Abstract
The p73 transcription factor is one of the members of the p53 family of tumor suppressors with unique biological functions in processes like neurogenesis, embryonic development and differentiation. For this reason, p73 activity is tightly regulated by multiple mechanisms, including transcription and post-translational modifications. Here, we identified a novel regulatory loop between TAp73 and the E3 ubiquitin ligase tripartite motif protein 32 (TRIM32). TRIM32, a new direct p73 transcriptional target in the context of neural progenitor cells, is differentially regulated by p73. Although TAp73 binds to the TRIM32 promoter and activates its expression, TAp73-induced TRIM32 expression is efficiently repressed by DNp73. TRIM32 in turn physically interacts with TAp73 and promotes its ubiquitination and degradation, impairing p73-dependent transcriptional activity. This mutual regulation between p73 and TRIM32 constitutes a novel feedback loop, which might have important implications in central nervous system development as well as relevance in oncogenesis, and thus emerges as a possible therapeutic target.
Keywords: p73, TRIM32, ubiquitination, transactivation, E3 ligase, neural differentiation
The p53 gene family of transcription factors has a role in the regulation of differentiation, and proliferation of somatic and stem cells, and it also has an important role in tumor suppression.1, 2 Emerging evidence suggests that while p73 share many functional properties with p53, it also has distinct and unique biological functions in processes such as neurogenesis and embryonic development. The TP73 gene exhibits a dual nature, which relies on the existence of TA and DNp73 variants. The TA proteins are transactivation competent,3, 4, 5, 6, 7 whereas the DN isoforms can have dominant-negative function towards p53 and TAp73,8, 9 acting as a major survival factor in postmitotic neurons,10 or transactivating target genes in a cell context-dependent manner.7, 11 Thus, the TA/ΔN differential expression is crucial for determining p73 function. This regulation can operate at transcriptional12 or post-transcriptional levels.13, 14, 15
The compilation of work regarding p73 function in neural development indicates that p73 has a complex multifunctional role in this process. There is strong evidence demonstrating TAp73 induction of neuronal differentiation. In this regard, the hippocampal defects also present in the TAp73−/−-specific mice, together with TAp73 regulation of miR-34a, support TAp73 function in neuronal differentiation.16 Furthermore, overexpression of TAp73 induces neurite outgrowth and expression of neuronal markers in neuroblastoma and pheochromocytoma cell lines3, 17 and spontaneous differentiation of oligodendrocytes.18 In contrast, recent reports have demonstrated p73 requirement, mainly TAp73, in the maintenance of neural stem/progenitor cells (NSC/NPC) self-renewal,19 where it blocks premature differentiation of embryonic neural progenitor cells (NPCs) in a p53-independent manner.20, 21
It is reasonable to consider that p73 would execute some of its multiple functions through the transcriptional regulation of cell context-specific target genes. In this regard, previous work using the human chronic myeloid leukemia cell line, K562, with ectopic ΔNp73 expression7 revealed tripartite motif protein 32 (TRIM32) as a putative p73 transcriptional target. Furthermore, published in silico analyses have also proposed TRIM32 as a TAp73 target.22, 23
TRIM32 belongs to the TRIM-NHL family24 and it has been involved in NPCs' regulation of differentiation and self-renewal during mouse embryonic brain development.25, 26 TRIM32 is asymmetrically distributed in NPC, and segregates with the daughter cell that undergoes neuronal differentiation.26 TRIM32 is retained in the cytoplasm of dividing progenitors and it is translocated to the nucleus to initiate neuronal differentiation.25 TRIM32 also regulates skeletal muscle stem cell differentiation27 as well as RARα-induced differentiation of acute promyelogenous leukemic cells.28 Furthermore, TRIM32 increased expression has been associated with enhanced epidermal carcinogenesis,29 cell growth and migration,30 linking TRIM32 altered regulation to the oncogenic process.
At the molecular level, TRIM32 has two functions:it mediates the polyubiquitination of c-Myc and it binds to the protein Argonaute-1, thereby increasing the activity of differentiation-inducing micro-RNAs like Let-7a.26, 31 Nevertheless, the mechanisms that regulate TRIM32 expression remain unclear, and while TRIM32 regulation via p73 has been predicted by in silico analysis,22 experimental evidence is still lacking.
In this article, we demonstrate for the first time that TRIM32 expression is differentially regulated by p73 at the transcriptional level. Although TAp73 activates TRIM32 expression, ΔNp73 efficiently represses TAp73 transactivation of this promoter. TRIM32 in turn, binds to TAp73, promotes its ubiquitination in a RING-finger-dependent manner and degrades it. Furthermore, TRIM32 overexpression can repress TAp73 transcriptional activation of the p21Cip promoter. Thus, we have identified a novel regulatory feedback loop between TP73 and TRIM32, similar to that of p53 and MDM2.
Results
P73 expression modulates TRIM32 levels
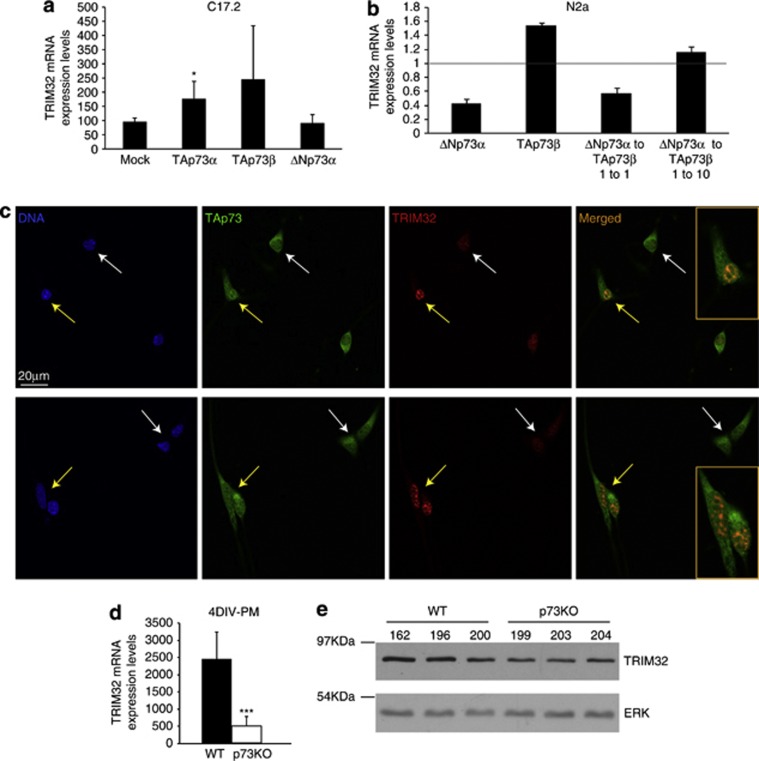
Previous genome-wide expression analysis7 revealed significant downregulation of the neuronal fate determinant TRIM32, suggesting that it could be a p73 transcriptional target. To address this, we first examined whether constitutive expression of TAp73 and/or DNp73 could modulate TRIM32 expression in immortalized mouse NPC C17.2,32 and in the mouse neuroblastoma cells, N2a (Figures 1a and b). TAp73 transfection resulted in a significant upregulation of TRIM32 expression in the case of TAp73α in C17.2 and TAp73β in N2a, by real-time quantitative PCR (qRT-PCR) analysis. Western blot analysis of C17.2 confirmed these results showing a moderate (threefold) but a statistically significant increase in TRIM32 protein level upon ectopic expression of TAp73α (Supplementary Figure 1A, P<0.005). DNp73 transfection alone had either no effect or decreased TRIM32 basal levels. Interestingly, when DNp73 and TAp73 were present at a 1 : 1 ratio, TRIM32 mRNA expression was repressed, indicating a dominant-negative effect of DNp73. Even in the presence of 10-fold levels of TAp73, DNp73 efficiently inhibits TAp73-induced upregulation (Figure 1b and Supplementary Figure 1A). Similar results were obtained in the murine embryonic stem cell line E14TG2a (Supplementary Figure 1B). Both isoforms of TAp73 (α and β) can regulate TRIM32 expression but their effect depends on the cell context. At the RNA level, TAp73α seems to have a more general and consistent effect than TAp73β. Therefore, we used TAp73α throughout this this study.
Figure 1.
P73 expression modulates TRIM32 levels. (a and b) Quantification of TRIM32 expression level by qRT-PCR analysis upon transfection with the indicated expression vectors in C17.2 (a) and N2a (b) cell lines. (c) Confocal microscopy images of WT NSs after 3 DIV-PM immunostained with antibodies against TRIM32 and TAp73. (d and e) qRT-PCR (d) and western blot analysis (e) of TRIM32 levels in NS cultures after 4 DIV-PM. Analysis was performed with data from three independent experiments with at least three embryos of each genotype (mean±standard deviation media (S.D.M.); *P<0.05, ***P<0.005)
To demonstrate the correlation between TAp73 and TRIM32 expression in a physiological in vitro context, we used NPC cultured with the neurosphere (NS) assay,33 as TRIM32 expression and regulation has been previously described in these cultures.25, 26 TAp73 and TRIM32 expression was analyzed by confocal microscopy in NS cultures obtained from olfactory bulbs (OBs) of wild-type (WT) E14.5 embryos. NPCs in the WT cultures presented low, but detectable, TRIM32. Interestingly, TRIM32 levels were lower in cells where TAp73 expression was mostly cytoplasmic, suggesting low or no TAp73 transcriptional activity (Figure 1c, white arrows). However, in cells with nuclear TAp73, TRIM32 expression was enhanced (Figure 1c, yellow arrows). Moreover, in those cells, TAp73 and TRIM32 colocalized within the nucleus.
We next analyzed endogenous TRIM32 levels on NS cultures from Trp73−/− mice (p73 knockout (KO) from now on).21 Comparative analysis by qRT-PCR and western blot revealed that TRIM32 expression levels were lower in p73KO cultures than in WT cells (Figures 1d and e, respectively). Consistently, confocal microscopy analysis showed that while TRIM32 expression in WT-NS was low, but detectable in most of the cells, in the p73KO cultures, only few cells presented detectable TRIM32 expression (Supplementary Figure 1C). Altogether, these data support the idea that TAp73 could be a transcriptional regulator of TRIM32. Furthermore, we found that partial TAp73 knockdown resulted in slight, but significant, downregulation of TRIM32 levels in HEK293 cells (Supplementary Figure 1D).
P73 differentially regulates TRIM32 expression at the transcriptional level
A number of p53/p73-binding sites have been reported in mouse and human TRIM32 promoters by in silico analysis,22 supporting the idea that TRIM32 could be a p73 transcriptional target. Consistently, in silico prediction of p53-responsive elements (p53REs) within the human TRIM32 locus using p53 Family Target Genes (FamTaG) database34 unveiled four p53-binding sites in this gene, two of them located in the 5′-flanking region (at positions −1551 to −1514 and −115 to −84, relative to the transcription start site at exon 1), whereas the other two were within intron 1. Thus, to demonstrate that p73 regulation of TRIM32 was at the transcriptional level, a fragment of the human TRIM32 promoter (−1351 to +270) was amplified (Supplementary Figure 2A) and subcloned into the pGL3-basic plasmid, generating the hTRIM32-luciferase (luc) reporter vector. This construct contains the p53RE identified in the proximal promoter (−115 to −84) (Supplementary Figure 2A, red box). In the murine TRIM32 promoter, another p53RE has been described in this region, which is quite conserved in the human promoter (Supplementary Figure 2A, orange box).
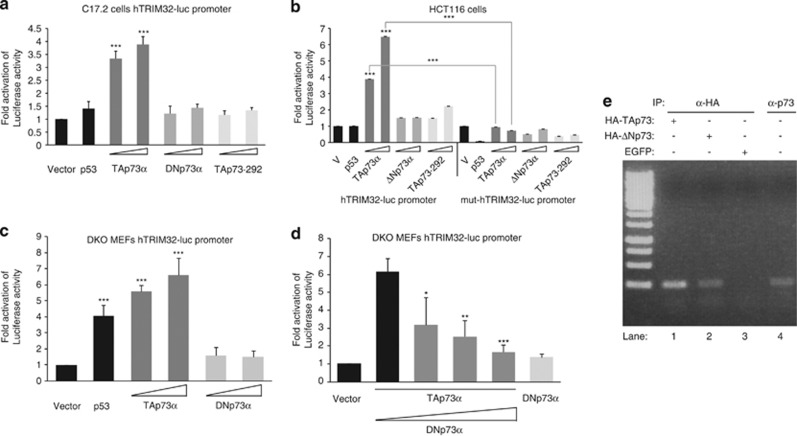
Transcriptional assays in C17.2 cells showed that TAp73α, but not the transcriptional-inactive mutant TAp73-292, significantly activated the hTRIM32-luc reporter, whereas p53 has no significant effect in this cell line (Figure 2a). To determine if this p73 function was limited to neural-like cells, transcriptional assays were performed in HCT116 human colon carcinoma cells, which have undetectable TRIM32 expression (Supplementary Figure 3A). TAp73, but not TAp73-292 nor DNp73, significantly activated the hTRIM32-luc reporter, demonstrating that this regulation was specific and not limited to neural-like cells (Figure 2b). TAp73α, but not TAP73α-292 or DNp73α, transactivates, but less efficiently, the commercially available (GeneCopoeia) murine TRIM32-luc promoter vector in the mouse cell lines, E14TG2a and C17.2 (Supplementary Figures 3B and C, respectively). Furthermore, transcriptional analysis with a mutant hTRIM32-luc, lacking the p53RE (Supplementary Figure 2), revealed a significant decrease in ectopic TAp73-induced activity (Figure 2b). Similar results were obtained in the C17.2 cell line (Supplementary Figure 3D). Altogether, our data demonstrate that TAp73 transcriptional activity over the TRIM32 promoter is specific and requires, at least in part, the p53RE. To address whether TAp73 could modulate TRIM32 promoter in the absence of endogenous p53, and to avoid TA versus DNp73 interactions, we used mouse embryonic fibroblasts deficient for both TP53 and TP73 genes (DKO MEFs). In these cells, both TAp73 and p53, but not DNp73, significantly transactivated the reporter, demonstrating that TRIM32 expression was regulated at the transcriptional level by these p53 family members (Figure 2c). To determine if DNp73 repression of TAp73-induced TRIM32 expression could be detected at the promoter level, we co-transfected DNp73 and TAp73 at various ratios in DKO MEFs and performed reporter assays. We observed a significant decrease of TAp73-induced promoter transactivation in the presence of increasing amounts of DNp73, even at <1 : 1 ratio. This supports the prediction that DNp73 efficiently repress TAp73 transactivation of TRIM32 promoter (Figure 2d). Finally, to unequivocally demonstrate that TRIM32 was a direct transcriptional target of TA and DNp73, we performed chromatin immunoprecipitation (ChIP) assays. Immunoprecipitation with anti-HA antibody (lanes 1 and 2) showed that both ectopic TA and DN p73 bound directly to the predicted p53/p73-binding site of the TRIM32 promoter. Furthermore, endogenous p73 immunoprecipitated with anti-p73 in untransfected cells (lane 4) also bound directly to this site. Altogether, these data demonstrated, for the first time, that TRIM32 is a direct transcriptional target of TAp73 and that TA and DN p73 differentially regulate this gene.
Figure 2.
P73 differentially regulates TRIM32 expression at the transcriptional level. (a–d) Transcriptional analysis with the reporter vector hTRIM32-luc (a, c and d) or mut-hTRIM32-luc (b) co-transfected with the indicated expression vectors in C17.2 (a), HCT116 (b) or p73KO/p53KO (DKO) MEFs (c and d) cells. Co-transfection of TA/DNp73 in (d) ranges from 1 : 0.6 to 1 : 2. (e) ChIP assay. DNA agarose gel of PCRs with primers spanning the putative p73-binding sides at positions −10 to −41 and −57 to −79 are shown. As templates immunoprecipitated chromatin was used. N2a cells were transfected either with expression constructs for hemagglutinin (HA)-TAp73 (lane 1), HA-DNp73 (lane 2), enhanced green fluorescent protein (EGFP) (mock control) (lane 3) or were left untransfected (lane 4). Chromatin was precipitated with an anti-HA antibody (lanes 1–3) or an anti-p73 antibody (lane 4). Association with the TRIM32 promoter is shown by PCR with primers specific for p73-binding site. The analyses were performed with data from at least three independent experiments (mean±S.D.M; *P<0.05, **P<0.01, ***P<0.005)
P73 deficiency results in lower TRIM32 levels
To corroborate that TA and ΔNp73 transcriptional regulation of TRIM32 promoter occurs under physiological conditions in vitro and in vivo, we sought to analyze the effect of p73 deficiency in differentiating NPC cultures and in the brain tissue of WT and p73KO mice.
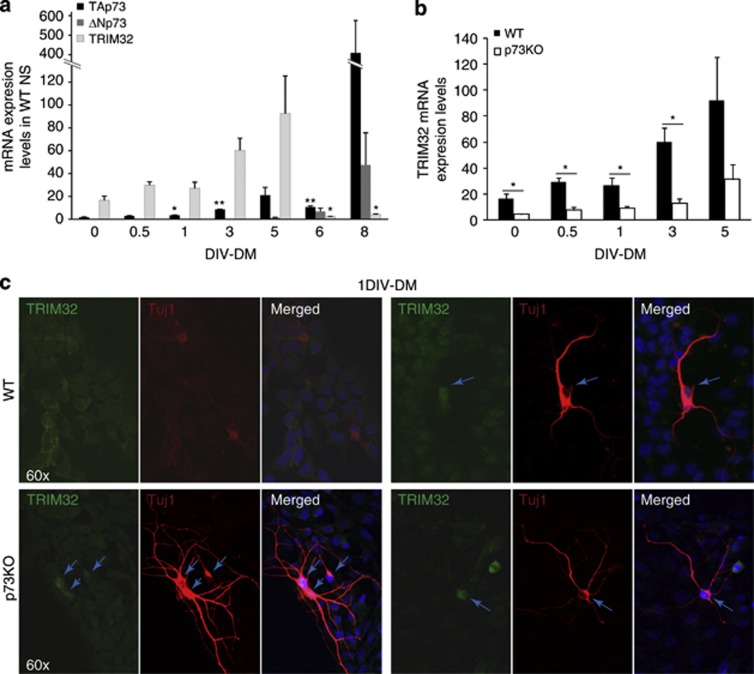
TRIM32 is involved in the regulation of NPC differentiation during mouse embryonic brain development.27 Thus, we analyzed whether p73 and TRIM32 expression kinetics correlated in a physiological and relevant system, such as the differentiation process of embryonic NPC (Figure 3a) and the effect of p73 deficiency on TRIM32 kinetics during this process (Figure 3b). TAp73 is significantly more abundant than DNp73 in NPCs.21 TAp73 expression is upregulated early during the differentiation process and increases steadily up to 5 days in vitro (DIV)-DM. At 6 DIV, TAp73 suffers a slight decrease and then reaches its maximal expression at 8 DIV (Figure 3a, black bars). DNp73 is undetectable during the first days of differentiation, beginning to be expressed at 5 DIV-DM and then increasing steadily up to 8 DIV-DM (Figure 3a, dark gray bars). TRIM32 expression (light gray bars) correlates with the initial upregulation of TAp73 during the first differentiation days, reaching maximal levels at 5 DIV-DM. TRIM32 upregulation coincides with DNp73 lowest expression levels, from 1 DIV-5DIV. Later on, its expression decreases concurrently with DNp73 upregulation, despite the elevated levels of TAp73 (8 DIV), probably reflecting DNp73 repression of TAp73 activity. This expression kinetic is consistent with TA- and DN-p73 differential regulation of TRIM32.
Figure 3.
P73 deficiency results in lower TRIM32 levels in vitro and in vivo. (a–c) NS cultures from p73KO E14.4 or WT E14.4 were analyzed after different DIV cultured under differentiating conditions (DIV-PM). (a and b) Quantitative analysis of TAp73 (black bars), DNp73 (dark gray bars) and TRIM32 (light gray bars) expression kinetics performed by qRT-PCR. (c) Confocal microscopy images of NSs after 1 DIV-DM, immunostained with antibodies against TRIM32 and the neuronal marker Tuj1. Blue arrows indicated cells with TRIM32 nuclear staining and Tuji-positive cell with long neurite extensions. Analysis was performed with data from three independent experiments with at least three embryos of each genotype (mean±standard error media (S.E.M.); *P<0.05, **P<0.01
Comparative analysis of TRIM32 expression in differentiating NPCs from WT and p73KO NS cultures revealed a significant decrease of TRIM32 levels in the absence of p73 at every stage analyzed (Figure 3b), highlighting the relevant role of TAp73 in TRIM32 upregulation during neuronal differentiation. Nevertheless, p73KO cultures express, and upregulate, TRIM32 during this process. Therefore, other p73-independent mechanisms are also regulating TRIM32 expression during NPC differentiation.
We next analyzed TRIM32 upregulation in individual cells in NPCs cultured under conditions supporting progressive neural differentiation and stained for TRIM32, and the neuronal marker, Tuj-1, after 24 h in culture (1 DIV-DM). At such early point of differentiation, WT cultures still showed detectable expression of TRIM32, with few cells beginning to express Tuj-1 presenting short lamellipodia and/or short unbranched processes (Figure 3c, upper left panel). In these cells, TRIM32 expression was mostly cytoplasmic, confirming the expected immature phenotype.25 At this stage of differentiation, we only rarely detected cells with neurite extensions longer than 4 × their soma in WT cultures. In these cases, TRIM32 expression was stronger in the nucleus indicating a mature phenotype (Figure 3, upper right panel, arrow). The majority of the cells in the p73KO cultures presented lower TRIM32 levels than in WT cells (Figure 3c, lower panel). However, as published previously,21 under identical mitotic culture conditions, we detected significantly higher number of Tuj-1-positive cells (2 versus 0.31%, P<0.001) with long and branched neurite extensions, highlighting the premature neuronal differentiation detected in p73KO cultures (Figure 3c, lower panel, arrows). Strikingly, despite the lack of p73, all Tuj-1-positive cells, with neurite extensions, had enhanced TRIM32 expression (24% of this was exclusively nuclear, whereas 52% was nuclear and cytoplasmic), all congruent with a mature neuronal phenotype and supporting the essential role of TRIM32 in neuronal differentiation. These data confirm the existence of p73-independent mechanisms of TRIM32 regulation. Furthermore, it supports the idea that lack of p73 leads to premature embryonic NPCs' differentiation in vitro and underlines the required role of TRIM32 in this process.
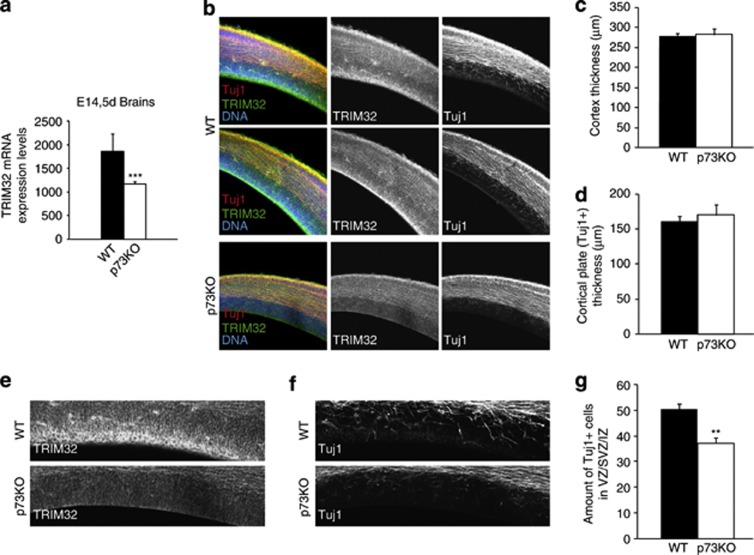
To confirm TRIM32 and TAp73 correlation in an in vivo context, we sought to study TRIM32 expression pattern in E14.5 embryo brains. This time coincides with the peak of mouse cortical neurogenesis, when TRIM32 is strongly expressed in the cortical plate and the ventricular zone, and colocalizes with the expression of Tuj-1.26 qRT-PCR analysis of whole E14.5 brains demonstrated that, whereas p73KO E14.5 embryos expressed TRIM32, there was a significant reduction in total TRIM32 expression levels compared with WT (Figure 4a). This reduction in TRIM32 expression is also observed, albeit diminished, in the brains of P7 young mice (Supplementary Figure S4). The gross anatomy of the cortex seems to be unaffected by the absence of p73, and neither neuronal differentiation per se seems to be strongly affected (Figures 4b–d). Furthermore, TRIM32 levels in the cortex seem to be normal, indicating that, as in vitro, there are p73-independent factors that ensure TRIM32 expression. Interestingly, TRIM32 levels were strongly reduced in the ventricular zone (VZ)/subventricular zone (SVZ) of p73KO animals (Figure 4e). These lower levels of TRIM32 in the VZ/SVZ correlate with the reduced amount of newly generated neurons (Tuj-1) in this region (Figures 4f and g), suggesting that p73 deficiency leads to TRIM32 downregulation affecting the population of NPCs that controls neurogenesis in the VZ/SVZ.
Figure 4.
TRIM32 expression levels are reduced in the brain of p73KO E14.5 mice. (a) Quantitative analysis of TRIM32 expression in E14.5 mice brains. (b–d) Immunostainings of brain sections from WT and p73KO mice labeled with the indicated antibodies and quantification of the thickness of the complete cortex (c) as well as the thickness of the TuJ1-positive cortical plate (d) based on the pictures shown in (b) (mean±S.E.M.). (e and f) Immunostainings of brain sections from WT and p73KO mice labeled with and anti-TRIM32 or TuJ-1 antibody are shown. Based on these pictures, the amount of TuJ1-positive cells in the VZ/SVZ was quantified (mean±S.E.M., **P<0.01, ***P<0.005)
TRIM32 interacts and promotes p73 ubiquitination and degradation, impairing TAp73 transcriptional activity
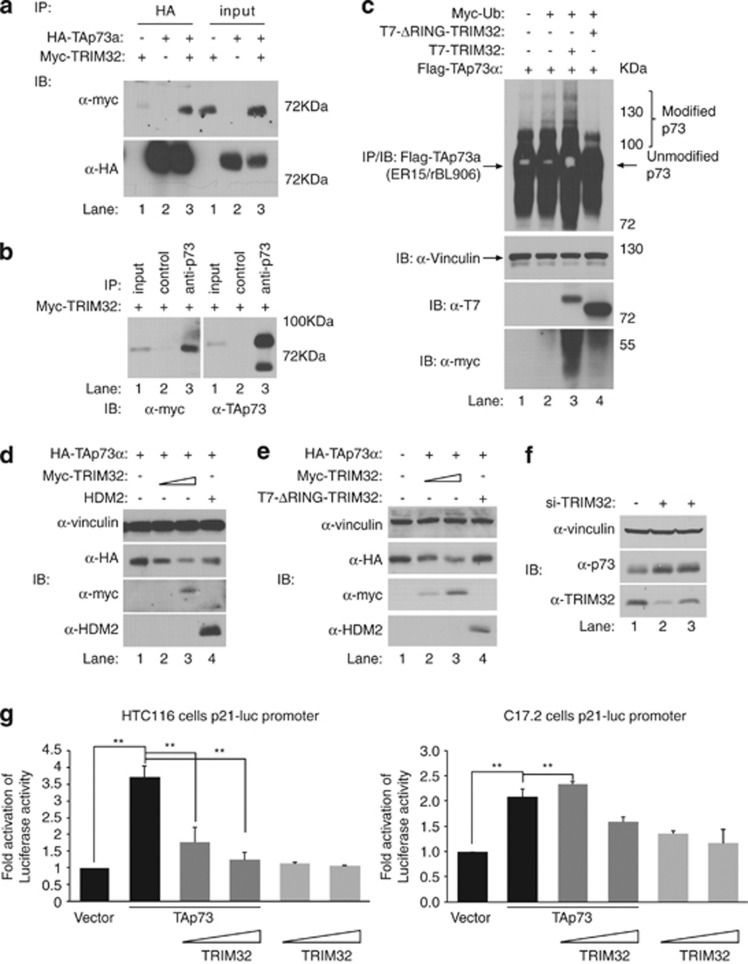
TRIM32 E3 ubiquitin ligase function35 leads us to hypothesize the possible existence of a regulatory loop between TRIM32 and its regulator, p73. To address this point, we first investigated whether p73 and TRIM32 could physically interact in vivo. HEK293 cells were transiently transfected with plasmids encoding human myc-TRIM32, with or without HA-TAp73α, treated with the proteasome inhibitor MG132 and immunoprecipitated with antibodies against the ectopic (Figure 5a) or the endogenous (Figure 5b) p73. Myc-TRIM32 specifically interacted with both overexpressed HA-TAp73α and endogenous TAp73α (Figure 5b). Similar results were seen with murine HA-TRIM32 (Supplementary Figure 5A). We also detected interaction between transfected DNp73α and HA-TRIM32 in HEK293 cells (Supplementary Figure 5B), suggesting that TRIM32 can form a stable complex with TAp73α and DNp73α.
Figure 5.
TRIM32 interacts and promotes p73 ubiquitination and degradation, impairing TAp73 transcriptional activity. (a) HEK293 cells were transfected with plasmids encoding myc-TRIM32 and hemagglutinin (HA)-TAp73α, and treated with MG132 (20 μM) for 6 h. Equal amounts of whole-cell extracts were immunoprecipitated with anti-HA (12CA5) antibody, resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotted (IB) with anti-myc antibody. The membrane was then stripped and reprobed with anti-HA (Y-11) antibody. (b) HEK293 cells were transfected with plasmid encoding myc-TRIM32 and treated with MG132 (20 μM) for 6 h. Equal amounts of whole-cell extracts were immunoprecipitated with control or anti-p73 (ER-15) antibody, resolved by SDS-PAGE and immunoblotted with anti-myc antibody. The membrane was then stripped and reprobed with anti-p73 (BL906) antibody. (c) HEK293 cells were transfected with plasmids encoding the indicated proteins and treated with MG132 (20 μM) for 6 h. Equal amounts of whole-cell extracts were immunoprecipitated with anti-p73 (ER-15) antibody, resolved by SDS-PAGE and immunoblotted with the indicated antibodies. (d and e) H1299 cells were transfected with constant amounts of plasmid encoding TAp73α, increasing amounts of plasmid encoding TRIM32 (p73/TRIM32 ratio=1 : 1, 1 : 10) and plasmid encoding MDM2 or ΔRING-TRIM32 (ratio=1 : 10). (f) HEK293 cells were transfected with two different small interfering (si)RNA oligonucleotides specific for TRIM32, or control oligonucleotides. Equal amounts of whole-cell extracts were resolved by SDS-PAGE and immunoblotted with the indicated antibodies. Equal amounts of whole-cell extracts were resolved by SDS-PAGE and immunoblotted with the indicated antibodies. (g) Transcriptional analysis with the reporter vector p21Cip1-FL-luc co-transfected with TAp73 alone or together with TRIM32 in a range from 1 : 0.6 to 1 : 2 or TRIM32 alone in HCT116 (left panel) or C17.2 (right panel) cells. Statistical analysis was performed with data from three independent experiments with at least three embryos of each genotype (mean±S.E.M.; **P<0.01)
We next asked whether p73 was a substrate for TRIM32-mediated ubiquitination. HEK293 cells were transfected with HA-TAp73α, with or without TRIM32 and ubiquitin, and treated with MG132. In cells in which TRIM32-WT was coexpressed, p73 slower-migrating ubiquitin-modified bands were detected (Figure 5c). In contrast, these modified forms of p73 were absent in the presence of Δ-RING-TRIM32-MUT, lacking the RING-finger domain. Consistent results were obtained with murine-TRIM32 and TAp73 α and β (Supplementary Figure 5C) altogether, suggesting that TRIM32 can promote p73 ubiquitination in a RING-finger-dependent manner. To determine whether this interaction resulted in TAp73 degradation, we transfected HCT116 cells with HA-TAp73α with or without GFP-TRIM32 and analyzed HA-TAp73 expression at different time points after transfection. Ectopic TAp73 expression is progressively lost in the presence of overexpressed TRIM32 (Supplementary Figure 5D). As expected, increasing amounts of TRIM32 in H1299 cells result in decreased steady-state levels of TAp73, in a dose-dependent manner (Figure 5d, lanes 2 and 3 versus lane 1). To determine whether this function was RING-finger-dependent, cells were transfected with HA-TAp73α alone or with either TRIM32-WT at different ratios (1 : 1, 1 : 10) or Δ-RING-TRIM32-MUT at the highest ratio (Figure 5e, lane 4). Although TRIM32-WT decreases TAp73 levels, the catalytic-inactive mutant transfected at the maximum amount does not degrade TAp73 (Figure 5e, compare lane 4 versus lane 1), indicating that TRIM32 degradation of TAp73 occurs, at least in part, in a RING-finger-dependent manner. To further demonstrate that endogenous TRIM32 maintains low endogenous TAp73 protein levels, we inhibited TRIM32 expression in HEK293 cells using two different siRNA oligonucleotides specific for TRIM32 and detected a clear increase in endogenous TAp73α protein upon TRIM32 knockdown (Figure 5f).
To gain insight into the functional consequences of the TRIM32/p73 interaction, we examined the effect of TRIM32 expression on TAp73-mediated transcriptional. We transfected the p21Cip1-full-length (FL) promoter-luc reporter plasmid (p21-FL promoter-luc), containing the p53/p73-binding sites, together with TAp73 with or without increasing amounts of TRIM32, or TRIM32 alone. Consistent with TRIM32 degradation of TAp73, increasing amounts of TRIM32 resulted in a significant reduction of TA-p73 transcriptional activation of the p21Cip1-FL promoter-luc in neural and non-neural cells (Figure 5g). Altogether, these results strongly support the idea that TRIM32 targets TAp73 and negatively regulates its protein levels hindering its transcriptional activity.
Discussion
We have identified a new p73 transcriptional target in the context of NPC differentiation and describe a novel regulatory feedback loop between TAp73 and TRIM32. We report, for the first time, that TRIM32 is differentially regulated by p73. Although TAp73 binds and activates TRIM32 promoter, this TAp73-induced TRIM32 expression is efficiently repressed by DNp73. TRIM32 in turn interacts with TAp73 and promotes its ubiquitination and degradation, impairing p73-dependent transcriptional activity.
TRIM32 levels in NPC are regulated by the ratio of TA/DN isoforms present in the cell in each particular moment. Consistent with TAp73 and TRIM32 functions as neural differentiation factors, TRIM32 is expressed at low but detectable levels in NPC under proliferating conditions. Upon induction of differentiation, TAp73 and TRIM32 levels rise sharply within the first hours. Meanwhile, DNp73 expression decreases to almost undetectable level to raise up only at late stages of differentiation.
Functional interaction between p73 and TRIM32 was confirmed by the lower TRIM32 steady-states levels in p73KO NPCs and the impaired TRIM32 expression in VZ/SVZ of p73KO E14.5 embryos, correlating with the reduced amount of newly generated neurons in this region. These data suggest that p73 deficiency results in a deregulation of TRIM32 levels that affects initial neuronal differentiation in the VZ in vivo. It is important to point out that TRIM32, although at lower levels, is expressed in p73-deficient cells, in vivo as well as in vitro, indicating that there are other p73-independent factors that support TRIM32 expression. This is consistent with TRIM32 essential role in neural differentiation, but the nature of these factors remains unknown.
TA and DN differential regulation of TRIM32 varies depending on the cell line and is not exclusive of neural-like cells, indicating that p73 transcriptional regulation of TRIM32 is cell context-dependent. This interpretation would be consistent with recent compilation of work regarding p73 function in central nervous system development, which indicates that p73 has a multifunctional role in this process.36 Although there is strong evidence demonstrating TAp73 induction of neuronal differentiation,17, 18, 37, 38 recent reports had unequivocally demonstrated p73 requirement, mainly TAp73, in the maintenance of NSC/NPC self-renewal.19, 20, 21, 39 This contradiction is also reflected in our data. While lower TRIM32 levels are detected in the absence of p73, which is consistent with p73 transcriptional regulation of TRIM32, prematurely differentiating Tuj-1-positive cells in p73KO NPC cultures had enhanced TRIM32 expression triggered by unknown mechanisms, despite the lack of p73. Thus, these data underline the crucial role of TRIM32 in neuronal differentiation and suggest that the appropriated ratios of TA/DNp73 are needed to control not only TRIM32 expression but also to maintain NPC stemness.
Owing to the role of p73 in cellular differentiation and tumor suppression, cells under physiological conditions require mechanisms to keep low-protein steady-state levels. This is achieved through degradation by the 26S proteasome, at least in part, by p73 interaction with E3 ubiquitin ligases like Itch or PIR2.14 In this regard, we also report a novel regulatory feedback loop between TAp73 and the E3 ligase TRIM32. This negative loop would be regulated by enhanced expression of DNp73, which efficiently inhibits TAp73-mediated TRIM32 expression. It has been proposed that p73 transcriptional activity is linked to its degradation40 and that regulation of p73 protein stability might be dependent on cell type.15 Consistently, we observed that TRIM32 overexpression induced TAp73 degradation and impaired its transcriptional activity. TRIM32 efficiency to affect TAp73 transcriptional activity varied depending on the cell line, supporting the role of a cell context factor. This feedback loop is not restricted to the neuronal context, and it may have special relevance in tumor cells. In this regard, TRIM32 has been proposed as a novel oncogene that promotes tumor growth, metastasis and resistance to anticancer drugs.30 Thus, the p73/TRIM32 loop could be similar to the p53/MDM2/ regulatory node where the Mdm2 oncogene binds to and inhibits the tumor suppressor protein, p53. Nevertheless, TRIM32 function and its interaction with p73 in tumorigenesis need to be further studied.
Materials and Methods
Mice husbandry and dissection of brain tissues
Animal experiments were conducted in agreement with European (Council Directive 86/609/CEE) and Spanish regulations (RD-1201/2005) for the protection of experimental animals with the appropriate institutional review committee approval. Mice heterozygous for Trp73 on a mixed background C57BL/6 × 129/svJae41 were backcrossed, at least five times, to C57BL/6 to enrich for C57BL/6 background. To obtain the double Trp73;p53KO mice (DKO), heterozygous animals for Trp73 were crossed with p53KO mice,42 and then Trp73+/−p53+/− mice were intercrossed to obtain the DKO animals. Mice were genotyped as described before.1, 41
To obtain embryo samples, pregnant females at 14.5 days postcoitum (E14.5d) were euthanized, embryos were extracted and brains were dissected.
To obtain 7 day-old postnatal mice samples, mice were deeply anesthetized by intraperitoneal injection of ketamine/medetomidine and euthanized. Brains were dissected and processed.
Primary culture of neural precursor cells
We utilized primary NS cultures obtained from OBs of Trp73−/− (p73KO from now on) and WT E14.5 embryos.21 NSCs cultured in the presence of epidermal growth factor (EGF) and basic fibroblast growth factor form NS that preserve NSC self-renewal and multipotency. These mitogenic culture conditions allow NPCs to behave like NSC giving rise to heterogeneous aggregates of NSC and NPC, in which the two populations are indistinguishable. To initiate each independent embryonic culture, the OBs of at least three different E14.5d embryos were dissected out and mechanically dissociated, as described previously.43 The methods for primary culture of neural precursor cells by the NS assay and differentiation have been described before.21
Immunostaining and confocal analysis
Immunocytochemistry was performed as described before44 using the following antibodies: mouse anti-TRIM32 (1 : 100; Abnova, Taipei City, Taiwan, China), rabbit anti-Tuj-1 (1 : 1000; Covance, Princeton, NJ, USA) and rabbit anti-p-His3 (1 : 100; Chemicon, Billerica, MA, USA). Images were obtained with Nikon EclipseTE2000 confocal microscope. Statistical analysis (Student's two-tailed t-test) was performed using triplicates from three independent experiments.
RNA isolation and real-time RT-PCR analysis
Total RNA was extracted with TRI reagent (Ambion, Austin, TX, USA) and cDNA was prepared using High Capacity RNA-to-cDNA kit (Applied Biosystems, Carlsbad, CA, USA) according to the manufacturer's instructions. P73 and TRIM32 expression was detected by qRT-PCR in a StepOnePlus Real-Time PCR System (Applied Biosystems) using FastStart Universal SYBR Green Master (Roche, Basel, Switzerland). Primers sequences and conditions were described before.16, 45 The expression levels of mRNA were expressed as 2ΔCt and normalized to 18S.
Samples and statistical analyses
The analyses were performed with data from three independent experiments with at least three embryos of each genotype. Statistical analyses were performed using the Student's two-tailed t-test (*P<0.05, **P<0.01, ***P<0.005), and the S.E. or S.D. of the samples is represented as indicated in each figure legend.
Plasmids and transfection
The cDNAs from human pcDNA3-HA-TAp73α, pcDNA3-HA-ΔNp73α and pcDNA3-HA-p53 were described previously.17 Cells were transfected with the indicated plasmids using Lipofectamine 2000 (Life Technologies, Carlsbad, CA, USA) following instructions from the manufacturer, and 24 h after transfection, the cells were harvested and processed.
Immunoprecipitation and ubiquitination assays
Cells treated with 20 μM MG132 (Sigma-Aldrich, St. Louis, MO, USA) were lysed and equal amounts of whole-cell extracts were subjected to immunoprecipitation and immunoblot as described previously.49 For the ubiquitination studies, a modified lysis method was used to preserve ubiquitin-modified forms as described in Watson and Irwin.46
Monoclonal antibodies ER15 anti-p7347 and 12CA5 anti-HA tag (Roche) were used in the immunoprecipitations.
Western blot analysis
Brains of 14.5 days embryos were homogenized, lysated and immunoblots were performed as described before.17 The immunoblots were incubated with the following primary antibodies: rabbit anti-TRIM32 (1 : 1000 (ref. 26), rabbit anti-ERK 2 (1 : 10 000; Santa Cruz Biotechnology, Dallas, TX, USA), rabbit BL906 anti-p73 (1 : 500; Bethyl Laboratories, Montgomery, TX, USA) or rabbit 2301 anti-p73 (0.75 μg/ml (ref. 48)), rabbit A14 anti-myc (1 : 500; Santa Cruz), mouse HA.11 anti-HA tag (1 : 1000; Covance), mouse anti-vinculin (1 : 20 000; Upstate-Millipore, Billerica, MA, USA), mouse anti-T7 (1 : 1000; Novagen, Billerica, MA, USA), followed by the appropriate HRP-conjugated secondary antibodies (Pierce, Waltham, MA, USA). The enhanced chemiluminescence was detected with Super Signal West-Pico Chemiluminiscent Substrate (Pierce), followed by the appropriated HRP-coupled secondary antibodies (Pierce).
Cloning of the human TRIM32 promoter and generation of a deletion mutant
Genomic DNA was isolated from human keratinocytes and a fragment of the TRIM32 promoter was PCR-amplified. Primers were designed according to the sequence in the genome databases (ENSEMBL Gene ID: ENSG00000119401). Primer sequences are indicated in Supplementary Figure 2. PCR amplification conditions are available upon request. The amplified fragment spans the region from −1351 to +270, and contains the p53 consensus binding site identified in the proximal promoter (−115 to −84), exon 1 and 218 bp from intron 1.The 1.6-kb PCR product was cloned using the TOPO TA Cloning kit (Invitrogen, Carlsbad, CA, USA). Afterwards, it was inserted as a HindIII–XhoI fragment into the pGL3-basic luciferase reporter vector (hTRIM32-luc promoter plasmid). Promoter sequence identity was confirmed before and after cloning into the reporter plasmid.
For generating mut-hTRIM32-luc promoter plasmid, the region spanning from −367 to −59, containing the putative p73-binding site, was deleted. A PCR with primers excluding this region was performed using hTRIM32-luc promoter plasmid as template. PCR product ends were polished by T4 DNA polymerase treatment and DNA was re-ligated. The deletion was confirmed by sequencing.
Luciferase assay
Cells (105) were transfected with 0.125 μg of the reporter luciferase pGL3-hTRIM32-luc or p21Cip1-FL promoter-luc plus 0.0625 μg pRLNull and 0.2–0.6 μg of the indicated expression vectors, using Lipofectamine 2000 (Life Technologies) following the manufacturer's protocol. Cellular extracts were prepared 24 h after transfection as described before.17 Measurements were taken in a Berthold's luminometer and normalized by the values of the Renilla luciferase of the same sample.
siRNA knockdown
siRNA transfection was performed as described previously.7, 49 Briefly, oligonucleotides (Dharmacon, Lafayatte, CO, USA) at a final concentration of 75 nM were transfected with oligofectamine (Invitrogen) according to the manufacturer's instructions. Oligonucleotide sequences for TRIM32 knockdown were as follows: si-TRIM32 1298: 5′-CAGTCAAGGTGAAGTACTA-3′ and si-TRIM32 2074: 5′-GCATCAAGATCTACAGCTA-3′. siGENOME RISC-Free Control siRNA (Dharmacon) was used as the negative control. Anti-p73 siRNA oligos has been described previously.7
Chromatin immunoprecipitation
For the ChIPs, N2a cells were transfected with expression constructs for HA-tagged TAp73, DNp73 or EGFP (negative control). Cells were lysed 48 h after transfection, and after crosslinking, the chromatin was precipitated with an anti-HA antibody (Roche). In addition, endogenous p73 was precipitated from untransfected cells with an anti-p73 antibody (Santa Cruz). The complete ChIP was conducted with the HighCell# ChIP kit (Diagenode, Denville, NJ, USA) according to the manufacturer's instructions. To investigate binding to the TRIM32 promoter, precipitated chromatin was used as a template in a PCR reaction with specific primers for TRIM32 promoter (primers were designed to amplify the predicted p73-binding sites).
An in silico prediction of p53REs within the human TRIM32 locus was performed with the p53FamTaG database.34
Immunohistochemistry of free-floating sections
Brains were fixed overnight at 4 °C in 4% PFA in phosphate-buffered saline. Sections of 40 μm were prepared using a vibratome (Leica, Wetzlar, Germany) and blocked for at least 1 h in TBS (0.1 M Tris,150 mM NaCl, pH 7.4) containing 0.5% Triton X-100, 0.1% Na-Azide, 0.1% Na-Citrate and 5% normal goat serum. Immunostainings were performed by incubation of the sections with primary antibodies diluted in the blocking solution for 48 h at 4 °C on a shaker, followed by incubation with the secondary antibody diluted in the blocking solution for 2 h at room temperature. Finally, sections were mounted in AquaMount (DAKO, Glostrup, Denmark). Images were collected by confocal microscopy using ZEN software (Zeiss, Jena, Germany); image analysis was performed with the ZEN software, Adobe Photoshop (San Jose, CA, USA), Image J software (Bethesda, MD, USA) and Imaris software (Zurich, Switzerland).
Acknowledgments
This work was supported by Grants SAF2009-07897 and SAF2012-36143 from Spanish Ministerio de Ciencia e Innovación (to MCM), Grant LE015A10-2 from the Junta de Castilla y Leon (to MCM), grants from the German Research Foundation (DFG: Emmy Noether Program, SCHW1392/2-1; SFB629 and SPP1356, SCHW1392/4-1) (to JCS) and grant from the Canadian Cancer Society Research Institute (CCSRI 2010-700580) (to MSI).
Glossary
- CNS
central nervous system
- NSC
neural stem cell
- NPC
neural progenitor cell
- NS
neurosphere
- WT
wild type
- DKO
p53−/−p73−/−
- MEF
mouse embryonic fibroblasts
- DIV
days in vitro
- DM
differentiation media
- PM
proliferation media
- VZ
ventricular zone
- SVZ
subventricular zone
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Disease website (http://www.nature.com/cddis)
Author contributions
L Gonzalez-Cano: conception and design, collection and/or assembly of data and manuscript writing; S Fuertes-Alvarez: collection and assembly of data; A-L Hillje: collection and assembly of data; A Blanch: collection and assembly of data; IR Watson: collection and assembly of data; MM Marques: conception and design, manuscript writing, data analysis and interpretation; MM Irwin: conception and design, manuscript writing, data analysis and interpretation, and financial support; JC Schwamborn: conception and design, manuscript writing, data analysis and interpretation, and financial support; MC Marin: conception and design, manuscript writing, data analysis and interpretation, and financial support.
Edited by G Melino
Supplementary Material
References
- Flores ER, Sengupta S, Miller JB, Newman JJ, Bronson R, Crowley D, et al. Tumor predisposition in mice mutant for p63 and p73: evidence for broader tumor suppressor functions for the p53 family. Cancer cell. 2005;7:363–373. doi: 10.1016/j.ccr.2005.02.019. [DOI] [PubMed] [Google Scholar]
- Lin Y, Cheng Z, Yang Z, Zheng J, Lin T. DNp73 improves generation efficiency of human induced pluripotent stem cells. BMC Cell Biol. 2012;13:9. doi: 10.1186/1471-2121-13-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Laurenzi V, Raschella G, Barcaroli D, Annicchiarico-Petruzzelli M, Ranalli M, Catani MV, et al. Induction of neuronal differentiation by p73 in a neuroblastoma cell line. J Biol Chem. 2000;275:15226–15231. doi: 10.1074/jbc.275.20.15226. [DOI] [PubMed] [Google Scholar]
- Fang L, Lee SW, Aaronson SA. Comparative analysis of p73 and p53 regulation and effector functions. J Cell Biol. 1999;147:823–830. doi: 10.1083/jcb.147.4.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jost CA, Marin MC, Kaelin WG., Jr. P73 is a simian [correction of human] p53-related protein that can induce apoptosis. Nature. 1997;389:191–194. doi: 10.1038/38298. [DOI] [PubMed] [Google Scholar]
- Jung MS, Yun J, Chae HD, Kim JM, Kim SC, Choi TS, et al. P53 and its homologues, p63 and p73, induce a replicative senescence through inactivation of NF-Y transcription factor. Oncogene. 2001;20:5818–5825. doi: 10.1038/sj.onc.1204748. [DOI] [PubMed] [Google Scholar]
- Marques-Garcia F, Ferrandiz N, Fernandez-Alonso R, Gonzalez-Cano L, Herreros-Villanueva M, Rosa-Garrido M, et al. P73 plays a role in erythroid differentiation through GATA1 induction. J Biol Chem. 2009;284:21139–21156. doi: 10.1074/jbc.M109.026849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishimoto O, Kawahara C, Enjo K, Obinata M, Nukiwa T, Ikawa S. Possible oncogenic potential of DeltaNp73: a newly identified isoform of human p73. Cancer Res. 2002;62:636–641. [PubMed] [Google Scholar]
- Zaika AI, Slade N, Erster SH, Sansome C, Joseph TW, Pearl M, et al. DeltaNp73, a dominant-negative inhibitor of wild-type p53 and TAp73, is up-regulated in human tumors. J Exp Med. 2002;196:765–780. doi: 10.1084/jem.20020179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs WB, Kaplan DR, Miller FD. The p53 family in nervous system development and disease. J Neurochem. 2006;97:1571–1584. doi: 10.1111/j.1471-4159.2006.03980.x. [DOI] [PubMed] [Google Scholar]
- Liu G, Nozell S, Xiao H, Chen X. DeltaNp73beta is active in transactivation and growth suppression. Mol Cell Biol. 2004;24:487–501. doi: 10.1128/MCB.24.2.487-501.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgunkova AA. The p53 gene family: control of cell proliferation and developmental programs. Biochemistry Biokhimiia. 2005;70:955–971. doi: 10.1007/s10541-005-0210-4. [DOI] [PubMed] [Google Scholar]
- Peschiaroli A, Scialpi F, Bernassola F, Pagano M, Melino G. The F-box protein FBXO45 promotes the proteasome-dependent degradation of p73. Oncogene. 2009;28:3157–3166. doi: 10.1038/onc.2009.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayan BS, Yang AL, Conforti F, Tucci P, Piro MC, Browne GJ, et al. Differential control of TAp73 and DeltaNp73 protein stability by the ring finger ubiquitin ligase PIR2. Proc Natl Acad Sci USA. 2010;107:12877–12882. doi: 10.1073/pnas.0911828107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Zeinab RA, Flores ER, Leng RP. Pirh2, a ubiquitin E3 ligase, inhibits p73 transcriptional activity by promoting its ubiquitination. Mol Cancer Res. 2011;9:1780–1790. doi: 10.1158/1541-7786.MCR-11-0157. [DOI] [PubMed] [Google Scholar]
- Tomasini R, Tsuchihara K, Wilhelm M, Fujitani M, Rufini A, Cheung CC, et al. TAp73 knockout shows genomic instability with infertility and tumor suppressor functions. Genes Dev. 2008;22:2677–2691. doi: 10.1101/gad.1695308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Garcia B, Vaque JP, Herreros-Villanueva M, Marques-Garcia F, Castrillo F, Fernandez-Medarde A, et al. P73 cooperates with Ras in the activation of MAP kinase signaling cascade. Cell Death Differ. 2007;14:254–265. doi: 10.1038/sj.cdd.4401945. [DOI] [PubMed] [Google Scholar]
- Billon N, Terrinoni A, Jolicoeur C, McCarthy A, Richardson WD, Melino G, et al. Roles for p53 and p73 during oligodendrocyte development. Development. 2004;131:1211–1220. doi: 10.1242/dev.01035. [DOI] [PubMed] [Google Scholar]
- Agostini M, Tucci P, Chen H, Knight RA, Bano D, Nicotera P, et al. P73 regulates maintenance of neural stem cell. Biochem Biophys Res Commun. 2010;403:13–17. doi: 10.1016/j.bbrc.2010.10.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujitani M, Cancino GI, Dugani CB, Weaver IC, Gauthier-Fisher A, Paquin A, et al. TAp73 acts via the bHLH Hey2 to promote long-term maintenance of neural precursors. Curr Biol. 2010;20:2058–2065. doi: 10.1016/j.cub.2010.10.029. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Cano L, Herreros-Villanueva M, Fernandez-Alonso R, Ayuso-Sacido A, Meyer G, Garcia-Verdugo JM, et al. P73 deficiency results in impaired self renewal and premature neuronal differentiation of mouse neural progenitors independently of p53. Cell Death Dis. 2010;1:e109. doi: 10.1038/cddis.2010.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boominathan L. The tumor suppressors p53, p63, and p73 are regulators of microRNA processing complex. PLoS One. 2010;5:e10615. doi: 10.1371/journal.pone.0010615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbluth JM, Pietenpol JA. The jury is in: p73 is a tumor suppressor after all. Genes Dev. 2008;22:2591–2595. doi: 10.1101/gad.1727408. [DOI] [PubMed] [Google Scholar]
- Reymond A, Meroni G, Fantozzi A, Merla G, Cairo S, Luzi L, et al. The tripartite motif family identifies cell compartments. EMBO J. 2001;20:2140–2151. doi: 10.1093/emboj/20.9.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillje AL, Worlitzer MM, Palm T, Schwamborn JC. Neural stem cells maintain their stemness through protein kinase C zeta-mediated inhibition of TRIM32. Stem Cells. 2011;29:1437–1447. doi: 10.1002/stem.687. [DOI] [PubMed] [Google Scholar]
- Schwamborn JC, Berezikov E, Knoblich JA. The TRIM-NHL protein TRIM32 activates microRNAs and prevents self-renewal in mouse neural progenitors. Cell. 2009;136:913–925. doi: 10.1016/j.cell.2008.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicklas S, Otto A, Wu X, Miller P, Stelzer S, Wen Y, et al. TRIM32 regulates skeletal muscle stem cell differentiation and is necessary for normal adult muscle regeneration. PLoS One. 2012;7:e30445. doi: 10.1371/journal.pone.0030445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, Okumura F, Iguchi A, Ariga T, Hatakeyama S. TRIM32 promotes retinoic acid receptor alpha-mediated differentiation in human promyelogenous leukemic cell line HL60. Biochem Biophys Res Commun. 2012;417:594–600. doi: 10.1016/j.bbrc.2011.12.012. [DOI] [PubMed] [Google Scholar]
- Albor A, Kulesz-Martin M. Novel initiation genes in squamous cell carcinomagenesis: a role for substrate-specific ubiquitylation in the control of cell survival. Mol Carcinogen. 2007;46:585–590. doi: 10.1002/mc.20344. [DOI] [PubMed] [Google Scholar]
- Kano S, Miyajima N, Fukuda S, Hatakeyama S. Tripartite motif protein 32 facilitates cell growth and migration via degradation of Abl-interactor 2. Cancer Res. 2008;68:5572–5580. doi: 10.1158/0008-5472.CAN-07-6231. [DOI] [PubMed] [Google Scholar]
- Hammell CM, Lubin I, Boag PR, Blackwell TK, Ambros V. Nhl-2 modulates microRNA activity in Caenorhabditis elegans. Cell. 2009;136:926–938. doi: 10.1016/j.cell.2009.01.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder EY, Deitcher DL, Walsh C, Arnold-Aldea S, Hartwieg EA, Cepko CL. Multipotent neural cell lines can engraft and participate in development of mouse cerebellum. Cell. 1992;68:33–51. doi: 10.1016/0092-8674(92)90204-p. [DOI] [PubMed] [Google Scholar]
- Reynolds BA, Weiss S. Clonal and population analyses demonstrate that an EGF-responsive mammalian embryonic CNS precursor is a stem cell. Dev Biol. 1996;175:1–13. doi: 10.1006/dbio.1996.0090. [DOI] [PubMed] [Google Scholar]
- Sbisa E, Catalano D, Grillo G, Licciulli F, Turi A, Liuni S, et al. P53FamTaG: a database resource of human p53, p63 and p73 direct target genes combining in silico prediction and microarray data. BMC Bioinform. 2007;8 (Suppl 1:S20. doi: 10.1186/1471-2105-8-S1-S20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn EJ, Albor A, Liu Y, El-Hizawi S, Vanderbeek GE, Babcock M, et al. RING protein Trim32 associated with skin carcinogenesis has anti-apoptotic and E3-ubiquitin ligase properties. Carcinogenesis. 2004;25:157–167. doi: 10.1093/carcin/bgh003. [DOI] [PubMed] [Google Scholar]
- Killick R, Niklison-Chirou M, Tomasini R, Bano D, Rufini A, Grespi F, et al. P73: a multifunctional protein in neurobiology. Mol Neurobiol. 2011;43:139–146. doi: 10.1007/s12035-011-8172-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agostini M, Tucci P, Steinert JR, Shalom-Feuerstein R, Rouleau M, Aberdam D, et al. MicroRNA-34a regulates neurite outgrowth, spinal morphology, and function. Proc Natl Acad Sci USA. 2011;108:21099–21104. doi: 10.1073/pnas.1112063108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Laurenzi V, Rossi A, Terrinoni A, Barcaroli D, Levrero M, Costanzo A, et al. P63 and p73 transactivate differentiation gene promoters in human keratinocytes. Biochem Biophys Res Commun. 2000;273:342–346. doi: 10.1006/bbrc.2000.2932. [DOI] [PubMed] [Google Scholar]
- Talos F, Abraham A, Vaseva AV, Holembowski L, Tsirka SE, Scheel A, et al. P73 is an essential regulator of neural stem cell maintenance in embryonal and adult CNS neurogenesis. Cell Death Differ. 2010;17:1816–1829. doi: 10.1038/cdd.2010.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Zhu H, Nie L, Maki CG. A link between p73 transcriptional activity and p73 degradation. Oncogene. 2004;23:4032–4036. doi: 10.1038/sj.onc.1207538. [DOI] [PubMed] [Google Scholar]
- Yang A, Walker N, Bronson R, Kaghad M, Oosterwegel M, Bonnin J, et al. P73-deficient mice have neurological, pheromonal and inflammatory defects but lack spontaneous tumours. Nature. 2000;404:99–103. doi: 10.1038/35003607. [DOI] [PubMed] [Google Scholar]
- Donehower LA, Harvey M, Slagle BL, McArthur MJ, Montgomery CA, Jr., Butel JS, et al. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 1992;356:215–221. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- Vicario-Abejon C, Yusta-Boyo MJ, Fernandez-Moreno C, de Pablo F. Locally born olfactory bulb stem cells proliferate in response to insulin-related factors and require endogenous insulin-like growth factor-I for differentiation into neurons and glia. J Neurosci. 2003;23:895–906. doi: 10.1523/JNEUROSCI.23-03-00895.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armesilla-Diaz A, Bragado P, Del Valle I, Cuevas E, Lazaro I, Martin C, et al. P53 regulates the self-renewal and differentiation of neural precursors. Neuroscience. 2009;158:1378–1389. doi: 10.1016/j.neuroscience.2008.10.052. [DOI] [PubMed] [Google Scholar]
- Kudryashova E, Wu J, Havton LA, Spencer MJ. Deficiency of the E3 ubiquitin ligase TRIM32 in mice leads to a myopathy with a neurogenic component. Hum Mol Genet. 2009;18:1353–1367. doi: 10.1093/hmg/ddp036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson IR, Irwin MS. Ubiquitin and ubiquitin-like modifications of the p53 family. Neoplasia. 2006;8:655–666. doi: 10.1593/neo.06439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin MC, Jost CA, Irwin MS, DeCaprio JA, Caput D, Kaelin WG., Jr Viral oncoproteins discriminate between p53 and the p53 homolog p73. Mol Cell Biol. 1998;18:6316–6324. doi: 10.1128/mcb.18.11.6316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson IR, Blanch A, Lin DC, Ohh M, Irwin MS. Mdm2-mediated NEDD8 modification of TAp73 regulates its transactivation function. J Biol Chem. 2006;281:34096–34103. doi: 10.1074/jbc.M603654200. [DOI] [PubMed] [Google Scholar]
- Lau LM, Nugent JK, Zhao X, Irwin MS. HDM2 antagonist Nutlin-3 disrupts p73-HDM2 binding and enhances p73 function. Oncogene. 2008;27:997–1003. doi: 10.1038/sj.onc.1210707. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.