Abstract
Genomic traits such as codon usage and the lengths of noncoding sequences may be subject to stabilizing selection rather than purifying selection. Mutations affecting these traits are often biased in one direction. To investigate the potential role of stabilizing selection on genomic traits, the effects of mutational bias on the equilibrium value of a trait under stabilizing selection in a finite population were investigated, using two different mutational models. Numerical results were generated using a matrix method for calculating the probability distribution of variant frequencies at sites affecting the trait, as well as by Monte Carlo simulations. Analytical approximations were also derived, which provided useful insights into the numerical results. A novel conclusion is that the scaled intensity of selection acting on individual variants is nearly independent of the effective population size over a wide range of parameter space and is strongly determined by the logarithm of the mutational bias parameter. This is true even when there is a very small departure of the mean from the optimum, as is usually the case. This implies that studies of the frequency spectra of DNA sequence variants may be unable to distinguish between stabilizing and purifying selection. A similar investigation of purifying selection against deleterious mutations was also carried out. Contrary to previous suggestions, the scaled intensity of purifying selection with synergistic fitness effects is sensitive to population size, which is inconsistent with the general lack of sensitivity of codon usage to effective population size.
Keywords: stabilizing selection, purifying selection, mutational bias, genetic drift, codon usage, indels
THERE is an increasing interest in the evolutionary factors that shape the properties of genomes. Weak purifying selection, together with mutation and genetic drift, has often been used as the basis for evolutionary models of genomic traits such as codon usage (Li 1987; Bulmer 1991; McVean and Charlesworth 1999), intron presence and size (Lynch 2002), and the mutation rate (Lynch 2011; Sung et al. 2012). This has led to the proposal that species with a low effective population size (Ne), in which selection is relatively ineffective in relation to genetic drift and mutation (Wright 1931; Kimura 1983), are more likely than species with a high Ne to evolve selectively disadvantageous properties, such as lower codon usage bias, larger genome size, and a higher mutation rate (Lynch 2002, 2007, 2011; Sung et al. 2012). But there is no a priori reason to exclude the possibility that at least some genomic traits are subject to stabilizing selection rather than purifying selection, so that individuals with extreme values of the trait are at a selective disadvantage compared with those with intermediate values (Kimura 1981; Johnson 1999; Parsch 2003; Wang and Yu 2011).
Evidence that quantitative traits can be subject to stabilizing selection started to accumulate over a century ago (Bumpus 1899; Weldon 1901; Di Cesnola 1907). Subsequently, Fisher (1930b, pp. 105–111) showed that mutation could maintain variability in a trait under stabilizing selection. This pioneering work stimulated many later theoretical studies, reviewed by Bürger (2000). Most applications to biological problems have concerned externally measurable phenotypes, which are known to experience relatively strong selection (Haldane 1954; Kingsolver et al. 2001), so that deterministic models have been commonly used in this context. Stabilizing selection on genomic traits is, however, likely to be very weak, and so it is important to examine the effects of genetic drift as well as mutation, if we are to understand their behavior under stabilizing selection.
Several models of stabilizing selection on quantitative traits in finite populations have shown that the probability distribution of the trait mean is held close to the optimal value for the trait, unless the population size is far lower than is usual for a natural population (Lande 1976; Campbell 1984; Barton 1989; Bürger and Lande 1994; Bürger 2000, pp. 268–282). This is because the dispersion of the mean around the optimum is controlled by the product of Ne and the net intensity of selection on the trait (Lande 1976). This is likely to be far larger than the product of Ne and the selection coefficient s for a mutation at a given site in the genome—it is Nes that controls the fixation probabilities of mutations and the distribution of variant frequencies within a population (Wright 1931; Kimura 1983). It is thus possible for selection to be in effective control of the mean of a quantitative trait, while mutation and drift have significant effects on the fates of individual variants affecting the trait (Kimura 1981, 1983; Campbell 1984; Barton 1989).
These models tell us that a trait mean is likely to stay close to the optimum, except for small artificial populations or species on the verge of extinction. However, the models mostly assume that mutational effects on the trait are unbiased, so that the trait mean is unchanged by mutation pressure alone. This is unlikely to be true of the genomic traits mentioned above; there is, for example, evidence for a mutational bias in favor of small deletions over small insertions (Petrov et al. 1996; Petrov and Hartl 1998; Comeron and Kreitman 2000; Ptak and Petrov 2002; Parsch 2003; Leushkin et al. 2013) and for unpreferred over preferred codons (Sharp et al. 2005; Hershberg and Petrov 2008; Zeng and Charlesworth 2009; Zeng 2010).
A theoretical investigation of stabilizing selection in an infinite population has shown that the trait mean may be maintained close to the optimal value in the face of mutational bias (Waxman and Peck 2003). In contrast, it is known from the theory of mutation, drift, and selection on codon usage that mutational bias can cause the equilibrium level of codon usage bias to depart substantially from its maximum value, if the population size is sufficiently small (Kimura 1981; Li 1987; Bulmer 1991; McVean and Charlesworth 1999). Similarly, Zhang and Hill (2008) used computer simulations of the combined effects of mutational bias, genetic drift, and stabilizing selection on a quantitative trait to show that substantial deviations of the trait mean from the optimum can be produced when Ne is sufficiently small in relation to the intensity of selection.
No systematic theoretical investigation of the interaction between stabilizing selection, mutational bias, and drift has previously been carried out. One purpose of this article is to fill this gap, by developing analytical approximations for the parameters of interest, using a method originally developed by Kimura (1981), which is related to that used in models of selection on codon usage (Li 1987; Bulmer 1991; McVean and Charlesworth 1999). The analytical results were checked by numerical modeling, using stochastic simulations as well as a matrix method similar to that employed in recent investigations of other modes of selection (Eyre-Walker and Keightley 2009; Zeng and Charlesworth 2009).
These methods can also be used to study the interaction between mutation pressure and weak purifying selection against deleterious mutations, allowing for the possibility of epistasis among the fitness effects of deleterious mutations at different sites. This has potential relevance to the evolution of sex and recombination (Kimura and Maruyama 1966; Feldman et al. 1980; Kondrashov 1982, 1984, 1988; Charlesworth 1990), but finite population effects have largely been neglected except for Kondrashov (1995). It has also been proposed that synonymous sites subject to selection on codon usage may be subject to synergistic fitness effects, thereby explaining the apparent lack of any strong relation between the effective population size of a species and its level of codon usage bias (Akashi 1995, 1996; Kondrashov et al. 2006). The reasoning is that the intensity of selection against unpreferred codons increases as the proportion of sites with unpreferred codons increases, eventually stabilizing the level of codon usage bias.
The main novel conclusion is that the scaled measure of the intensity of selection for individual sites under stabilizing selection (defined here as γ = 4 Nes) is approximately independent of Ne over a wide range of parameters, provided that there is sufficient mutational bias to perturb the population mean from the optimal value by a small amount, thereby creating an overall pressure of selection in the opposite direction to the effect of mutational bias. This is because a larger departure of the mean from the optimum due to a lower Ne causes an increase in the magnitude of the selection coefficient s, which can almost precisely counteract the effect of the change of Ne on γ.
These results mean that studies utilizing data such as the frequency spectra of DNA sequence variants may find it virtually impossible to distinguish stabilizing from purifying selection. Furthermore, there is no reason to expect a noticeable positive relation between the trait mean and Ne for most biologically realistic values of Ne, since the deviation of the trait mean from the optimum is usually extremely small, in contrast to the results of models of purifying selection that have been used to model various aspects of genome evolution, e.g., Lynch (2002, 2007, 2011). In contrast, when there is synergistic selection against deleterious mutations, γ is a strongly increasing function of Ne; in addition, the mean number of deleterious mutations per individual declines as Ne increases, contradicting the idea that synergistic selection can counteract a reduction in codon usage bias caused by lower Ne.
Models of Stabilizing Selection
Basic assumptions
A model of selection and mutation acting on a quantitative trait in a randomly mating diploid population is used. This assumes a large number of exchangeable, independent sites, with a pair of additively acting, biallelic variants at each site, as first proposed and analyzed by Fisher (1930b, pp. 105–111). Essentially the same model can also be applied to haploids. Consider a large number m of sites, each with a variant of type A1 decreasing the trait value z and a variant of type A2 that increases it. The scale of measurement is such that individuals homozygous for type A1 alleles at every site have a value of –ma, and individuals homozygous for type A2 alleles at every site have a value of ma. In the context of genome evolution, for example, the trait could be the length of a collection of introns or intergenic sequences, with each variant representing an insertion or deletion of a set of bases.
It is convenient to assume a quadratic deviations model, such that the fitness of individuals with phenotypic value z is
| (1) |
where zo is the optimal value of the trait, and S is the intensity of selection (Fisher 1930b, p. 105).
For the case of very weak selection analyzed here, this is an excellent approximation to the commonly used nor-optimal selection model, where the natural logarithm of fitness is a quadratic function of the deviation from the optimum, e.g., Haldane (1954) and Lande (1976). The general conclusions should thus apply to this case.
Under the assumption that each site evolves independently within a population (i.e., there is no linkage disequilibrium), the change due to selection in the frequency qi of the A2 allele at the ith site in the system, neglecting higher-order terms in Sa2, is given by
| (2) |
This equation or its equivalent has been used repeatedly in the literature (e.g., Wright 1935; Barton 1986; Bürger 2000, p. 217). The parameter δ measures the deviation of the population mean from the optimum, relative to the effect of a single variant, such that
| (3) |
The term in 2δ in the equation represents the effect of directional selection due to the deviation of the mean from the optimum, while the term in 2qi – 1 represents the effect of stabilizing selection, which tends to push the allele frequency toward the nearest extreme when the mean coincides with the optimum.
We also have the useful relation
| (4) |
where is the mean of q over all sites (Bürger 2000, p. 217).
Following Zhang and Hill (2008), an additional “pleiotropic” directional selection coefficient, sd, could be added to 2δ + 2qi – 1 in Equation 2. All of the analyses described below can be carried out with this extension of the model, with 2Sa2δ being replaced by 2Sa2δ + sd in the calculations. If sd > 0, the expected equilibrium value of δ, as given by Equations A.3 and 12b, is reduced, but the value of the scaled selection coefficient, as given by Equations 9 and 12a, is unchanged. This extension is not considered further.
Mutational models
Two extreme possibilities for representing the effects of mutation are considered here. First, mutations in both directions may occur at each site, with a frequency u of mutations from A1 to A2 and a frequency κu in the opposite direction, where κ represents the extent of mutational bias. Mutation rates are thus dependent on the allelic states of sites, as is appropriate for nucleotide substitutions, such as those at sites affecting a quantitative trait. The mutational contribution to the change in frequency of type A2 alleles is then
| (5) |
If both mutation and selection terms are small, this can be added to Equation 2 to obtain an expression for the net allele frequency change at the ith site, Δqi = Δqis + Δqim. If the mutation rate at each site is sufficiently low, the mutational terms need not be applied to segregating sites, yielding a model that is similar to the modifications of the infinite sites model described by Kimura (1981), McVean and Charlesworth (1999), and Charlesworth and Charlesworth (2010, pp. 268–279). This allows simple analytical approximations to be obtained (see below).
Alternatively, the state of a site may not restrict the direction of a future mutation. This probably applies to indels, where the presence of an insertion or deletion does not preclude a further change of the same kind. Any site can then mutate to an insertion with probability u and to a deletion with probability κu, regardless of any previous mutational events at that site. Since in reality successive additions or deletions of sets of bases are unlikely to involve precisely the same nucleotide site, in this model it is probably best to regard m as referring to the number of representatives of a defined class of sequences that can be affected by the indels in question, such as short introns in Drosophila. This case is referred to as the “state-independent model,” in contrast to the alternative “state-dependent model.” It is similar to the ladder model of mutation used for electrophoretic and microsatellite loci (Ohta and Kimura 1973; Slatkin 1995), since the state of a particular location in the genome can evolve indefinitely in either direction by successive insertions or deletions.
Analysis of the state-dependent model: Further assumptions
The method for including the effects of finite population size will first be developed in relation to the state-dependent model, since it uses procedures previously developed for modeling selection on codon usage bias (Kimura 1981; Li 1987; Bulmer 1991; McVean and Charlesworth 1999). The number of sites is assumed to be sufficiently large that the distribution of variant frequencies over sites (including the two fixed classes) is close to the probability distribution of variant frequencies generated by drift, mutation, and selection. This implies that the mean value () of the frequencies of A2 variants across all sites for a given population is close to the expected value (q*) of the frequency at a random site, taken over the probability distribution of frequencies under drift, mutation, and selection. This assumption can be used to develop both numerical and analytical results. Its accuracy is evaluated in the Supporting Information, File S1.
In addition, linkage equilibrium is assumed; simulations have shown that this provides a good approximation to exact multilocus models of stabilizing selection, provided that selection is weak in relation to recombination, and the population size is sufficiently large (Bürger 2000). For simplicity, a Wright–Fisher population of size N is assumed; more generally, N can be replaced by the effective population size, Ne (Wright 1931; Charlesworth and Charlesworth 2010, Chap. 5).
Approximate analytical results for the state-dependent model
When there is sufficient mutational bias to perturb the population mean away from the optimum, the term in 2δ in Equation 2 dominates over 2qi – 1 (Kimura 1981). This means that selection on individual allele frequencies is effectively directional, so that Equation 2 can be replaced by an equation of the form
| (6a) |
where
| (6b) |
A difficulty is that δ depends on the population mean, and hence on . However, this problem can be solved by assuming that the mutation rate is sufficiently low that an infinite-sites model can be used (i.e., we have 4Nu << 1). In this case, the approximate expected value of , q*, at mutation–selection–drift equilibrium is determined by the probabilities of sites being fixed for A1 and A2, defined as f(0) and f(1), respectively (Kimura 1981; Bulmer 1991).
The ratio (1 – q*)/q* is then ≈f(0)/f(1); this is also close to the ratio (1 – )/ for a given population (File S1). At equilibrium, the rate of flow of probability from sites with q = 1 to sites with q = 0 must be equal to the flow in the opposite direction; these flows are proportional to κuQ1f(1) and uQ2f(0), respectively, where Q1 and Q2 are the probabilities of fixation of new A1 and A2 mutations, respectively (Kimura 1981; Bulmer 1991; Charlesworth and Charlesworth 2010, p. 272). We then have
| (7a) |
With a fixed selection coefficient s, the standard formula for fixation probability implies that Q1/Q2 ≈ exp(–γ), where γ = 4Ns is the scaled selection coefficient (Fisher 1930a; Charlesworth and Charlesworth, Chap. 6, p. 262).
If fluctuations in δ around its expectation over the entire probability distribution of q, denoted by δ*, are sufficiently small in relation to δ*, δ can be equated to δ*, and s in Equations 6 can be treated as fixed and equal to 2Sa2 δ*. The conditions under which this is valid are examined in File S1. From Equation 6b, this assumption implies that γ = 8NSa2δ*. Substituting into Equation 7a, we have
| (7b) |
This is equivalent to the Li–Bulmer equation used in the theory of selection on codon usage (Li 1987; Bulmer 1991).
Furthermore, using Equations 3 and 4, and writing b = zo/ma, we have
| (8) |
Useful approximations for γ and δ* can then be obtained, as shown in the Appendix. In particular, when 4NSma2 >> 1, Equations 6 and A.3b imply that
| (9) |
This analysis brings out the remarkable fact that γ at mutation–selection–drift equilibrium is approximately independent of the population size for a given set of mutation and selection parameters, at least over some of the range of possible parameter values. If b is small, so that the optimum is in the mid-range of possible values, γ ≈ ln(κ). Furthermore, the argument leading to Equations A.7 of the Appendix shows that Equations A3 for δ* can also be used for the case when there is a mixture of stabilizing and directional selection, but γ = 8NSa2δ* can no longer be interpreted as the product of 4N and a fixed selection coefficient and is therefore not sufficient to determine the fixation probabilities of A1 and A2 mutations.
Equations A.3b and A.7b imply that, as NSa2 increases, δ* becomes indefinitely small, consistent with previous results for very different models of mutation, which found that mutational bias has only a small effect on the population mean in a large population (Waxman and Peck 2003; Zhang and Hill 2008). Alternatively, approximate expressions for δ* and γ under the above conditions can be derived using the model of the joint effects of drift and selection on a quantitative trait developed by Lande (1976); they give qualitatively similar results to those obtained here, although quantitatively there is a disagreement, reflecting the different assumptions made in the two cases (see File S1). As described in File S1, this approach also can be used to show that the expected value of the trait mean is expected to approach the optimum as NSa2 increases, even when the conditions needed for the validity of the above results are violated.
Equation 9 also suggests, somewhat counterintuitively, that the values of δ* and γ are nonzero if b is nonzero, even if there is no mutational bias at the level of individual nucleotide sites (i.e., when κ = 1). This arises because a nonzero b implies that the mean frequency of A2 at equilibrium under selection departs from one-half; in a finite population, the frequencies of sites fixed for A1 and A2 are then unequal, so that there will be a higher net frequency of mutations from the allelic type favored by selection, effectively creating a mutational bias.
Knowing δ* and hence γ, we can use standard results from diffusion theory to obtain the overall probability distribution of q when Equations 6 hold. With reversible mutation and a Wright–Fisher population of size N, the probability density of q is
| (10) |
where C is a constant ensuring that the integral of φ(q) between 0 and 1 is equal to 1 (Wright 1931).
The distribution with the infinite sites assumption is represented by the limiting value of Equation 10 as 4Nu tends to zero, which is useful for calculating the theoretical value of the site frequency spectrum of variants affecting the trait in a sample from the population (McVean and Charlesworth 1999). This is described in more detail below. In addition, approximate expressions for the expected genetic variance in z (Vg*) can then be derived, as shown in the Appendix.
Numerical results for the state-dependent model
Some representative numerical results for equilibrium populations, generated as described in the Appendix, are shown in Table 1, together with approximate values of δ, γ, and Vg, generated by both the first-order approximations described above (the App. 1 columns), and the more exact method described in the Appendix (the App. 2 columns), which allows for both stabilizing and directional selection effects on allele frequencies. In all cases shown, the net intensity of selection on the trait, S, was 0.01, representing weak selection in relation to the population sizes used here: the maximal value of NS was 4 (for N = 400). If we were to scale up to a population size of 100,000 with this NS value, S would be only 4 × 10−5, corresponding to extremely weak selection compared with that normally expected for externally measurable traits (Haldane 1954; Turelli 1984; Kingsolver et al. 2001). As shown above, only the product NSa2 is relevant to the values of δ and γ, but Vg* is proportional to ma2 (see Appendix).
Table 1. Simulation and theoretical values of parameters under stabilizing selection with state-dependent mutations.
| N | δ |
γ (× 102) |
Vg (× 102) |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sim. |
Mat. | App. 1 | App. 2 | Sim. |
Mat. | App. 1 | App. 2 | Sim. |
Mat. | App. 1 | App. 2 | Stab. sel. | |||||
| Mean | S.D. | Mean | S.D. | Mean | S.D. | ||||||||||||
| κ = 2 | 50 | 14.9 (0.3) | 6.76 | 16.6 | 16.5 | 16.5 | 0.602 (0.012) | 0.270 | 0.672 | 0.660 | 0.660 | 2.89 (0.04) | 0.976 | 2.83 | 2.88 | 2.87 | 2.97 |
| z0 = 0 | 100 | 7.54 (0.21) | 4.77 | 8.57 | 8.45 | 8.40 | 0.610 (0.017) | 0.381 | 0.691 | 0.676 | 0.685 | 6.14 (0.06) | 1.42 | 5.64 | 5.77 | 5.73 | 5.88 |
| 200 | 4.37 (0.16) | 3.62 | 4.41 | 4.28 | 4.22 | 0.718 (0.026) | 0.580 | 0.715 | 0.685 | 0.702 | 12.7 (0.1) | 2.04 | 10.1 | 11.8 | 11.4 | 11.5 | |
| 400 | 2.11 (0.11) | 2.42 | 2.24 | 2.15 | 2.10 | 0.711 (0.035) | 0.774 | 0.736 | 0.688 | 0.724 | 25.4 (0.1) | 2.91 | 21.5 | 23.1 | 22.5 | 22.2 | |
| z0 = 20 | 50 | 26.2 (0.3) | 6.79 | 26.5 | 26.0 | 26.0 | 1.06 (0.01) | 0.273 | 1.06 | 1.04 | 1.05 | 2.94 (0.04) | 0.958 | 2.86 | 2.91 | 2.90 | 3.17 |
| 100 | 13.3 (0.2) | 4.90 | 13.6 | 13.3 | 13.3 | 1.08 (0.02) | 0.393 | 1.09 | 1.07 | 1.08 | 6.05 (0.06) | 1.41 | 5.70 | 5.83 | 5.79 | 6.27 | |
| 200 | 7.13 (0.15) | 3.41 | 6.95 | 6.75 | 6.69 | 1.17 (0.02) | 0.547 | 1.13 | 1.08 | 1.11 | 12.5 (0.1) | 2.04 | 11.2 | 11.7 | 11.5 | 12.3 | |
| 400 | 4.02 (0.11) | 2.42 | 3.58 | 3.39 | 3.32 | 1.34 (0.04) | 0.780 | 1.17 | 1.09 | 1.14 | 23.8 (0.1) | 2.74 | 21.7 | 23.3 | 22.7 | 23.7 | |
| κ = 4 | 50 | 31.9 (0.3) | 6.91 | 33.6 | 33.0 | 33.3 | 1.29 (0.01) | 0.277 | 1.35 | 1.32 | 1.32 | 4.40 (0.06) | 1.42 | 4.20 | 4.29 | 4.19 | 4.95 |
| z0 = 0 | 100 | 16.6 (0.2) | 5.11 | 17.3 | 16.9 | 16.8 | 1.34 (0.02) | 0.411 | 1.39 | 1.35 | 1.37 | 9.00 (0.08) | 1.75 | 8.38 | 8.62 | 8.75 | 9.80 |
| 200 | 8.54 (0.16) | 3.62 | 8.97 | 8.56 | 8.45 | 1.39 (0.03) | 0.580 | 1.44 | 1.37 | 1.40 | 18.6 (0.1) | 2.54 | 16.4 | 17.3 | 17.1 | 19.2 | |
| 400 | 4.70 (0.11) | 2.49 | 4.63 | 4.31 | 4.20 | 1.57 (0.36) | 0.793 | 1.51 | 1.39 | 1.44 | 35.0 (0.1) | 3.26 | 31.4 | 34.6 | 33.7 | 37.0 | |
| z0 = 20 | 50 | 42.0 (0.3) | 7.18 | 43.5 | 42.5 | 42.5 | 1.68 (0.01) | 0.287 | 1.74 | 1.70 | 1.71 | 4.42 (0.05) | 1.21 | 4.35 | 4.44 | 4.43 | 5.54 |
| 100 | 21.6 (0.2) | 5.02 | 22.4 | 21.8 | 21.8 | 1.75 (0.02) | 0.402 | 1.80 | 1.74 | 1.76 | 9.31 (0.07) | 1.62 | 8.65 | 8.91 | 8.86 | 10.1 | |
| 200 | 11.8 (0.2) | 3.61 | 11.5 | 11.0 | 10.9 | 1.94 (0.03) | 0.575 | 1.85 | 1.76 | 1.80 | 18.7 (0.1) | 2.53 | 16.9 | 17.8 | 17.6 | 21.5 | |
| 400 | 4.90 (0.11) | 2.48 | 5.98 | 5.55 | 5.43 | 1.63 (0.04) | 0.801 | 1.96 | 1.78 | 1.85 | 33.0 (0.1) | 3.30 | 32.4 | 35.7 | 34.9 | 41.5 | |
N is the population size; δ is the difference between the optimal and mean values of the trait, divided by a; γ is the scaled selection coefficient 4Ns for an A2 variant; Vg is the trait variance; κ is the mutational bias parameter; b is the ratio of the optimal value of the trait to ma. The mutation rate is u = 1 × 10−5; the selection parameters are S = 0.01, m = 1000, a = 0.1 (Sa2 = 0.0001). The columns headed by Sim. are values obtained from 500 stochastic simulations of each parameter set, with the mean in the top part of each cell, and the standard error (in parentheses); the standard deviations are given to the right of the mean. The value of γ is obtained by averaging 4NSa2(2δ + 2qi – 1) over all segregating sites. The columns headed by Mat. give the expected values for the relevant parameters obtained from iterations of the matrix equation for the probability distribution vector, f, to near to its equilibrium. The columns headed by App. 1 show the approximations for δ*, γ, and Vg* given by Equations A.3a, A.1, and A.4b, respectively. The columns headed by App. 2 show the values of these parameters using Newton–Raphson iteration of Equation A.2, as described in the Appendix, where γ is estimated as (1 – q*)γ2– q*γ1 . The column headed by Stab. sel. are the approximate equilibrium genetic variances under pure stabilizing selection (δ* = 0), given by Barton (1989, Equation 8).
As expected from the approximate analytical results, despite very weak selection on the trait, the equilibrium value of δ* is always fairly small, even for the smallest population size modeled here (N = 50; NS = 0.5). Both the matrix method values of δ* and the mean values obtained from the stochastic simulations agree quite closely with the simple approximations in the parameter range shown in Table 1; as predicted by Equation A.3b, δ* is roughly inversely proportional to N; and the eightfold ratio between the smallest and largest N values is reflected in a roughly 7.4-fold ratio of δ* values. In contrast, the equilibrium γ values show only modest increases as N increases. While the simple approximation for γ works best for the smaller N values, where the correspondingly larger δ* values mean that directional selection is the dominant force in Equation 2, the maximum differences from the observed values are still only of the order of 10% of the observed value even for N = 400. The expression for γ given by Equation 9 gives results very close to the App. 1 values in Table 1. Large standard deviations of δ and γ are observed in the stochastic simulation results, which agree quite closely with the values predicted by Equation S1.1 in File S1. Comparisons of numerical results for m = 1000 and 4000 show little differences for the values of these parameters, as expected theoretically (data not shown).
The variance of the trait, Vg, provides a further test of the utility of the approximations. These work quite well over the entire parameter range in Table 1, as does Equation 8 of Barton (1989) for the case of pure stabilizing selection. This generally good agreement, regardless of the specific assumptions, probably reflects the fact that selection is very weak in all these cases, so that the variants are close to neutrality. The expected variance in the neutral case is then the product of ma2 and the expected neutral nucleotide diversity π; the latter is equal to 8Nuκ/(1 + κ) (Charlesworth and Charlesworth 2010, p. 274). The values for Vg* in Table 1 are quite close to 8Nuκma2/(1 + κ); for example, with N = 100, κ = 2, and b = 0, the neutral value is 0.053, compared with the observed value of 0.057. The standard deviations of Vg are generally much smaller in relation to the expected values than those for the other variables, as is expected for the neutral approximation.
Approximate analytical results for the state-independent model
It is simpler to obtain approximate analytical results for this model than for the state-dependent model. When 2δ is the dominant term in Equation 2, we can once again treat the problem as one of directional selection, with a scaled selection coefficient γ = 8NSa2δ in favor of A2 vs. A1 variants. The condition for statistical equilibrium is that the expected number of new A1 mutations arising each generation that become fixed, 2NκuQ1, is equal to the expected number of new A2 mutations that become fixed, 2NuQ2. Equations 7 are thus replaced by
| (11) |
so that we obtain
| (12a) |
| (12b) |
As was shown to be the case for the state-dependent model (see Equation A.7), these expressions can also be used when there is a mixture of stabilizing and directional selection, although γ can no longer be interpreted as the product of 4N and a fixed selection coefficient.
Numerical results for the state-independent model
Some numerical results for this case, using the matrix and stochastic simulations methods described in the Appendix, are shown in Table 2, for parameter values that are similar to those in Table 1. The overall picture is similar to that for the state-dependent case, except that the γ values are close to ln(κ) (App. 1). The approximate results for δ* and γ provide a better fit to the simulation results than do the matrix method results for N = 100 and 400. The more exact values for γ, based on Newton–Raphson numerical solutions of the equation for equilibrium (see Appendix) fit somewhat better, except for N = 400. All the methods confirm that δ * is smaller with larger N, and the ratios of δ* values for different N values are close to the corresponding ratios of N.
Table 2. Simulation and theoretical values of parameters under stabilizing selection with state-independent mutations.
| N | δ |
γ (× 102) |
Vg (× 102) |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sim. |
Mat. | App. 1 | App. 2 | Sim. |
Mat. | App. 1 | App. 2 | Sim. |
Mat. | App. 1 | App. 2 | Stab. sel. | |||||
| Mean | S.D. | Mean | S.d. | Mean | S.D. | ||||||||||||
| κ = 2 | 50 | 17.7 (0.3) | 7.31 | 17.5 | 17.3 | 17.3 | 0.711 (0.013) | 0.294 | 0.706 | 0.693 | 0.698 | 5.78 (0.07) | 1.48 | 5.55 | 5.77 | 5.75 | 5.94 |
| z0 = 0 | 100 | 8.57 (0.21) | 4.70 | 8.61 | 8.66 | 8.61 | 0.694 (0.017) | 0.376 | 0.698 | 0.693 | 0.702 | 11.4 (0.1) | 2.06 | 10.7 | 11.5 | 11.5 | 11.8 |
| 200 | 4.20 (0.16) | 3.49 | 4.65 | 4.33 | 4.28 | 0.691 (0.025) | 0.557 | 0.762 | 0.693 | 0.711 | 22.6 (0.1) | 2.80 | 21.1 | 23.1 | 22.8 | 23.1 | |
| 400 | 2.06 (0.11) | 2.50 | 1.69 | 2.17 | 2.11 | 0.689 (0.036) | 0.801 | 0.579 | 0.693 | 0.729 | 41.5 (0.2) | 3.54 | 36.3 | 46.2 | 45.0 | 44.4 | |
| z0 = 20 | 50 | 17.2 (0.3) | 6.64 | 17.5 | 17.3 | 17.3 | 0.693 (0.01) | 0.267 | 0.706 | 0.693 | 0.698 | 5.70 (0.06) | 1.41 | 5.55 | 5.77 | 5.75 | 5.94 |
| 100 | 8.97 (0.23) | 5.07 | 8.61 | 8.66 | 8.61 | 0.724 (0.02) | 0.408 | 0.698 | 0.693 | 0.702 | 11.5 (0.08) | 1.88 | 10.7 | 11.5 | 10.8 | 11.5 | |
| 200 | 4.43 (0.15) | 3.41 | 4.65 | 4.33 | 4.28 | 0.726 (0.02) | 0.550 | 0.762 | 0.693 | 0.710 | 22.3 (0.1) | 2.81 | 21.1 | 23.1 | 22.8 | 23.1 | |
| 400 | 2.05 (0.10) | 2.23 | 1.69 | 2.17 | 2.11 | 0.687 (0.03) | 0.717 | 0.579 | 0.693 | 0.729 | 40.3 (0.2) | 3.46 | 36.3 | 46.2 | 45.0 | 44.4 | |
| κ = 4 | 50 | 35.1 (0.3) | 7.46 | 35.0 | 34.7 | 34.5 | 1.41 (0.01) | 0.299 | 1.41 | 1.39 | 1.39 | 8.24 (0.07) | 1.63 | 8.19 | 8.66 | 8.63 | 9.90 |
| z0 = 0 | 100 | 17.4 (0.2) | 4.65 | 17.3 | 17.3 | 17.2 | 1.41 (0.02) | 0.371 | 1.40 | 1.39 | 1.40 | 16.3 (0.1) | 2.31 | 15.5 | 17.3 | 17.2 | 19.6 |
| 200 | 8.66 (0.16) | 3.46 | 9.20 | 8.66 | 8.55 | 1.43 (0.02) | 0.550 | 1.51 | 1.39 | 1.42 | 30.0 (0.1) | 2.98 | 29.1 | 34.6 | 34.2 | 38.5 | |
| 400 | 4.22 (0.12) | 2.70 | 3.99 | 4.33 | 4.22 | 1.42 (0.38) | 0.857 | 1.35 | 1.39 | 1.45 | 53.7 (0.2) | 3.81 | 47.1 | 69.2 | 67.6 | 74.1 | |
| z0 = 20 | 50 | 34.7 (0.3) | 7.27 | 35.0 | 34.7 | 34.5 | 1.40 (0.01) | 0.290 | 1.41 | 1.39 | 1.39 | 8.23 (0.08) | 1.78 | 8.19 | 8.66 | 8.63 | 9.90 |
| 100 | 17.3 (0.2) | 5.09 | 22.4. | 21.8 | 21.8 | 1.40 (0.02) | 0.409 | 1.80 | 1.74 | 1.76 | 16.3 (0.1) | 2.35 | 8.65 | 8.91 | 8.86 | 10.1 | |
| 200 | 8.69 (0.16) | 3.53 | 9.21 | 8.66 | 8.55 | 1.42 (0.03) | 0.565 | 1.51 | 1.39 | 1.42 | 30.6 (0.1) | 2.99 | 29.1 | 34.6 | 34.2 | 38.5 | |
| 400 | 4.49 (0.12) | 2.59 | 3.99 | 4.33 | 4.22 | 1.51 (0.04) | 0.830 | 1.35 | 1.39 | 1.45 | 53.4 (0.2) | 4.05 | 47.1 | 69.2 | 67.6 | 74.1 | |
The composition of the table is as described for Table 1, except that the approximations for δ*, γ, and Vg* are those given by Equations 12 and A.9.
The predictions for Vg* are accurate for N = 50 and N = 100, but the first two approximations tend to overestimate Vg* for the larger N values, especially for the higher level of mutational bias, reflecting the increasing importance of the stabilizing selection component, which these approximations ignore. The prediction based on Barton’s (1989) Equation 8 performs even worse for these larger N values when κ = 4, presumably because it does not take the directional selection component into account. In this case, the state independence implies that the net mutation rate is (1 + κ)u, so that the neutral value of Vg* is 4Numa2(1 + κ). This fits the values in Table 1 quite well for N = 50 and 100, but tends to overestimate Vg* for the higher values of N, reflecting the increasing effectiveness of selection in eliminating deleterious variants.
Site frequency spectra
Estimates of the strength of selection on individual variants affecting a trait can be obtained from DNA sequence polymorphism data, by comparing the distribution over sites of the frequencies of individual variants (the site frequency spectrum) with the predictions of models of selection, mutation, and drift of the type described above. This has been particularly useful for estimating selection on codon usage in organisms such as bacteria and Drosophila, where alternative synonymous codons for a given amino acid can be clearly defined as a priori candidates for being preferred or disfavored by directional selection; a fixed value of γ can then be estimated (Hartl et al. 1994; Akashi 1999; Comeron and Guthrie 2005; Zeng and Charlesworth 2009; Sharp et al. 2010). In this case, the results described in the previous section suggest that essentially similar results will be obtained over a wide range of parameter space of mutation and selection parameters if the trait in question is subject to stabilizing rather than directional selection, as was originally proposed by Kimura (1981, 1983, pp. 143–148).
Site frequency spectra have also been used for estimating selection on indel mutations, especially in Drosophila noncoding sequences, with mixed results (Comeron and Kreitman 2000; Schaeffer 2002; Ometto et al. 2005; Presgraves 2006; Leushkin et al. 2013). This raises the question of whether the model of state-independent mutations with stabilizing selection could be used for this purpose, since this mutational model is probably the simplest one that is appropriate for indels. A detailed investigation of how to estimate the parameters of this model from polymorphism data will be the subject of another article. Here, I simply point out that, under the state-independent model, a mutational bias toward deletions is consistent with a stable statistical equilibrium for sequence length only if there is stabilizing selection; this can be seen from the fact that even a small difference between the products of the respective mutation rates and fixation probabilities between insertions and deletions will lead to indefinite evolution in the direction of the class with the higher product. Furthermore, if the population is at a statistical equilibrium under this model, an excess of polymorphisms that are inferred to be deletion mutations by comparison with an outgroup, relative to polymorphisms inferred to be insertion mutations, necessarily implies a mutational bias toward deletions. Such an excess is, for example, commonly observed in data from natural populations of Drosophila (Comeron and Kreitman 2000; Schaeffer 2002; Ometto et al. 2005; Presgraves 2006; Leushkin et al. 2013).
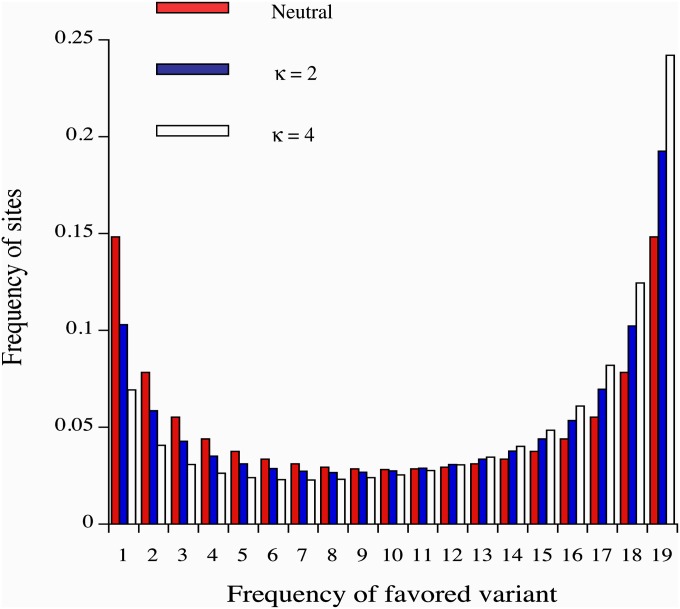
Mutational bias toward deletions should thus create a directional selection pressure in favor of insertions over deletions, so that we can use the “pooled” frequency spectrum of indels, in which the longer variant at a given location in a noncoding sequence is treated as the variant favored by selection (A2 in the above model), to estimate γ, ignoring the stabilizing selection component of selection in Equation 2 as a first approximation. In addition, under the infinite-sites assumption, the mean frequency of the A2-type variants in a sample will exceed 0.5, if they are favored by selection, providing a nonparametric test for selection (e.g., McVean and Charlesworth 1999). Figure 1 shows the frequency spectra generated by the matrix calculations for two of the parameter sets shown in Table 2, for the case of a sample of 20 alleles. The departure from neutrality in the direction of a higher abundance of high-frequency A2-type variants is clearly visible, with a greater departure from symmetry around a frequency of 0.5 with the higher level of mutational bias.
Figure 1.
Site frequency spectra in a sample of 20 alleles with a predominance of directional selection, from matrix iterations. The histograms show the spectra for the cases of neutrality (red bars) and stabilizing selection with state-independent mutations and two different levels of mutational bias (blue and white bars) with N = 400, S = 0.01, m = 1000, a = 0.1, z0 = 0, and u = 1 × 10−5.
These spectra are very close to those obtained by pooling the results of replicate stochastic simulations (data not shown), as expected from the results in File S1. It should be noted, however, that there are differences between the frequency spectra for individual replicate runs of the stochastic simulations, indicating that the same population observed at different times, or independently evolving populations subject to the same evolutionary forces, could be inferred to have significantly different γ values if standard methods for inferring selection from frequency spectra are applied. With low mutational bias, there is a substantial chance that a population would yield a spectrum that fails to provide significant evidence of selection; for example, in the case in Figure 1 with κ = 2, the mean frequency of A2 among segregating sites is 0.606, and its standard deviation among 500 replicates is 0.046 (for more details, see File S1). Such failure is less likely with a larger population size and higher mutational bias, since the ratio of the mean frequency of A2 to its standard deviation becomes larger. This effect raises some interesting questions concerning the estimation of γ, which will be considered in a subsequent article.
Purifying Selection
Assumptions of the model
A similar approach can be used to study the interaction between mutation pressure and weak purifying selection in a finite population, when selection, mutation, and drift each significantly influence allele frequencies. Let the A1 and A2 variants at a site represent the disfavored and favored alternatives, respectively. If there is semidominance with respect to mutational effects on fitness, we can assume that the fitness of an individual is determined by the number of A1 variants, n = n11 + 0.5n12, where n11 is the number of sites homozygous for A1 variants and n12 is the number of sites that are heterozygous. If mating is random and qi is the frequency of A2 at the ith site, the mean of n, , is equal to the sum of 2(1 – qi) over all sites. Equivalently, = 2m(1 – ), where is the mean of q over all m sites.
To include the possibility of epistatic interactions among the fitness effects of mutations, it is convenient to use the quadratic form
| (13) |
where α and β are constants (Kondrashov 1982; Charlesworth 1990).
When β = 0, fitnesses are multiplicative, and epistasis is absent on the logarithmic scale. “Synergistic epistasis” is represented by a positive value of β; “diminishing returns” epistasis is represented by a negative value of β. Since synergistic epistasis is of the most interest in relation to the topics discussed here, numerical results are presented for this case and for multiplicative fitnesses. The latter case is essentially equivalent to the standard models of selection on codon usage (Li 1987; Bulmer 1991; McVean and Charlesworth 1999).
Approximate analytic results are derived in the Appendix for the expected values at statistical equilibrium of the mean number of mutations per individual, the scaled selection coefficient on a mutation, and the genetic variance.
Numerical results for purifying selection
The numerical methods for this case are very similar to those used for stabilizing selection, except that now the selection coefficient s in the equivalent of Equation 6a is calculated from Equation A.11b, where n* is the product of 2m and the mean value of (1 – q) across the probability distribution of allele frequencies, f. Some results for this model are shown in Table 3, with 500 sites under selection, corresponding to the number of third coding positions in a typical human or Drosophila gene. The upper set of results is for β = 0, corresponding to multiplicative fitnesses (no epistasis); the middle set is for weak synergistic selection, with a value of β chosen so that the net selection coefficient s is approximately the same as for β = 0; and the bottom set is for purely synergistic selection (α = 0).
Table 3. Parameter values with purifying selection.
|
n* |
γ |
Vg |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| N | Mat. | App. 1 | App. 2 | Mat. | App. 1 | App. 2 | Mat. | App. 1 | App. 2 | |
| α = 0.0015, | 50 | 600 | 598 | 597 | 0.300 | 0.300 | 0.300 | 1.37 | 1.39 | 1.39 |
| β = 0 | 100 | 525 | 523 | 523 | 0.600 | 0.600 | 0.600 | 2.82 | 2.87 | 2.87 |
| 200 | 389 | 373 | 376 | 1.20 | 1.20 | 1.20 | 5.67 | 5.81 | 5.81 | |
| 400 | 177 | 73.3 | 154 | 2.40 | 2.40 | 2.40 | 9.94 | 10.3 | 10.6 | |
| α = 0.0010, | 50 | 581 | 581 | 580 | 0.368 | 0.369 | 0.368 | 1.38 | 1.40 | 1.40 |
| β = 1.45 | 100 | 503 | 501 | 501 | 0.692 | 0.690 | 0.690 | 2.84 | 2.88 | 2.88 |
| × 10−6 | 200 | 379 | 367 | 369 | 1.24 | 1.23 | 1.23 | 5.66 | 5.81 | 5.81 |
| 400 | 212 | 173 | 202 | 2.09 | 2.00 | 2.07 | 10.4 | 10.9 | 10.8 | |
| α = 0.0, | 50 | 493 | 490 | 490 | 0.740 | 0.734 | 0.734 | 1.42 | 1.45 | 1.45 |
| β = 7.5 | 100 | 390 | 385 | 386 | 1.17 | 1.15 | 1.16 | 2.86 | 2.91 | 2.91 |
| × 10−6 | 200 | 281 | 269 | 276 | 1.69 | 1.62 | 1.66 | 5.52 | 5.68 | 5.66 |
| 400 | 189 | 168 | 183 | 2.27 | 2.02 | 2.19 | 10.1 | 10.9 | 10.6 | |
The columns headed by Mat. come from the matrix iterations; those headed by App. 1 come from the approximations for n*, γ, and Vg obtained from Equations A.14 and A.15, and the columns headed by App. 2 give the values of these quantities obtained from Newton–Raphson iteration of Equation A.13. The mutation parameters were u = 1 × 10−5 and κ =2; m = 500.
In all cases, the numerical results generated by the matrix results are quite well predicted by the two types of approximation (stochastic simulation results are not shown, due to their agreement with the other results); this is, of course, not surprising for the multiplicative case, in which γ is fixed and equal to 4Nα. As expected from the approximations given above, γ increases with N for the same set of selection and mutation parameters, although the ratio of γ values for successive pairs of N values is somewhat less than two with epistatic selection and becomes smaller with increasing N, suggesting that an asymptotic value may eventually be approached. From the analysis of the derivative with respect to N of the approximation for γ given by Equation A.14b, it would be expected that this will happen only when the expected frequency of the favored allele A2, q* = 1 – n*/(2m), is close to one (this corresponds to the frequency of optimal codons for a model of selection on codon usage).
This was tested by running matrix iterations with α = 0 (giving the maximum effect of synergism) and β = 7.5 ×10−5, 10 times the value in Table 3, bottom. By using mutation rates that are also 10 times the values in Table 3, the system behaves as though N is 10 times the Table 3 values, as far as the mean and variance of n, and the value of γ, are concerned. For N values equivalent to 200, 400, 800, and 1600, the equilibrium values of q* are 0.843, 0.902, 0.937, and 0.940; the corresponding values of γ are 2.35, 2.93, 3.76, and 5.51. Thus, as N increases, q* approaches 1, and γ continues to increase, at an accelerating rate. This acceleration is expected from Equation A.13; if we put q* = 1 – υ in this equation, when υ is very small, we have γ ≈ ln(κ) + υ – ln(υ), which increases very fast as υ approaches zero. This reinforces the conclusion based on the approximations of Equations A.14 that synergism does not prevent a large increase in the scaled strength of selection as N increases.
Using a similar argument to that employed for the state-dependent model of stabilizing selection, the expected variance in the number of mutations in the neutral case in this case is 8Nmuκ/(1 + κ). Comparison with the values for Vg in Table 3 shows that this formula tends to slightly underpredict the variance; e.g., for the case with β = 0 and N = 100, the neutral value is 2.67, compared with the observed value of 2.82. This probably reflects the fact that selection leads to a higher frequency of sites fixed for A2 than under neutrality, and these have a higher rate of mutation than A1 sites.
Discussion
Biological implications of the results
A major conclusion from this work is that, in a finite population, weak stabilizing selection with mutational bias can cause a sufficient deviation of the population mean from the optimum to induce a net pressure of directional selection on the trait (see Tables 1 and 2). This agrees with the results of Zhang and Hill (2008), obtained from simulations of a reflected gamma distribution of mutational effects on a quantitative trait. While the magnitude of this deviation declines with the population size, it is generally extremely small compared with the total possible range of trait values (2ma). For example, in Table 2 with κ = 4 and N = 50, the deviation of the expected mean from the optimum (aδ*) is ∼0.1 × 35 = 3.5, whereas the range is 200.
Another way of looking at the size of the deviation from the optimum is to consider a genomic trait that may be under stabilizing selection, such as the total length of a set of noncoding sequences like short introns in Drosophila, for which a conservative value is 50,000 bp (Misra et al. 2002). If small insertions or deletions are associated with a typical a value of 5 bp, consistent with what is seen in Drosophila polymorphism data (Leushkin et al. 2013), a δ* of 35 (as above) would imply that the expected mean value of the trait departs from the optimum by 5 × 35 bp = 175 bp (this uses the fact that the value of δ* for a fixed value of Sa2 is invariant with respect to m and a, for sufficiently large m). Using Equation S1.1, File S1, δ has a standard deviation due to drift of 5 × 7.1 = 35 bp. The maximum probable departure of the mean from the optimum is thus ∼175 + 2 × 35 = 245 bp. Clearly, a deviation of this magnitude would be undetectable relative to a trait value of 5 × 104. It would then be difficult to detect any relation between mean trait values and N in comparisons across species with different N values, as is done in efforts to test evolutionary hypotheses concerning genomic traits (Lynch 2007).
Nevertheless, such departures can cause a significant pressure of directional selection on individual variants, provided that δ exceeds one-half. This is because they then dominate the expression for allele frequency change relative to the contribution from the stabilizing selection term that would exist if the mean and optimum coincided. This effect allows the use of established theoretical results on the interaction between purifying selection, mutation, and drift, yielding useful approximate expressions for the expected values of the population mean and its departure from the optimum, the scaled directional selection parameter for an individual variant (γ = 4Ns), and the genetic variance in the trait, Vg. The results can also be used to investigate the important issue of the genetic load generated by pervasive weak selection on sites distributed throughout the genome (Kondrashov 1995), as is discussed in Charlesworth (2013).
Perhaps the most striking conclusion is that γ can remain remarkably constant over a wide range of parameter space (see Tables 1 and 2), even though the difference between the population mean and the optimum declines sharply with the population size. In the case of the state-independent mutation model, which is likely to be relevant to mutations such as insertions and deletions, the approximate predicted equilibrium value of γ is given by ln(κ); this prediction matches the values generated by the matrix iterations and stochastic simulations over a wide range of parameter space (Table 2). The same is true of the slightly more complex expectation for the state-dependent model, which is more appropriate for quantitative phenotypic traits or for codon usage (Table 1). This lack of sensitivity of γ to N reflects the fact that the directional selection component is generated by the deviation of the population mean from the optimal trait value, driven by mutational bias and drift. This deviation is expected to increase with smaller N (see Equations 9 and 12b and Tables 1 and 2), causing an increase in the intensity of stabilizing selection that approximately compensates for the reduction in N in the product Ns over a wide range of parameter space. A similar result was obtained by Cherry (1998), for a model of amino-acid sequence evolution under directional selection, but that uses an equilibrium condition similar to that in Equation 11. The validity of this is, however, not clear for a system where a state-dependent mutational model is required.
This behavior contrasts sharply with what happens with mutation and purifying selection with a constant s, where Ns is proportional to N, as in the standard Li–Bulmer model of selection on codon usage (Table 3, top). It also contrasts with what is found with the synergistic selection model (Equations A.14 and Table 3), where the magnitude of γ increases with N over a wide range of parameters. Indeed, q* in Equation A.13 is forced toward one as γ increases. These results imply that synergistic selection is unlikely to account for the general lack of a relationship between the codon usage for a species and its inferred species effective population size, as has been previously proposed (Akashi 1995, 1996; Kondrashov et al. 2006).
The results also have the important implication that weak stabilizing selection may be extremely difficult to distinguish from purifying selection, because the directional component of selection introduced by drift and mutational bias can be dominant, even when these forces have only very minor effects in pushing the population mean away from the optimum. The main difference from purifying selection lies in the insensitivity of γ to N for equilibrium populations. For mutations affecting genomic traits such as codon usage, GC content, or the lengths of nonfunctional noncoding DNA sequences such as short introns, κ is likely to be in the range 2–4 in organisms such as Drosophila (Petrov and Hartl 1998; Comeron and Kreitman 2000; Haag-Liautard et al. 2007; Keightley et al. 2009; Zeng 2010). Since γ is strongly determined by ln(κ) in the range of parameter space considered here, this sets bounds on the magnitude of γ that should be detected from surveys of DNA sequence variation within species, if these traits are indeed subject to stabilizing selection. The data becoming available from genome-wide resequencing studies will allow tests of these predictions, and statistical methods for fitting models of the kind described here to such data are in the process of development.
Since it is usually unclear a priori whether stabilizing selection or purifying selection is operating on genomic traits, these results raise the question of whether there may be alternative explanations for the correlations with Ne of such properties as genome size and mutation rate, which have been used as evidence in favor of the hypothesis that the reduced efficiency of selection with relatively Ne values may drive larger genome sizes and mutation rates (Lynch 2002, 2007, 2011; Sung et al. 2012). As an example of such an alternative, many studies of species differences suggest that faster development time disfavors larger genome size (Cavalier-Smith 1985; Pagel and Johnstone 1992). Development rate is correlated with smaller body size, which in turn is correlated with larger Ne (Lynch 2007), so that an apparent relation between Ne and the genome size of a species can be generated indirectly. It is therefore interesting to note that A. thaliana has a reduced genome size relative to its outcrossing relative, A. lyrata (Hu et al. 2011). This difference in genome size is consistent with the hypothesis that the evolution of rapid development in relation to the colonizing lifestyle of A. thaliana has caused a shift toward a lower optimal value for the size of noncoding sequences. It is in the opposite direction to that predicted by Lynch’s models from the difference in Ne between the two species, since A. thaliana has the smaller Ne (Qiu et al. 2011).
The results also suggest that a role for stabilizing selection in causing codon usage bias, originally proposed by Kimura (1981, 1983, pp. 183–193), should be reexamined. Kimura assumed that translational efficiency is optimized by matching the frequencies of use of specific codons to the abundance of corresponding tRNAs, without providing a specific mechanism to justify this assumption. A similar idea has recently been proposed by Qian et al. (2012), who suggested that tRNA availability during translation may be rate limiting. Under these conditions, they show that Kimura’s conjecture is justified, using an argument that is equivalent to the ideal free distribution for foraging strategy used in behavioral ecology (Fretwell and Lucas 1970), and discuss experimental evidence that supports this model. Agashe et al. (2013) report results of experimental manipulation of codon frequencies on fitness in a bacterium that also suggest stabilizing selection, but with a different mechanistic basis.
Such a model does not, however, provide an obvious explanation for the well-documented relation between the level of expression of a gene and its codon usage bias (Hershberg and Petrov 2008). Several other biochemical mechanisms may influence selection on codon usage (Plotkin and Kudla 2011), and their relative importance is unclear. One possibility for explaining the expression–codon usage bias relation on the basis of a stabilizing selection model is that the GC content of a message may increase its folding energy, so that GC-rich messages are tightly folded and hence obstruct translation initiation (Plotkin and Kudla 2011). Harrison and Charlesworth (2011) suggested that this process may account for the fact that, in budding yeast, highly expressed genes in GC-rich regions of the genome tend to have lower optimal codon usage than genes with equivalent levels of expression elsewhere in the genome. This effect could provide a mechanism for stabilizing selection on codon usage in taxa such as Drosophila, where GC-ending codons appear to preferred by selection in opposition to mutational bias favoring AT over GC basepairs. The selective forces favoring GC codons, such as greater translational efficiency, would probably be stronger for more highly expressed genes, overcoming the negative effects of GC content on the folding of the message, and hence moving the optimal GC content upward.
Limitations and extensions of the models
The models described here are clearly very limited in many respects, and more general models need to be examined to determine the generality of the major conclusions. In particular, the assumption of the same mutational effects at all sites is unrealistic, as is the assumption of equal effects of variants in each direction. Similarly, the effects of relaxing assumption of a completely symmetrical selection, quadratic selection function, and the effects of close linkage among the sites under selection (e.g., Charlesworth et al. 2010) need to be examined. These investigations will almost certainly require computer simulations, due to the mathematical complexities involved.
Supplementary Material
Acknowledgments
I thank Nick Barton, Reinhard Bürger, Deborah Charlesworth, Isabel Gordo, Joachim Hermisson, Alexey Kondrashov, Russell Lande, Michael Lynch, David Waxman, Xu-Sheng Zhang, and an anonymous reviewer for their comments on this article. I am especially grateful to Nick Barton for pressing me to justify the approach leading to Equations 7 and 11 and for suggesting the method leading to Equations A7; I am also grateful to Reinhard Bürger and Xu-Sheng Zhang for correcting many confusing small errors. This work was supported by research grant BB/H006028/1 from the Biotechnology and Biological Sciences Research Council of United Kingdom.
Appendix
Approximate solution for state-dependent mutation when directional selection dominates
It is reasonable to assume that δ*/m is close to zero, unless Ns is very small. Using Equation 6b, writing ε = δ*/m and taking logarithms of the expressions in Equations 7b and 8, this implies that
| (A.1) |
For a given value of b, both δ*and γ can be determined from Equation A.1, e.g. by Newton–Raphson iteration. A good approximation to γ can be found simply by putting ε = 0 in this expression, giving
| (A.2) |
First-order approximations to the logarithmic terms on the left-hand side of Equation A.2 yield
| (A.3a) |
For many biologically realistic situations, 4NSma2 >> 1; in this case, Equation A.3a reduces to
| (A.3b) |
The expected genetic variance in z, Vg*, is equal to ma2π, where π is the pairwise diversity at a single nucleotide site. Substituting the expression for s from Equation 6b into the expression for π given by McVean and Charlesworth (1999, Equation 15), the genetic variance contributed by a single site is
| (A.4a) |
where γ and ε = δ*/m are given by Equations 6b and A.3a.
Summing over all sites, and using Equation A.3a, this gives
| (A.4b) |
Vg* in Equation A.4b increases without limit as N increases, which is clearly incorrect. This reflects the fact that δ* approaches zero as N increases, so that the approximations used above break down. The approximation described in the next section avoids this problem.
Approximate solution for state-dependent mutations, with both stabilizing and directional selection components
An approximate solution can be obtained in the following way for the case when δ is so small that the stabilizing selection component of Equation 2 becomes significant. Provided that NSa2 >> 1, so that the frequency of A2 is close to zero or one with high probability, Equation 2 implies that the change in qi due to selection is ≈ qi(1 – qi) Sa2(1+ 2δ) for sites where new mutations to A1 have been introduced, and –qi (1 – qi) Sa2 (1 – 2 δ) for sites where new mutations to A2 have been introduced. We can therefore obtain approximations to the fixation probabilities Q1 and Q2 of new mutations corresponding to these cases, by using the standard formula for fixation probability under directional selection (e.g., Charlesworth and Charlesworth 2010, p. 261). These are given by two scaled selection coefficients γ1 ≈ – 4N Sa2 (2δ * + 1) (for rare A1 mutations) and γ2 ≈ 4NSa2(2δ* – 1) (for rare A2 mutations). These expressions are not exact, since they ignore the dependence on allele frequency of the term (2δ + 2qi – 1) in Equation 2, but numerical studies indicate that they provide a fairly good approximation when selection is weak, as assumed here (data not shown). Equation 7b is then replaced by
The same argument that led to Equation A.2 yields the following expression, which allows determination of δ*, and hence γ1 and γ2, by Newton–Raphson iteration
| (A.5) |
Given a value of δ*, the two γ values can be obtained from the above expressions for γ1 and γ2. The equilibrium variance can then be calculated by combining the variances contributed by sites mutating to A1 and A2, respectively, in the proportions q* and 1 – q*, using the corresponding diversities given by Equation B6.7.3 of Charlesworth and Charlesworth (2010, p. 278). This yields the expression
| (A.6a) |
When the population size is sufficiently large that 2δ * ≪ 1, the system approaches that of pure stabilizing selection, with both γ1 and γ2 becoming equal to γ = – 4NSa2. Under these conditions, ε can be set to 0, and Equation A.6a simplifies to
| (A.6b) |
The limit of this as N approaches infinity is similar to the standard expression for the variance under stabilizing selection and mutation in a infinite population (e.g., Charlesworth and Charlesworth 2010, p. 190).
Alternatively, an expression for the ratio Q1/Q2 in the general case considered here can be found without making the above approximation, as suggested to me by Nick Barton. The fixation probability for a new A2 mutation in a system that obeys selection gradient dynamics, as is true in the present case (Barton 1989), is proportional to the integral between 0 and 1/(2N) of
where is the mean fitness of the population as a function of the allele frequency at the locus in question, holding all other allele frequencies constant (e.g., Charlesworth and Charlesworth 2010, p. 299, Equation 6A.6b). Since 1/(2N) is close to zero, it follows that the integral for a new A2 mutation is proportional to –2N. Similarly, the integral for a new A1 mutation is proportional to –2N. From the full equation for fixation probability (Charlesworth and Charlesworth 2010, p. 299), the expressions for A1 and A2 share common constants of proportionality, so that we have
| (A.7a) |
In the present case, application of Equation 1 to a population with a specified mean and variance of z shows that, for a single locus with small effect a, we have ln – ln ≈ 4Sa2δ, so that
| (A.7b) |
This is identical in form to the expression for the special case with constant selection coefficient used to obtain Equation 7b, which assumed δ* < 1/2. This means that the argument used for this case to obtain an expression for δ* can be generalized, so that Equations A.3 can be used even for cases when δ* < 1/2. However, in these cases, it is no longer the case that the fixation probabilities of A1 and A2 can be found from a single γ parameter, nor can the distribution of variant frequencies be found from Equation 10.
Numerical methods for analyzing the state-dependent model
Under a Wright–Fisher model of drift, it is possible to represent the transition from one generation to the next by a matrix of the type described by Zeng and Charlesworth (2009), of size (2N + 1) × (2N + 1), where N is the population size. The only substantial difference from their model is the form of the expression for Δqis in Equation 2. The state of the population at the start of a generation is represented by a column vector f of dimension 2N + 1, such that the element fj represents the probability of frequency q(j) = j/(2N), where j = 0, 1, 2, ..., 2N. The q(j) values are then modified by selection, according to Equation 2, where the expected value of z is obtained from Equation 4 by averaging [2q(j) – 1]a over all values of q(j). With reversible mutation, they are further modified for segregating sites according to Equation 4; the value of q at sites fixed for A1 is changed from 0 to u, and the value of q at sites fixed for A1 is changed from 0 to 1– κu (Zeng and Charlesworth 2009).
This procedure assumes that the value of δ for a single population out of the ensemble of populations generated by the probability distribution of allele frequencies can be replaced by the expected value of δ over this ensemble, δ*. A justification of this assumption as a first-order approximation with respect to the strength of selection on an individual variant is given in File S1.
Using binomial sampling from the allele frequencies after selection and mutation, we can write down a transition matrix A, whose element aij describes the probability that a site with frequency j/(2N) at the start of the generation produces an allele frequency i/(2N) in the next generation. This is used to premultiply f to get a new vector, f′ (for details, see Zeng and Charlesworth 2009). Successive iterations are conducted until the system approaches equilibrium.
It is only feasible to use a vector with a dimension of a few hundred to generate numerical results in a reasonable amount of time. To represent a biologically realistic population size, it is necessary to appeal to the fact that, when all deterministic evolutionary forces are sufficiently weak that second-order terms in their magnitude are negligible, implying that diffusion equation approximations are justified, it is the products of 2N and these magnitudes that determine the rate of change of f when time is rescaled to units of 2N generations (Ewens 2004, p. 137). Thus, provided that values of NS, Nu, etc., that are comparable with those for a natural population are used, we can make accurate inferences about the outcome of evolution from the much smaller populations that can be easily modeled. This principle has been used successfully in several previous studies, e.g., Li (1987), Dolgin and Charlesworth (2006), Keightley and Eyre-Walker (2007), Eyre-Walker and Keightley 2009), and Zeng and Charlesworth (2009); tests of several examples using both the matrix method and the stochastic simulations described below show that it also works well here (data not shown).
The distribution of the numbers of A2 vs. A1 variants in a sample of k alleles, taken from a population with vector f of probabilities of allele frequencies, can be obtained by applying binomial sampling conditioned on a population frequency q of A2, weighting by the corresponding element of f, and summing over all possible q values, as described by Zeng and Charlesworth (2009). This gives the pooled site frequency spectrum for the sample, which can be used to test for selection (Cutter and Charlesworth 2006; Galtier et al. 2006; Zeng and Charlesworth 2009; Sharp et al. 2010).
Stochastic simulations of a set of m freely recombining sites were carried out using a similar model. In this case, however, a single population was followed to provide a replicate run. Each generation, deterministic changes in allele frequencies at each site were calculated according to Equations 2 and 5. The effect of drift was modeled using a binomial random number generator (Press et al. 1992), to generate a new allele frequency at each site from a binomial deviate with sample size 2N, and the postselection and mutation allele frequency as the parameter. Care was taken to ensure that populations were run for sufficient time to approach statistical equilibrium. For the smaller population sizes, this could take 4000 generations. The means, standard deviations, and standard errors of the statistics described in the main text were calculated, as well as the fractions of sites fixed for A1 and A2 in a sample of 20 alleles, and the frequency of A2 in the sample, to determine the extent to which the properties of the sample frequency spectrum varied between replicates. In addition, the frequency spectrum across pooled replicate runs was computed, for comparison with the matrix and analytical results.
Approximate solution for state-independent mutations, with both stabilizing and directional selection components
A very similar procedure to that for the state-dependent model can be used. The condition for equilibrium analogous to Equation A.5, but without the state dependence, yields the following expression
| (A.8) |
This can be solved for δ* by Newton–Raphson iteration. Using the same approach as above, but noting that A1 to A2 and A2 to A1 mutations now occur at rates u and κu, respectively, the equilibrium genetic variance is given by
| (A.9) |
By putting γ2 = – γ1 = ln(κ) in Equation A.9, an expression for Vg* can be obtained for the case when directional selection is dominant.
State-independent mutation model of stabilizing selection: Numerical methods
This model presents some bookkeeping difficulties, because the state of an individual site is not as clearly assignable as in the previous model, since a site fixed for an A1 type allele can mutate with probability u to an A2 allele (associated with an effect on the trait of a) or with probability κu to another A1 type allele with effect –a (and similarly for a site fixed for an A2 allele). An individual site can therefore evolve indefinitely in either direction. This problem can be overcome as follows. Assume arbitrarily that the ensemble of initial populations is fixed for a collection of A1 and A2 type alleles at each site, such that the expected value of z, z*, is equal to the optimum, z0. By Equation 4, the proportion of sites fixed for A2 alleles, f2N = q*, must thus satisfy the relation z0 = z* = (ma)(2f2N – 1). Since the proportion of sites fixed for A1 alleles is f0 = 1 – q*, this expression can also be written as z0 = z* = (ma)(f2N – f0).
At a site fixed for A1, a mutation to A2 can be treated in the exactly the same way as with the state-dependent model; if it eventually becomes fixed, the value of f2N is increased accordingly. But a mutation to another A1-type variant means that we now have a site in which a derived A1 mutation is segregating, so that the state of the ancestral variant at the site must be switched from A1 to A2. Each generation, therefore, an expected number of 2Nuκ f0 new A1 mutations arise at sites previously fixed for A1; the switching of the ancestral state implies an initial frequency of an A2-type allele at these sites of q0 = 1 – 1/(2N), so that f2N–1 is increased by 2Nκu f0. According to Equation 4, these sites would contribute an amount 2Nκuf0ma(2q0 – 1) = 2Nκu f0ma(1 – 1/N) to z*. However, in reality they contribute –2Nκuf0ma (2q0 – 1). To compensate for this, if we are to continue to use Equation 4, z* must be adjusted by –4Nκuf0ma (1 – 1/N) ≈ –4Nκuf0ma. The same argument can be applied to sites fixed for A2, where new mutations to other A2-type variants (with q0 = 1/(2N) and 2q0 – 1 = –(1 – 1/N) must be compensated for by adjusting z* by 4Nuf0ma (1 – 1/N) ≈ 4Nuf0ma, to correct for the switch of the ancestral state from A2 to A1. This procedure can be repeated in each subsequent generation. In addition, the two types of mutational event at each class of site mean that, each generation, new mutations cause f0 to decrease by 2Nu(1 + κ) f0, and f2N to decrease by 2Nu(1 + κ) f2N. (Use of these terms removes the need to include mutational changes in the values of q for formerly fixed sites, as was done with the state-dependent model; the two methods are equivalent, to the order of the approximations used here.)
With these changes, the matrix method described for the state-dependent model can be used to generate numerical results. However, it should be noted that f0 and f2N must be interpreted differently from the previous model, since their values at a given time now simply describe the proportions of sites where A1 and A2 types of variant, respectively, were the latest to become fixed. This reflects the fact that the designation as A1 or A2 merely describes variants associated with effects of –a vs. a at a segregating site, so that only the probability distribution of frequencies at segregating sites has any biological meaning.
Stochastic simulations were carried out in a similar way to those described for the state-dependent mutational model. The only difference is that the infinite-sites assumption was made, so that there were no deterministic mutational contributions to allele frequency changes. Instead, at nonsegregating sites, new A1-type mutations were introduced each generation with a probability 2Nκu per site, and new A2-type mutations with a probability 2Nu, regardless of the allelic state of the site. Corrections to the population mean, to take into account switching of the ancestral allelic state with A1 to A1 or A2 to A2 mutations, were made as described for the matrix calculations.
Approximate analytical results for purifying selection
When is sufficiently large, the central limit theorem implies that the distribution of n among individuals in the population is approximately normal, and the log mean fitness of the population is given by
| (A.10) |
where Vg is now the variance of n among individuals within the population (Charlesworth 1990, Equation A2).
The selection coefficient in the equivalent of Equation 6a is given by
| (A.11a) |
(e.g., Charlesworth and Charlesworth 2010, p. 123).
In an infinitely large population with free recombination, there is an approximately Poisson distribution of n among individuals within the population, so that (Charlesworth 1990). In a finite population, drift generates a substantial variance in allele frequencies, Vq, among sites; we then have Vg = 2m[], so that Vg < . For large m, we can replace and by their expected values over the probability distributions of allele frequencies, q* and n*, and hence obtain an expected value for Vg. For small values of α and βn*, Equation A.11a can be well approximated by
| (A.11b) |
With mutations occurring at rate u from A1 to A2 at each site, and at rate κu in the reverse direction, we can write down an expression that is similar in form to Equation 7b. In this case, Equation A.11b implies that
| (A.12) |
This can be substituted into the equilibrium equation corresponding to Equation 7b, yielding the following expression
| (A.13) |
An exact solution for q* for assigned values of κ, α, β, and N can be obtained from Equation A.13 by Newton-Raphson iteration, and substituted into Equation A.12 to find the equilibrium value of γ.
Some insights into the behavior of γ as a function of N can be found by assuming that selection is weak compared with mutational bias, so that q* is close to one-half at equilibrium. We can then write (1 – q*)/q* = 1 + ζ, where second-order terms in ζ are negligible. This implies that ln(1 – q*) – ln(q*) ≈ ζ. Substituting this into Equation A.13, we obtain
| (A.14a) |
and
| (A.14b) |
These expressions allow n* to be found, using the results discussed above.
Differentiation of Equation A.14b with respect to N gives dγ/dN = 4m{α + mβ[1 + ln(κ)]}/(1 + 2Nmβ)2, so that γ is an increasing function of N even when β > 0. This shows that synergistic epistasis among deleterious mutations does not immunize the scaled selection coefficient against changes in population size, unless 2Nmβ >> 1, in which case Equation A.13 implies that q* approaches 1, an unrealistically high value compared with the frequency of usage of preferred codons obtained in most studies. This contrasts with the behavior of γ with stabilizing selection.
Given the values of n* = 2m(1 – q*) and hence of s in Equation A.11b, Vg* is given approximately by using the equivalent of McVean and Charlesworth’s (1999) Equation 15:
| (A.15) |
Footnotes
Communicating editor: J. Wakeley
Literature Cited
- Agashe D., Martinez-Gomez N. C., Drummond D. A., Marx C. J., 2013. Good codons, bad transcript: large reductions in gene expression and fitness arising from synonymous mutations in a key enzyme. Mol. Biol. Evol. 30: 549–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akashi H., 1995. Inferring weak selection from patterns of polymorphism and divergence at “silent” sites in Drosophila DNA. Genetics 139: 1067–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akashi H., 1996. Molecular evolution between Drosophila melanogaster and D. simulans: reduced codon bias, faster rates of amino-acid substitution, and larger proteins in D. melanogaster. Genetics 144: 1297–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akashi H., 1999. Inferring the fitness effects of DNA polymorphisms and divergence data: statistical power to detect directional selection under stationarity and free recombination. Genetics 151: 221–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton N. H., 1986. The maintenance of polygenic variation through a balance between mutation and stabilizing selection. Genet. Res. 47: 209–216 [DOI] [PubMed] [Google Scholar]
- Barton N. H., 1989. The divergence of a polygenic system subject to stabilizing selection and drift. Genet. Res. 54: 59–77 [DOI] [PubMed] [Google Scholar]
- Bulmer M. G., 1991. The selection-mutation-drift theory of synonomous codon usage. Genetics 129: 897–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bumpus H. C., 1899. The elimination of the unfit as illustrated by the introduced sparrow, Passer domesticus. Biol. Lectures Woods Hole Marine Biol. Station 6: 209–226 [Google Scholar]
- Bürger R., 2000. The Mathematical Theory of Selection, Recombination and Mutation, Wiley, Chichester, UK [Google Scholar]
- Bürger R., Lande R., 1994. On the distribution of the mean and variance of a quantitative trait under mutation-selection-drift balance. Genetics 138: 901–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell R. B., 1984. The manifestation of phenotypic evolution at constituent loci. I. Stabilizing selection. Evolution 38: 1033–1038 [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T., 1985. Eukaryote gene numbers, non-coding DNA and genome size, pp. 69–103 in The Evolution of Genome Size, edited by Cavalier-Smith T. Wiley, Chichester, UK. [Google Scholar]
- Charlesworth B., 1990. Mutation-selection balance and the evolutionary advantage of sex and recombination. Genet. Res. 55: 199–221 [DOI] [PubMed] [Google Scholar]
- Charlesworth, B., 2013 Why we are not dead 100 times over. Evolution. (in press). [DOI] [PubMed]
- Charlesworth B., Charlesworth D., 2010. Elements of Evolutionary Genetics. Roberts, Greenwood Village, CO [Google Scholar]
- Charlesworth B., Betancourt A. J., Kaiser V. B., Gordo I., 2010. Genetic recombination and molecular evolution. Cold Spring Harb. Symp. Quant. Biol. 74: 177–186 [DOI] [PubMed] [Google Scholar]
- Cherry J. L., 1998. Should we expect substitution rate to depend on population size? Genetics 150: 911–919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comeron J. M., Kreitman M., 2000. The correlation between intron length and recombination in Drosophila: equilibrium between mutational and selective forces. Genetics 156: 1175–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comeron J. M., Guthrie T. B., 2005. Intragenic Hill–Robertson interference influences selection on synonymous mutations in Drosophila. Mol. Biol. Evol. 22: 2519–2530 [DOI] [PubMed] [Google Scholar]
- Cutter A. D., Charlesworth B., 2006. Selection intensity on preferred codons correlates with overall codon usage bias in Caenorhabitis remanei. Curr. Biol. 16: 2053–2057 [DOI] [PubMed] [Google Scholar]
- Di Cesnola A. P., 1907. A first study of natural selection in Helix arbustorum (Helicogena). Biometrika 5: 387–399 [Google Scholar]
- Dolgin E. S., Charlesworth B., 2006. The fate of transposable elements in asexual populations. Genetics 174: 817–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewens W. J., 2004. Mathematical Population Genetics. 1. Theoretical Introduction. Springer, New York [Google Scholar]
- Eyre-Walker A., Keightley P. D., 2009. Estimating the rate of adaptive mutations in the presence of slightly deleterious mutations and population size change. Mol. Biol. Evol. 26: 2097–2108 [DOI] [PubMed] [Google Scholar]
- Feldman M. W., Christiansen F. B., Brooks L. D., 1980. Evolution of recombination in a constant environment. Proc. Natl. Acad. Sci. USA 77: 4824–4827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher R. A., 1930a The distribution of gene ratios for rare mutations. Proc. R. Soc. Edinb. 50: 205–220 [Google Scholar]
- Fisher R. A., 1930b The Genetical Theory of Natural Selection. Oxford University Press, Oxford, UK. [Google Scholar]
- Fretwell S. D., Lucas H. J., 1970. On territorial behavior and other factors influencing habitat distribution in birds. Acta Biotheor. 19: 16–36 [Google Scholar]
- Galtier N., Bazin E., Bierne N., 2006. GC-biased segregation of non-coding polymorphisms in Drosophila. Genetics 172: 221–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haag-Liautard C., Dorris M., Maside X., Macaskill S., Halligan D. L., et al. , 2007. Direct estimation of per nucleotide and genomic deleterious mutation rates in Drosophila. Nature 445: 82–85 [DOI] [PubMed] [Google Scholar]
- Haldane J. B. S., 1954. The measurement of natural selection. Caryologia 6 (Suppl.): 480–487 [Google Scholar]
- Harrison R. J., Charlesworth B., 2011. Biased gene conversion affects patterns of codon usage and amino acid usage in the Saccharomyces sensu stricto group of yeasts. Mol. Biol. Evol. 28: 117–129 [DOI] [PubMed] [Google Scholar]
- Hartl D. L., Moriyama E. N., Sawyer S. A., 1994. Selection intensity for codon bias. Genetics 138: 227–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershberg R., Petrov D. A., 2008. Selection on codon bias. Annu. Rev. Genet. 42: 287–299 [DOI] [PubMed] [Google Scholar]
- Hu T. T., Pattyn P., Bakker E. G., Cao J., Cheng J.-F., et al. , 2011. The Arabidopsis lyrata genome sequence and the basis of rapid genome size change. Nat. Genet. 43: 476–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson T., 1999. Beneficial mutations, hitchhiking and the evolution of mutation rates in sexual populations. Genetics 151: 1621–1631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keightley P. D., Eyre-Walker A., 2007. Joint inference of the distribution of fitness effects of deleterious mutations and population demography based on nucleotide polymorphism frequencies. Genetics 177: 2251–2261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keightley P. D., Trivedi M., Thomson M., Oliver F., Kumar S., et al. , 2009. Analysis of the genome sequences of three Drosophila melanogaster spontaneous mutation accumulation lines. Genome Res. 19: 1195–1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M., 1981. Possibility of extensive neutral evolution under stabilizing selection with special reference to non-random usage of synonymous codons. Proc. Natl. Acad. Sci. USA 78: 454–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M., 1983. The Neutral Theory of Molecular Evolution. Cambridge University Press, Cambridge, UK [Google Scholar]
- Kimura M., Maruyama T., 1966. The mutational load with epistatic gene interactions in fitness. Genetics 54: 1303–1312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsolver J. G., Hoekstra H. E., Hoekstra J. M., Berrigan D., Vignieri S. N., et al. , 2001. The strength of phenotypic selection in natural populations. Am. Nat. 157: 245–261 [DOI] [PubMed] [Google Scholar]
- Kondrashov A. S., 1982. Selection against harmful mutations in large sexual and asexual populations. Genet. Res. 44: 325–332 [DOI] [PubMed] [Google Scholar]
- Kondrashov A. S., 1984. Deleterious mutations as an evolutionary factor. 1. The advantage of recombination. Genet. Res. 44: 199–217 [DOI] [PubMed] [Google Scholar]
- Kondrashov A. S., 1988. Deleterious mutations and the evolution of sexual reproduction. Nature 336: 435–440 [DOI] [PubMed] [Google Scholar]
- Kondrashov A. S., 1995. Contamination of the genome by very slightly deleterious mutations: Why have we not died 100 times over? J. Theor. Biol. 175: 583–594 [DOI] [PubMed] [Google Scholar]
- Kondrashov F. A., Ogurtsov A. Y., Kondrashov A. S., 2006. Selection in favor of nucleotides G and C diversifies evolution rates and levels of polymorphism at mammalian synonymous sites. J. Theor. Biol. 240: 616–626 [DOI] [PubMed] [Google Scholar]
- Lande R., 1976. Natural selection and random genetic drift in phenotypic evolution. Evolution 30: 314–334 [DOI] [PubMed] [Google Scholar]
- Leushkin E. V., Bazykin G. A., Kondrashov A. S., 2013. Strong mutational bias towards deletions in the Drosophila melanogaster genome is compensated by selection. Genome Biol. Evol. 5: 514–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W.-H., 1987. Models of nearly neutral mutations with particular implications for non-random usage of synonymous codons. J. Mol. Evol. 24: 337–345 [DOI] [PubMed] [Google Scholar]
- Lynch M., 2002. Intron evolution as a population-genetic process. Proc. Natl. Acad. Sci. USA 99: 6118–6123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M., 2007. The Origins of Genome Architecture. Sinauer, Sunderland, MA [Google Scholar]
- Lynch M., 2011. The lower bound to the evolution of mutation rates. Genome Biol. Evol. 3: 1107–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVean G. A. T., Charlesworth B., 1999. A population genetic model for the evolution of synonymous codon usage: patterns and predictions. Genet. Res. 74: 145–158 [Google Scholar]
- Misra S., Crosby M. A., Mungall C. J., Matthews B. B., Campbell K. S., et al. , 2002. Annotation of the Drosophila melanogaster euchromatic genome: a systematic review. Gen. Biol. 3: Research0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta T., Kimura M., 1973. A model of mutation appropriate to estimate the number of electrophoretically detectable alleles in a finite population. Genet. Res. 22: 201–204 [DOI] [PubMed] [Google Scholar]
- Ometto L., Stephan W., De Lorenzo D., 2005. Insertion/deletion and nucleotide polymorphism reveal constraints in Drosophila melanogaster introns and intergenic regions. Genetics 169: 1521–1527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagel M., Johnstone R. A., 1992. Variation across species in the size of the nuclear genome supports the junk-DNA explanation for the C-value paradox. Proc. Biol. Sci. 249: 119–124 [DOI] [PubMed] [Google Scholar]
- Parsch J., 2003. Selective constraints on intron evolution in Drosophila. Genetics 165: 1843–1851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov D. A., Hartl D. L., 1998. High rate of DNA loss in the Drosophila melanogaster and Drosophila virilis species groups. Mol. Biol. Evol. 15: 1562–1567 [DOI] [PubMed] [Google Scholar]
- Petrov D. A., Lozovskaya E. R., Hartl D. L., 1996. High rate of DNA loss in Drosophila. Nature 384: 346–349 [DOI] [PubMed] [Google Scholar]
- Plotkin J. B., Kudla G., 2011. Synonymous but not the same: the causes and consequencs of codon bias. Nat. Rev. Genet. 12: 33–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presgraves D., 2006. Intron length evolution in Drosophila. Mol. Biol. Evol. 23: 2203–2213 [DOI] [PubMed] [Google Scholar]
- Press W. H., Teukolsky S. A., Vetterling W. T., Flannery B. P., 1992. Numerical Recipes in Fortran: The Art of Scientific Computing. Cambridge University Press, Cambridge, UK. [Google Scholar]
- Ptak S. E., Petrov D. A., 2002. How intron splicing affects the deletion and insertion profile in Drosophila melanogaster. Genetics 162: 1233–1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian W., Yang J.-R., Pearson N. M., Maclean C., Zhang J., 2012. Balanced codon usage optimizes translational efficiency. PLoS Genet. 8: e1002603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu S., Zeng K., Slotte T., Wright S. I., Charlesworth D., 2011. Reduced efficacy of natural selection on codon usage bias in selfing Arabidopsis and Capsella species. Genome Biol. Evol. 3: 868–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffer S. W., 2002. Molecular population genetics of sequence length diversity in the Adh region of Drosophila pseudoobscura. Genet. Res. 80: 163–175 [DOI] [PubMed] [Google Scholar]
- Sharp P. M., Bailes E., Grocock R. J., Peden J. F., Sockett R. E., 2005. Variation in the strength of selected codon usage bias among bacteria. Nucleic Acids Res. 33: 1141–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P. M., Emery L. R., Zeng K., 2010. Forces that influence the evolution of codon bias. Phil. Trans. R. Soc. B. 365: 1203–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slatkin M., 1995. A measure of population subdivision based on microsatellite allele frequencies. Genetics 139: 457–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung W., Ackerman M. S., Miller S. F., Doag T. G., Lynch M., 2012. Drift-barrier hypothesis and mutation-rate evolution. Proc. Natl. Acad. Sci. USA 109: 18488–18492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turelli M., 1984. Heritable variation via mutation-selection balance: Lerch’s zeta meets the abdominal bristle. Theor. Popul. Biol. 25: 138–193 [DOI] [PubMed] [Google Scholar]
- Wang D. P., Yu J., 2011. Both size and GC-content of mimimal introns are selected in human populations. PLoS ONE 6: e17945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waxman D., Peck J. R., 2003. The anomalous effects of biased mutation. Genetics 164: 1615–1626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weldon W. F. R., 1901. A first study of natural selection in Clausilia laminata (Montagu). Biometrika 1: 109–124 [Google Scholar]
- Wright S., 1931. Evolution in Mendelian populations. Genetics 16: 97–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S., 1935. Evolution in populations in approximate equilibrium. J. Genet. 30: 257–266 [Google Scholar]
- Zeng K., 2010. A simple multiallele model and its application to identifying preferred–unpreferred codons using polymorphism data. Mol. Biol. Evol. 27: 1327–1337 [DOI] [PubMed] [Google Scholar]
- Zeng K., Charlesworth B., 2009. Estimating selection intensity on synonymous codon usage in a non-equilibrium population. Genetics 183: 651–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.-S., Hill W. G., 2008. The anomalous effects of biased mutation: mean-optimum deviation and apparent directional selection under stabilizing selection. Genetics 179: 1135–1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.