Abstract
Previous studies have shown that core leptosporangiates, the most species-rich group of extant ferns (monilophytes), have a distinct plastid genome (plastome) organization pattern from basal fern lineages. However, the details of genome structure transformation from ancestral ferns to core leptosporangiates remain unclear because of limited plastome data available. Here, we have determined the complete chloroplast genome sequences of Lygodium japonicum (Lygodiaceae), a member of schizaeoid ferns (Schizaeales), and Marsilea crenata (Marsileaceae), a representative of heterosporous ferns (Salviniales). The two species represent the sister and the basal lineages of core leptosporangiates, respectively, for which the plastome sequences are currently unavailable. Comparative genomic analysis of all sequenced fern plastomes reveals that the gene order of L. japonicum plastome occupies an intermediate position between that of basal ferns and core leptosporangiates. The two exons of the fern ndhB gene have a unique pattern of intragenic copy number variances. Specifically, the substitution rate heterogeneity between the two exons is congruent with their copy number changes, confirming the constraint role that inverted repeats may play on the substitution rate of chloroplast gene sequences.
Keywords: chloroplast genome, core leptosporangiates, schizaeoid ferns, heterosporous ferns, ndhB, intragenic rate heterogeneity
Leptosporangiates are the most diverse group of nonflowering vascular plants, with more than 11,000 species and a total of seven orders identified in the most recent classification system of living ferns (monilophytes) (Pryer et al. 2004; Smith et al. 2006; Christenhusz et al. 2011). Three most derived leptosporangiate orders, that is, Salviniales (heterosporous ferns), Cyatheales (tree ferns), and Polypodiales (polypods), comprise a large monophyletic clade called core leptosporangiates (Pryer et al. 2004). Current knowledge of leptosporangiate plastid genomes (plastomes) is mostly derived from the available complete chloroplast (cp) genome data of tree ferns (Gao et al. 2009) and polypods (Wolf et al. 2003, 2011). The plastomes of the two core leptosporangiate lineages display same gene order, nearly constant gene component, and similar GC content (hereafter the core type plastomes) (Gao et al. 2009). On the other hand, the core type plastomes differ considerably from those of basal fern lineages, including whisk ferns, ophioglossoids, marattioids, and horsetails (hereafter the basal type plastomes) (Roper et al. 2007; Karol et al. 2010; Grewe et al. 2013). For example, in comparison to the basal type plastomes, the core type plastomes have rearranged inverted repeats (IRs) and rpoB to psbZ (BZ) regions, lack five tRNA genes, and contain higher GC contents (Gao et al. 2009, 2011). To trace the evolutionary pathway of gene order transformation between the basal and core type plastomes, two in-depth studies have recently been carried out by generating partial genome sequences from selected representative species (Wolf et al. 2010; Gao et al. 2011), but more complete plastome data from other leptosporangiate lineages are needed to fully understand the evolutionary processes.
In this study, the complete plastome sequences of Lygodium japonicum (Lygodiaceae, a schizaeoid fern) (supplementary fig. S1, Supplementary Material online) (GenBank accession number: KC536645) and Marsilea crenata (Marsileaceae, a heterosporous fern) (supplementary fig. S2, Supplementary Material online) (GenBank accession number: KC536646) have been determined. These two species were selected because they represent the sister and the basal lineage of core leptosporangiates, respectively, but whose plastome sequences are currently unavailable. Among ferns, the 157,260-bp L. japonicum plastome is the largest sequenced to date, whereas Alsophila spinulosa (a tree fern, 156,661 bp) and Cheilanthes lindheimeri (a polypod fern, 155,770 bp) rank second and third (supplementary table S1, Supplementary Material online). Except for Angiopteris evecta (a marattioid fern), the expanded genome sizes of Lygodium, Alsophila, and Cheilanthes are due to relatively larger IRs and noncoding sequences compared with other ferns. The A. evecta plastome has the longest noncoding sequences because of the pseudogenization of a large gene ycf1. Among leptosporangiates, the shortest IRs are found in the plastome of M. crenata, leading to its relatively small genome size.
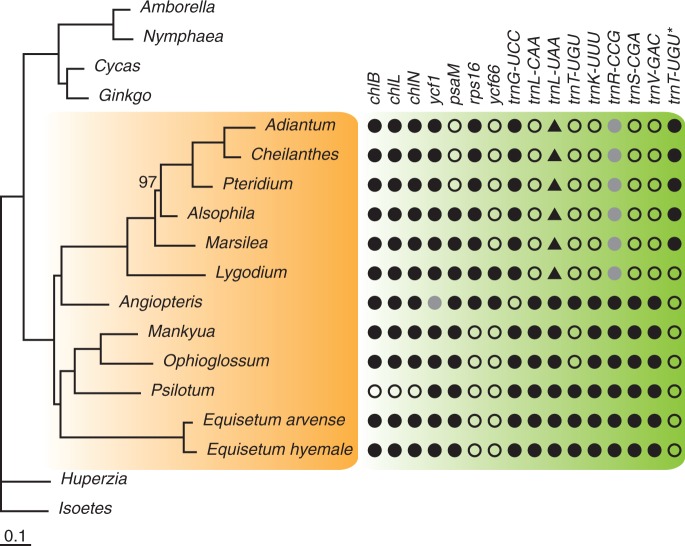
A total of 86 protein genes are encoded in the L. japonicum plastome, which is the only one that contains all the protein-coding genes previously found in ferns. All other sequenced fern plastomes were found to lose certain genes with numbers ranging from 1 to 5 (fig. 1). Some gene loss events seem to be phylogenetically informative, whereas others are not. The three chlorophyll biosynthesis genes (chlB, chlL, and chlN) disappear in Psilotum nudum (a whisk fern), ycf1 is pseudogenized in Angiopteris, and psaM is lost in all sequenced polypods. All these genes have also been parallelly lost in some or all angiosperm plastomes (Jansen et al. 2007). Our previous study has indicated that ycf66 is highly unstable in ferns and has been independently lost at least four times (Gao et al. 2011). The presence of rps16 in Angiopteris and leptosporangiates and absence in other three fern lineages (whisk ferns, ophioglossoid ferns, and horsetails) (Grewe et al. 2013) support a basal dichotomy within ferns (fig. 1). It is unexpected that the intron of rpoC1 gene has completely disappeared in the L. japonicum plastome. This intron has undergone multiple independent losses in angiosperms (Downie et al. 1996; Jansen et al. 2007).
Fig. 1.—
ML tree of 18 taxa of 87 plastid genes. Support values are only shown for nodes with bootstrap values less than 100%. The changes of gene contents among ferns are shown on the left. The novel intron-containing trnT-UGU is marked by a star. The filled and open circles indicate the genes present and absent, respectively, in the corresponding species. The gray circles denote pseudogenes. The filled triangles indicate that the trnL-UAA appears to be trnL-CAA in these species.
The cp tRNA contents are variable in ferns. All the sequenced plastomes of leptosporangiates have lost five tRNAs in comparison to the other four fern lineages (fig. 1). It is noteworthy that, coupled with the loss of trnL-CAA, the intron-containing trnL-UAA has turned to trnL-CAA via a one-base anticodon mutation. This tRNA gene alteration may have been caused by the pressure of codon translation. The trnR-CCG has decayed in both of the L. japonicum and M. crenata plastomes. Our previous studies have detected degraded trnR-CCG in tree ferns and polypods but intact one in Gleicheniales, Hymenophyllales, and Osmundales (Gao et al. 2009). Therefore, the degradation of trnR-CCG represents a common feature shared by both the core leptosporangiates and their sister group. An open question is why the degraded trnR-CCG still exists in these groups. One possible explanation is that the degraded versions of trnR-CCG have developed some new structural or biochemical functions related or unrelated to tRNA-Arg. For instance, the degraded trnR-CCG in the Adiantum plastome (trnSeC-UCA) has been suggested to be a selenocysteine tRNA (Wolf et al. 2003), whereas the one in tree ferns (trnR-UCG) appears to still be an arginine tRNA (Gao et al. 2009). The relict of the trnR-CCG gene contains short palindromic sequences that may form stem-loop structures (Gao et al. 2010), known to be hot spots for mutations and associated with short inversions (Kim and Lee 2004; Gao et al. 2011).
There is an intron-free trnT-UGU located between rps4 and trnL-UAA that is widespread in land plant plastomes but absent in all sequenced plastomes of leptosporangiates and ophioglossoids (fig. 1). Notably, a novel intron-containing trnT-UGU has been annotated between ndhB and trnR-ACG in the Adiantum plastome (Wolf et al. 2003). Similar sequences have also been found in other polypods (Wolf et al. 2011) and tree ferns (Gao et al. 2009). Nevertheless, the sequences of the intron-free and intron-containing trnT-UGU share no apparent similarity (supplementary fig. S3, Supplementary Material online). For the L. japonicum and M. crenata plastomes, no intron-containing trnT-UGU can be detected by using tRNA prediction programs. We have manually identified two exons in the corresponding region of the M. crenata plastome based on sequence similarity (supplementary fig. S3, Supplementary Material online). Thus, this gene appears to be a new component specific to core leptosporangiates. However, because no convincing experimental evidence is available, we are unsure whether the novel trnT-UGU is functional in nature.
The L. japonicum plastome has a unique gene order differing from either the core- or basal-type plastomes. Its IR regions show a complete synteny with that of the core type, whereas BZ regions have the same order as the basal type (Wolf et al. 2010; Gao et al. 2011). Hence, the L. japonicum plastome reflects an intermediate between the core and basal types.
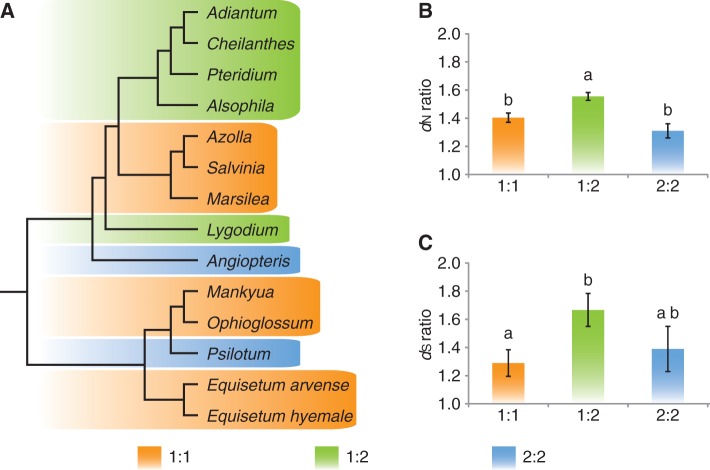
The M. crenata plastome gene order is generally syntenic with the core type plastome (tree ferns and polypods). Their only difference is that the ndhB exists as a complete single copy gene in the M. crenata plastome but whose second exon is duplicated in the IRs of L. japonicum, tree fern, and polypod plastomes. To test whether this feature is specific to M. crenata, we have determined the sequences covering the two boundaries between IRs and the large single copy (LSC) region (rrn23 to rpl2 and rrn23 to matK) in Salvinia molesta (GenBank accession number: KC626075) and Azolla caroliniana (GenBank accession number: KC626076), which represent two other genera in heterosporous ferns. Our results indicate that in all heterosporous ferns, ndhB is single copied and located in the LSC. Of note, the copy status of ndhB in the four basal fern lineages is variable. ndhB is complete and resides in IRs in whisk ferns and marattioids. In contrast, it is translocated to LSC due to IR contractions in ophioglossoids and horsetails. In summary, there are three types of ndhB copies in ferns: one copy of exon 1 plus one copy of exon 2 (1:1 type), one copy of exon 1 plus two copies of exon 2 (1:2 type), and two copies of exon 1 plus two copies of exon 2 (2:2 type) (fig. 2).
Fig. 2.—
The copy number variance of the two exons of ndhB gene in ferns and its impact on evolutionary rate. 1:1, one copy exon 1 plus one copy exon 2; 1:2, one copy of exon 1 plus two copies of exon 2; and 2:2, two copies of exon 1 plus two copies of exon 2. (A) The distribution of copy status of different ndhB exons among ferns. (B) The ratio of nonsynonymous (dN) substitution rates between exon 1 and exon 2. (C) The ratio of synonymous (dS) substitution rates between exon 1 and exon 2. (B) and (C), means labeled with different letters are significantly different (Tukey HSD test, P < 0.05).
For each ndhB exon of ferns, we have calculated the nonsynonymous (dN) and synonymous (dS) substitution rates separately. Then the ratio of dN and of dS between the two exons in a given species has been determined (fig. 2). Our results indicate that for both dN and dS ratios, the 1:1 and 2:2 types show no significant difference, because the copy status of exon 1 and exon 2 is the same for each of the two types. However, the dN and dS ratios of the 1:2 type are higher than the 1:1 and 2:2 types, although the difference is not statistically significant for the dS ratio. Therefore, genomic locations (LSC or IR region) and copy number ratios (1:1, 1:2, and 2:2) of the two exons form the main sources of their evolutionary rate divergence.
The impact of IRs on the evolutionary rate of cp genes has been evaluated by examining genes that exhibit copy number changes caused by IR loss or rearrangement (Perry and Wolfe 2002; Sen et al. 2012). However, the results may be plagued by lineage-specific rate heterogeneity (Muse and Gaut 1997; Lopez et al. 2002), because high heterotachous genes are common in the plastome (Lockhart et al. 2006; Wu et al. 2011). For this reason, the variance of evolutionary rate for a given gene among different species may be affected by the lineage effect rather than the gene’s location. In this respect, the fern ndhB gene provides a unique opportunity with its intragenic location change between the two exons. Comparison of the ratio of substitution rates between exon 1 and exon 2 within the same gene should avoid the lineage effect.
Materials and Methods
Genome Sequencing, Assembly, and Annotation
Young leaves of L. japonicum and M. crenata were collected from the plants growing in Wuhan Botanical Garden, Chinese Academy of Sciences. Total DNA was extracted using DNeasy Plant Mini Kit (QIAGEN) and sequenced by HiSeq 2000 Sequencing System (Illumina). A total of 15.2 and 13.7 million paired-end sequence reads of 100 bp were generated for L. japonicum and M. crenata, respectively. After culling the low-quality and too short (<50 bp) sequences, 2.9 and 2.4 GB sequences were acquired for L. japonicum and M. crenata, respectively. For each species, the reads were de novo assembled with Velvet (Zerbino and Birney 2008). To enrich the high-coverage data from cpDNAs, the coverage cutoff value was set as 30 (-cov_cutoff 30). To identify the chloroplast contigs, all the returned contigs were blasted to previously reported fern cp genomes. A total of six and nine contigs with lengths of 132,060 and 132,339 bp were found to be derived from cpDNAs for L. japonicum and M. crenata, respectively. The putative gaps were filled by polymerase chain reaction (PCR) amplification based on the cp contig sequences. The PCR and DNA sequencing were as described previously (Gao et al. 2009).
The L. japonicum and M. crenata plastomes were annotated by performing Dual Organellar GenoMe Annotator (DOGMA) (Wyman et al. 2004). From this initial annotation, putative starts, stops, and intron positions were determined by comparisons with homologous genes in other cp genomes and by considering the possibility of RNA editing, which can modify the start and stop positions. The tRNA genes were detected by two online programs, ARAGORN (Laslett and Canback 2004) and tRNAscan-SE (Lowe and Eddy 1997). The circular gene maps were drawn using OGDRAW (Lohse et al. 2007).
Young leaves of S. molesta and Azo. caroliniana were acquired from Wuhan Botanical Garden, Chinese Academy of Sciences. Total DNAs were extracted using DNeasy Plant Mini Kit. Primers were designed based on the rrn23, rpl2, ndhB, and matK sequences of the M. crenata plastomes and other heterosporous ferns in GenBank. The PCR and DNA sequencing were as described previously (Gao et al. 2009).
Phylogenomic Analysis
Eighty-three protein-coding and four rRNA sequences were extracted from 12, 4, and 2 sequenced plastomes of ferns, seed plants, and lycophytes, respectively. Multiple sequence alignments for each gene were conducted using MUSCLE (Edgar 2004) as implemented in MEGA5 (Tamura et al. 2011). Three independent maximum likelihood (ML) runs were conducted in GARLI (Zwickl 2006), using the automated stopping criterion, and terminating the search when the -ln score remained constant for 20,000 consecutive generations. A total of 100 ML bootstrap replicates were also performed using GARLI with the same parameters.
Nucleotide Substitution Rates of the Two Exons of ndhB
To obtain an estimate of the values of dN and dS in the two exons of ndhB gene in ferns, we made pairwise comparisons between 14 ferns and 3 lycophyte outgroups (NC_006861, NC_014675, and NC_013086). For each exon, the sequences were aligned by reverse translation of MUSCLE (Edgar 2004) alignments of the corresponding protein sequences. The codeml program from the PAML package (Yang 2007) was used to calculate dN and dS using the ML method (using the options seqtype = 1, runmode = -2 and CodonFreq = 2 in the codeml.ctl files). The Tukey HSD test (JMP 9.0) was used to evaluate differences of the ratios of dN and dS between the two exons among different exon copy number types.
Supplementary Material
Supplementary figures S1–S3 and table S1 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
The authors thank Jia Li and Chuan-De Wang for experimental assistance. They are deeply indebted to the anonymous reviewer for his/her valuable comments to improve the manuscript. This work was supported by National Natural Science Foundation of China (nos. 30970290, 31070594, and 31000171), Knowledge Innovation Program of the Chinese Academy of Sciences (nos. KSCX2-EW-J-20 and KSCX2-YW-Z-0940), Basic Research Project of the Department of Science and Technology of Zhuhai city, China (No. 2012D0401990031), and the Key Laboratory of Plant Germplasm Enhancement and Specialty Agriculture, Wuhan Botanical Garden, Chinese Academy of Sciences.
Literature Cited
- Christenhusz MJM, Zhang X-C, Schneider H. A linear sequence of extant families and genera of lycophytes and ferns. Phytotaxa. 2011;19:7–54. [Google Scholar]
- Downie SR, Llanas E, Katz-Downie DS. Multiple independent losses of the rpoC1 intron in angiosperm chloroplast DNA’s. Syst Bot. 1996;21:135–151. [Google Scholar]
- Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L, Su Y-J, Wang T. Plastid genome sequencing, comparative genomics, and phylogenomics: current status and prospects. J Syst Evol. 2010;48:77–93. [Google Scholar]
- Gao L, Yi X, Yang Y-X, Su Y-J, Wang T. Complete chloroplast genome sequence of a tree fern Alsophila spinulosa: insights into evolutionary changes in fern chloroplast genomes. BMC Evol Biol. 2009;9:130. doi: 10.1186/1471-2148-9-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L, Zhou Y, Wang Z-W, Su Y-J, Wang T. Evolution of the rpoB-psbZ region in fern plastid genomes: notable structural rearrangements and highly variable intergenic spacers. BMC Plant Biol. 2011;11:64. doi: 10.1186/1471-2229-11-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewe F, Guo W, Gubbels EA, Hansen AK, Mower JP. Complete plastid genomes from Ophioglossum californicum, Psilotum nudum, and Equisetum hyemale reveal an ancestral land plant genome structure and resolve the position of Equisetales among monilophytes. BMC Evol Biol. 2013;13:8. doi: 10.1186/1471-2148-13-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen RK, et al. Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc Natl Acad Sci U S A. 2007;104:19369–19374. doi: 10.1073/pnas.0709121104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karol K, et al. Complete plastome sequences of Equisetum arvense and Isoetes flaccida: implications for phylogeny and plastid genome evolution of early land plant lineages. BMC Evol Biol. 2010;10:321. doi: 10.1186/1471-2148-10-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KJ, Lee HL. Complete chloroplast genome sequences from Korean ginseng (Panax schinseng Nees) and comparative analysis of sequence evolution among 17 vascular plants. DNA Res. 2004;11:247–261. doi: 10.1093/dnares/11.4.247. [DOI] [PubMed] [Google Scholar]
- Laslett D, Canback B. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 2004;32:11–16. doi: 10.1093/nar/gkh152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockhart P, et al. Heterotachy and tree building: a case study with plastids and eubacteria. Mol Biol Evol. 2006;23:40–45. doi: 10.1093/molbev/msj005. [DOI] [PubMed] [Google Scholar]
- Lohse M, Drechsel O, Bock R. OrganellarGenomeDRAW (OGDRAW): a tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr Genet. 2007;52:267–274. doi: 10.1007/s00294-007-0161-y. [DOI] [PubMed] [Google Scholar]
- Lopez P, Casane D, Philippe H. Heterotachy, an important process of protein evolution. Mol Biol Evol. 2002;19:1–7. doi: 10.1093/oxfordjournals.molbev.a003973. [DOI] [PubMed] [Google Scholar]
- Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muse SV, Gaut BS. Comparing patterns of nucleotide substitution rates among chloroplast loci using the relative ratio test. Genetics. 1997;146:393–399. doi: 10.1093/genetics/146.1.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry AS, Wolfe KH. Nucleotide substitution rates in legume chloroplast DNA depend on the presence of the inverted repeat. J Mol Evol. 2002;55:501–508. doi: 10.1007/s00239-002-2333-y. [DOI] [PubMed] [Google Scholar]
- Pryer KM, et al. Phylogeny and evolution of ferns (monilophytes) with a focus on the early leptosporangiate divergences. Am J Bot. 2004;91:1582–1598. doi: 10.3732/ajb.91.10.1582. [DOI] [PubMed] [Google Scholar]
- Roper JM, et al. The complete plastid genome sequence of Angiopteris evecta (G. Forst.) Hoffm. (Marattiaceae). Am Fern J. 2007;97:95–106. [Google Scholar]
- Sen L, Fares M, Su Y-J, Wang T. Molecular evolution of psbA gene in ferns: unraveling selective pressure and co-evolutionary pattern. BMC Evol Biol. 2012;12:145. doi: 10.1186/1471-2148-12-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AR, et al. A classification for extant ferns. Taxonomy. 2006;55:705–731. [Google Scholar]
- Tamura K, et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf PG, Roper JM, Duffy AM. The evolution of chloroplast genome structure in ferns. Genome. 2010;53:731–738. doi: 10.1139/g10-061. [DOI] [PubMed] [Google Scholar]
- Wolf PG, Rowe CA, Sinclair RB, Hasebe M. Complete nucleotide sequence of the chloroplast genome from a leptosporangiate fern, Adiantum capillus-veneris L. DNA Res. 2003;10:59–65. doi: 10.1093/dnares/10.2.59. [DOI] [PubMed] [Google Scholar]
- Wolf PG, et al. The evolution of chloroplast genes and genomes in ferns. Plant Mol Biol. 2011;76:251–261. doi: 10.1007/s11103-010-9706-4. [DOI] [PubMed] [Google Scholar]
- Wu CS, Wang YN, Hsu CY, Lin CP, Chaw SM. Loss of different inverted repeat copies from the chloroplast genomes of Pinaceae and Cupressophytes and influence of heterotachy on the evaluation of gymnosperm phylogeny. Genome Biol Evol. 2011;3:1284–1295. doi: 10.1093/gbe/evr095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyman SK, Jansen RK, Boore JL. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 2004;20:3252–3255. doi: 10.1093/bioinformatics/bth352. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwickl DJ. Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion [dissertation] [Austin (TX)]: University of Texas at Austin; 2006. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.