Abstract
In Schizosaccharomyces pombe, over 90% of transcription factor genes are nonessential. Moreover, the majority do not exhibit significant growth defects under optimal conditions when deleted, complicating their functional characterization and target gene identification. Here, we systematically overexpressed 99 transcription factor genes with the nmt1 promoter and found that 64 transcription factor genes exhibited reduced fitness when ectopically expressed. Cell cycle defects were also often observed. We further investigated three uncharacterized transcription factor genes (toe1+–toe3+) that displayed cell elongation when overexpressed. Ectopic expression of toe1+ resulted in a G1 delay while toe2+ and toe3+ overexpression produced an accumulation of septated cells with abnormalities in septum formation and nuclear segregation, respectively. Transcriptome profiling and ChIP-chip analysis of the transcription factor overexpression strains indicated that Toe1 activates target genes of the pyrimidine-salvage pathway, while Toe3 regulates target genes involved in polyamine synthesis. We also found that ectopic expression of the putative target genes SPBC3H7.05c, and dad5+ and SPAC11D3.06 could recapitulate the cell cycle phenotypes of toe2+ and toe3+ overexpression, respectively. Furthermore, single deletions of the putative target genes urg2+ and SPAC1399.04c, and SPBC3H7.05c, SPACUNK4.15, and rds1+, could suppress the phenotypes of toe1+ and toe2+ overexpression, respectively. This study implicates new transcription factors and metabolism genes in cell cycle regulation and demonstrates the potential of systematic overexpression analysis to elucidate the function and target genes of transcription factors in S. pombe.
Keywords: Schizosaccharomyces pombe, fission yeast, transcription factor, overexpression, microarray, cell cycle
TRANSCRIPTIONAL regulatory networks establish the gene expression programs responsible for normal growth and disease states. These networks are composed of direct interactions between transcription factors and the promoters of their target genes. Deletion mutant collections in model organisms have the potential to rapidly map transcriptional regulatory networks by systematic characterization of transcription factors. However, in Saccharomyces cerevisiae, almost 90% of transcription factor deletion strains do not exhibit growth defects in rich medium, complicating the use of this approach (Chua et al. 2004; Yoshikawa et al. 2011). One explanation for this occurrence is that most transcription factors are not active under optimal growth conditions. Transcriptome profiling of more than half of transcription factor deletion strains in rich medium have not been productive in identifying their direct target genes (Chua et al. 2004, 2006). Moreover, condition-specific transcription factors do not occupy promoters of their target genes when ChIP-chip experiments are conducted in rich medium (Lee et al. 2002; Chua et al. 2004; Harbison et al. 2004). Chemical genetic profiling has uncovered environmental perturbations that reduce the growth rate of deletion mutants, thereby identifying conditions in which gene activity may be required (Winzeler et al. 1999; Giaever et al. 2002; Hillenmeyer et al. 2008). However, the correlation between reduced fitness of the deletion strain and increased messenger RNA expression of the gene in wild type under the same conditions is surprisingly low, suggesting that growth phenotypes of deletion mutants may not indicate gene activity (Winzeler et al. 1999; Giaever et al. 2002). Alternatively, the lack of obvious phenotypes of transcription factor deletion strains in optimal conditions could be caused by a high level of functional redundancy among transcription factors. This is not likely the primary reason as the frequency of negative genetic interactions among transcription factor genes appears substantially lower than genes encoding other types of proteins (Costanzo et al. 2010; Zheng et al. 2010).
Systematic gene overexpression circumvents the difficulties associated with deletion studies and identifying the activating conditions of Saccharomyces cerevisiae transcription factors (Chua et al. 2006). Global analysis revealed that genes causing reduced fitness when overexpressed resulted mostly in gain-of-function phenotypes and were functionally enriched in transcription factor genes (Gelperin et al. 2005; Sopko et al. 2006; Yoshikawa et al. 2011). The reduced fitness was attributed to the induction of transcription factor activity by ectopic expression and the inappropriate expression of their target genes (hence the term “phenotypic activation”). Transcriptome profiling of 55 overexpression strains with reduced fitness identified putative target genes and binding specificities for most known and several uncharacterized transcription factors (Chua et al. 2006). These results reveal the potential of systematic overexpression to characterize transcription factors in organisms amenable to transgenic technologies.
The transcriptional regulatory network of the fission yeast Schizosaccharomyces pombe consists of ∼100 sequence-specific DNA-binding transcription factors regulating ∼5000 genes in the genome. Despite being an extensively studied model organism, its transcriptional regulatory network remains substantially incomplete. Approximately two-thirds of S. pombe transcription factors have been characterized to some degree with biological roles focused mainly on cell cycle control, meiosis, mating, iron homeostasis, stress response, and flocculation (Fujioka and Shimoda 1989; Miyamoto et al. 1994; Sugiyama et al. 1994; Nakashima et al. 1995; Takeda et al. 1995; Watanabe and Yamamoto 1996; Ribar et al. 1997; Horie et al. 1998; Labbe et al. 1999; Ohmiya et al. 1999, 2000; Abe and Shimoda 2000; Mata et al. 2002; Buck et al. 2004; Cunliffe et al. 2004; Alonso-Nunez et al. 2005; Mata and Bahler 2006; Mercier et al. 2006, 2008; Mata et al. 2007; Rustici et al. 2007; Aligianni et al. 2009; Prevorovsky et al. 2009; Ioannoni et al. 2012; Matsuzawa et al. 2012). However, for many of these, few bona fide target genes have been identified. The remaining one-third of transcription factors are poorly characterized with unknown functions, target genes, and binding specificity.
In this study, we constructed transcription factor deletion and overexpression strains to advance the mapping of the S. pombe transcriptional regulatory network. Most transcription factor deletion strains did not exhibit defects in generation time when grown in rich medium. Consequently, we constructed and characterized an array consisting of 99 strains, each overexpressing a unique transcription factor gene. Sixty-four of 99 S. pombe transcription factor genes caused a decrease in fitness when ectopically expressed with the nmt1 promoter. Of these transcription factor overexpression strains, 76.6% exhibited an elongated cell morphology relative to the control strain with some displaying various cell cycle defects. We further investigated three previously uncharacterized genes encoding fungal-specific Zn (2)-Cys (6) transcription factors that exhibited reduced fitness and cell elongation when ectopically expressed. These genes were named toe1+–toe3+ (toe1+/SPAC1399.05c, toe2+/SPAC139.03, toe3+/SPAPB24D3.01) for transcription factor overexpression elongated. Ectopic expression of toe1+ caused a G1 delay while overexpression of toe2+ and toe3+ resulted in an accumulation of septated cells with aberrant septum deposition and nuclear missegregation, respectively. Transcriptome profiling and ChIP-chip analysis of HA-tagged Toe1–3 under control of the nmt41 promoter revealed that Toe1-regulated genes were involved in the pyrimidine-salvage pathway, while Toe3 target genes likely functioned in polyamine synthesis. Ectopic expression of several putative target genes could recapitulate the phenotype of toe2+ and toe3+ overexpression, while the deletion of certain putative target genes could suppress the phenotypes of toe1+ and toe2+ overexpression.
Materials and Methods
Yeast strains, media, and general methods
Strains were grown on rich (YES) or minimal (EMM) medium and supplemented with G418, nourseothricin, and thiamine hydrochloride at a concentration of 150 mg/liter, 100 mg/liter, and 15 μM, respectively. Chlorpromazine hydrochloride (Sigma Aldrich, St. Louis) was added to YES medium at 100 and 300 µg/ml for hypersensitivity assays and transcriptome profiling, respectively. The strains used in this study are listed in Supporting Information, Table S1. Matings were performed on sporulation medium (SPA). For EMM minus nitrogen supplemented with uracil medium (EMM-N+U), NH4Cl was substituted with 200 mg/liter of uracil. ORFs driven by nmt1/41 promoters were ectopically expressed by culturing the overexpression strains in EMM lacking thiamine medium for 18–24 hr unless indicated otherwise. Standard genetics and molecular and cell biology techniques were carried out as described in Moreno et al. (1991).
Construction of deletion and overexpression strains
The oligonucleotides used to construct the transcription factor deletion and overexpression strains are listed in Table S2. Genes regulated by the nmt1 promoter were cloned into the pREP1 vector. For ChIP-chip experiments, toe1+, toe2+, and toe3+ were cloned into pSLF272 to generate C-terminal triple HA fusions (Forsburg and Sherman 1997). All clones were confirmed by sequencing, and lithium acetate was transformed to generate the overexpression strains. Western blotting with anti-HA F-7 antibody (Santa Cruz Biotechnology, Santa Cruz, CA) was used to verify the expression of the HA-tagged transcription factors. For deletion of putative target genes, the open reading frame was deleted by a PCR stitching method as described in detail in Kwon et al. (2012). The gene deletions were confirmed by colony PCR.
Fitness and cell-length scoring of transcription factor overexpression strains
All transcription factor overexpression strains were induced on solid EMM medium without thiamine for 48 hr and then microscopically examined. Each strain was initially patched on EMM medium supplemented with thiamine and incubated overnight at 30°. The strains were then transferred to EMM medium lacking thiamine, incubated for 24 hr at 30° to induce the nmt1 promoter, and then transferred again onto EMM medium lacking thiamine. After 24 hr at 30°, the strains were examined for colony and cell morphologies with a Zeiss AxioScope A1 tetrad microscope (Zeiss, Thornwood, NY). Because the nmt1 promoter does not reach maximum induction until ∼18 hr, the second transfer of the strains onto EMM medium lacking thiamine was required to accurately observe the colony and cell morphologies caused by overexpression of the transcription factor. Reduced fitness was identified by a decrease in colony size and scored as slight (1), moderate (2), and severe (3) consisting of approximately 30–100 cells/colony, 10–30 cells/colony, and <10 cells/colony, respectively, relative to the empty vector control strain (>100 cells/colony). Cell elongation was scored as mild (1), moderate (2), and severe (3) with cell lengths ∼1.5, 2, and 3 times of the control strain, respectively. A score of −1 was assigned to cells that appeared shorter than the control strain. The fitness and cell length of the control strain were scored as 0.
Fluorescence microscopy
Transcription factor overexpression strains were grown in liquid EMM lacking thiamine medium for 24 hr at 30°. Cells were methanol-fixed and stained with DAPI (1 µg/ml) and calcofluor white (50 µg/ml) to visualize nuclei and cell-wall material, respectively. Images were acquired with a Zeiss Axioskop 2 microscope (Zeiss) and Scion CFW Monochrome CCD Firewire Camera (Scion, Frederick, MD). Cell cycle defects detected in transcription factor overexpression strains were classified as aberrant septal deposition and/or multisepta, abnormal nuclear morphology reminiscent of condensed chromosomes and chromosome missegregation.
Microarray expression profiling and ChIP-chip experiments
Strains containing nmt41-driven HA-tagged Toe1–3 were cultured and induced in 200 ml EMM medium lacking thiamine for 20–24 hr at 30°. Half of the culture was utilized for microarray expression profiling in which the transcriptome of the transcription factor overexpression strain was compared to an empty vector control, while the other half was subjected to ChIP-chip analysis. Culturing, sample preparation, hybridization, normalization, and data analysis of the transcriptome and ChIP-chip experiments were carried out as described with detail in Kwon et al. (2012). Labeled complementary DNA samples were hybridized to Agilent S. pombe 8X15K expression and 4X44K Genome ChIP-on-chip microarrays and washed according to the manufacturer’s instructions (Agilent Technology, Santa Clara, CA). The microarrays were scanned with a GenePix4200A scanner (Molecular Devices, Sunnyvale, CA), and the transcriptome and ChIP-chip data were normalized with the R Bioconductor Limma package. The ChIP-chip data were analyzed by ChIPOTle Peak Finder Excel Macro (Buck et al. 2005). Cluster 3.0 (Eisen et al. 1998) and Java Treeview 1.1.6r2 (Saldanha 2004) were used to create heat-map images of microarray expression and ChIP-chip data. The microarray expression and ChIP-chip data have been submitted to the NCBI Gene Expression Omnibus Database (GSE46811).
Quantitative PCR
Strains containing nmt41-driven HA-tagged Toe1–3 were cultured and induced in 100 ml EMM medium without thiamine for 20–24 hr at 30°. The expression level of putative target genes in the nmt41-driven toe-HA strains were compared against an empty vector control. Culturing and total RNA extractions were performed as in the expression microarray experiments. Reverse transcription was performed on total RNA using SuperScript II Reverse Transcriptase (Life Technologies, Carlsbad, CA) and Oligo(dT)23 anchored primers (Sigma-Aldrich, St Louis), following the manufacturers’ instructions. Quantitative PCR (qPCR) reactions were set up in MicroAmp Fast Optical 48-Well Reaction Plates using 5–50 ng cDNA, 1.2 ul of 0.5 uM forward and reverse primers, and 10 μl SYBR green master mix (Life Technologies). The act1+ gene was used as a reference for determining the relative expression of putative target genes. qPCR was performed on a StepOne Real-Time PCR system (Life Technologies) using the following program: 95° for 10 min, 40 cycles of 95° for 15 sec, and 58° for 1 min, followed by a melting curve program of 58°–95° with a heating rate of 0.3°/sec. Three replicates were carried out for each combination of query gene and strain. Fold changes were determined by the ΔΔCt method according to the manufacturer’s recommendations (Life Technologies).
Flow cytometry
A strain containing chromosomal-integrated pREP1-toe1+ was cultured in 100 ml EMM medium with and without thiamine for 20–24 hr at 30°. This strain was used to reduce the phenotypic heterogeneity caused by variations in plasmid copy number. Approximately 1 × 107 cells were fixed in 1 ml of 95% EtOH, resuspended in 50 mM sodium citrate (pH 7.0), and treated with 250 µg/ml RNAse A (Roche Applied Science, Indianapolis) at 50° for 2 hr and 2 µg/ml Proteinase K (Promega, Madison, WI) at 37° for 1 hr. Cells were then washed and resuspended in 50 mM sodium citrate (pH 7.0) containing propidium iodide (8 µg/ml) and sonicated briefly to minimize doublets. Flow cytometry was carried out with a FACSCalibur Flow Cytometer and FACSDiva 6.0 software (BD Biosciences, Franklin Lakes, NJ).
Results
Construction and phenotypic characterization of the transcription factor overexpression array
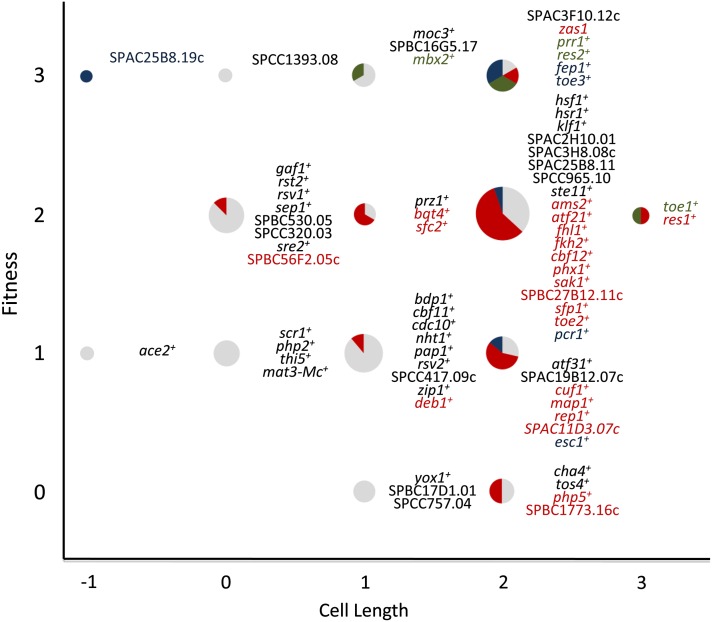
The transcription factors were derived from a list of 129 S. pombe candidate proteins that contained bona fide DNA-binding domains and other domains known to be associated with transcriptional regulation (Beskow and Wright 2006). This list was reduced to 99 candidate sequence-specific transcription factors after removal of proteins involved in chromatin remodeling, general transcription, and nontranscriptional roles. Among the 99 genes that encode these transcription factors, 62 have gene names and are primarily implicated in cell cycle control, meiosis, mating, iron homeostasis, stress response, and flocculation (Fujioka and Shimoda 1989; Miyamoto et al. 1994; Sugiyama et al. 1994; Nakashima et al. 1995; Takeda et al. 1995; Watanabe and Yamamoto 1996; Ribar et al. 1997; Horie et al. 1998; Labbe et al. 1999; Ohmiya et al. 1999, 2000; Abe and Shimoda 2000; Mata et al. 2002; Buck et al. 2004; Cunliffe et al. 2004; Alonso-Nunez et al. 2005; Mata and Bahler 2006; Mercier et al. 2006, 2008; Mata et al. 2007; Rustici et al. 2007; Aligianni et al. 2009; Prevorovsky et al. 2009; Ioannoni et al. 2012; Matsuzawa et al. 2012). The remaining 37 transcription factors have not been characterized and most contain the fungal-specific Zn (2)-Cys (6) DNA-binding domain. This transcription factor family is most predominant in S. pombe and S. cerevisiae containing 32 and 56 members, respectively, and has been implicated in diverse functions such as metabolism, meiosis, and flocculation (Todd and Andrianopoulos 1997; Kwon et al. 2012; Matsuzawa et al. 2013). We measured the generation times of 91 nonessential transcription factor haploid gene deletions in rich medium and found that only 10 displayed significant differences in their generation times compared to wild type (L. Vachon and G. Chua, unpublished data. The remaining eight transcription factor genes were either essential or previously published as nonessential but not able to be deleted from our study. We next constructed an overexpression array containing 99 strains of nmt1-driven transcription factor genes and microscopically examined their colony morphology to detect reduced fitness. Most transcription factor genes (64/99) resulted in a fitness defect when ectopically expressed (Figure 1). Among these 64 strains, the relative fitness decrease compared to the empty vector control was scored as mild (32.8%), moderate (50.0%), and severe (17.2%). Additionally, cell elongation and reduced fitness appeared to be correlated in the transcription factor overexpression strains (Figure 1). In fact, 76.6% of the strains with a fitness defect also displayed increased cell lengths relative to the empty vector control. Seven transcription factor overexpression strains displayed an abnormal cell length but no fitness defect (Figure 1). The remaining transcription factors (28.3%) did not exhibit reduced fitness or abnormal cell lengths when ectopically expressed.
Figure 1.
Phenotypic characterization of the S. pombe transcription factor overexpression array. Graph showing the phenotypes associated with ectopic expression of transcription factors in S. pombe. Strains containing an nmt1-driven transcription factor gene were scored for fitness defects (y-axis) and cell elongation (x-axis) on EMM lacking thiamine plates after 48 hr. To observe cell cycle phenotypes, transcription factor overexpression strains were grown in EMM lacking thiamine liquid medium for 24 hr and stained with DAPI and calcofluor white to visualize nuclei and cell-wall material, respectively. Transcription factors that did not result in a phenotype when ectopically expressed were not included. Fitness defects were scored as the following: (1) slight (∼30–100 cells per colony), (2) moderate (∼10–30 cells per colony), and (3) severe (<10 cells per colony). Cell elongation was scored as the following: (1) mild (∼1.5 times longer than control), (2) moderate (about twice the length of control), (3) severe (about three times longer than control), and (−1) short (shorter than control). Cell cycle phenotypes were classified as (red) aberrant septal deposition and/or multisepta, (green) abnormal nuclear morphology reminiscent of condensed chromosomes, and (blue) chromosome missegregation. The proportion of transcription factor overexpression strains with no cell cycle phenotypes were shown as gray sectors The relative fitness and cell length of the empty vector control were scored as 0. Transcription factor overexpression strains that do not exhibit any fitness and cell length defects were not shown.
The cell elongation phenotype suggested that ectopic expression of these transcription factors may cause defects in the cell cycle. Microscopic examination of these overexpression strains revealed that several exhibited cell cycle phenotypes such as multiseptation, multinucleation, nuclear missegregation, and aberrant septum deposition (Figure 1). We proceeded to investigate three uncharacterized Zn (2)-Cys (6) transcription factor genes that exhibited cell elongation when ectopically expressed. These three transcription factor genes were named toe+ for transcription factor overexpression elongated. Additional cell cycle phenotypes were detected from the ectopic expression of toe2+/SPAC139.03 (abnormally heavy septum deposition that often appeared lengthwise) and toe3+/SPAPB24D3.01 (nuclear missegregation). In contrast, the single-deletion strains of all three toe+ genes did not exhibit any detectable mutant phenotype in rich medium (data not shown).
Toe1 is a novel transcriptional regulator of the pyrimidine-salvage pathway
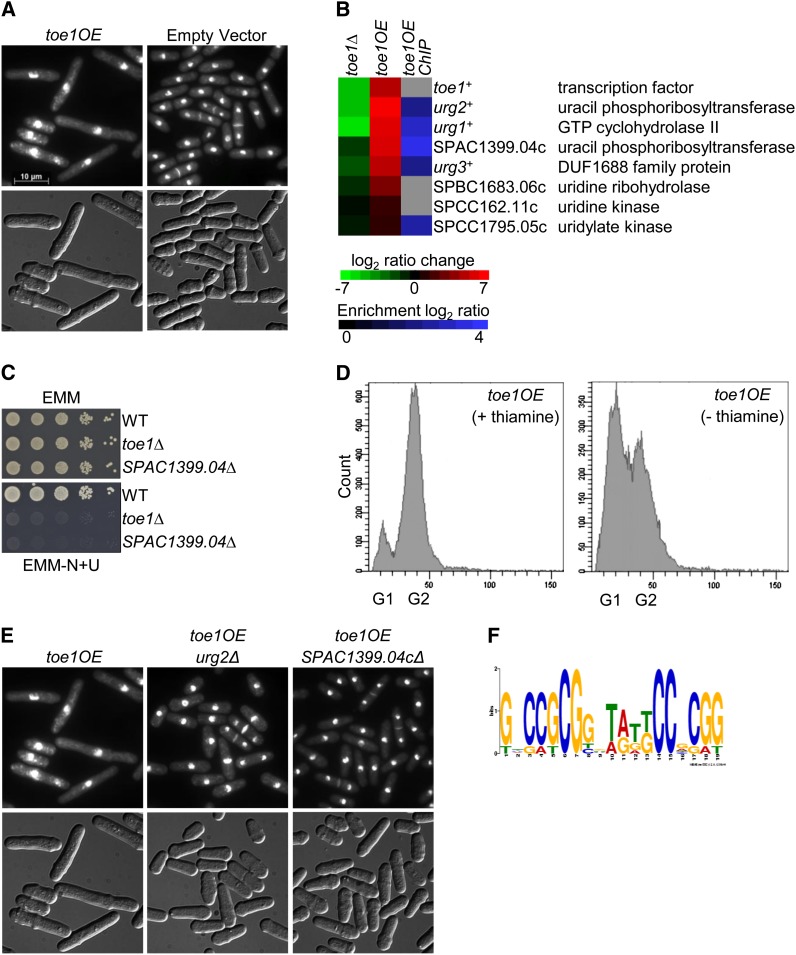
The ectopic expression of toe1+ causes a cell elongation phenotype (Figure 2A). Transcriptome profiling of S. cerevisiae transcription factor overexpression strains that exhibit reduced fitness have successfully identified their target genes and binding specificity (Chua et al. 2006; Chua 2009). We took a similar approach to characterize transcription factors in S. pombe, but also incorporated ChIP-chip experiments to better distinguish the target genes. An nmt41-driven toe1-HA strain was grown in medium lacking thiamine for 20–24 hr to induce the transcription factor gene, and then the culture was divided in two for transcriptome and ChIP-chip analyses. The moderate-strength nmt41 promoter was chosen over the strong nmt1 promoter to reduce secondary transcriptional effects in the microarray experiments. Similar to the toe1OE (nmt1) strain, cells containing the nmt41-driven toe1-HA were elongated after grown for 24 hr in medium lacking thiamine (data not shown).
Figure 2.
Identification of toe1 putative target genes by phenotypic activation. (A) Overexpression of toe1+ by the nmt1 promoter produces elongated cells. The toe1OE and empty vector strains were grown for 24 hr in EMM lacking thiamine medium at 30°. Cells were fixed with methanol and stained with DAPI and calcofluor white to visualize nuclei and cell-wall material, respectively (top panels). Cells are shown with Normarski in the bottom panels. (B) Putative target genes of toe1 involved in pyrimidine salvage are down-regulated in the toe1Δ strain, induced in the nmt41-toe1OE-HA strain, and bound by Toe1 at their promoters. The heat map shows the relative expression of seven putative target genes in the toe1Δ strain compared to wild type (left column) and the nmt41-driven toe1-HA strain compared to an empty vector control (middle column) by transcriptome profiling with dye reversal. The right column shows promoter occupancy of the putative target genes by toe1 with ChIP-chip analysis of an nmt41-driven toe1-HA strain. The color bars indicate the relative expression and ChIP enrichment ratios between experimental and control strains. (C) Loss of toe1+ and its putative target gene SPAC1399.04c prevents growth in medium containing uracil as the sole nitrogen source (EMM-N+U). Strains were spot-diluted on EMM and EMM lacking ammonium chloride with uracil (200 mg/liter) and incubated for 4 days at 30°. (D) Ectopic expression of toe1+ causes a G1 delay. Flow cytometric analysis of a chromosomal-integrated nmt1-driven toe1-HA strain under inducing (thiamine absent) and non-inducing (thiamine present) conditions. The histograms depict an increase in the percentage of cells in G1 and a reduction of cells in G2 in the toe1OE strain under inducing conditions compared to non-inducing conditions. (E) The cell elongation phenotype of the toe1OE strain is suppressed by the single deletion of the putative target genes urg1+ and SPAC1399.04c. An nmt1-driven toe1+ was ectopically expressed in each of the two corresponding deletion backgrounds. These strains were prepared and stained as described above. The presence of the pREP1-toe1+ vector in these strains was confirmed by growth on selective medium as well as by PCR. (F) A putative DNA motif resembling the binding specificity of Zn (2)-Cys (6) transcription factors was retrieved by promoter analysis of the toe1 putative target genes found in the heat map. The promoter regions (1000 bp upstream of the start codon) of the toe1 putative target genes were analyzed by MEME (Bailey et al. 2006).
Transcriptome profiling of the nmt41-driven toe1-HA strain revealed that 97 genes were induced at least twofold (Table S3). Gene ontology analysis of the top 50 most induced genes with the Princeton GO Term Finder (http://go.princeton.edu/cgi-bin/GOTermFinder) showed functional enrichment for the pyrimidine salvage pathway (P = 4.6e-5). Strikingly, the four most highly induced genes (ranging from 35.5- to 113.8-fold relative to the empty vector control) consisted of the uracil-regulatable genes urg1+, urg2+, and urg3+ (Watt et al. 2008) and an uncharacterized gene (SPAC1399.04c) predicted to encode a uracil phosphoribosyltransferase (Figure 2B). Moreover, these four genes were the most downregulated in the toe1Δ strain (ranging from 3.0- to 76.4-fold relative to wild type) (Figure 2B). These four genes contained protein sequence homology to the URC genes of Saccharomyces kluyveri, which function in the pyrimidine-salvage pathway through degradation of uracil (Andersen et al. 2008). Loss-of-function alleles of the URC genes result in growth inhibition on medium containing uracil as the sole nitrogen source (Andersen et al. 2008).
Interestingly, one of the URC genes encodes a Zn (2)-Cys (6) transcription factor, suggesting that Toe1 could be a putative regulator of the homologous genes in S. pombe. To determine if this was the case, we tested whether the toe1Δ strain and deletion of its putative target genes would be sensitive to medium containing uracil as the sole nitrogen source. Indeed, loss of toe1+ and SPAC1399.04c prevented growth under this condition (Figure 2C). In addition, several putative genes functioning in the pyrimidine-salvage pathway such as SPBC1683.06c (uridine ribohydrolase), SPCC162.11c (uridine kinase), and SPCC1795.05c (uridylate kinase were upregulated (10.7-, 2.8-, and 2.5-fold, respectively) in the toe1OE strain (Figure 2B).
ChIP-chip analysis of the nmt41-driven toe1-HA strain showed Toe1 association with 15 promoters (Table S4). Of the seven highly up-regulated pyrimidine-salvage pathway genes in the toe1+ overexpression data, five were detected with ChIP-chip, indicating that these genes are likely direct target genes of Toe1 (Figure 2B). Because urg2+ and urg3+ are adjacent divergent genes, Toe1 binding in the intergenic region may result in the regulation of both these genes. The seven most highly induced putative target genes were also validated by qPCR (Table S5).
The cell elongation phenotype of the toe1OE strain suggests a defect in the cell cycle. Examination of the septation index between toe1OE and wild-type strains revealed no significant difference (data not shown). However, overexpression of toe1+ appeared to cause an accumulation of cells in G1, indicating a delay in this cell cycle phase (Figure 2D). We also constructed single overexpressions of the pyrimidine-salvage pathway genes and examined the strains for cell elongation. None of these overexpression strains resulted in cell elongation (data not shown). Interestingly, single deletions of urg2+ and SPAC1399.04c could suppress the cell elongation phenotype of ectopic toe1+ expression (Figure 2E).
The promoter regions (1000 bp upstream of the start codon) of the pyrimidine-salvage pathway genes were subjected to Multiple Em for Motif Elicitation (MEME) analysis to elucidate the binding specificity of Toe1 (Bailey et al. 2006). The highest-scoring DNA motif for Toe1 contained inverted terminal CCG/GGC trinucleotides flanking a predominantly degenerate region of 11 nucleotides (P = 5.5e-14; Figure 2F). This DNA motif most resembled the known binding specificity (CGGN11CCG) of the Zn (2)-Cys (6) transcription factors Gal4p (S. cerevisiae) and Lac9p (Kluyveromyces lactis) (Carey et al. 1989; Halvorsen et al. 1991; Todd and Andrianopoulos 1997).
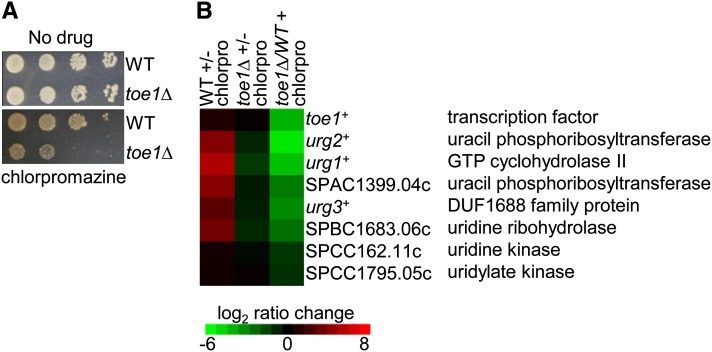
From screening our transcription factor deletion array to several drug compounds, we discovered that the toe1Δ strain was hypersensitive to the phenothiazine antipsychotic drug chlorpromazine (Figure 3A; L. Vachon and G. Chua, unpublished data). Chlorpromazine may inhibit uridine kinase, a key enzyme in pyrimidine salvage (Tseng et al. 1986). The hypersensitivity could indicate that the activity of toe1+ is required for adapting to chloropromazine, and thus Toe1 target genes may be induced by chlorpromazine treatment. Indeed, most of the Toe1 putative target genes functioning in the pyrimidine-salvage pathway were induced in chlorpromazine-treated wild type, but not in the toe1Δ strain (Figure 3B; left and middle columns, respectively). Consistently, the transcript levels of these target genes were lower in the toe1Δ strain relative to wild type when both strains were treated with chlorpromazine (Figure 3B; right column). We also investigated whether overexpression and deletion of the putative target genes could confer resistance and sensitivity, respectively, to chlorpromazine. However, none of these strains exhibited altered responses to chloropromazine treatment, possibly because many of the enzymes in the pyrimidine-salvage pathway are encoded by multiple genes with overlapping gene function (data not shown). Altogether, these results indicate that Toe1 transcriptionally activates genes functioning in the pyrimidine-salvage pathway and has a role in regulating cell cycle progression.
Figure 3.
Response of toe1+ to chlorpromazine treatment. (A) Loss of toe1+ results in sensitivity to chlorpromazine. Exponentially growing toe1Δ and wild-type strains were spot diluted on YES medium lacking or containing chlorpromazine (100 µg/ml) and incubated for 3 days at 30°. (B) Transcriptome profiling of toe1Δ and wild-type strains treated with chlorpromazine. toe1 putative target genes were induced in the wild type but not in the toe1Δ strain upon chlorpromazine treatment (left and middle columns, respectively). As a result, the expression of the putative target genes is lower in the toe1Δ strain relative to wild type when treated with chlorpromazine (right column). The microarray experiments were performed with dye reversal. Relative expression ratios are indicated in the color bar. Chloropromazine treatment for the transcriptome profiling experiments was 300 µg/ml for 1.5 hr.
Putative target genes of Toe2 are required for proper septum formation
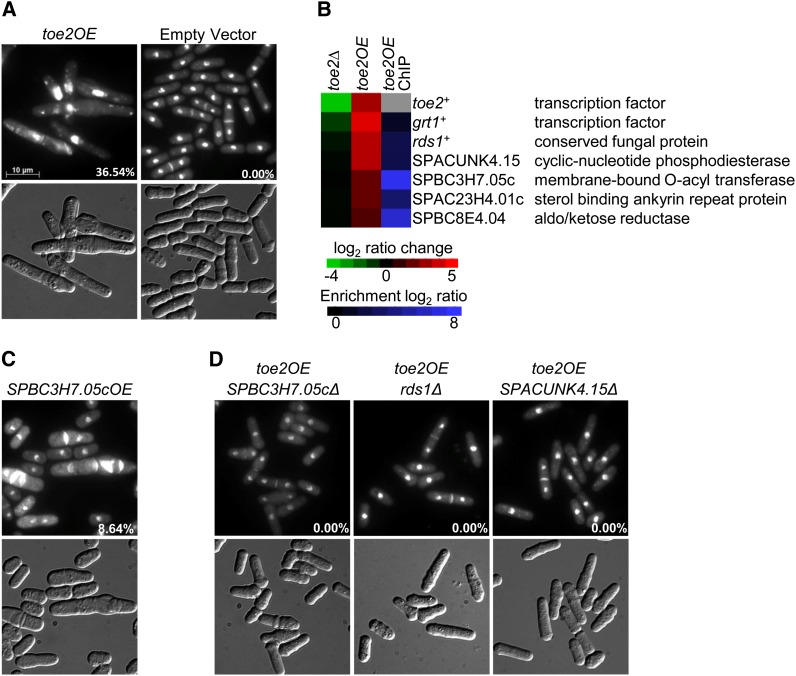
The ectopic expression of toe2+ under control of the nmt1 promoter causes defects in septum formation with abnormally heavy and often longitudinal septal deposition (Figure 4A). The proportion of cells exhibiting this aberrant phenotype was ∼36%. Ectopic expression of toe2+ under control of the nmt41 promoter also caused similar defects, although to a lesser degree (data not shown). In addition, the percentage of septated cells in the toe2OE strain was significantly higher than in the empty vector control (58.8% vs. 9.5%; two-tailed t-test; P-value < 0.002), indicating a stage-specific defect in the cell cycle. The nmt41-driven toe2-HA strain was analyzed by transcriptome profiling and ChIP-chip to uncover putative target genes. We found 114 genes that were upregulated at least twofold and 71 genes in which their promoters were associated with Toe2 (Table S6 and Table S7). The application of the Princeton GO Term Finder to the 114 genes showed functional enrichment for amino acid catabolism (P = 3.9e-4) while no functional enrichment was observed with the ChIP-chip data. Only 11 genes in the ChIP-chip data showed upregulation at least twofold in response to toe2+ overexpression (Table S6 and Table S7). These genes appeared to primarily function in metabolism and ion transport, and their involvement in septum formation was not obvious.
Figure 4.
Identification of Toe2 putative target genes by phenotypic activation. (A) Overexpression of toe2+ by the nmt1 promoter produces elongated cells that exhibit aberrant septal deposition. The toe2OE and empty vector strains were grown for 24 hr in EMM lacking thiamine medium at 30°. Cells were fixed with methanol and stained with DAPI and calcofluor white to visualize nuclei and cell-wall material, respectively (top panels). Cells are shown with Normarski in the bottom panels. (B) Putative target genes of Toe2 are induced in the nmt41-toe2OE-HA strain and are bound by Toe2 at their promoters. The heat map shows the relative expression of six putative target genes in the toe2Δ strain compared to wild type (left column) and the nmt41-driven toe2-HA strain compared to an empty vector control (middle column) by transcriptome profiling with dye reversal. The right column shows promoter occupancy of the putative target genes by Toe2 with ChIP-chip analysis of an nmt41-driven toe2-HA strain. The color bars indicate the relative expression and ChIP enrichment ratios between experimental and control strains. (C) Ectopic expression of the sequence orphan SPBC3H7.05c results in a similar aberrant septal deposition phenotype as seen in the toe2OE strain. The SPBC3H7.05cOE strain (nmt1-regulated) was cultured and prepared as described above. (D) The aberrant septal deposition phenotype of the toe2OE strain is abrogated by the single deletion of the putative target genes SPBC3H7.05c, rds1+, and SPACUNK4.15c. An nmt1-driven toe2+ was ectopically expressed in each of the three corresponding deletion backgrounds. These strains were prepared and stained as described above. The presence of the pREP1-toe2+ vector in these strains was confirmed by growth on selective medium as well as by PCR. Percentages indicate the proportion of cells exhibiting the septal defect.
Of these 11 genes, we decided to focus on the 6 most induced genes (3- to 21-fold induction) when toe2+ was overexpressed (Figure 4B). The induction of these 6 genes in the nmt41-driven toe2-HA strain was validated by qPCR (Table S5). These 6 genes appeared to not be differentially expressed in the toe2Δ strain (Figure 4B). Ectopic expression of these 6 genes singly revealed that only SPBC3H7.05c, which encodes a membrane-bound O-acyl transferase, resulted in aberrant septal deposition similar to the toe2OE strain although a lower proportion of cells exhibited this phenotype (Figure 4C). In addition, a few cells showing multiseptation and nuclear missegregation were observed in the SPBC3H7.05cOE strain (data not shown). The putative target gene SPAC23H4.01c that encodes a sterol-binding ankyrin repeat protein did not replicate the septal phenotype of the toe2OE strain when overexpressed, but produced elongated multiseptated cells (data not shown). To further validate the Toe2 putative target genes, toe2+ was overexpressed in strains containing single deletions of these genes. We found that loss of SPBC3H7.05c, as well as of SPACUNK4.15 and rds1+ that encode a predicted 2′,3′-cyclic-nucleotide 3′-phosphodiesterase and conserved fungal protein, respectively, could suppress the septal phenotype of the toe2OE strain (Figure 4D). These results identify several putative target genes of Toe2, including SPBC3H7.05c, that appear to play a role in septation in S. pombe.
Toe3 activates putative target genes involved in arginine catabolism and nuclear segregation
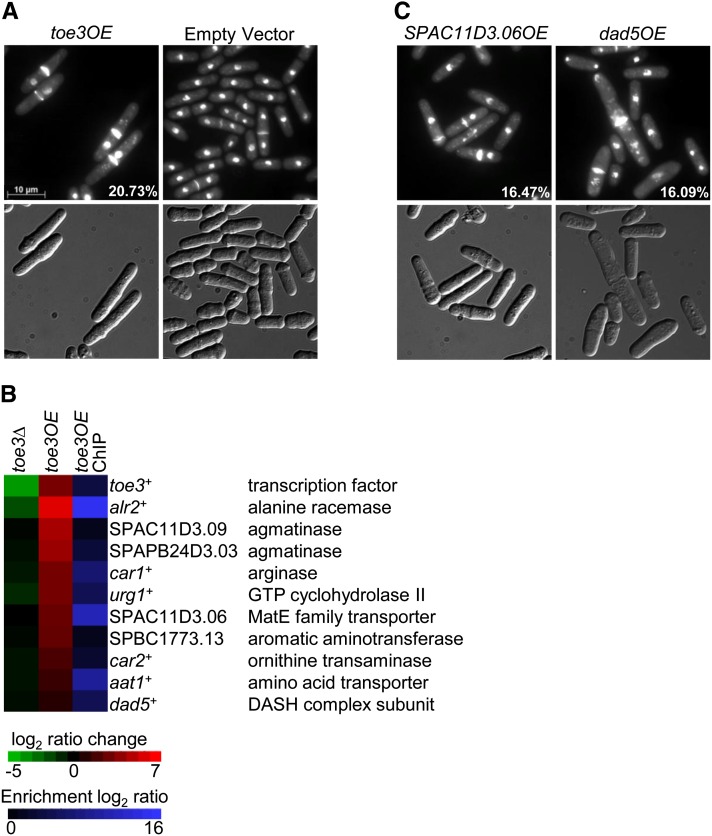
The ectopic expression of toe3+ under control of the nmt1 promoter results in a defect in nuclear segregation, where ∼20% of cells are observed with a septum and a single nucleus positioned distally (Figure 5A). The nmt41-driven toe3-HA strain exhibited a similar phenotype, although with reduced penetrance (data not shown). The percentage of septated cells in the toe3OE strain was also significantly higher than the empty vector control (20.6% vs. 9.5%; two-tailed t-test; P-value < 0.03), indicating a stage-specific defect in the cell cycle. Among the septated cells, over 80% exhibited the nuclear missegregation phenotype. To identify the Toe3 target genes, we performed transcriptome and ChIP-chip analyses on the nmt41-driven toe3-HA strain. We found that 95 genes were induced at least twofold relative to the control strain while the promoters of 174 genes were associated with Toe3 (Table S8 and Table S9). The 95 genes induced at least twofold by toe3+ overexpression were subjected to the Princeton GO Term Finder and found to be functionally enriched in arginine catabolic process (P = 2.4e-5). The same functional enrichment was observed in the 10 genes identified by ChIP-chip and upregulated at least twofold when toe3+ was ectopically expressed (P = 4.8e-6). The genes implicated in arginine catabolism and potentially influencing polyamine intracellular levels included car1+, car2+, SPAPB24D3.03, and SPAC11D3.09 (Figure 5B). SPAC11D3.06 may have a role in polyamine transport as MatE transporters have been reported to transport agmatine in human embryonic kidney (HEK293) cells (Winter et al. 2011). In addition, Toe3 bound to its own promoter, suggesting the possibility of autoregulation (Figure 5B). The top 10 highly induced putative target genes identified by microarray expression profiling and ChIP-chip of the nmt41-driven toe3-HA strain were validated by qPCR (Table S5). Among these putative target genes, only alr2+ and urg1+ were downregulated at least twofold in the toe3Δ strain (Figure 5B).
Figure 5.
Identification of Toe3 putative target genes by phenotypic activation. (A) Overexpression of toe3+ by the nmt1 promoter produces elongated cells that exhibit a nuclear missegregation phenotype. The toe3OE and empty vector strains were grown for 24 hr in EMM lacking thiamine medium at 30°. Cells were fixed with methanol and stained with DAPI and calcofluor white to visualize nuclei and cell-wall material, respectively (top panels). Cells are shown with Normarski in the bottom panels. (B) Putative target genes of Toe3 are induced in the nmt41-toe3OE-HA strain and are bound by Toe3 at their promoters. The heat map shows the relative expression of 10 putative target genes in the toe3Δ strain compared to wild type (left column) and the nmt41-driven toe3-HA strain compared to an empty vector control (middle column) by transcriptome profiling with dye reversal. The right column shows promoter occupancy of the putative target genes by Toe3 with ChIP-chip analysis of an nmt41-driven toe3-HA strain. The color bars indicate the relative expression and ChIP enrichment ratios between experimental and control strains. (C) Ectopic expression of either SPAC11D3.06 or dad5+, which encodes a MATE family transporter and a DASH complex subunit, respectively, results in a nuclear missegregation phenotype that is similar to the toe3OE strain. The SPAC11D3.06OE and dad5OE strains (both nmt1-regulated) were cultured and prepared as described above. The presence of the pREP1-toe3+ vector in these strains was confirmed by growth on selective medium as well as by PCR. Percentages indicate the proportion of cells exhibiting the nuclear missegregation phenotype.
We next determined whether overexpression of the putative target genes could produce the nuclear missegregation phenotype of the toe3OE strain. Eight of the 10 putative target genes were overexpressed singly with the nmt1 promoter (aat1+, alr2+, car1+, car2+, dad5+, SPAC11D3.06, SPAPB24D3.03, and SPBC1773.13). Among these genes, ectopic expression of dad5+ and SPAC11D3.06 resulted in a nuclear missegregation phenotype with penetrance comparable to the toe3OE strain (Figure 5C). These results were consistent with the known essential role of Dad5 as a component of the Dam1/Duo1, Ask1, Spc34/Spc19, Hsk1 (DASH) complex in chromosome segregation (Sanchez-Perez et al. 2005). However, we did not observe suppression of the nuclear missegregation phenotype caused by toe3+ overexpression when these putative target genes were deleted singly (data not shown). Altogether, these results suggest that Toe3 may play a role in nuclear segregation by regulating dad5+, SPAC11D3.06, and potentially other genes involved in polyamine biosynthesis.
Discussion
The transcriptional regulatory network in S. pombe remains substantially incomplete. The target genes have not been identified for the majority of sequence-specific transcription factors and over one-third of them have not been investigated at all. Here, we employed systematic genetics to analyze all the transcription factors by overexpression.
Systematic overexpression analysis revealed that 65% of S. pombe transcription factors exhibited reduced fitness, approximately twice the frequency in S. cerevisiae (Sopko et al. 2006). This difference could be attributed to variations in scoring for reduced fitness and promoter strength. Interestingly, ∼75% of S. pombe transcription factor overexpression strains that showed reduced fitness also exhibited cell elongation, suggesting a potential role in the cell cycle. Approximately 8–15% of S. pombe genes exhibit moderate-to-strong periodic expression during the cell cycle, and thus a considerable number of transcription factors would probably be required for their transcriptional control (Rustici et al. 2004; Oliva et al. 2005; Peng et al. 2005). Moreover, approximately one-third of S. pombe transcription factors have been detected to display strong periodic expression during the cell cycle (Bushel et al. 2009). Furthermore, in S. cerevisiae, genes causing reduced fitness when ectopically expressed were functionally enriched for transcription factor and cell cycle regulator genes, which could be similar in S. pombe (Gelperin et al. 2005; Sopko et al. 2006; Yoshikawa et al. 2011).
Another possible explanation for transcription factor overexpression toxicity is the occurrence of transcriptional squelching (Gill and Ptashne 1988). Ectopic expression of a strong transcriptional activator has been shown to sequester general transcription factors of RNA polymerase II (Liu and Berk 1995; Tavernarakis and Thireos 1995; McEwan and Gustafsson 1997). The inhibition of cell growth usually associated with squelching is likely caused by the transcriptional repression of essential genes or a lethal combination of nonessential genes. These genes could potentially encode ribosomal proteins and cell cycle activators, which are found to be predominantly repressed in a hypomorphic allele encoding the RNA polymerase II component Rpb11p (Mnaimneh et al. 2004). Although we cannot rule out squelching, down-regulated genes in our toeOE strains were not enriched for ribosomal and cell cycle genes.
We discovered that the transcription factor Toe1 activates genes implicated in the pyrimidine-salvage pathway. The putative target genes urg1+, urg3+, and urg2+/SPAC1399.04c appear to be homologous to URC1, URC4, and URC6, respectively, in S. kluyveri, while toe1+ is probably the homolog of URC2 (Andersen et al. 2008). The URC genes function in the catabolism of uracil in S. kluyveri (Andersen et al. 2008). Similar to the URC genes, deletion of toe1+ and SPAC1399.04c prevented growth on medium containing uracil as the sole nitrogen source (Figure 2C). Moreover, several other genes involved in the pyrimidine-salvage pathway, such as SPBC1683.06c and SPCC162.11c, which encode a uridine ribohydrolase and uridine kinase, respectively, were induced by toe1+ overexpression (Figure 2B). We also detected chlorpromazine sensitivity in the toe1Δ strain, suggesting that Toe1 activity and activation of its target genes may be required for the proper cellular response to this drug (Figure 3A). Chlorpromazine has been reported to possibly inhibit uridine kinase activity in murine sarcoma cells (Tseng et al. 1986). If this is also the case in S. pombe, then inhibition of uridine kinase by chlorpromazine treatment could compromise overall pyrimidine-salvage capacity, thereby triggering a compensatory response by activating other genes of similar function. Indeed, the uracil catabolic genes were induced in chlorpromazine-treated wild type but not in the chlorpromazine-treated toe1Δ strain (Figure 3B). Furthermore, we discovered that toe1+ overexpression causes a G1 delay (Figure 2D). It may be that induction of pyrimidine-salvage genes could represent a signal for insufficient levels of nucleotides, thus preventing cells from undergoing a round of DNA replication.
The toe2OE strain exhibits a delay in cytokinesis with thickened and misplaced septa, indicating that this transcription factor functions in the proper formation of the division septum for cytokinesis. The uncharacterized gene SPBC3H7.05c is most likely a target gene of Toe2. Ectopic expression of SPBC3H7.05c replicated the septal phenotype of the toe2OE strain, while the septal phenotype of toe2+ overexpression was rescued in the SPBC3H7.05c deletion background. The SPBC3H7.05c gene encodes a membrane-bound O-acyl transferase (MBOAT), suggesting a function in lysophospholipid synthesis, but its exact role in septation remains unclear (Benghezal et al. 2007; Riekhof et al. 2007; Matsuda et al. 2008). In S. cerevisiae, loss of the MBOAT-encoding gene GUP1 causes defects in the cell wall and bipolar budding while loss of the homologous gene in Candida albicans showed misplaced septa and compromised hyphae formation (Ni and Snyder 2001; Ferreira et al. 2006, 2010). In addition, the single deletion of the putative target genes rds1+ and SPACUNK4.15, which encode a conserved fungal protein and predicted 2′,3′-cyclic-nucleotide 3′-phosphodiesterase, respectively, could also suppress the septation phenotype of the toe2OE strain. The rds1+ gene appears to be stress-responsive and a putative target gene of the iron and copper starvation transcription factor Cuf1, while the SPACUNK4.15 product has been implicated in transfer RNA splicing in other organisms (Culver et al. 1994; Ludin et al. 1995; Rustici et al. 2007; Schwer et al. 2008). How these genes actually function in septation remains unknown.
Ectopic expression of toe3+ results in an accumulation of septated cells containing a single nucleus in one compartment. The putative target genes of Toe3 were functionally enriched in arginine catabolism, including five that are likely to play a direct role in influencing polyamine levels. These include genes encoding for agmatinase (SPAC11D3.09 and SPAPB24D3.03), arginase (Car1), ornithine transaminase (SPBC1773.13), and a MatE transporter (SPAC11D3.06), which may be involved in transporting polyamines (Winter et al. 2011). These results indicate a possible role for toe3+ in proper nuclear segregation through the regulation of polyamine levels in the cell. Indeed, we observed that ectopic expression of SPAC11D3.06 recapitulates the nuclear missegregation phenotype of the toe3OE strain. In addition, the nuclear missegregation phenotype was also seen when another putative target gene, dad5+, was ectopically expressed. Dad5 is a subunit of the DASH complex involved in sister-chromatid segregation during anaphase by linking spindle fibers to the kinetochore (Miranda et al. 2005; Sanchez-Perez et al. 2005). Increased expression of dad5+ in the toe3OE strain might perturb the DASH complex by altering the stoichiometry of its components, thereby resulting in nuclear missegregation. However, deletion of dad5+ and SPAC11D3.06 singly could not suppress the nuclear missegregation phenotype of the toe3OE strain. This may be due to a functional redundancy in nuclear segregation by dad5+ and SPAC11D3.06.
In summary, we have utilized systematic overexpression to characterize transcription factors in S. pombe. Our analyses of three Zn (2)-Cys (6) transcription factors, which are commonly associated with metabolic regulation, have implicated several metabolites in cell cycle regulation. Metabolism genes are periodically expressed in the fission yeast cell cycle during maximal growth (Rustici et al. 2004). Because the majority of transcription factor genes cause reduced fitness when ectopically expressed, further analysis of these overexpression strains with approaches from this study have the potential to significantly contribute to the complete mapping of the transcriptional regulatory network in S. pombe.
Supplementary Material
Acknowledgments
This work was supported by grants from the Canadian Institutes of Health Research to G.C., National Sciences and Engineering Research Council of Canada to J.K., and Canada Foundation for Innovation to G.C. and J.K.
Footnotes
Communicating editor: K. M. Arndt
Literature Cited
- Abe H., Shimoda C., 2000. Autoregulated expression of Schizosaccharomyces pombe meiosis-specific transcription factor Mei4 and a genome-wide search for its target genes. Genetics 154: 1497–1508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aligianni S., Lackner D. H., Klier S., Rustici G., Wilhelm B. T., et al. , 2009. The fission yeast homeodomain protein Yox1p binds to MBF and confines MBF-dependent cell-cycle transcription to G1-S via negative feedback. PLoS Genet. 5: e1000626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso-Nunez M. L., An H., Martin-Cuadrado A. B., Mehta S., Petit C., et al. , 2005. Ace2p controls the expression of genes required for cell separation in Schizosaccharomyces pombe. Mol. Biol. Cell 16: 2003–2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen G., Bjornberg O., Polakova S., Pynyaha Y., Rasmussen A., et al. , 2008. A second pathway to degrade pyrimidine nucleic acid precursors in eukaryotes. J. Mol. Biol. 380: 656–666 [DOI] [PubMed] [Google Scholar]
- Bailey T. L., Williams N., Misleh C., Li W. W., 2006. MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 34: W369–W373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benghezal M., Roubaty C., Veepuri V., Knudsen J., Conzelmann A., 2007. SLC1 and SLC4 encode partially redundant acyl-coenzyme A 1-acylglycerol-3-phosphate O-acyltransferases of budding yeast. J. Biol. Chem. 282: 30845–30855 [DOI] [PubMed] [Google Scholar]
- Beskow A., Wright A. P., 2006. Comparative analysis of regulatory transcription factors in Schizosaccharomyces pombe and budding yeasts. Yeast 23: 929–935 [DOI] [PubMed] [Google Scholar]
- Buck V., Ng S. S., Ruiz-Garcia A. B., Papadopoulou K., Bhatti S., et al. , 2004. Fkh2p and Sep1p regulate mitotic gene transcription in fission yeast. J. Cell Sci. 117: 5623–5632 [DOI] [PubMed] [Google Scholar]
- Buck M. J., Nobel A. B., Lieb J. D., 2005. ChIPOTle: a user-friendly tool for the analysis of ChIP-chip data. Genome Biol. 6: R97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushel P. R., Heard N. A., Gutman R., Liu L., Peddada S. D., et al. , 2009. Dissecting the fission yeast regulatory network reveals phase-specific control elements of its cell cycle. BMC Syst. Biol. 3: 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey M., Kakidani H., Leatherwood J., Mostashari F., Ptashne M., 1989. An amino-terminal fragment of GAL4 binds DNA as a dimer. J. Mol. Biol. 209: 423–432 [DOI] [PubMed] [Google Scholar]
- Chua G., 2009. Identification of transcription factor targets by phenotypic activation and microarray expression profiling in yeast. Methods Mol. Biol. 548: 19–35 [DOI] [PubMed] [Google Scholar]
- Chua G., Robinson M. D., Morris Q., Hughes T. R., 2004. Transcriptional networks: reverse-engineering gene regulation on a global scale. Curr. Opin. Microbiol. 7: 638–646 [DOI] [PubMed] [Google Scholar]
- Chua G., Morris Q. D., Sopko R., Robinson M. D., Ryan O., et al. , 2006. Identifying transcription factor functions and targets by phenotypic activation. Proc. Natl. Acad. Sci. USA 103: 12045–12050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo M., Baryshnikova A., Bellay J., Kim Y., Spear E. D., et al. , 2010. The genetic landscape of a cell. Science 327: 425–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culver G. M., Consaul S. A., Tycowski K. T., Filipowicz W., Phizicky E. M., 1994. tRNA splicing in yeast and wheat germ. A cyclic phosphodiesterase implicated in the metabolism of ADP-ribose 1”,2”-cyclic phosphate. J. Biol. Chem. 269: 24928–24934 [PubMed] [Google Scholar]
- Cunliffe L., White S., McInerny C. J., 2004. DSC1-MCB regulation of meiotic transcription in Schizosaccharomyces pombe. Mol. Genet. Genomics 271: 60–71 [DOI] [PubMed] [Google Scholar]
- Eisen M. B., Spellman P. T., Brown P. O., Botstein D., 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95: 14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira C., Silva S., van Voorst F., Aguiar C., Kielland-Brandt M. C., et al. , 2006. Absence of Gup1p in Saccharomyces cerevisiae results in defective cell wall composition, assembly, stability and morphology. FEMS Yeast Res. 6: 1027–1038 [DOI] [PubMed] [Google Scholar]
- Ferreira C., Silva S., Faria-Oliveira F., Pinho E., Henriques M., et al. , 2010. Candida albicans virulence and drug-resistance requires the O-acyltransferase Gup1p. BMC Microbiol. 10: 238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsburg S. L., Sherman D. A., 1997. General purpose tagging vectors for fission yeast. Gene 191: 191–195 [DOI] [PubMed] [Google Scholar]
- Fujioka H., Shimoda C., 1989. A mating-type-specific sterility gene map1 is required for transcription of a mating-type gene mat1-Pi in the fission yeast Schizosaccharomyces pombe. FEMS Microbiol. Lett. 51: 45–48 [DOI] [PubMed] [Google Scholar]
- Gelperin D. M., White M. A., Wilkinson M. L., Kon Y., Kung L. A., et al. , 2005. Biochemical and genetic analysis of the yeast proteome with a movable ORF collection. Genes Dev. 19: 2816–2826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaever G., Chu A. M., Ni L., Connelly C., Riles L., et al. , 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418: 387–391 [DOI] [PubMed] [Google Scholar]
- Gill G., Ptashne M., 1988. Negative effect of the transcriptional activator GAL4. Nature 334: 721–724 [DOI] [PubMed] [Google Scholar]
- Halvorsen Y. D., Nandabalan K., Dickson R. C., 1991. Identification of base and backbone contacts used for DNA sequence recognition and high-affinity binding by LAC9, a transcription activator containing a C6 zinc finger. Mol. Cell. Biol. 11: 1777–1784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbison C. T., Gordon D. B., Lee T. I., Rinaldi N. J., Macisaac K. D., et al. , 2004. Transcriptional regulatory code of a eukaryotic genome. Nature 431: 99–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillenmeyer M. E., Fung E., Wildenhain J., Pierce S. E., Hoon S., et al. , 2008. The chemical genomic portrait of yeast: uncovering a phenotype for all genes. Science 320: 362–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horie S., Watanabe Y., Tanaka K., Nishiwaki S., Fujioka H., et al. , 1998. The Schizosaccharomyces pombe mei4+ gene encodes a meiosis-specific transcription factor containing a forkhead DNA-binding domain. Mol. Cell. Biol. 18: 2118–2129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioannoni R., Beaudoin J., Lopez-Maury L., Codlin S., Bahler J., et al. , 2012. Cuf2 is a novel meiosis-specific regulatory factor of meiosis maturation. PLoS ONE 7: e36338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon E. J., Laderoute A., Chatfield-Reed K., Vachon L., Karagiannis J., et al. , 2012. Deciphering the transcriptional-regulatory network of flocculation in Schizosaccharomyces pombe. PLoS Genet. 8: e1003104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labbe S., Pena M. M., Fernandes A. R., Thiele D. J., 1999. A copper-sensing transcription factor regulates iron uptake genes in Schizosaccharomyces pombe. J. Biol. Chem. 274: 36252–36260 [DOI] [PubMed] [Google Scholar]
- Lee T. I., Rinaldi N. J., Robert F., Odom D. T., Bar-Joseph Z., et al. , 2002. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science 298: 799–804 [DOI] [PubMed] [Google Scholar]
- Liu X., Berk A. J., 1995. Reversal of in vitro p53 squelching by both TFIIB and TFIID. Mol. Cell. Biol. 15: 6474–6478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludin K. M., Hilti N., Schweingruber M. E., 1995. Schizosaccharomyces pombe rds1, an adenine-repressible gene regulated by glucose, ammonium, phosphate, carbon dioxide and temperature. Mol. Gen. Genet. 248: 439–445 [DOI] [PubMed] [Google Scholar]
- Mata J., Bahler J., 2006. Global roles of Ste11p, cell type, and pheromone in the control of gene expression during early sexual differentiation in fission yeast. Proc. Natl. Acad. Sci. USA 103: 15517–15522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mata J., Lyne R., Burns G., Bahler J., 2002. The transcriptional program of meiosis and sporulation in fission yeast. Nat. Genet. 32: 143–147 [DOI] [PubMed] [Google Scholar]
- Mata J., Wilbrey A., Bahler J., 2007. Transcriptional regulatory network for sexual differentiation in fission yeast. Genome Biol. 8: R217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda S., Inoue T., Lee H. C., Kono N., Tanaka F., et al. , 2008. Member of the membrane-bound O-acyltransferase (MBOAT) family encodes a lysophospholipid acyltransferase with broad substrate specificity. Genes Cells 13: 879–888 [DOI] [PubMed] [Google Scholar]
- Matsuzawa T., Yoritsune K., Takegawa K., 2012. MADS box transcription factor Mbx2/Pvg4 regulates invasive growth and flocculation by inducing gsf2+ expression in fission yeast. Eukaryot. Cell 11: 151–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzawa T., Kageyama Y., Ooishi K., Kawamukai M., Takegawa K., 2013. The zinc finger protein Gsf1 regulates Gsf2-dependent flocculation in fission yeast. FEMS Yeast Res. 13: 259–266 [DOI] [PubMed] [Google Scholar]
- McEwan I. J., Gustafsson J., 1997. Interaction of the human androgen receptor transactivation function with the general transcription factor TFIIF. Proc. Natl. Acad. Sci. USA 94: 8485–8490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercier A., Pelletier B., Labbe S., 2006. A transcription factor cascade involving Fep1 and the CCAAT-binding factor Php4 regulates gene expression in response to iron deficiency in the fission yeast Schizosaccharomyces pombe. Eukaryot. Cell 5: 1866–1881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercier A., Watt S., Bahler J., Labbe S., 2008. Key function for the CCAAT-binding factor Php4 to regulate gene expression in response to iron deficiency in fission yeast. Eukaryot. Cell 7: 493–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda J. J., De Wulf P., Sorger P. K., Harrison S. C., 2005. The yeast DASH complex forms closed rings on microtubules. Nat. Struct. Mol. Biol. 12: 138–143 [DOI] [PubMed] [Google Scholar]
- Miyamoto M., Tanaka K., Okayama H., 1994. res2+, a new member of the cdc10+/SWI4 family, controls the ‘start’ of mitotic and meiotic cycles in fission yeast. EMBO J. 13: 1873–1880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mnaimneh S., Davierwala A. P., Haynes J., Moffat J., Peng W. T., et al. , 2004. Exploration of essential gene functions via titratable promoter alleles. Cell 118: 31–44 [DOI] [PubMed] [Google Scholar]
- Moreno S., Klar A., Nurse P., 1991. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194: 795–823 [DOI] [PubMed] [Google Scholar]
- Nakashima N., Tanaka K., Sturm S., Okayama H., 1995. Fission yeast Rep2 is a putative transcriptional activator subunit for the cell cycle ‘start’ function of Res2-Cdc10. EMBO J. 14: 4794–4802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni L., Snyder M., 2001. A genomic study of the bipolar bud site selection pattern in Saccharomyces cerevisiae. Mol. Biol. Cell 12: 2147–2170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohmiya R., Kato C., Yamada H., Aiba H., Mizuno T., 1999. A fission yeast gene (prr1(+)) that encodes a response regulator implicated in oxidative stress response. J. Biochem. 125: 1061–1066 [DOI] [PubMed] [Google Scholar]
- Ohmiya R., Yamada H., Kato C., Aiba H., Mizuno T., 2000. The Prr1 response regulator is essential for transcription of ste11+ and for sexual development in fission yeast. Mol. Gen. Genet. 264: 441–451 [DOI] [PubMed] [Google Scholar]
- Oliva A., Rosebrock A., Ferrezuelo F., Pyne S., Chen H., et al. , 2005. The cell cycle-regulated genes of Schizosaccharomyces pombe. PLoS Biol. 3: e225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng X., Karuturi R. K., Miller L. D., Lin K., Jia Y., et al. , 2005. Identification of cell cycle-regulated genes in fission yeast. Mol. Biol. Cell 16: 1026–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prevorovsky M., Grousl T., Stanurova J., Rynes J., Nellen W., et al. , 2009. Cbf11 and Cbf12, the fission yeast CSL proteins, play opposing roles in cell adhesion and coordination of cell and nuclear division. Exp. Cell Res. 315: 1533–1547 [DOI] [PubMed] [Google Scholar]
- Ribar B., Banrevi A., Sipiczki M., 1997. sep1+ encodes a transcription-factor homologue of the HNF-3/forkhead DNA-binding-domain family in Schizosaccharomyces pombe. Gene 202: 1–5 [DOI] [PubMed] [Google Scholar]
- Riekhof W. R., Wu J., Jones J. L., Voelker D. R., 2007. Identification and characterization of the major lysophosphatidylethanolamine acyltransferase in Saccharomyces cerevisiae. J. Biol. Chem. 282: 28344–28352 [DOI] [PubMed] [Google Scholar]
- Rustici G., Mata J., Kivinen K., Lio P., Penkett C. J., et al. , 2004. Periodic gene expression program of the fission yeast cell cycle. Nat. Genet. 36: 809–817 [DOI] [PubMed] [Google Scholar]
- Rustici G., van Bakel H., Lackner D. H., Holstege F. C., Wijmenga C., et al. , 2007. Global transcriptional responses of fission and budding yeast to changes in copper and iron levels: a comparative study. Genome Biol. 8: R73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldanha A. J., 2004. Java Treeview: extensible visualization of microarray data. Bioinformatics 20: 3246–3248 [DOI] [PubMed] [Google Scholar]
- Sanchez-Perez I., Renwick S. J., Crawley K., Karig I., Buck V., et al. , 2005. The DASH complex and Klp5/Klp6 kinesin coordinate bipolar chromosome attachment in fission yeast. EMBO J. 24: 2931–2943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwer B., Aronova A., Ramirez A., Braun P., Shuman S., 2008. Mammalian 2’,3′ cyclic nucleotide phosphodiesterase (CNP) can function as a tRNA splicing enzyme in vivo. RNA 14: 204–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sopko R., Huang D., Preston N., Chua G., Papp B., et al. , 2006. Mapping pathways and phenotypes by systematic gene overexpression. Mol. Cell 21: 319–330 [DOI] [PubMed] [Google Scholar]
- Sugiyama A., Tanaka K., Okazaki K., Nojima H., Okayama H., 1994. A zinc finger protein controls the onset of premeiotic DNA synthesis of fission yeast in a Mei2-independent cascade. EMBO J. 13: 1881–1887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda T., Toda T., Kominami K., Kohnosu A., Yanagida M., et al. , 1995. Schizosaccharomyces pombe atf1+ encodes a transcription factor required for sexual development and entry into stationary phase. EMBO J. 14: 6193–6208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavernarakis N., Thireos G., 1995. Transcriptional interference caused by GCN4 overexpression reveals multiple interactions mediating transcriptional activation. Mol. Gen. Genet. 247: 571–578 [DOI] [PubMed] [Google Scholar]
- Todd R. B., Andrianopoulos A., 1997. Evolution of a fungal regulatory gene family: the Zn(II)2Cys6 binuclear cluster DNA binding motif. Fungal Genet. Biol. 21: 388–405 [DOI] [PubMed] [Google Scholar]
- Tseng A., Jr, Brooks M., Cadman E., 1986. Modulation of fluoropyrimidine metabolism by chlorpromazine. Biochem. Biophys. Res. Commun. 138: 1009–1014 [DOI] [PubMed] [Google Scholar]
- Watanabe Y., Yamamoto M., 1996. Schizosaccharomyces pombe pcr1+ encodes a CREB/ATF protein involved in regulation of gene expression for sexual development. Mol. Cell. Biol. 16: 704–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt S., Mata J., Lopez-Maury L., Marguerat S., Burns G., et al. , 2008. urg1: a uracil-regulatable promoter system for fission yeast with short induction and repression times. PLoS ONE 3: e1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter T. N., Elmquist W. F., Fairbanks C. A., 2011. OCT2 and MATE1 provide bidirectional agmatine transport. Mol Pharm 8: 133–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzeler E. A., Shoemaker D. D., Astromoff A., Liang H., Anderson K., et al. , 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285: 901–906 [DOI] [PubMed] [Google Scholar]
- Yoshikawa K., Tanaka T., Ida Y., Furusawa C., Hirasawa T., et al. , 2011. Comprehensive phenotypic analysis of single-gene deletion and overexpression strains of Saccharomyces cerevisiae. Yeast 28: 349–361 [DOI] [PubMed] [Google Scholar]
- Zheng J., Benschop J. J., Shales M., Kemmeren P., Greenblatt J., et al. , 2010. Epistatic relationships reveal the functional organization of yeast transcription factors. Mol. Syst. Biol. 6: 420. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.