation of endogenous FoxOs by immunohistochemistry and only upon fasting and not loss of IIS (Kitamura et al., 2006).
Loss of p63 or epidermal expression of FoxO1-ADA phenotypically mimic each other and result in much more severe phenotype than combined epidermal loss of IR and IGF-1R. Although this in part may be explained by the developmental timing of deletion versus expression, this does suggest that other signals than IIS control FoxO and p63 in epidermal morphogenesis. Potential candidates are TGFβ-superfamily members, as several TGFβ downstream transcription targets, some of which are also regulated by p63, require the cooperative activity of Smad4 and FoxOs in keratinocytes (Gomis et al., 2006). Perturbed FoxO-p63 interactions may also contribute to a range of common and rare skin diseases characterized by disturbed epidermal differentiation, such as psoriasis, ichthyosis, atopic dermatitis, and skin cancer.
Our data provide evidence for a model in which IIS in keratinocytes controls epidermal morphogenesis through exclusion of FoxO from the nucleus thereby releasing its inhibitory action on p63. This in turn allows p63 to exert its transcriptional control of genes that regulate proliferative potential, cell cycle progression, and spindle orientation. Our data suggest that 14-3-3σ, an important regulator of epidermal differentiation, might be one of the key targets of IR/IGF-1R/FoxO/p63 axis. P63 promoter binding inhibits Sfn transcription, which encodes 14-3-3σ (Westfall et al., 2003). This is relieved upon loss of insulin/IGF-1 accompanied by increased FoxO binding to the p63 site in the endogenous Sfn promoter resulting in more RNA and protein expression. In line with our findings, overexpression of 14-3-3σ in murine epidermis results in a hypomorphic phenotype (Cianfarani et al., 2011), similar to the dkoepi mice. It was also shown that 14-3-3σ can induce G2/M arrest in cell culture (Reinhardt and Yaffe, 2009).
Although insulin/IGFs and FoxO have not yet been directly implicated in the regulation of mitosis and ACD, AKT, the upstream inhibitor of FoxO, regulates ACD in cancer cells (Dey-Guha et al., 2011) and combined loss of Akt1 and Akt2 results in less suprabasal layers similar to epidermal loss of IR and IGF-1R (Peng et al., 2003). The small GTPase Rac, previously shown to mediate IIS regulation on epidermal morphogenesis likely upstream of Akt (Stachelscheid et al., 2008), has also been implicated in control of ACD (Halet and Carroll, 2007; Lu et al., 2012). How insulin/IGF-mediated regulation of FoxO and p63 regulates ACD and mitotic progression to control the balance between proliferation and differentiation will be an important question for the near future.
Experimental Procedures
Mice
Epidermis-specific deletion of insulin receptor (IR), IGF-1-receptor (IGF-1R), or both using K14-Cre-mediated deletion in mice have been described (Stachelscheid et al., 2008). Conditional FoxO knockin mice carrying a CACG promoter, a loxP-flanked stop cassette followed by the cDNA of constitutively nuclear FoxO1-ADA or dominant negative FoxO1-DN in the Rosa26 locus (Belgardt et al., 2008; Stöhr et al., 2013) or a loxP-flanked stop cassette followed by FoxO1-GFP in the Rosa26 locus (Fukuda et al., 2008) were crossed to K14-Cre transgenic mice (Hafner et al., 2004) to induce epidermal-specific expression. All mice are in C57Bl/6 background and experiments were performed according to institutional guidelines and animal license of the State Office North Rhine-Westphalia, Germany.
Immunohistochemistry and Immunofluorescence
For histology, embryos or mouse skin were fixed in 4% PFA and embedded in paraffin. Paraffin sections (5 μm) were deparaffinized and stained with hematoxylin and eosin (H&E) and imaged with an Olympus BX51 microscope. For immunofluorescence, paraffin sections were deparaffinized, antigens were retrieved with buffer A, UG, or AG (EMS), and sections were blocked in PBS containing 5% normal goat serum and 0.1% Triton X-100. Slides were incubated with primary antibody followed by washing and incubation with the appropriate secondary antibodies coupled to Alexa 488, Alexa 594, or Cy3 (Invitrogen). Nuclei were counterstained with DAPI (Sigma) and sections examined using Olympus IX81 fluorescence or Olympus FV1000 confocal microscopes. Primary antibodies are listed in Supplemental Experimental Procedures.
Division Axis Orientation Determination
To analyze the angle of divisions, the axis of divisions in E16.5 embryos was determined in anaphase/telophase cells using survivin staining as described by Williams et al. (2011). The angle of division was determined by measuring the angle of the plane transecting two daughter cells relative to the plane of the basement membrane. The angles of divisions were quantified and angle orientation was plotted with Oriana 4 (KCS). The different divisions were then categorized as described with asymmetric divisions having an angle of 60°–90°, random 30°–60°, and symmetric 0°–30° (Lechler and Fuchs, 2005). Each of the total number of asymmetric, random, or symmetric divisions of the control were then set to 100% to compared the relative loss within each of the division categories to either knockouts or transgene.
Isolation and Transfection of Primary Keratinocytes and CHO Cells
Primary keratinocytes were isolated and cultured in minimal Ca2+ medium (50 μM Ca2+) as described (Stachelscheid et al., 2008). For FoxO1 and p63 overexpression, primary mouse keratinocytes were transfected using Lipofectamine2000 (Invitrogen) at a confluence of 70%–90% according to manufacturer’s protocol. CHO cells were transfected using Lipofectamine (Invitrogen). For transfection 250 ng of the following plasmids were used: pcDNA3-FoxO1-ADA, pcDNA3-FoxO1-ADA-DBD, and pCMV-FLAG-FoxO1 (Kitamura et al., 2007) and p63 plasmids pCMV-FAG-ΔNp63α (Ferone et al., 2012), pEGFP-ΔNp63α, or pEGFP empty vector as control. For luciferase reporter assays, cells were cotransfected with 25 ng pTK-Renilla (Promega) and 250 ng of one of the following p63 luciferase reporters: 250 ng of pGL3-BDS-2 (3x) and pG13-Luc (Hermeking et al., 1997; Yang et al., 1998) or the 6xDBE-Fkhre FoxO reporter (Potente et al., 2005). Experiments were performed in triplicates and luciferase activity was determined 24 to 48 hr after transfection with the dual-luciferase reporter assay kit (Promega) and a Berthold TriStar Luminometer. Firefly luciferase activity was normalized for transfection efficiency to Renilla luciferase activity. The results were presented as the average of at least three independent experiments.
FACS Analysis
Cells were harvested by trypsin/EDTA treatment, washed in 5 ml PBS and resuspended in 3 ml PBS. For fixation, cells were incubated with 70% ethanol/PBS at 4°C for minimum 2 hr and then centrifuged for 7 min at 1,000 rpm. The pellet was 2× washed with PBS, filtered with a 70 μm cell strainer and resuspended in PBS containing 0.1% Triton X-100, RNase A (10 mg/ml) and propidium iodide (2 mg/ml; Sigma). Cells were stained at 37°C for 15 min and analyzed by FACS (FACScalibur, BD). FACS data were analyzed using FlowJo software (Tree Star).
RNA Interference
To silence FoxO gene expression, keratinocytes were transfected with ON-TARGET plus SMARTpool siRNAs (Thermo Fisher Scientific) targeting FoxO1, FoxO3, FoxO4, and FoxO6. Subconfluent primary mouse keratinocytes were transfected using 50 nM of each SMARTpool or nontargeting control pool and Lipofectamine2000 Reagent (Invitrogen) according to manufacturer’s protocol. Efficient knockdown was observed 48 hr posttransfection in RT-PCR and western blot analysis.
Chromatin and Coimmunoprecipitation
For ChIP assays ∼3 × 106 primary mouse keratinocytes or newborn epidermis were crosslinked with 1% formaldehyde. Crosslinking was stopped and cells lysed. Lysates were subjected to sonication on ice to obtain DNA fragments ranging from 200 to 1,000 bp in length. The supernatant was diluted for immunoprecipitation in IP buffer (150 mM NaCl, 20 mM Tris pH 8, 2 mM EDTA) and precleared with protein A beads (Roche). Supernatant was incubated with 4 μg antibody overnight at 4°C, and beads were sequentially washed as previously described (Ferone et al., 2012). Chromatin was eluted, DNA was purified and analyzed by quantitative real-time PCR, or protein was analyzed by western blot. Real-time PCR was performed using the SYBR Green PCR master mix (Applied Biosystems) in an ABI StepOne light cycler. Primer sets for analysis are listed in Supplemental Experimental Procedures.
For Co-IP, CHO cells were lysed in HEPES buffer (50 mM HEPES-KOH pH 7.8; 140 mM NaCl; 1 mM EDTA; 10% glycerol; 0.25% Triton X-100; 1% NP-40) for 20 min at 4°C, subsequently nuclei were disrupted by mild sonication and lysate after preclearing incubated with 4 μg of antibody overnight. Antibodies were precipitated with Protein A/G (Roche) beads and after washing in IP buffer, samples were analyzed by western blot.
Quantitative Real-Time PCR and Global Gene Expression Analysis
Gene expression was analyzed using quantitative real-time PCR. RNA was extracted from keratinocytes and epidermis using Trizol (Invitrogen) and RNeasy Minikit (QIAGEN). RNA was reversely transcribed with Quantitect Reverse Transcriptase (QIAGEN) and amplified using the TaqMan Universal PCR Master Mix (Applied Biosystems). PCR was performed on an ABI StepONE Plus machine. Calculations were performed by comparative cycle threshold (ΔΔCt) method with data normalized relative to 18S and Hprt1. Probes for target genes were ordered from TaqMan Assay-on-Demand Kits (Applied Biosystems). TaqMan probes are listed in Supplemental Experimental Procedures. For microarray analysis, RNA was isolated from control and dkoepi−/− epidermis (n = 4 each) and sent to the DNA Sciences Core at the University of Virginia for labeling, amplification, and hybridization to the Affymetrix 430-2.0 platform. For global gene expression analysis, significantly regulated genes sets (p < 0.05) from the different microarrays were analyzed for overlap using VENNY Tool by Oliveros (http://bioinfogp.cnb.csic.es/tools/venny/index.html). The significance of overlapping and nonoverlapping genes was determined using hypergeometric distribution algorithm. Overlapping and nonoverlapping gene sets were then annotated using DAVID functional annotation tool (Dennis et al., 2003).
Protein Isolation and Immunoblotting
Epidermis was separated from dermis and dissociated with a MixerMill homogenizer and lysed in 1% SDS lysis buffer. Primary keratinocytes were scraped of culture dishes and lysed in 1% SDS lysis buffer. Lysates were separated on SDS-PAGE gels (Novex), transferred to a PVDF membrane, blocked in 5% western blot blocking solution (Roche) and incubated with primary antibodies followed by incubation with the appropriate horseradish peroxidase coupled secondary antibodies and detected using ECL (Thermo Fisher Scientific).
Statistical Methods
Data were analyzed for statistical significance using two-tailed unpaired Student’s t test unless otherwise stated. Relative loss of ACD, random divisions, and SCD were tested using one-way ANOVA in Prism 5 (GraphPad). The asterisks shown in graphs correspond to the p values as stated in the figure legends. The results were presented as the average of at least three independent experiments unless otherwise stated in the legends and arrow bars indicate SD or SEM.
Acknowledgments
We would like to thank B. Habermann and A. Yeroslaviz for their help with the statistical analysis of the hypergeometric distribution and our colleagues in the Niessen laboratory for stimulating discussion. Funding was provided by the DFG-funded SFBs 829 (to J.C.B. and C.M.N.) and 832 (to C.M.N.), the German Cancer Aid (to C.M.N.), Telethon, GGP09230, Italy (to C.M.), and the European ERA-Net Research Program on Rare Diseases (E-Rare-2; Skindev; to C.M.)
Published: July 29, 2013
Footnotes
Supplemental Information includes Supplemental Experimental Procedures and six figures and can be found with this article online at http://dx.doi.org/10.1016/j.devcel.2013.05.017.
Contributor Information
Caterina Missero, Email: missero@ceinge.unina.it.
Jens C. Brüning, Email: jens.bruening@uni-koeln.de.
Carien M. Niessen, Email: carien.niessen@uni-koeln.de.
Accession Numbers
Microarray data have been deposited into the GEO database with accession number GSE47065.
Supplemental Information
References
- Accili D., Arden K.C. FoxOs at the crossroads of cellular metabolism, differentiation, and transformation. Cell. 2004;117:421–426. doi: 10.1016/s0092-8674(04)00452-0. [DOI] [PubMed] [Google Scholar]
- Bakkers J., Hild M., Kramer C., Furutani-Seiki M., Hammerschmidt M. Zebrafish DeltaNp63 is a direct target of Bmp signaling and encodes a transcriptional repressor blocking neural specification in the ventral ectoderm. Dev. Cell. 2002;2:617–627. doi: 10.1016/s1534-5807(02)00163-6. [DOI] [PubMed] [Google Scholar]
- Belgardt B.F., Husch A., Rother E., Ernst M.B., Wunderlich F.T., Hampel B., Klöckener T., Alessi D., Kloppenburg P., Brüning J.C. PDK1 deficiency in POMC-expressing cells reveals FOXO1-dependent and -independent pathways in control of energy homeostasis and stress response. Cell Metab. 2008;7:291–301. doi: 10.1016/j.cmet.2008.01.006. [DOI] [PubMed] [Google Scholar]
- Calnan D.R., Brunet A. The FoxO code. Oncogene. 2008;27:2276–2288. doi: 10.1038/onc.2008.21. [DOI] [PubMed] [Google Scholar]
- Cianfarani F., Bernardini S., De Luca N., Dellambra E., Tatangelo L., Tiveron C., Niessen C.M., Zambruno G., Castiglia D., Odorisio T. Impaired keratinocyte proliferative and clonogenic potential in transgenic mice overexpressing 14-3-3σ in the epidermis. J. Invest. Dermatol. 2011;131:1821–1829. doi: 10.1038/jid.2011.137. [DOI] [PubMed] [Google Scholar]
- Della Gatta G., Bansal M., Ambesi-Impiombato A., Antonini D., Missero C., di Bernardo D. Direct targets of the TRP63 transcription factor revealed by a combination of gene expression profiling and reverse engineering. Genome Res. 2008;18:939–948. doi: 10.1101/gr.073601.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis G., Jr., Sherman B.T., Hosack D.A., Yang J., Gao W., Lane H.C., Lempicki R.A. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:3. [PubMed] [Google Scholar]
- Dey-Guha I., Wolfer A., Yeh A.C., G Albeck J., Darp R., Leon E., Wulfkuhle J., Petricoin E.F., 3rd, Wittner B.S., Ramaswamy S. Asymmetric cancer cell division regulated by AKT. Proc. Natl. Acad. Sci. USA. 2011;108:12845–12850. doi: 10.1073/pnas.1109632108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferone G., Thomason H.A., Antonini D., De Rosa L., Hu B., Gemei M., Zhou H., Ambrosio R., Rice D.P., Acampora D. Mutant p63 causes defective expansion of ectodermal progenitor cells and impaired FGF signalling in AEC syndrome. EMBO Mol. Med. 2012;4:192–205. doi: 10.1002/emmm.201100199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferone G., Mollo M.R., Thomason H.A., Antonini D., Zhou H., Ambrosio R., De Rosa L., Salvatore D., Getsios S., van Bokhoven H. p63 control of desmosome gene expression and adhesion is compromised in AEC syndrome. Hum. Mol. Genet. 2013;22:531–543. doi: 10.1093/hmg/dds464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda M., Jones J.E., Olson D., Hill J., Lee C.E., Gautron L., Choi M., Zigman J.M., Lowell B.B., Elmquist J.K. Monitoring FoxO1 localization in chemically identified neurons. J. Neurosci. 2008;28:13640–13648. doi: 10.1523/JNEUROSCI.4023-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomis R.R., Alarcón C., He W., Wang Q., Seoane J., Lash A., Massagué J. A FoxO-Smad synexpression group in human keratinocytes. Proc. Natl. Acad. Sci. USA. 2006;103:12747–12752. doi: 10.1073/pnas.0605333103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner M., Wenk J., Nenci A., Pasparakis M., Scharffetter-Kochanek K., Smyth N., Peters T., Kess D., Holtkötter O., Shephard P. Keratin 14 Cre transgenic mice authenticate keratin 14 as an oocyte-expressed protein. Genesis. 2004;38:176–181. doi: 10.1002/gene.20016. [DOI] [PubMed] [Google Scholar]
- Halet G., Carroll J. Rac activity is polarized and regulates meiotic spindle stability and anchoring in mammalian oocytes. Dev. Cell. 2007;12:309–317. doi: 10.1016/j.devcel.2006.12.010. [DOI] [PubMed] [Google Scholar]
- Hermeking H., Lengauer C., Polyak K., He T.C., Zhang L., Thiagalingam S., Kinzler K.W., Vogelstein B. 14-3-3 sigma is a p53-regulated inhibitor of G2/M progression. Mol. Cell. 1997;1:3–11. doi: 10.1016/s1097-2765(00)80002-7. [DOI] [PubMed] [Google Scholar]
- Jacobs F.M., van der Heide L.P., Wijchers P.J., Burbach J.P., Hoekman M.F., Smidt M.P. FoxO6, a novel member of the FoxO class of transcription factors with distinct shuttling dynamics. J. Biol. Chem. 2003;278:35959–35967. doi: 10.1074/jbc.M302804200. [DOI] [PubMed] [Google Scholar]
- Jensen K.S., Binderup T., Jensen K.T., Therkelsen I., Borup R., Nilsson E., Multhaupt H., Bouchard C., Quistorff B., Kjaer A. FoxO3A promotes metabolic adaptation to hypoxia by antagonizing Myc function. EMBO J. 2011;30:4554–4570. doi: 10.1038/emboj.2011.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King K.E., Ponnamperuma R.M., Yamashita T., Tokino T., Lee L.A., Young M.F., Weinberg W.C. deltaNp63alpha functions as both a positive and a negative transcriptional regulator and blocks in vitro differentiation of murine keratinocytes. Oncogene. 2003;22:3635–3644. doi: 10.1038/sj.onc.1206536. [DOI] [PubMed] [Google Scholar]
- Kitamura T., Feng Y., Kitamura Y.I., Chua S.C., Jr., Xu A.W., Barsh G.S., Rossetti L., Accili D. Forkhead protein FoxO1 mediates Agrp-dependent effects of leptin on food intake. Nat. Med. 2006;12:534–540. doi: 10.1038/nm1392. [DOI] [PubMed] [Google Scholar]
- Kitamura T., Kitamura Y.I., Funahashi Y., Shawber C.J., Castrillon D.H., Kollipara R., DePinho R.A., Kitajewski J., Accili D. A Foxo/Notch pathway controls myogenic differentiation and fiber type specification. J. Clin. Invest. 2007;117:2477–2485. doi: 10.1172/JCI32054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloet D.E., Burgering B.M. The PKB/FOXO switch in aging and cancer. Biochim. Biophys. Acta. 2011;1813:1926–1937. doi: 10.1016/j.bbamcr.2011.04.003. [DOI] [PubMed] [Google Scholar]
- Koster M.I., Roop D.R. Mechanisms regulating epithelial stratification. Annu. Rev. Cell Dev. Biol. 2007;23:93–113. doi: 10.1146/annurev.cellbio.23.090506.123357. [DOI] [PubMed] [Google Scholar]
- Koster M.I., Kim S., Huang J., Williams T., Roop D.R. TAp63alpha induces AP-2gamma as an early event in epidermal morphogenesis. Dev. Biol. 2006;289:253–261. doi: 10.1016/j.ydbio.2005.10.041. [DOI] [PubMed] [Google Scholar]
- Kouwenhoven E.N., van Heeringen S.J., Tena J.J., Oti M., Dutilh B.E., Alonso M.E., de la Calle-Mustienes E., Smeenk L., Rinne T., Parsaulian L. Genome-wide profiling of p63 DNA-binding sites identifies an element that regulates gene expression during limb development in the 7q21 SHFM1 locus. PLoS Genet. 2010;6:e1001065. doi: 10.1371/journal.pgen.1001065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechler T., Fuchs E. Asymmetric cell divisions promote stratification and differentiation of mammalian skin. Nature. 2005;437:275–280. doi: 10.1038/nature03922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefkimmiatis K., Caratozzolo M.F., Merlo P., D’Erchia A.M., Navarro B., Levrero M., Sbisa’ E., Tullo A. p73 and p63 sustain cellular growth by transcriptional activation of cell cycle progression genes. Cancer Res. 2009;69:8563–8571. doi: 10.1158/0008-5472.CAN-09-0259. [DOI] [PubMed] [Google Scholar]
- Lu W., Casanueva M.O., Mahowald A.P., Kato M., Lauterbach D., Ferguson E.L. Niche-associated activation of rac promotes the asymmetric division of Drosophila female germline stem cells. PLoS Biol. 2012;10:e1001357. doi: 10.1371/journal.pbio.1001357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills A.A., Zheng B., Wang X.J., Vogel H., Roop D.R., Bradley A. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature. 1999;398:708–713. doi: 10.1038/19531. [DOI] [PubMed] [Google Scholar]
- Nakae J., Barr V., Accili D. Differential regulation of gene expression by insulin and IGF-1 receptors correlates with phosphorylation of a single amino acid residue in the forkhead transcription factor FKHR. EMBO J. 2000;19:989–996. doi: 10.1093/emboj/19.5.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemoto S., Fergusson M.M., Finkel T. Nutrient availability regulates SIRT1 through a forkhead-dependent pathway. Science. 2004;306:2105–2108. doi: 10.1126/science.1101731. [DOI] [PubMed] [Google Scholar]
- Nguyen B.C., Lefort K., Mandinova A., Antonini D., Devgan V., Della Gatta G., Koster M.I., Zhang Z., Wang J., Tommasi di Vignano A. Cross-regulation between Notch and p63 in keratinocyte commitment to differentiation. Genes Dev. 2006;20:1028–1042. doi: 10.1101/gad.1406006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partridge L., Brüning J.C. Forkhead transcription factors and ageing. Oncogene. 2008;27:2351–2363. doi: 10.1038/onc.2008.28. [DOI] [PubMed] [Google Scholar]
- Peng X.D., Xu P.Z., Chen M.L., Hahn-Windgassen A., Skeen J., Jacobs J., Sundararajan D., Chen W.S., Crawford S.E., Coleman K.G., Hay N. Dwarfism, impaired skin development, skeletal muscle atrophy, delayed bone development, and impeded adipogenesis in mice lacking Akt1 and Akt2. Genes Dev. 2003;17:1352–1365. doi: 10.1101/gad.1089403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potente M., Urbich C., Sasaki K., Hofmann W.K., Heeschen C., Aicher A., Kollipara R., DePinho R.A., Zeiher A.M., Dimmeler S. Involvement of Foxo transcription factors in angiogenesis and postnatal neovascularization. J. Clin. Invest. 2005;115:2382–2392. doi: 10.1172/JCI23126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulson N.D., Lechler T. Robust control of mitotic spindle orientation in the developing epidermis. J. Cell Biol. 2010;191:915–922. doi: 10.1083/jcb.201008001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt H.C., Yaffe M.B. Kinases that control the cell cycle in response to DNA damage: Chk1, Chk2, and MK2. Curr. Opin. Cell Biol. 2009;21:245–255. doi: 10.1016/j.ceb.2009.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senoo M., Pinto F., Crum C.P., McKeon F. p63 Is essential for the proliferative potential of stem cells in stratified epithelia. Cell. 2007;129:523–536. doi: 10.1016/j.cell.2007.02.045. [DOI] [PubMed] [Google Scholar]
- Stachelscheid H., Ibrahim H., Koch L., Schmitz A., Tscharntke M., Wunderlich F.T., Scott J., Michels C., Wickenhauser C., Haase I. Epidermal insulin/IGF-1 signalling control interfollicular morphogenesis and proliferative potential through Rac activation. EMBO J. 2008;27:2091–2101. doi: 10.1038/emboj.2008.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stöhr O., Schilbach K., Moll L., Hettich M.M., Freude S., Wunderlich F.T., Ernst M., Zemva J., Brüning J.C., Krone W. Insulin receptor signaling mediates APP processing and β-amyloid accumulation without altering survival in a transgenic mouse model of Alzheimer’s disease. Age (Dordr) 2013;35:83–101. doi: 10.1007/s11357-011-9333-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tothova Z., Gilliland D.G. FoxO transcription factors and stem cell homeostasis: insights from the hematopoietic system. Cell Stem Cell. 2007;1:140–152. doi: 10.1016/j.stem.2007.07.017. [DOI] [PubMed] [Google Scholar]
- Truong A.B., Khavari P.A. Control of keratinocyte proliferation and differentiation by p63. Cell Cycle. 2007;6:295–299. doi: 10.4161/cc.6.3.3753. [DOI] [PubMed] [Google Scholar]
- Vanbokhoven H., Melino G., Candi E., Declercq W. p63, a story of mice and men. J. Invest. Dermatol. 2011;131:1196–1207. doi: 10.1038/jid.2011.84. [DOI] [PubMed] [Google Scholar]
- Westfall M.D., Mays D.J., Sniezek J.C., Pietenpol J.A. The Delta Np63 alpha phosphoprotein binds the p21 and 14-3-3 sigma promoters in vivo and has transcriptional repressor activity that is reduced by Hay-Wells syndrome-derived mutations. Mol. Cell. Biol. 2003;23:2264–2276. doi: 10.1128/MCB.23.7.2264-2276.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams S.E., Beronja S., Pasolli H.A., Fuchs E. Asymmetric cell divisions promote Notch-dependent epidermal differentiation. Nature. 2011;470:353–358. doi: 10.1038/nature09793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang A., Kaghad M., Wang Y., Gillett E., Fleming M.D., Dötsch V., Andrews N.C., Caput D., McKeon F. p63, a p53 homolog at 3q27-29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol. Cell. 1998;2:305–316. doi: 10.1016/s1097-2765(00)80275-0. [DOI] [PubMed] [Google Scholar]
- Yang A., Schweitzer R., Sun D., Kaghad M., Walker N., Bronson R.T., Tabin C., Sharpe A., Caput D., Crum C., McKeon F. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature. 1999;398:714–718. doi: 10.1038/19539. [DOI] [PubMed] [Google Scholar]
- Yang A., Zhu Z., Kapranov P., McKeon F., Church G.M., Gingeras T.R., Struhl K. Relationships between p63 binding, DNA sequence, transcription activity, and biological function in human cells. Mol. Cell. 2006;24:593–602. doi: 10.1016/j.molcel.2006.10.018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.