Abstract
RNA interference is a post transcriptional gene silencing mechanism that is triggered by double-stranded RNA (dsRNA). Various attributes of the 3′ end structure, including overhang length and sequence composition, plays a primary role in determining the position of the Dicer cleavage in both dsRNA and unimolecular, short hairpin RNA (shRNA). The specificity and robustness of RNAi have triggered an immense interest in using RNAi as a tool in various settings. RNAi is a mechanism for controlling normal gene expression which has recently began to be employed as a potential therapeutic agent for a wide range of disorders, including cancer, infectious diseases and metabolic disorders. Clinical trials with RNAi have now begin, but major obstacles, such as off-target effects, toxicity and unsafe delivery methods, have to be overcome before RNAi can be considered as a conventional drug. It is also used as a tool to improve crops by providing resistance against parasites and modified versions of siRNA that are directed against disease causing genes are being developed, some of which are already tested in clinical trials. In this paper, we first reviewed the RNAi mechanism and then focussed on some of its applications in biomedical research such as treatment for HIV, viral hepatitis and several other diseases.
Abbreviations: SiRNAs, small interfering RNAs; RISC, RNA induced silencing complex; MiRNAs, microRNAs
Keywords: RNAi, Gene silencing, Antiviral, Dicer
1. Introduction
RNA interference (RNAi) is an evolutionary regulatory mechanism of most cells that uses ∼21–25 long siRNA transcripts to effectively control the expression of desired genes. By inhibiting the expression of mRNA transcripts through degrading or binding sequence specifically thus hindering translation into proteins. It is a conserved biological mechanism controlling normal gene expression. The silencing mechanism occurs at the levels of transcription, post-transcription and translation. RNAi can also cause augmentation of gene expression due to direct effects on the translation (Ebbesen et al., 2008). Two types of small ribonucleic acid (RNA) molecules – micro-RNA (miRNA) and small interfering RNA (siRNA) – are central to RNA interference.
1.1. DNA-directed RNAi
DNA-directed RNA interference (ddRNAi) uses DNA templates to synthesize si/shRNA in vivo. ddRNAi depends on U6 or H1 [RNA polymerase III], or U1 [RNA polymerase II] promoters for the expression of siRNA target sequences that have been transfected into mammalian cells (Miyagishi and Taira, 2002). si/shRNA target sequences can be generated by PCR, creating “expression cassettes” that can be transfected directly into cells (Castanotto et al., 2002) or cloned into expression vectors (Sui et al., 2002).ddRNAi technology involves inserting a DNA construct into a cell, triggering the production of double stranded RNA (dsRNA), which is then cleaved into small interfering RNA (siRNA) as part of the RNAi process, resulting in the destruction of the target mRNA and knocking-down or silencing the expression of the target gene. It is not just the size that complicates the cellular uptake and release of the nucleic acid, but the requirement for getting the DNA from the cytoplasm to the nucleus is the major rate-limiting step differentiating it from synthetic siRNA delivery.
1.2. Mechanism of RNA interference
RNA interference (RNAi) is a phenomenon in which double-stranded RNA (dsRNA) suppresses the expression of a target protein by stimulating the specific degradation of the target mRNA. Long dsRNA is processed to short interfering RNAs (siRNAs) by the action of a dsRNA-specific endonuclease known as Dicer (Fig. 1) (Bernstein et al., 2001 and Hammond et al., 2000). The resultant siRNAs usually 21–24 nt in length, are double stranded, and have 3 overhangs of 2 nt. Exogenous synthetic siRNAs or endogenously expressed siRNAs can also be incorporated into the RNA-induced silencing complex (RISC), thereby bypassing the requirement for dsRNA processing by Dicer. siRNAs are incorporated into the multi protein RISC. A helicase in RISC unwinds the duplex siRNA, which then pairs by means of its unwound antisense strand to messenger RNAs (mRNAs) that bear a high degree of sequence complementarities to the siRNA (Freda, 2004). Cleavage of the target mRNA begins at a single site 10 nt upstream of the 5-most residue of the siRNA-target mRNA duplex (Elbashir et al., 2001). Although the composition of RISC is not completely known, it includes members of the Argonaute family (Angaji et al., 2010) that have been implicated in processes directing post transcriptional silencing (Freda, 2004). Argonaute proteins are essential components of the RNAi machinery that are associated with distinct classes of small RNAs to exert their effector functions. Argonaute proteins are associated with small interfering RNAs (siRNAs) or microRNAs (miRNAs), and silence gene expression by either siRNA guided cleavage of the target mRNA transcript, or by miRNA mediated post transcriptional repression involving both translational inhibition and/or mRNA degradation.
Figure 1.
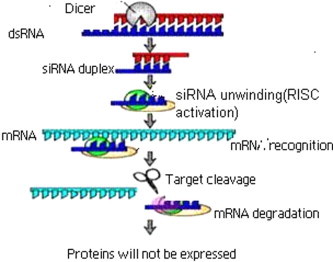
Mechanism of RNAi.
Functional specialization extends to the biogenesis pathways associated with these small RNAs; miRNAs are processed from endogenous hairpin precursors by cleavage events involving the RNAse III enzymes Drosha and Dicer1 (Dcr-1) with its partner loquacious (Loqs). There are two small RNAs in the RNAi pathway: small interfering RNAs (siRNAs) and micro-RNAs (miRNAs) that are generated via processing of longer dsRNA and stem loop precursors. Dicer enzymes play a critical role in the formation of these two effectors of RNAi (Tijsterman et al., 2002). They can cleave long dsRNAs and stem-loop precursors into siRNAs and miRNAs in an ATP-dependent manner, respectively.
1.3. Influence of overhang structure and sequence on Dicer specificity and efficiency
Dicer processes long double-stranded RNA (dsRNA) and pre-microRNAs to generate the functional intermediates (short interfering RNAs and microRNAs) of the RNA interference pathway. Although overhang composition has no impact on cleavage product sizes, nucleotide sequence does play a role in determining Dicer efficiency. Overhangs that contain a C and A in the penultimate and terminal positions (respectively) are processed most efficiently, while those containing an A in the penultimate position and a U in the terminal position are less optimal substrates. Overall, ranking the Dicer cleavage efficiency of substrates with different overhangs reveals a hierarchy with C > U = G > A being the preference at the penultimate position and A > G = U > C being the order of terminal nucleotides (Annaleen et al., 2005).
Some studies of substrates with varying 3′ overhang lengths showed that the number of nucleotides in the overhang was a critical factor in Dicer specificity. Substrates with overhang lengths of 1, 2, and 3 nt showed concomitant shifts in the primary Dicer cleavage position. Increasing the overhang length further reduced the diversity of cleavage products generating primarily a 21-nt product. Thus, when the overhang length is 0–3 nt it appears as though Dicer cleaves dsRNA by “counting” a distance of approximately 23 nt from the 3′ end of the overhang to cleave both strands. However, when the overhang length is ⩾4 nt, the primary cleavage product is similar to that observed in duplexes containing 3-nt overhangs. Therefore, in cases where the overhang length extends beyond 3 nt, Dicer no longer uses the 3′ end of the overhang to determine cleavage position.
Therefore, RNA end structure, particularly 3′ overhang length, plays a critical role in determining the position and efficiency of Dicer cleavage. (Annaleen et al., 2005) This knowledge has a profound implication for siRNA and shRNA design. shRNA expression by the Pol III promoter results in shRNA termini with variable overhang lengths (1–5 Us). And this variation drastically affects the specificity of Dicer cleavage and consequently the functionality of cleaved siRNA.
1.4. Effect of siRNA end structure on silencing efficiency
It has recently been suggested that Dicer is involved in siRNA RISC loading (Lee et al., 2004). In vitro Dicer cleavage assays demonstrate that end structure influences Dicer efficiency. This could be due to the fact that siRNA end structure impacts overall silencing efficiency. In general, siRNAs with asymmetric overhangs on the antisense strand (i.e., a 3′ overhang on the antisense strand) performed better than siRNAs with asymmetric sense strand overhangs. When compared, the difference in functionality was more profound between structurally asymmetric siRNA (blunt–overhang, overhang–blunt) than symmetric molecules (overhang–overhang, blunt–blunt). This indicates that the effect on functionality may be the result of a shift in equilibrium between sense and antisense loading rather than overall RISC loading efficiency (Annaleen et al., 2005).
2. Designing siRNAs
Small interfering RNAs (siRNAs), the guides that direct RNA interference (RNAi), provide a powerful tool to reduce the expression of a single gene in human cells. (Schwarz et al., 2008) The ability of an siRNA to silence the gene expression is determined by its sequence and not all target sites are equal (Peek and Behlke, 2007; Amarzguioui et al., 2006). Other considerations such as cross hybridization and chemical modification can also alter the effectiveness of siRNA in addition to the actual sequence (Peek and Behlke, 2007).
2.1. Location of siRNA
The location of siRNA within the entire gene is less of a concern for potency than the localization of siRNA within a particular gene exon structure. (Peek and Behlke, 2007) Targets should be located 50–100 nt downstream of the start codon (ATG). Therefore, the knowledge of when specific splice variants are used is essential to determine how to most effectively target the desired isoform(s) of the gene.
2.2. Modification
siRNAs must have phosphate groups at the 5′ end in order to have activity so it is important not to block the 5′ end of the antisense strand with any modifications other than the phosphate group. (Peek and Behlke, 2007) However, transfected unmodified 5′OH ends are rapidly phosphorylated by cellular kinases, indicating not to phosphorylate synthetic siRNAs. Regardless of the target sequence, DNA ‘TT’ dinucleotide overhangs are often added to the 3′ ends; however some evidence suggests that using RNA bases may lead to greater potency (Peek and Behlke, 2007).
2.3. Thermodynamic stability
The thermodynamic stability of siRNAs has a significant influence on their potency; thermodynamic asymmetry is fundamentally important for siRNA function and strand loading into RISC. The most effective siRNA has a relatively low Tm and duplex stability (less stable, more A/U rich) toward the 5′ end of the guide strand and a relatively high Tm (more stable, more G/C rich) toward the 5′ end of passenger strand. For situations in which selecting thermodynamically favorable regions is impossible, introducing mismatches (to lower Tm) or adding modified bases (to increase Tm) to the siRNA duplex can create thermodynamic asymmetry. If non-complementary bases must be introduced, it is important that it is at the 3′ end of the sense strand (passenger strand) rather than the 5′ end of the AS strand (guide strand) to avoid impairing the ability of the AS strand to anneal to the active target (Peek and Behlke, 2007).
2.4. Sequence characteristics and specificity
To maintain specificity, certain sequence characteristics should be avoided in the guide strand such as homopolymeric runs (those with four or more identical nucleotides) and nine –base or greater segments of G/C bases in addition, the secondary structure of the target and the site accessibility are important factors in the activity of siRNAs. A moderate to low GC content (30–52%) tends to be a feature of functional siRNA. Due to the high specificity of nucleic acid base pairing, even a single mismatch in a 19-base sequence can prevent duplex formation; however, cross hybridization can and does occur. Therefore, it is very important to screen all candidate siRNAs for homology to other targets and exclude those with significant complementarity.
Like targeted effects, off-target effects (OTEs) are dose dependent. Therefore, it is important to establish dose response profiles for all siRNAs in use and always use the lowest concentration of siRNA that will provide adequate target knockdown. An additional measure to prevent OTE bias is to ensure that at least two, and ideally three, independent siRNAs against a target give the same result (Peek and Behlke, 2007).
3. Applications of RNA interference in various areas
Besides being an area of intense, upfront basic research, the RNAi process holds the key to future technological applications in various sectors, such as in functional genomics, therapeutic intervention, agriculture and other areas. Since the discovery of RNAi, the idea of RNAi therapeutics has blown into a creative and highly competitive field that has attracted some of the most brilliant minds and it is one of the most highly invested in fields in biotechnology research. Some studies have demonstrated that it is safe to systemically deliver therapeutically effective doses of siRNAs to primates, leading the way for other future systemic applications of RNAi. Some potential diseases that may be therapeutic targets for RNAi in the near future include viral diseases, genetic diseases and cancer. However, the major obstacles, such as off-target effects, toxicity and unsafe delivery methods, have to be overcome before RNAi can be considered as a conventional drug.
3.1. RNAi as a potential therapeutic for humans; genetic diseases
Dominant negative genetic disorders, in which a mutant allele of a gene causes disease in the presence of a second, normal copy, have been challenging since there is no cure and treatments are only to alleviate the symptoms. Current therapies involving pharmacological and biological drugs are not suitable to target mutant genes selectively due to structural indifference of the normal variant of their targets from the disease-causing mutant ones (Table 1). In instances when the target contains single nucleotide polymorphism (SNP), whether it is an enzyme or structural or receptor protein are not ideal for treatment using conventional drugs due to their lack of selectivity. Although there is a cooling trend by the pharmaceutical industry for the potential of RNA interference (RNAi), RNAi and other RNA targeting drugs (antisense, ribozyme, etc.) still hold their promise as the only drugs that provide an opportunity to target genes with SNP mutations found in dominant negative disorders, genes specific to pathogenic tumor cells, and genes that are critical for mediating the pathology of various other diseases (Seyhan, 2011).
Table 1.
Applications of RNA interference (RNAi) and genes involved.
| Genes | Fungi | Animals | Plants |
|---|---|---|---|
| RdRp | QDE1 | EGO1 | SGS2/SDE1 |
| Elf2c | QDE2 | RDE1 | AGO1 |
| Rnase D | – | MUT7 | – |
| RNA Helicase | – | – | MUT6 |
| Coiled Coil | – | – | SGS3 |
Compiled from: http://www.ejb.org accessed on 3.08.2012.
A promising lead toward using RNAi for the treatment of genetic diseases has been provided by preliminary studies demonstrating how single nucleotide polymorphisms (SNPs) in mutant allele transcripts can be used as selective targets for RNAi. Disease causing polyglutamine proteins encoded by CAG repeat containing transcripts found in several neurological diseases present especially challenging targets because CAG repeats are common to many normal transcripts as well, and cannot be selectively targeted by siRNAs. Alternatively, single nucleotide polymorphisms are very often found in mutant allele transcripts, and represent potential selective targets. Systematic analyses of siRNAs in which the polymorphic nucleotide is complementary to the mid region of the siRNA provides an siRNA/SNP combination that is highly selective. In certain examples, the siRNAs direct selective degradation of only the mutant transcripts, leaving the wild type transcripts intact despite having only a single mismatch with the wild type sequence (Miller et al., 2004a, Miller et al., 2004b). Particular purine–purine mismatches at positions 10 and 16 relative to 5′ end of the guide strand provide selectivity (Schwarz et al., 2008). Since the wild-type SOD1 performs important functions it is important to selectively eliminate the expression of only the mutant allelic transcript. Many SOD1 mutations are single nucleotide changes. Since delivery of siRNAs and viral vectors expressing siRNAs to affected regions of the brain is technically feasible (McCaffrey et al., 2003), the promise of clinical use of RNAi for treatment of degenerative, neurological diseases may approach reality soon. Despite the excitement and promise of therapeutic RNAi, there are many obstacles, the greatest of which is delivery. Systemically delivered siRNAs face degradation by nucleases, and the use of viral vectors to target organs of interest is still in its infancy.
3.2. RNAi in gene regulation and antiviral responses
RNAi and RNAi-related mechanisms play essential roles in the regulation of cellular gene expression, as well as in innate antiviral immune responses. RNAi is regarded as a natural defense mechanism against mobile endogenous transposons and invasion by exogenous viruses which have dsRNA as an intermediate product. With this defense mechanism, organisms maintain genetic integrity and hinder infection (Ebbesen et al., 2008). For many applications, it may be complicated to introduce short dsRNAs directly into cells. However, many groups have now shown that appropriately designed DNA molecules containing inverted repeat sequences can be transcribed into RNA molecules that form RNA hairpins. If the sequences are chosen correctly, these are processed by the Dicer nuclease to form siRNAs. Thus all the methods derived for delivering genes into cells can in principle be used to deliver siRNAs as well. This made the application of RNAi therapy for the prevention and treatment of viral infection convenient (Fig. 2).
Figure 2.
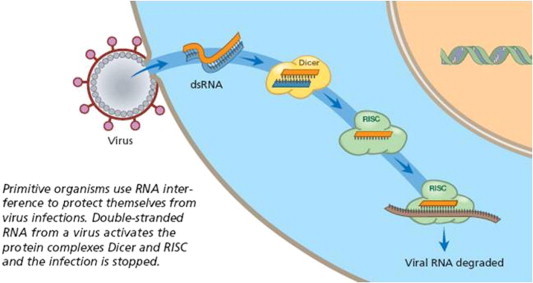
Antiviral mechanism of RNA interference.
RNAi has got a potential for the treatment of viral diseases such as those caused by the hepatitis C virus (HCV) and the human immunodeficiency virus (HIV). However, there are important issues and concerns about the therapeutic application of this technology, including difficulties with delivery and uncertainty about potential toxicity that needs to be solved. The HCV genome is a positive-strand RNA molecule with a single open reading frame encoding a polyprotein that is processed post-translationally to produce at least ten proteins. The only therapy currently available uses combined interferon (IFN) and ribavirin. Subgenomic and full-length HCV replicons that replicate and express HCV proteins in stably transfected human hepatoma cell-derived Huh-7 cells have been used to study viral replication and the effects of various antiviral drugs.
Small inhibitory RNAs targeting the internal ribosome entry site (IRES) and non-structural protein NS3 and NS5b encoding mRNAs were shown to inhibit HCV replicon function in cell culture (Wilson et al., 2003). Furthermore, anti-HCV siRNAs were shown to “cure” Huh-7.5 cells bearing persistently replicating HCV replicons. Delivery of the siRNAs or vectors that carry siRNA expression cassettes is the major challenge for treatment of HCV. The method of delivery used in a number of in vivo studies, hydrodynamic intravenous injection, is not feasible for the treatment of human hepatitis. Delivery is a problem that must be confronted for any therapeutic application of RNAi. A recent report demonstrates that it is feasible to introduce genetic material into hepatocytes using catheters or even localized hydrodynamic procedures (Eastman et al., 2002).
Numerous studies have established the proof of concept that diseases can be targeted by therapeutic RNAi, and several small interfering RNAs (siRNAs) are currently being tested in clinical trials (Grimm and Kay, 2007). Despite these rapid advances, significant hurdles still need to be overcome for the widespread therapeutic application of siRNAs. Perhaps the greatest challenge is the delivery of effective quantities of siRNAs into the cytoplasm of relevant target cells in vivo (Dykxhoorn and Lieberman, 2006).
HIV was the first infectious agent targeted by RNAi perhaps owing to the fact that the life cycle of HIV is well understood as is its pattern of gene expression. Synthetic and expressed siRNAs have been used to target a number of early and late HIV-encoded RNAs including the TAR element, tat, rev, gag, env, vif, nef (Jacque et al., 2002) and reverse transcriptase. Cellular cofactors, such as NFκβ, the HIV receptor CD4 and co-receptors CXCR4 and CCR5 have also been successfully down regulated by RNAi resulting in an inhibition of HIV replication. Moreover, inhibition of HIV replication has been achieved in numerous human cell lines and primary cells including T lymphocytes and hematopoietic stem cell derived macrophages.
Despite the success of in vitro RNAi-mediated inhibition of HIV-1, for future clinical applications, targeting the virus directly represents a substantial challenge since the high viral mutation rate will certainly lead to escape mutants (Boden et al., 2003). RNAi-mediated down regulation of cellular co-factors required for HIV infection is an attractive alternative or complementary approach.
Delivery of siRNAs or shRNA encoding genetic units to HIV infected cells is also a challenging problem. The target cells are primarily T lymphocytes, monocytes and macrophages. Since synthetic siRNAs will not persist for long periods in cells, delivery would have to be done repetitively for years to effectively treat the infection. Systemic delivery of siRNAs to T lymphocytes is a major barrier and probably not feasible. T-cell isolation from patients followed by transduction, expansion of the transduced cells and re-infusion has been found to be the preferred path (Dropulic, 2001; Davis et al., 2004).
There are additional challenges for using siRNAs in the treatment of HIV-1 infection, including validating the approach in a relevant animal model and preventing the emergence of variants resistant to treatment because of the high sequence diversity of the virus. In this issue, Kumar et al. (2008) exploit a series of recent technical advances to overcome these obstacles. They demonstrate that targeting a combination of host and viral proteins with siRNAs can efficiently inhibit HIV-1 infection in a humanized mouse model. It has been shown previously that antibodies can be used to deliver siRNAs into the cytoplasm of specific target cells (Song et al., 2005). This approach decreases the amount of siRNA that is needed, thereby minimizing the risk of undesired effects in bystander cells. In their new work, Kumar et al. use a single-chain antibody to the CD7 receptor conjugated to a nonamer arginine peptide (9R) (Fig. 1). The CD7-specific antibody is well suited for siRNA delivery because CD7 is expressed by most T cells and is rapidly internalized. Moreover, this antibody has already been used in clinical studies to target toxins to T cell lymphomas and leukemias. In previous work, Kumar et al. (2007) demonstrated that the positively charged 9R peptide binds to polyanionic nucleic acids and can be used to deliver siRNA to neuronal cells. They now show that these techniques can be used to suppress HIV-1 replication and prevent CD4+ T cell depletion in vivo in a humanized mouse model of acquired immune deficiency syndrome (AIDS). This is a significant advance, not only because these findings enhance the prospect of a new HIV-1/AIDS therapy but also because this study introduces an siRNA delivery system that could be adapted to target different receptors and hence other cell types. Moreover, given that the binding of the siRNA to the 9R tag is noncovalent, this approach should make it possible to easily compare the efficacy of different siRNAs.
3.3. Cancer
The use of RNAi for cancer therapeutics could transform treatment of this devastating disease. The strong appeal of RNAi in therapeutics is the potency and specificity with which gene expression can be inhibited. What makes RNAi so exciting to many researchers is its potential for knocking out a protein without harming the cell. By comparison, chemotherapy invariably kills tumors by destroying cancerous cells as well as healthy cells nearby. The possible targets for various diseases range from oncogenes to growth factors and single nucleotide polymorphisms (SNPs).
Animal models are widely used to investigate the therapeutic efficiency of RNAi. In vivo utilization of siRNA was effectively performed by targeting the colorectal cancer-associated gene beta-catenin. Decreased proliferation and diminished invasiveness were observed following siRNA-mediated silencing of this gene in human colon cancer cells. Additionally, when treated cancer cells were placed in a nude mouse, prolonged survival was seen compared with mice receiving unmanipulated tumors. Similarly, silencing the oncogene H-ras led to the inhibition of in vivo tumor growth of human ovarian cancer in a SCID mouse model (Ebbesen et al., 2008). The challenges for cancer are similar to those faced for other diseases which include finding good targets, delivery and minimizing toxicity. Perhaps the most significant work utilized transferin containing nano-particles to target Ewing’s sarcoma cells in a mouse xenograph model (Boden et al., 2003). This study demonstrated the feasibility of using non-lipid based nano-particles for the targeted delivery of siRNAs in a cancer model, and provides a powerful proof of principle for systemic delivery of siRNAs to a metastatic cancer.
Interestingly, RNAi may also be exploited to silence pathways that facilitate the effects of traditional cancer drugs. This includes targeting of the multidrug resistance gene (MDR1) for re-sensitization to chemotherapy and silencing of double-strand break repair enzymes for enhanced effects of radio- and chemotherapy (Olivier et al., 2008). Although the trials are in their early phase, the promises they are showing now are only indicative of the potential RNAi has for future therapeutic process. There has been progress over the last 4 years in terms of different delivery system technologies and the movement of RNAi therapeutics from pre-clinical into human trials. With the first substantial patient data from these RNAi-based therapies on the near-term horizon, and new studies likely to begin soon, it is too early to dismiss RNAi therapeutics for cancer treatment.
4. RNAi technology
Developments like the 2′-acetoxyethyl (ACE) RNA chemistry and the incorporation of modified, especially cationic, nucleotides form the basis for the synthesis of highly stable effective siRNAs. One of the major problems with synthetic siRNAs is their low stability in serum. A study was conducted with much different chemistry at the 2′O ribose position such as aminoethyl and guanidinoethyl (Odadzic et al., 2008), and it was shown that siRNA half life and efficacy can be greatly enhanced by introducing modifications at specific positions both in the passenger and the guide strand of the siRNA. In this study off-target effects caused by the incorporation of the passenger strand in RISC were effectively avoided by design of a nicked passenger strand in the so called small internally segmented interfering RNA (sisiRNA) design. Furthermore, off-target effects could be avoided by incorporation of specific modifications in the guide strand of the siRNA. Nano-particles based on chitosan are highly effective for siRNA delivery, particularly in organs like the lungs.
RNAu, is a new technique based on expression of U1 small nuclear RNA (snRNA) of which the 5′ nucleotides 2–11 are modified to base-pair with a 10 nucleotide target within the 3′ terminal exon of a gene of interest. Binding of the modified U1 snRNA inhibits polyadenylation, resulting in degradation of the transcript and gene knockdown. The U1 snRNA mechanism tolerates a single mismatch at positions 1, 2, 9 and 10, the central 6 nucleotides require perfect base-pairing but do allow a single G–U base-pair. The presence of multiple target sites within the 3′ exon enhances inhibition, and a knockdown of gene expression of up to 700-fold can be achieved.
4.1. Technological applications of RNA interference in Gene knockdown
The RNA interference pathway is often used to study the function of genes in cell culture and in vivo in model organisms (Daneholt, 2007). Double-stranded RNA is synthesized with a sequence complementary to a gene of interest and introduced into a cell or organism, where it is recognized as exogenous genetic material and activates the RNAi pathway. Using this mechanism, researchers can cause a drastic decrease in the expression of a targeted gene. Studying the effects of this decrease can show the physiological role of the gene product. Since RNAi may not totally abolish expression of the gene, this technique is sometimes referred as a “knockdown”, to distinguish it from “knockout” procedures in which expression of a gene is entirely eliminated (Voorhoeve and Agami, 2003).
Extensive efforts in computational biology have been directed toward the design of successful dsRNA reagents that maximize gene knockdown but minimize “off-target” effects. Off-target effects arise when an introduced RNA has a base sequence that can pair with and thus reduce the expression of multiple genes at a time. Such problems are more frequent when the dsRNA contains repetitive sequences.
Depending on the organism and experimental system, the exogenous RNA may be a long strand designed to be cleaved by Dicer, or short RNAs designed to serve as siRNA substrates. In most mammalian cells, shorter RNAs are used because long double-stranded RNA molecules induce the mammalian interferon response, a form of innate immunity that reacts nonspecifically to foreign genetic material (Naito et al., 2006). Mouse oocytes and cells from early mouse embryos lack this reaction to exogenous dsRNA and are therefore a common model system for studying gene-knockdown effects in mammals. Specialized laboratory techniques have also been developed to improve the utility of RNAi in mammalian systems by avoiding the direct introduction of siRNA, for example, by stable transfection with a plasmid encoding the appropriate sequence from which siRNAs can be transcribed, (Reynolds et al., 2006) or by more elaborate lentiviral vector systems allowing the inducible activation or deactivation of transcription, known as conditional RNAi (Stein et al., 2005).
5. Current trends in clinical trials using RNAi
Since the discovery of RNAi, the idea of RNAi therapeutics has blown into a creative and highly competitive field that has attracted some of the most brilliant minds and it is one of the most highly invested in fields in biotechnology research. Many pharmaceutical companies devoted to RNAi research are coming up with interesting insights into how human ailments can be cured using the RNAi mechanism. Although the trials are in their early phase, the promises they are showing now are only indicative of the potential RNAi has for future therapeutic process. For example, Nucleonics Inc., a pharmaceutical company, has initiated a phase l clinical trial of the systemically administrated RNAi-based therapeutic NUCB-1000 for the potential treatment of HBV infection. NUCB-1000 consists of a plasmid DNA construct designed to produce four different shRNAs, targeting different sequences of the HBV genome, under the control of an RNA polymerase III promoter. The plasmid DNA was formulated with a proprietary cationic-lipid delivery system. Another antiviral strategy that has received much attention is currently under development in a phase l clinical trial for the potential treatment of HIV. In this approach, CD34+ cells were collected from patients after induction and treated ex vivo. A mixture of shRNA, ribozyme and RNA decoy targeting three different HIV-related genes: trans-activator of transcription/regulator of virion (tat/rev), CCR5, and transactivation-response genes, respectively, were delivered by a lentiviral vector. The trial combined gene therapy with RNAi, and involved transfecting CD34+ hematopoietic progenitor cells ex vivo and then returning the cells to the patients; RNAi treatment thereby influences all the CD34+ hematopoietic progenitor cells’ future progeny, including T-cells. This represents an important trial since it is the first in which a lentivirus was used as a vector in combination with DNA-directed shRNAs. Preclinical safety and efficacy parameters were encouraging and the pluripotent precursor cells were able to differentiate normally after lentiviral transduction (Thu Ngyuen et al., 2008).
6. Prospects of utilizing RNAi techniques
Since the onset of its discovery, few breakthroughs in history can match the level of understanding and research progress made in RNAi in such a short time. Already, the time when RNAi research in vitro cell cultures used to attract an awe stricken attention is long gone as in vivo trials using RNAi dominate the biotechnology scene.
In a remarkably short time since its discovery in model organisms, the RNAi pathway has emerged as a powerful tool for the study of gene function in mammals. The future of RNAi researches is exciting and there are many applications to be considered in this field. An instance where RNAi technology can be used to silence the gene(s) responsible for the production of β-oxalylaminoalanine-L-alanine (BOAA), a neurotoxin found in a leafy vegetable known as Lathyrus sativus, which is commonly used as a food by the people in the lower socioeconomic class of the poor nations. Another case where RNAi may be successfully applied is in the production of banana varieties resistant to the Banana Bract Mosaic Virus (BBrMV), currently devastating the banana population in Southeast Asia and India (Rodoni et al., 1999).
With the use of RNAi in whole animals increasing, a growing enthusiasm for using RNAi triggers in therapy is anticipated. Despite considerable hurdles to overcome it seems likely that RNAi will find a place along side of many conventional approaches in the treatment of diseases, although it is unclear as how long we have to wait to witness the first RNAi based drug. The future studies of RNAi will pertain to it for being investigated for more applications of human, animal and plant therapeutics. There is great hope that in the near future RNAi based treatment for diseases such as Huntington’s disease, HIV, cancer and other genetic based or infectious diseases will be available.
7. Limitations of RNAi
Despite the proliferation of promising cell culture studies for RNAi-based drugs, some concern has been raised regarding the safety of RNA interference, especially the potential for “off-target” effects, in which a gene with a coincidentally similar sequence to the targeted gene is also repressed. A computational genomics study estimated that the error rate of off-target interactions is about 10%. One major study of liver disease in mice led to high death rates in the experimental animals, suggested by researchers to be the result of “oversaturation” of the dsRNA pathway (SrinivasaRao et al., 2011).
8. Conclusion
Clinical trials with RNAi have now begun, but major obstacles, such as off-target effects, toxicity and unsafe delivery methods, have to be overcome before RNAi can be considered as a conventional drug. Generally, the success of the therapeutic use of RNAi relies on lack of toxicity, specificity of silencing effects and efficacy in vitro and in vivo. So if RNAi is to be used therapeutically one should weigh the possible harms against the possible benefits of this method and perform a risk-benefit analysis. Moreover, as discussed already, delivery methods and clinical trials are nowhere complete and are under rigorous developmental programs. With the increase in technological advancements and novel breakthroughs though it can only be but anticipated that the future of RNAi therapeutics is going to be a bright one.
Acknowledgement
The authors are really thankful to those who contributed toward the completion of this manuscript.
Footnotes
Peer review under responsibility of King Saud University

References
- Amarzguioui M., Lundberg P. Rational design and in vitro and in vivo delivery of Dicer substrate siRNA. Nat. Protoc. 2006;1(2):508–517. doi: 10.1038/nprot.2006.72. [DOI] [PubMed] [Google Scholar]
- Angaji S.A., Hedayati S.S., Poor R.H., Madani S., Poor S.S., Panahi S. Application of RNA interference in treating human diseases. J. Gene. 2010;89:xx. doi: 10.1007/s12041-010-0073-3. [DOI] [PubMed] [Google Scholar]
- Annaleen Vermeulen, Linda Behlen, Angela Reynolds, Alexey Wolfson, William S. Marshall, Jon Karpilow, Anastasia Khvorova. Dicer specificity and efficiency. RNA J. 2005 [Google Scholar]
- Bernstein E., Caudy A.A., Hammond S.M., Hannon G.J. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- Boden D., Pusch O., Lee F., Tucker L., Ramratnam B. Human immunodeficiency virus type 1 escape from RNA interference. J. Virol. 2003;77:11531–11535. doi: 10.1128/JVI.77.21.11531-11535.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castanotto D. Functional siRNA expression from transfected PCR products. RNA. 2002;8:1454–1460. doi: 10.1017/s1355838202021362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daneholt Bertil. Advanced information: RNA interference. Nobel Prize Physiol. Med. 2007 2006 01, 25. 89–97. [Google Scholar]
- Davis B.M., Humeau L., Dropulic B. In vivo selection for human and murine hematopoietic cells transduced with a therapeutic MGMT lentiviral vector that inhibits HIV replication. Mol. Ther. 2004;9:160–172. doi: 10.1016/j.ymthe.2003.11.003. [DOI] [PubMed] [Google Scholar]
- Dropulic B. Lentivirus in the clinic. Mol. Ther. 2001;4:511–512. doi: 10.1006/mthe.2001.0501. [DOI] [PubMed] [Google Scholar]
- Dykxhoorn D.M., Lieberman J. PLoS Med. 2006;3:e242. doi: 10.1371/journal.pmed.0030242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastman S.J., Baskin K.M., Hodges B.L., Chu Q., Gates A., Dreusicke R., Anderson S., Scheule R.K. Development of catheter-based procedures for transducing the isolated rabbit liver with plasmid DNA. Hum. Gene Ther. 2002;13:2065–2077. doi: 10.1089/10430340260395910. [DOI] [PubMed] [Google Scholar]
- Ebbesen M., Jensen T.G., Andersen S., Pedersen F.S. Ethical perspectives on RNA interference therapeutics. Int. J. Med. Sci. 2008;5(3):159–168. doi: 10.7150/ijms.5.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir S.M., Harborth J., Lendecknel W., Yalcin A., Weber K., Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in mammalian cell culture. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- Freda Stevenson K. DNA vaccines and adjuvants. Immunol. Rev. 2004;199(1):5–8. doi: 10.1111/j.0105-2896.2004.00146.x. [DOI] [PubMed] [Google Scholar]
- Grimm D., Kay M.A. J. Clin. Invest. 2007;117:3633–3641. doi: 10.1172/JCI34129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond S.M., Berstein E., Beach D., Hannon G.J. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature. 2000;404:293–296. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]
- Jacque J.M., Triques K., Stevenson M. Modulation of HIV-1 replication by RNA interference. Nature. 2002;418:435–438. doi: 10.1038/nature00896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P., Wu H., McBride J.L., Jung K.E., Kim M.H., Davidson B.L., Lee S.K., Shankar P., Manjunath N. Nature. 2007;448:39–43. doi: 10.1038/nature05901. [DOI] [PubMed] [Google Scholar]
- Kumar, P., Ban, H.S., Kim, S.-S., Wu, H., Pearson, T., Greiner, D.L., Laouar, A., Yao, J., Haridas, V., Habiro, K., et al. (this issue). Cell.
- Lee Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell. 2004;117(1):69–81. doi: 10.1016/s0092-8674(04)00261-2. [DOI] [PubMed] [Google Scholar]
- McCaffrey A.P., Nakai H., Pandey K., Huang Z., Salazar F.H., Xu H., Wieland S.F., Marion P.L., Kay M.A. Inhibition of hepatitis B virus in mice by RNA interference. Nat. Biotechnol. 2003;21:639–644. doi: 10.1038/nbt824. [DOI] [PubMed] [Google Scholar]
- Miller V.M., Gouvion C.M., Davidson B.L., Paulson H.L. Targeting Alzheimer’s disease genes with RNA interference: an efficient strategy for silencing mutant alleles. Nucl. Acids Res. 2004;32:661–668. doi: 10.1093/nar/gkh208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagishi M., Taira K. U6 promoter driven siRNAs with four uridine 3′ overhangs efficiently suppress targeted gene expression in mammalian cells. Nat. Biotechnol. 2002;20:497–500. doi: 10.1038/nbt0502-497. [DOI] [PubMed] [Google Scholar]
- Naito Y., Ui-Tei K., Nishikawa T., Takebe Y., Saigo K. SiVirus: web-based antiviral siRNA design software for highly divergent viral sequences. Nucl. Acids Res. 2006;54:W448–50. doi: 10.1093/nar/gkl214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odadzic D., Bramsen J.B., Smicius R., Bus C., Kjems J., Engels J.W. Synthesis of 2′-O-modified adenosine building blocks and application for RNA interference. Bioorg. Med. Chem. 2008;16:518–529. doi: 10.1016/j.bmc.2007.09.019. [DOI] [PubMed] [Google Scholar]
- Olivier ter Brake, Joost Haasnoot, Jens Kurreck, Ben Berkhout. Antiviral applications of RNA interference. Retrovirology. 2008;5:81. doi: 10.1186/1742-4690-5-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peek A.S., Behlke M.A. Design of active small interfering RNAs. Curr. Opin. Mol. Ther. 2007;9(2):110–118. [PubMed] [Google Scholar]
- Reynolds A., Anderson E., Vermeulen A., Fedorov Y., Robinson K., Leake D., Karpilow J., Marshall W., Khvorova A. Induction of the interferon response by siRNA is cell type- and duplex length-dependent. RNA. 2006;12(6):988–993. doi: 10.1261/rna.2340906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodoni B.C., Dale J.L., Harding R.M. Characterization and expression of the coat protein-coding region of the banana bract mosaic potyvirus, development of diagnostic assays and detection of the virus in banana plants from five countries in Southeast Asia. Arch. Virol. 1999;144:1725–1737. doi: 10.1007/s007050050700. [DOI] [PubMed] [Google Scholar]
- Schwarz D.S., Ding H., Kennington L., Moore J.T., Schelter J., Burchard J., Linsley P.S., Aronin N., Xu Z., Zamore P.D. Designing siRNA that distinguish between genes that differ by a single nucleotide. Adv. Drug Deliv. Rev. 2008;2006(2):14. doi: 10.1371/journal.pgen.0020140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seyhan A.A. RNAi: a potential new class of therapeutic for human genetic disease. Hum. Genet. 2011;130(5):583–605. doi: 10.1007/s00439-011-0995-8. [DOI] [PubMed] [Google Scholar]
- Song E., Zhu P., Lee S.K., Chowdhury D., Kussman S., Dykxhoorn D.M., Feng Y., Palliser D., Weiner D.B., Shankar P. Nat. Biotechnol. 2005;23:709–717. doi: 10.1038/nbt1101. [DOI] [PubMed] [Google Scholar]
- SrinivasaRao T., SrinivasaPrasad C., Shah S.A., Rather M.A. RNA interference and its therapeutic applications. Vet. World. 2011;4(5):225229. [Google Scholar]
- Stein P., Zeng F., Pan H., Schultz R. Absence of non-specific effects of RNA interference triggered by long double-stranded RNA in mouse oocytes. Dev. Biol. 2005;286(2):464–471. doi: 10.1016/j.ydbio.2005.08.015. [DOI] [PubMed] [Google Scholar]
- Sui G. A DNA vector-based RNAi technology to suppress gene expression in mammalian cells. Proc. Natl. Acad. Sci. USA. 2002;99:5515–5520. doi: 10.1073/pnas.082117599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thu Ngyuen RNAi therapeutics: an update on delivery. Curr. Opin. Mol. Ther. 2008;10(2):158–167. [PubMed] [Google Scholar]
- Tijsterman M., Ketting R.F., Okihara K.L., Sijen T., Plasterk R.H.A. RNA helicase MUT-14-dependent gene silencing triggered in Caenorhabditis elegans by short antisense RNAs. Science. 2002;295:694–697. doi: 10.1126/science.1067534. [DOI] [PubMed] [Google Scholar]
- Voorhoeve P.M., Agami R. Knockdown stands up. Trends Biotechnol. 2003;21(1):2–4. doi: 10.1016/s0167-7799(02)00002-1. [DOI] [PubMed] [Google Scholar]
- Wilson J.A., Jayasena S., Khvorova A., Sabatinos S., Rodrigue-Gervais I.G., Arya S., Sarangi F., Harris- Brandts M., Beaulieu S., Richardson C.D. RNA interference blocks gene expression and RNA synthesis from hepatitis C replicons propagated in human liver cells. Proc. Natl. Acad. Sci. USA. 2003;100:2783–2788. doi: 10.1073/pnas.252758799. [DOI] [PMC free article] [PubMed] [Google Scholar]


