Abstract
We have adapted a bacterial CRISPR RNA/Cas9 system to precisely engineer the Drosophila genome and report that Cas9-mediated genomic modifications are efficiently transmitted through the germline. This RNA-guided Cas9 system can be rapidly programmed to generate targeted alleles for probing gene function in Drosophila.
Keywords: CRISPR RNA, Cas9, homologous recombination, genome engineering, Drosophila
GENETIC and molecular techniques to manipulate the genomes of model organisms are invaluable tools for understanding gene function. In Drosophila, chemical and insertional mutagenesis are powerful and widely utilized methods for disrupting gene function (St Johnston 2002; Venken and Bellen 2005). Imprecise excision of a transposable element inserted near a gene of interest can result in deletion of all or part of the locus. More recently, techniques that stimulate homologous recombination (HR) using an exogenous template have made precisely targeted genome modifications possible (Gloor et al. 1991; Banga and Boyd 1992; Nassif et al. 1994; Rong and Golic 2000; Gong and Golic 2003; Huang et al. 2009). The most widely used methods rely on double-strand breaks (DSBs) in the donor template to trigger HR, but these can be time consuming and labor intensive (Gao et al. 2008; Huang et al. 2009). Zinc-finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs) have been successfully employed in Drosophila to generate DSBs in genomic targets that trigger repair by either HR or error-prone nonhomologous end joining (NHEJ) (Bibikova et al. 2002; Liu et al. 2012). However, ZFNs and TALENs, which both comprise a nuclease joined to a site-specific DNA-binding domain, require the generation of a unique protein for each genomic manipulation (Wood et al. 2011). In the past few months, the simplified CRISPR RNA-guided Cas9 nuclease has shown broad potential for genome engineering in metazoans.
CRISPRs (clustered regularly interspaced short palindromic repeats) were first identified in Escherichea coli in 1987 and later shown to function as part of an adaptive immune system in bacteria and archaea (Ishino et al. 1987; Makarova et al. 2006; Barrangou et al. 2007). In type II CRISPR systems, a CRISPR RNA (crRNA), which contains sequence complementary to invading virus or plasmid DNA, and a trans-activating CRISPR RNA (tracrRNA) interact with a CRISPR-associated nuclease (Cas9) to direct sequence-specific cleavage of exogenous DNA. Recognizing the potential of harnessing this system for precise genome engineering in other organisms, Jinek and colleagues (Jinek et al. 2012) identified a minimal two-component system required for the site-specific cleavage of DNA—Cas9 and a chimeric RNA (chiRNA) comprising the crRNA and tracrRNA from Streptococcus pyogenes. Thus, in contrast to ZFNs and TALENs, this modified CRISPR RNA/Cas9 system directs a common nuclease to specific DNA sequences by a short, readily generated RNA. This obviates the need to create a unique chimeric nuclease for each genome manipulation and raises the possibility and promise of routine genome engineering.
In recent months, a number of laboratories have demonstrated that this heterologously expressed two-component CRISPR RNA/Cas9 system can induce site-specific DSBs in eukaryotic genomes, including cultured mammalian cells, human stem cells, and yeast (Cho et al. 2013; Cong et al. 2013; Dicarlo et al. 2013; Ding et al. 2013; Jinek et al. 2013; Mali et al. 2013). In vivo experiments in zebrafish and mice yielded mosaic animals, demonstrating that the system has the capacity to manipulate a variety of eukaryotic genomes (Chang et al. 2013; Hwang et al. 2013; Shen et al. 2013). Most recently, mice carrying homozygous mutations in two genes were generated through embryo injections (Wang et al. 2013). Nonetheless, to date the CRISPR RNA/Cas9 system has not been adapted for Drosophila, a key genetic model, and germline transmission of Cas9-induced changes has not been achieved in any organism. Here we demonstrate that the CRISPR RNA/Cas9 system can (1) mediate efficient genome engineering in Drosophila and (2) induce targeted genome modifications in the Drosophila germline that are transmitted to progeny.
Results
Site-directed cleavage of Drosophila genes
To test whether the CRISPR RNA/Cas9 system could induce site-specific DSBs in Drosophila, we targeted the yellow gene, which is on the X chromosome and commonly used for genome engineering studies (Rong and Golic 2000; Bibikova et al. 2002; Liu et al. 2012). Efficient target recognition by the CRISPR RNA/Cas9 system requires 20 nucleotides (nt) of homology between the chiRNA and its genomic target. Cleavage also requires that the 3′ end of the genomic target sequence contains a 3-basepair (bp) proto-spacer adjacent motif (PAM) sequence, NGG, which differentiates self from invading DNA in the endogenous system (Jinek et al. 2012). Thus, selection of a 20-nt target sequence is limited only by the requirement for an adjacent PAM sequence. In our plasmid-based system, the ideal target sequence also begins with a G that allows for precise transcriptional initiation of the chiRNA from the U6 promoter (Wakiyama et al. 2005, Materials and Methods).
Plasmids encoding Cas9 and a chiRNA targeting the first exon of yellow (YE1), disruption of which is expected to abolish yellow function, were co-injected into preblastoderm embryos (Figures 1A and Supporting Information, Figure S1, Figure S2, and Table S1. To assess Cas9-mediated cleavage, we isolated DNA from individual embryos 24-hr post-injection. Following PCR amplification of the targeted genomic site, we used the SURVEYOR mutation detection assay, which employs an endonuclease that recognizes and cuts at the site of mismatches, to detect small insertions or deletions (indels) that result when DSBs are imperfectly repaired by NHEJ. DSBs that are repaired by HR using either the sister chromatid or homologous chromosome as a template do not result in an altered locus and are not detectable. The SURVEYOR assay indicated site-specific indels in one of 13 embryos injected with Cas9 and the YE1 chiRNA (Figure 1B). Consistent with the loss of yellow in a subset of somatic cells, we observed mosaic yellow patches in adult male cuticles after YE1 injection (6%, n = 67). Based on the size of the cleavage products produced in the SURVEYOR assay, the induced cleavage occurred at the YE1 target site indicating that the chiRNA successfully directed Cas9 to the yellow locus where it induced DSBs in the targeted DNA.
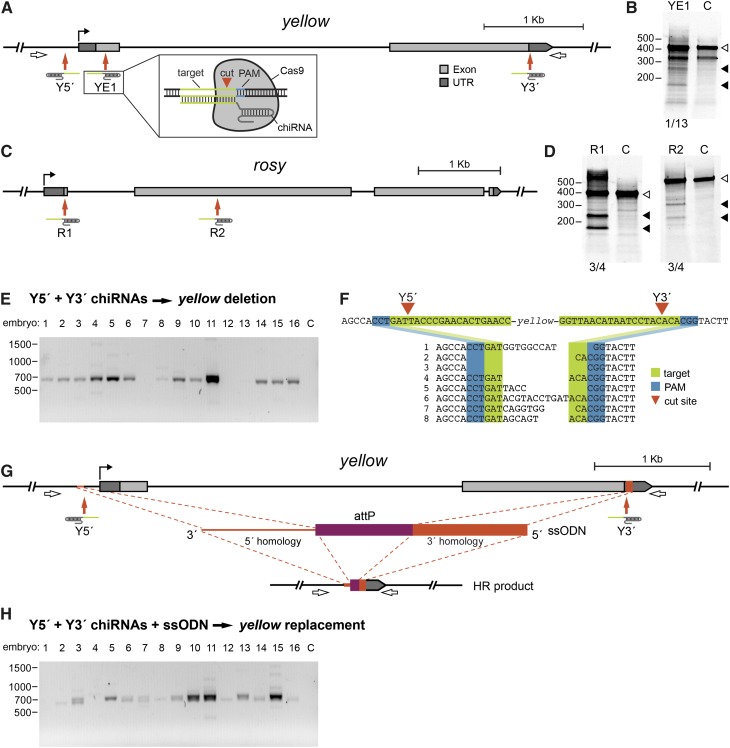
Figure 1.
Cas9-generated DSBs are repaired by NHEJ and HR in Drosophila embryos. (A) Schematic of the yellow locus indicating YE1, Y5′, and Y3′ chiRNA target sites. The predicted cut site for each chiRNA is indicated (red arrows). Inset shows schematic of CRISPR RNA/Cas9 system components interacting with target DNA. For additional detail, see Figure S1A. (B) Results of the SURVEYOR assay indicate that targeted cleavage occurred in yellow at the predicted YE1 cut site (left, embryos injected with YE1 chiRNA; right, control, C, embryos). Duplexes lacking indels were uncut (open arrowhead), whereas duplexes containing indels were cut asymmetrically at the targeted site (solid arrowheads). One of 13 embryos injected with YE1 chiRNA produced duplexes with indels. (C) Schematic of the rosy locus indicating R1 and R2 chiRNA target sites (red arrows). (D) Results from SURVEYOR assays indicate R1 and R2 chiRNAs generated indels at the targeted cut sites (embryos injected with R1 chiRNA, R2 chiRNA, or control, C, are as indicated). Uncut duplexes (open arrowhead) and cut duplexes (solid arrowhead) are indicated. Three of four embryos injected with either R1 chiRNA or R2 chiRNA produced duplexes with indels. (E) The yellow locus was amplified from embryos injected with Cas9, Y5′ chiRNA, and Y3′ chiRNA using the primers indicated in A (open arrows). Targeted deletions result in a 650-bp PCR product (C, uninjected control embryo). (F) Products from nine PCRs were pooled for sequencing. Sequence alignments reveal breakpoints and imprecise repair between predicted cut sites (red arrowheads). One repair event that resulted in the 4.6-kb deletion of yellow plus an indel deleting an additional 27 bp and inserting 93 bp is not shown. (G) Schematic of the yellow ssODN design. The ssODN includes 60-nt homology arms (red) flanking a 50-nt attP docking site (purple). The homology arms correspond to the sequences immediately adjacent to the predicted cut sites (red arrows). For additional detail see Figure S1C. (H) The yellow locus was amplified from embryos injected with Cas9, Y5′ chiRNA, Y3′ chiRNA, and ssODN using the primers indicated in A. HR events in which the locus is deleted and replaced with attP will yield a 700-bp product. Products from three reactions were pooled for sequence analysis, which confirmed the replacement of yellow with attP in five of eight clones.
To determine if co-injection of Cas9 and a chiRNA could generate indels at other loci, we designed two chiRNAs to target the rosy gene (Figure 1C, R1 and R2). Again, the SURVEYOR assay identified Cas9-induced site-specific cleavage at the targeted sites in a subset of embryos (Figure 1D, 3/4 embryos for each chiRNA). Thus, the RNA-guided Cas9 system can be readily programmed to induce DSBs in multiple Drosophila genomic targets.
Engineering defined deletions
Encouraged that the CRISPR RNA/Cas9 system functioned in Drosophila, we next asked whether Cas9-mediated DSBs could be harnessed to generate defined deletions. We reasoned that introducing two chiRNAs, Y5′ targeting the 5′ end of yellow and Y3′ targeting the 3′ end, might guide Cas9 to precisely delete the 4.6-kilobase (kb) yellow locus (Figure 1A). We isolated DNA from embryos 24 hr after co-injection of Cas9, Y5′ chiRNA, and Y3′ chiRNA and performed PCR to determine whether yellow was deleted. Using primers flanking the targeted sites (Figure 1A, open arrows), a 650-bp product was amplified in 13/16 embryos, as expected if a targeted deletion occurred. (Figure 1E). Moreover, as adults, 66% of injected males (n= 52) displayed yellow mosaicism, consistent with a high level of targeted deletion in somatic cells.
To investigate individual repair events, the DNA amplified from the targeted locus was pooled, subcloned, and sequenced. In nine of nine clones analyzed, the 4.6-kb yellow locus was deleted and the junction created by repair of the two cut sites showed the small indels typical of NHEJ (Figure 1F). These results extend findings from mammalian cell culture in which 19- and 118-bp sequences were deleted by cotransfection of Cas9 and two chiRNAs (Cong et al. 2013; Mali et al. 2013) and demonstrate that such “multiplex” genome editing is an effective tool for precisely deleting large genomic regions in organisms.
Cas9-mediated homologous recombination
The ability to edit genomic sequence by introducing specific mutations or exogenous sequences to monitor or manipulate protein activity provides a critical tool for dissecting gene function. One means of achieving this goal is to replace a gene with a sequence that enables site-specific integration of engineered genes into the targeted locus, such as the attP ΦC31 phage recombination site. Such gene replacement can be accomplished by stimulating HR in the presence of a donor template that includes the site-specific integration sequence (Gao et al. 2008; Huang et al. 2009). To this end, we designed a single-strand oligodeoxynucleotide (ssODN) repair template that includes a 50-nt attP site flanked by 60-nt homology arms corresponding to sequences 5′ and 3′ of yellow (Figure 1G and Figure S1C). We co-injected Cas9 and the Y5′ and Y3′ chiRNAs with this ssODN, which we predicted would serve as a template for repair of the deleted locus by HR resulting in the replacement of yellow with attP. Using the same primers as above (Figure 1A, open arrows), we performed PCR on DNA extracted from single injected embryos. In 13/16 embryos, we detected the 700-bp product that is expected to result from replacement of yellow with attP (Figure 1H). In agreement with highly efficient disruption of yellow, 62% of injected males exhibited a mosaic cuticle as adults (n = 35).
To confirm that the ssODN donor served as a template for repair by HR, the PCR products amplified from injected embryos were pooled and subcloned. Sequence analysis of individual repair events revealed that the yellow gene was replaced by attP from the ssODN template in five of eight clones. Two clones contained the 4.6-kb deletion but lacked the attP sequence, indicating site-specific induction of DSBs repaired by NHEJ rather than HR. This mirrors our PCR results, in which doublets of ∼700 and 650 bp were detected, likely representing the inclusion or lack of the 50-bp attP sequence (Figure 1H, lanes 3, 10 and 11). The higher-molecular-weight bands faintly visible in some lanes may reflect aberrant repair events (Figure 1H, lanes 11 and 15). We observed such an event in one of eight clones. In this clone, the 4.6-kb yellow locus was deleted and part of the attP sequence was incorporated along with 40 bp of novel sequence. This unusual event may reflect repair via a synthesis-dependent strand annealing (SDSA) mechanism in which the two broken DNA strands independently select repair templates (Nassif et al. 1994). Previous studies of HR induced by DSBs in genomic targets have also identified SDSA as a primary mechanism of repair and reported similar events (Nassif et al. 1994; Bozas et al. 2009).
Germline transmission of Cas9-induced genome modifications
Our results demonstrate that the readily programmed CRISPR RNA/Cas9 system can be employed to precisely mutate, delete, or replace genes in vivo, so we next sought to determine whether Cas9-induced mutations could be transmitted through the germline to create stable mutant strains. We crossed adult flies injected as embryos with Cas9 and YE1 chiRNA to flies carrying a null mutation in yellow (y1), allowing us to phenotypically screen female progeny of injected males and all progeny of injected females. A total of 6.4% of injected males and 4.8% of injected females yielded at least one offspring exhibiting the yellow null phenotype (Table 1, injected flies that yield yellow offspring are referred to as “founders”). To determine if yellow progeny resulted from Cas9-induced mutations, the target locus was amplified and sequenced from at least one yellow progeny per founder. Indels were present at the predicted Cas9 cut site in each case (Figure 2A). Interestingly, we recovered a 4-bp deletion in the progeny of multiple founders. This is likely due to preferential repair by microhomology-mediated end joining (MMEJ) facilitated by the presence of a short repeat adjacent to the YE1 cut site (Figure 2A, underlined in wild-type sequence) (McVey and Lee 2008). Thus, the rate of mutations induced by Cas9 and a single chiRNA is comparable to that obtained with a pair of ZFNs targeting the yellow locus (6% transmission for males and 0% for females) and lower than that reported for two TALENs (17% for males and 45% for females) (Bibikova et al. 2002; Liu et al. 2012).
Table 1. Germline transmission rates.
| Injected males |
Injected females |
||||||
|---|---|---|---|---|---|---|---|
| chiRNA(s) | ssODN donor | % (no.) founders | % (no.) progeny | % (no.) founders | % (no.) progeny | % (no.) founders yielding targeted event | % (no.) overall germline transmission |
| YE1 | — | 6.4 (3/47) | 0.23 (5/2128) | 4.8 (1/21) | 0.26 (4/1525) | 100 (4/4) | 5.9 (4/68) |
| Y5′, Y3′ | — | 25 (13/52) | 1.3 (61/4608) | 14.3 (5/35) | 1.5 (35/2421) | 5.6 (1/18) | 1.1 (1/87) |
| Y5′, Y3′ | + | 8.6 (3/35) | 1.0 (24/2336) | 23.1 (6/26) | 1.3 (34/2655) | 22.2 (2/9) | 3.3 (2/61) |
Flies injected with Cas9 and the indicated chiRNAs and ssODN template were crossed to yellow (y1) and progeny screened for yellow cuticles. The percentage of injected flies producing one or more yellow progeny (founders) is indicated along with the percentage of total progeny exhibiting yellow cuticle. At least one progeny per founder was sequenced to determine if the targeted event had occurred. The percentage of founders in which the targeted event occurred in one or more progeny is reported, as is the overall germline transmission rate (percentage injected flies yielding expected event).
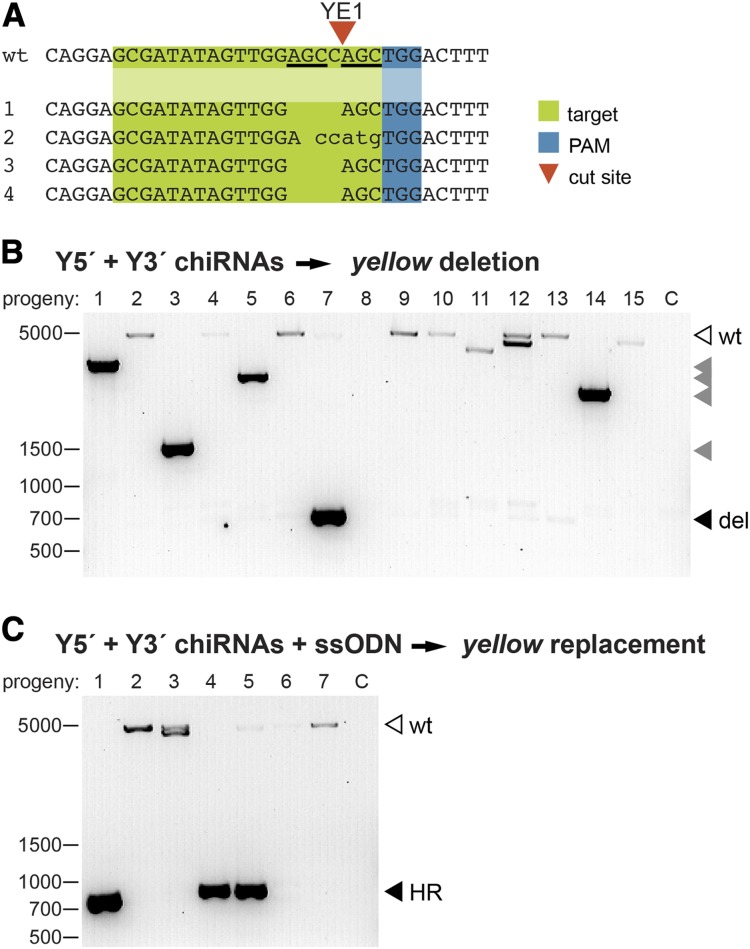
Figure 2.
CRISPR-induced modifications are efficiently transmitted through the germline. (A) Representative sequences of independent yellow mutants generated by Y1E-guided Cas9-induced DSBs. The predicted YE1 cleavage site is indicated with a red arrowhead. Short sequence repeats flanking the cleavage site (underlined in the wild-type sequence) may have facilitated the preferential generation of the same 4-bp deletion in three independent founders by MMEJ. (B) The yellow locus was amplified from yellow progeny of males and females injected with Cas9, Y5′ chiRNA, and Y3′ chiRNA using the primers indicated in Figure 1A. Fifteen independent progeny are represented (C, no template control). A 650-bp PCR product is expected (solid arrowhead) if a targeted deletion of yellow has been transmitted. PCR products corresponding to partial deletions (shaded arrowheads), and wild-type (open arrowhead) yellow are indicated. (C) The yellow locus was amplified from yellow progeny of flies injected with Cas9, Y5′ chiRNA, Y3′ chiRNA, and ssODN using the primers indicated in Figure 1A. Seven independent progeny are represented (C, no template control). A 700-bp product is expected following the targeted replacement of yellow with attP. PCR products corresponding to correct HR events are indicated (solid arrowheads) and were confirmed by Sanger sequencing.
We also investigated whether multiplex-mediated genome editing was transmitted through the germline. We first analyzed the progeny of flies that had been injected with Cas9, Y5′ chiRNA, and Y3′ chiRNA to precisely delete yellow. A total of 25% of injected males and 14.3% of injected females produced yellow progeny (Table 1, founders). PCR and sequence analysis of mutant progeny identified the precise deletion of yellow in the progeny of one founder (Figure 2B and Table 1). Partial deletions of yellow, up to ∼3.7 kb in size, were also detected by PCR and sequence analysis, consistent with imperfect repair of Cas9-induced deletions (Figure 2B).
We next analyzed flies that had been injected with Cas9, Y5′ chiRNA, Y3′ chiRNA, and the ssODN donor template to determine if the replacement of yellow with attP was also transmitted to progeny. A total of 8.6% of injected males and 23.1% of injected females produced one or more yellow offspring (Table 1, founders). Sequence analysis showed that two (22%) of the founders transmitted the precisely engineered replacement of yellow with attP (Figure 2C and Table 1). By comparison, depending on the specific donor, 1–19% of mutant progeny of flies injected with ZFNs and ssODN donor templates designed to introduce stop mutations in a rosy exon reflected HR-mediated repair (Beumer et al. 2013). These data demonstrate that Cas9-mediated HR events are effectively transmitted through the germline.
A common concern with sequence-guided nucleases is the potential for off-target cleavage. Cong et al. (2013) demonstrated that cleavage by Cas9 requires a PAM and a perfect match of at least 12 nt at the 3′ end of the targeting sequence, while single mismatches in the remaining 8 nt of targeting sequence are tolerated. Given the high frequency at which PAM sites occur in the Drosophila genome, we were able to readily select targets with minimal potential for off-target cleavage. Of the three chiRNAs used to generate germline transformants, only the Y5′ chiRNA has a 12-nt match to sequence adjacent to a PAM elsewhere in the genome. PCR and sequence analysis of this site in flies carrying Y5′-mediated deletion or replacement of yellow demonstrated the presence of wild-type sequence, consistent with the absence of off-target cleavage (Figure S3). Moreover, engineered flies developed into viable and fertile adults. The lack of detectable off-target effects is in accordance with findings in other systems and suggests high specificity of targeting by the CRISPR RNA/Cas9 system (Wang et al. 2013).
In summary, overall germline transmission rates, defined as the percentage of injected flies that yielded at least one offspring carrying the targeted event, ranged from 1.1% for multiplex-mediated deletion of yellow to 5.9% for single chiRNA-induced NHEJ. Perfect replacement of yellow with attP was transmitted through the germline of 3.3% of injected flies (2/61). Importantly, transformed flies were healthy and fertile and showed no evidence of off-target cleavage.
Discussion
Our results demonstrate the efficient generation of genome modifications via the CRISPR RNA/Cas9 system in Drosophila and for the first time show germline transmission of Cas9-mediated modifications. A single chiRNA can guide Cas9 to a specific genomic sequence to induce DSBs that are imperfectly repaired by NHEJ, while multiplex targeting can be used to generate large defined deletions. By combining multiplex targeting of yellow with the introduction of an ssODN donor template containing exogenous sequence, we successfully replaced yellow with an attP docking site. Interestingly, while germline transmission rates for each of these manipulations were similar, injection of two chi-RNAs (with or without the ssODN donor) yielded ∼10 times more mosaic males than a single chiRNA. This may be due to the fact that multiplex injections provide multiple routes for mutating the target locus, including single DSBs induced by either chiRNA and repaired by NHEJ. The lower percentage of founders yielding the precise targeted event in the multiplex injection paradigms supports this interpretation. Our results also suggest that the frequency of events in somatic tissue does not directly correlate with the frequency of events in the germline, consistent with previous observations with ZFNs and TALENs in Drosophila (Bibikova et al. 2002; Liu et al. 2012).
It is likely that the CRISPR RNA/Cas9 system can be further optimized. Studies in vertebrates suggest that expression levels of CRISPR RNAs are a key determinant of efficiency (Jinek et al. 2013). Transgenic lines expressing Cas9 and tracrRNA under the control of endogenous germline-specific promoters might yield increased efficiency. Moreover, it may be possible to bias repair to HR over NHEJ by exploiting a Cas9 mutant that induces single-strand DNA breaks (Jinek et al. 2012; Cong et al. 2013; Mali et al. 2013) or conducting Cas9-mediated genome engineering in a genetic background that reduces NHEJ (Beumer et al. 2008). Because the components of the CRISPR RNA/Cas9 system can be injected into embryos of any genotype, it should also be possible to use a marked transposable element residing in the targeted locus to identify defined deletions or replacements by loss of the positive marker, obviating the need for molecular screening when screening by mutant phenotype is not possible. Finally, combining the CRISPR RNA/Cas9 system with dsDNA donors containing larger homology regions may also increase efficiency (Nassif et al. 1994; Beumer et al. 2013). While the generation of such templates is more labor intensive, the ability to incorporate a selectable marker would add to the utility of the CRISPR RNA/Cas9 system for some applications.
The ease of producing sequence-specific chiRNAs makes the CRISPR RNA/Cas9 system an appealing method for genome editing. From the initial cloning steps to identifying transformants, stable lines with targeted genome alterations can be generated within a month. Our demonstration that the CRISPR RNA/Cas9 system can be used to create and transmit precise genomic modifications in Drosophila opens the door to rapid engineering of the genome to investigate gene function and regulation.
Supplementary Material
Acknowledgments
We thank the Bloomington Drosophila Stock Center for fly stocks and Michael Cox, Carlos Flores, Judith Kimble, and Andrew Mehle for invaluable discussions and critical comments on the manuscript. We are grateful to Feng Zhang for sharing reagents and an unpublished manuscript with us, Kevin Eliceiri for sharing injection equipment, and Kathy Vaccaro for sharing her expertise on embryo injections. This work was funded by startup funds from the University of Wisconsin to M.M.H., J.W., and K.O.C.G. and grants from the National Institute of Neurological Disorders and Stroke, National Institutes of Health to J.W. (R00 NS072252) and K.O.C.G. (R00 NS060985). All plasmids used in this study will be made available to academic laboratories through the nonprofit distributor Addgene along with detailed guidance at http://FlyCRISPR.molbio.wisc.edu.
Footnotes
Communicating editor: J. Sekelsky
Literature Cited
- Banga S. S., Boyd J. B., 1992. Oligonucleotide-directed site-specific mutagenesis in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 89: 1735–1739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrangou R., Fremaux C., Deveau H., Richards M., Boyaval P., et al. , 2007. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315: 1709–1712 [DOI] [PubMed] [Google Scholar]
- Beumer K. J., Trautman J. K., Bozas A., Liu J. L., Rutter J., et al. , 2008. Efficient gene targeting in Drosophila by direct embryo injection with zinc-finger nucleases. Proc. Natl. Acad. Sci. USA 105: 19821–19826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beumer K. J., Trautman J. K., Mukherjee K., Carroll D., 2013. Donor DNA Utilization during gene targeting with zinc-finger nucleases. Genes Genomes Genet. 3: 657-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibikova M., Golic M., Golic K. G., Carroll D., 2002. Targeted chromosomal cleavage and mutagenesis in Drosophila using zinc-finger nucleases. Genetics 161: 1169–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozas A., Beumer K. J., Trautman J. K., Carroll D., 2009. Genetic analysis of zinc-finger nuclease-induced gene targeting in Drosophila. Genetics 182: 641–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang N., Sun C., Gao L., Zhu D., Xu X., et al. , 2013. Genome editing with RNA-guided Cas9 nuclease in zebrafish embryos. Cell Res. 23: 465–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S. W., Kim S., Kim J. M., Kim J. S., 2013. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat. Biotechnol. 31: 230–232 [DOI] [PubMed] [Google Scholar]
- Cong L., Ran F. A., Cox D., Lin S., Barretto R., et al. , 2013. Multiplex genome engineering using CRISPR/Cas systems. Science 339: 819–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dicarlo J. E., Norville J. E., Mali P., Rios X., Aach J., et al. , 2013. Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Res. 41: 4336–4343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Q., Regan S. N., Xia Y., Oostrom L. A., Cowan C. A., et al. , 2013. Enhanced efficiency of human pluripotent stem cell genome editing through replacing TALENs with CRISPRs. Cell Stem Cell 12: 393–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao G., McMahon C., Chen J., Rong Y. S., 2008. A powerful method combining homologous recombination and site-specific recombination for targeted mutagenesis in Drosophila. Proc. Natl. Acad. Sci. USA 105: 13999–14004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gloor G. B., Nassif N. A., Johnson-Schlitz D. M., Preston C. R., Engels W. R., 1991. Targeted gene replacement in Drosophila via P element-induced gap repair. Science 253: 1110–1117 [DOI] [PubMed] [Google Scholar]
- Gong W. J., Golic K. G., 2003. Ends-out, or replacement, gene targeting in Drosophila. Proc. Natl. Acad. Sci. USA 100: 2556–2561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Zhou W., Dong W., Watson A. M., Hong Y., 2009. From the cover: directed, efficient, and versatile modifications of the Drosophila genome by genomic engineering. Proc. Natl. Acad. Sci. USA 106: 8284–8289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang W. Y., Fu Y., Reyon D., Maeder M. L., Tsai S. Q., et al. , 2013. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat. Biotechnol. 31: 227–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishino Y., Shinagawa H., Makino K., Amemura M., Nakata A., 1987. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J. Bacteriol. 169: 5429–5433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J. A., et al. , 2012. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337: 816–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M., East A., Cheng A., Lin S., Ma E., et al. , 2013. RNA-programmed genome editing in human cells. Elife 2: e00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Li C., Yu Z., Huang P., Wu H., et al. , 2012. Efficient and specific modifications of the Drosophila genome by means of an easy TALEN strategy. J. Genet. Genomics 39: 209–215 [DOI] [PubMed] [Google Scholar]
- Makarova K. S., Grishin N. V., Shabalina S. A., Wolf Y. I., Koonin E. V., 2006. A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol. Direct 1: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P., Yang L., Esvelt K. M., Aach J., Guell M., et al. , 2013. RNA-guided human genome engineering via Cas9. Science 339: 823–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVey M., Lee S. E., 2008. MMEJ repair of double-strand breaks (director’s cut): deleted sequences and alternative endings. Trends Genet. 24: 529–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nassif N., Penney J., Pal S., Engels W. R., Gloor G. B., 1994. Efficient copying of nonhomologous sequences from ectopic sites via P-element-induced gap repair. Mol. Cell. Biol. 14: 1613–1625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong Y. S., Golic K. G., 2000. Gene targeting by homologous recombination in Drosophila. Science 288: 2013–2018 [DOI] [PubMed] [Google Scholar]
- Shen B., Zhang J., Wu H., Wang J., Ma K., et al. , 2013. Generation of gene-modified mice via Cas9/RNA-mediated gene targeting. Cell Res. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Johnston D., 2002. The art and design of genetic screens: Drosophila melanogaster. Nat. Rev. Genet. 3: 176–188 [DOI] [PubMed] [Google Scholar]
- Venken K. J., Bellen H. J., 2005. Emerging technologies for gene manipulation in Drosophila melanogaster. Nat. Rev. Genet. 6: 167–178 [DOI] [PubMed] [Google Scholar]
- Wakiyama M., Matsumoto T., Yokoyama S., 2005. Drosophila U6 promoter-driven short hairpin RNAs effectively induce RNA interference in Schneider 2 cells. Biochem. Biophys. Res. Commun. 331: 1163–1170 [DOI] [PubMed] [Google Scholar]
- Wang H., Yang H., Shivalila C. S., Dawlaty M. M., Cheng A. W., et al. , 2013. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 153: 910–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood A. J., Lo T. W., Zeitler B., Pickle C. S., Ralston E. J., et al. , 2011. Targeted genome editing across species using ZFNs and TALENs. Science 333: 307. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.