Abstract
Current tandem mass spectral libraries for lipid annotations in metabolomics are limited in size and diversity. We provide a freely available computer generated in-silico tandem mass spectral library of 212,516 MS/MS spectra covering 119,200 compounds from 26 lipid compound classes, including phospholipids, glycerolipids, bacterial lipoglycans and plant glycolipids. Platform independence is shown by using tandem mass spectra from 40 different mass spectrometer types including low-resolution and high-resolution instruments.
Hundreds of metabolite signals with tandem mass spectra (MS/MS) are detected in metabolomic applications from complex biological matrices1. While library matches for some of those spectra may be found in MS/MS databases of pure chemical standards, the identification rates are usually very low, because such libraries like NIST, Metlin and MassBank cover less than 20,000 compounds. In comparison, known chemical structures deposited in PubChem, ChemSpider and CAS (Chemical Abstracts), account for more than 100 million structures combined. In addition, the complexity of metabolism in nature implies that there are many more compounds for which no pure reference standards can be purchased. Unlike genes or peptides, metabolites cover a diverse structural space and show large variations in mass spectral fragmentations; therefore, de-novo methods cannot be used with high confidence. We here propose an in-silico generation of tandem mass spectra from small molecule compound structures by means of cheminformatics. This approach works analogous to annotation for peptide MS/MS sequencing, where experimental tandem mass spectra are matched against theoretically predicted mass spectral fragmentations obtained from known amino acid sequences. As first instance of such an in-silico MS/MS metabolite library, we chose lipids as target structures because these compounds are ubiquitous in nature and represent a well investigated class of molecules with consistent mass spectral fragmentations. Online databases and computational tools have been developed for mass spectral lipid analysis2-8, but they do not provide stand-alone MS/MS libraries. We close this gap by providing LipidBlast as a large and platform independent MS/MS database, freely available for commercial and non-commercial use at http://fiehnlab.ucdavis.edu/projects/LipidBlast/.
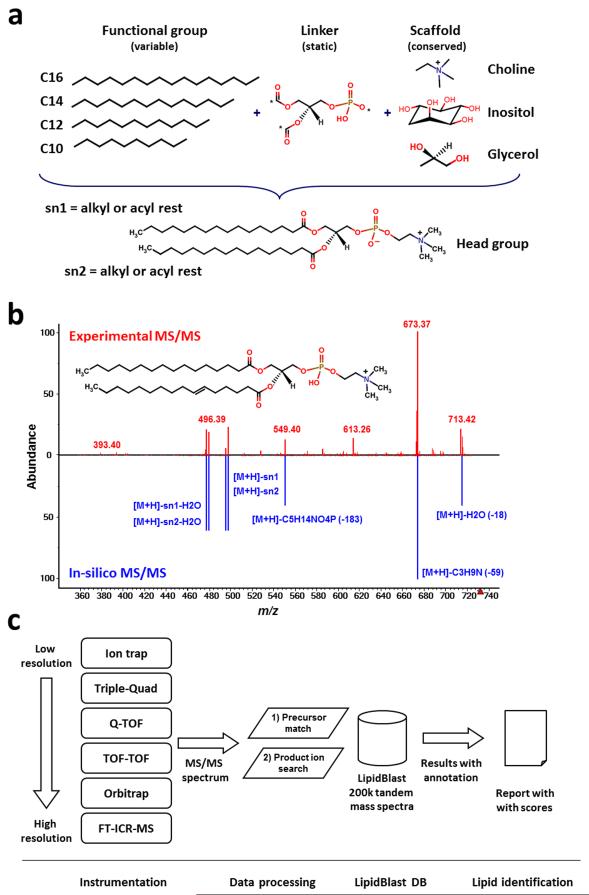
In order to generate an in-silico MS/MS library several steps are required: (i) the definition of structures to be included, the definition of structural boundaries to exclude biologically very improbable compounds and the subsequent exhaustive in-silico generation of all possible structures using combinatorial methods (Fig. 1a); (ii) the experimental acquisition of MS/MS spectra on different platforms and theoretical interpretation of structural class-specific fragmentations and rearrangements; (iii) the rule-based generation of characteristic fragmentations and heuristic modeling of ion abundances for each lipid class covering a series of observed adduct ions (Fig. 1b); (iv) the rigorous validation of the in-silico generated tandem mass spectra including decoy database search and false positive and false negative identification rate investigations and finally (v) the application for high-throughput lipid identification (Fig. 1c).
Figure 1. Creation, validation and application of in-silico generated tandem mass spectra in LipidBlast.
(a) New lipid compound structures were created using combinatorial chemistry approaches. A scaffold of the lipid core structure and linker are connected to fatty acyls with different chain lengths and different degrees of unsaturation. (b) The reference tandem spectra (upper panel) are used to simulate the mass spectral fragmentations and ion abundances of the in-silico spectra (lower panel). The compound shown here is phosphatidylcholine PC(16:0/16:1) at precursor m/z=732.55 [M+H]+. (c) Tandem mass spectra are obtained from LC-MS/MS or direct-infusion experiments. The MS/MS spectra are submitted to LipidBlast MS/MS search. An m/z precursor ion filter serves as first powerful filter and a subsequent product ion match creates a library hit score that is related to the level of confidence for the compound annotation.
Around half of all LipidBlast compound structures were imported from the LIPID MAPS database or generated using the LIPID MAPS Tools9, covering 13 lipid classes of the most common glycerophospholipids and glycerolipids10. Many bacterial and plant lipids were not covered in LIPID MAPS. Therefore we created additional 54,805 compounds from 13 additional lipid classes using the combinatorial chemistry algorithms provided by ChemAxon Reactor11, 12 and SmiLib13 to give a total of 119,200 compounds (see Table 1). Structure examples from each lipid class can be found in Supplementary Figure 1.
Table 1. Lipid classes of common phospholipids, plant and bacterial lipids and number of lipid species and tandem mass spectra in the LipidBlast in-silico MS/MS database.
Positive and negative mode ionization and several adducts including [M+H]+; [M+Na]+; [M+NH4]+; [M-H]−; [M–2H](2−); [M+NH4–CO]+; [M+2Na–H]+; [M]+; [M–H+Na]+; [M+Li]+ are covered. Many of the complex glycolipids structures and MS/MS spectra are enumerated for the first time and were not covered in existing databases.
| Num | Lipid class | Short Name | Number compounds |
Number MS/MS spectra |
Number MS/MS libraries |
|---|---|---|---|---|---|
| 1 | Phosphatidylcholines | PC | 5,476 | 10,952 | 2 |
| 2 | Lysophosphatidylcholines | lysoPC | 80 | 160 | 2 |
| 3 | Plasmenylphosphatidylcholines | plasmenyl-PC | 222 | 444 | 2 |
| 4 | Phosphatidylethanolamines | PE | 5,476 | 16,428 | 3 |
| 5 | Lysophosphatidylethanolamines | lysoPE | 80 | 240 | 3 |
| 6 | Plasmenylphosphatidylethanolamines | plasmenyl-PE | 222 | 666 | 3 |
| 7 | Phosphatidylserines | PS | 5,123 | 15,369 | 3 |
| 8 | Sphingomyelines | SM | 168 | 336 | 2 |
| 9 | Phosphatidic acids | PA | 5,476 | 16,428 | 3 |
| 10 | Phosphatidylinositols | PI | 5,476 | 5,476 | 1 |
| 11 | Phosphatidylglycerols | PG | 5,476 | 5,476 | 1 |
| 12 | Cardiolipins | CL | 25,426 | 50,852 | 2 |
| 13 | Ceramide-1 -phosphates | CerP | 168 | 336 | 2 |
| 14 | Sulfatides | ST | 168 | 168 | 1 |
| 15 | Gangliosides | [glycan]-Cer | 880 | 880 | 1 |
| 16 | Monoacylglycerols | MG | 74 | 148 | 2 |
| 17 | Diacylglycerols | DG | 1,764 | 3,528 | 2 |
| 18 | Triacylglycerols | TG | 2,640 | 7,920 | 3 |
| 19 | Monogalactosyldiacylglycerols | MGDG | 5,476 | 21,904 | 4 |
| 20 | Digalactosyldiacylglycerols | DGDG | 5,476 | 10,952 | 2 |
| 21 | Sulfoquinovosyldiacylglycerols | SQDG | 5,476 | 5,476 | 1 |
| 22 | Diacylated phosphatidylinositol monomannoside |
Ac2PIM1 | 144 | 144 | 1 |
| 23 | Diacylated phosphatidylinositol dimannoside |
Ac2PIM2 | 144 | 144 | 1 |
| 24 | Triacylated phosphatidylinositol dimannoside |
Ac3PIM2 | 1,728 | 1,728 | 1 |
| 25 | Tetraacylated phosphatidylinositol dimannoside |
Ac4PIM2 | 20,736 | 20,736 | 1 |
| 26 | Diphosphorylated hexaacyl Lipid A | LipidA-PP | 15,625 | 15,625 | 1 |
|
| |||||
| Total | All libraries | 119,200 | 212,516 | 50 | |
For lipid fragmentation analysis we performed over 500 experimental measurements of phospholipid and glycerolipid standard reference compounds; a high diversity set of authentic reference compounds with different carbon and double bond numbers per lipid class is preferable as development set. Experiments were performed under 0-55V CID voltage in positive and negative ionization mode. We selected tandem mass spectra from approximately 300 published literature reports for those lipid classes that were unavailable to us as pure reference standards (Supplementary Note 1). Subsequently we have studied which fragmentations and rearrangements were observed for each single lipid class, starting from the precursor ions, including [M+H]+, [M+Na]+, [M+NH4]+, [M-H]−, [M-2H](2−), [M]+, [M+Li]+ (Supplementary Table 1) and continuing to the detailed analysis of product ions, including their relative ion abundances (Supplementary Figure 2). Lipids show predictable MS/MS spectra with dominant fragmentations being the loss of the polar head groups, the acyl or alkyl chain losses from precursor ions (M-sn1, M-sn2) and product ions of the fatty acid (FA) fragments (sn1, sn2; best observed in negative ionization as [FA-H]−). We observed many other specific fragments and rearrangements that were subsequently added to the rule-based generation of tandem mass spectra in LipidBlast (Supplementary Figure 3).
The creation of the in-silico MS/MS libraries itself was performed by transforming the obtained knowledge about fragmentations and ion abundances from the reference lipids to the thousands of lipid structures that were created with combinatorial methods. We used heuristic methods to model precursor and product ions including their relative ion abundances for each of the unique lipid classes (see Online methods). Structure files were imported into the Instant-JChem chemical database and subsequently exported into Microsoft EXCEL. For each individual precursor ion, the characteristic losses and specific fragment ions together with their accurate masses and molecular formulas were calculated. Specific types of mass spectrometers may yield different relative ion intensities; for best MS/MS matching results, we therefore created libraries according to the observed ion intensities from reference spectra acquired by the corresponding instruments. Finally, all MS/MS spectra with lipid species name, adduct name, lipid class, accurate precursor mass, accurate mass fragment, heuristic modeled abundance and fragment annotation were generated as electronic files. Overall 212,516 tandem mass spectra for 119,200 different lipids in 26 lipid classes (see Table 1) have been created.
The validation of LipidBlast was performed by (i) false positive and false negative evaluations, (ii) by using decoy database searches and (iii) by MS/MS analysis of authentic lipid standards measured in-house and from the literature. Search parameters and detailed statistics are given at http://fiehnlab.ucdavis.edu/projects/LipidBlast/.
We first searched all LipidBlast MS/MS spectra against the full LipidBlast library itself using the NIST MS Search program, assuring that there were no technical or systematic errors and validating that each specific lipid would only correspond to their unique counterpart. With very few exceptions (<1%) this test succeeded. Subsequently, LipidBlast was validated against the NIST08 tandem mass spectral library. Using LipidBlast we determined a true positive rate (sensitivity) of 89%, a specificity of 96% and a false positive rate of 4%. We performed an additional and independent validation using 325 accurate mass QTOF MS/MS spectra from the NIST11 database that were not included in the LipidBlast development. 87% of these validation MS/MS spectra were correctly annotated by the true lipid class, number of carbons and double bonds. LipidBlast also correctly identified the correct acyl chains in 76% of all cases. Annotation of double bond positions, stereospecificity and regiospecificity is currently not possible with LipidBlast searches.
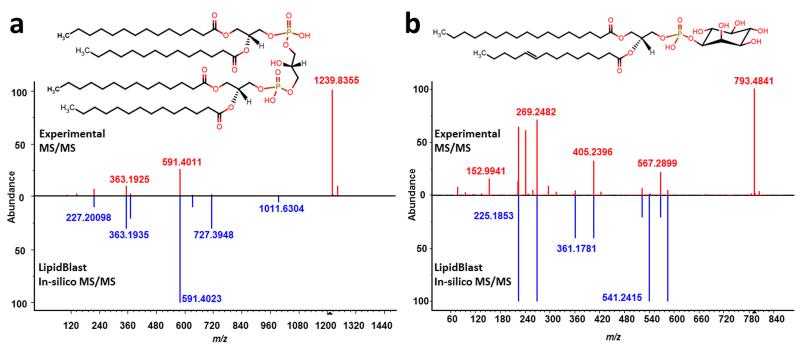
As next validation step, we manually extracted MS/MS spectra from the peer-reviewed literature (see Supplementary Note 1) and converted the printed spectra into digitized formats. We found 134 MS/MS lipid mass spectra of 110 different ionized lipid species of 26 lipid classes covering 40 different types of mass spectrometers (Supplementary Table 2). Falsely annotated spectra and spectra from compound mixtures were excluded. Due to the broad range of instruments and ionization modes, the library search hit scores differed widely. The MS/MS search of these literature spectra using the LipidBlast libraries revealed that in 91% of the 117 remaining cases the correct lipid class was detected including the correct number of carbon atoms and double bonds (Supplementary Figure 4). Next, we used decoy database searches to determine false positive rates. The decoy search determined that all reverse hit scores below 300 should be discarded. As a final validation step we measured an additional twenty-seven authentic reference standards on our QTOF instrument (Fig 2). The compounds included eight different lipid classes with varying chain lengths and different degrees of unsaturation. All compounds except one were correctly identified as first hit. More importantly, the correct carbon number and degree of unsaturation for each specific fatty acyl side chain was determined (see Supplementary Figure 5).
Figure 2. LipidBlast was mostly developed with ion trap tandem mass spectra but can be used with data from other platforms such as QTOF mass spectrometers.
a) The Cardiolipin example shows that even in the in the case of the non-matching but abundant precursor ion at m/z 1239.8355 [M-H]−, the correct result is obtained with LipidBlast. b) The standard reference compound with precursor m/z=793.4841 [M-H]− is correctly identified as phosphatidylinositol PI(17:0/14:1) as first hit in a library search with LipidBlast.
As an example application, we analyzed lipid extracts of the NIST SRM 1050 standard reference human plasma14 using a low resolution mass spectrometer. A total of 264 lipids were structurally annotated of which 90 peaks required manual inspection with scores lower than 600. The dataset was cross-checked with manual peak annotations and data available from LIPID MAPS. With accurate mass LC-MS/MS, a total of 523 molecular lipid species were annotated. Similar numbers of plasma lipids were reported in other methods14, 15. Differences can be attributed to variations in analytical approaches.
Without MS/MS investigation, lipids cannot be unambiguously annotated. When searching the accurate mass of the lipid precursor ion alone with up to 100,000 resolving power, 10-14% of all lipids in LipidBlast would be wrongly annotated with respect to the total number of carbons and double bonds due to isobaric overlaps of lipid adducts. Using LipidBlast, even low-resolution MS/MS spectra can be successfully used, yielding lipid annotations including biochemically meaningful specifications of their accurate acyl chain lengths and double bond counts. With the advent of the LipidBlast library we propose a paradigm shift in metabolomics towards the use of in-silico libraries. Analogous to proteomics, it is not feasible to chemically synthesize all analytical targets (metabolites or natural products) as authentic standards and use these for library generation or quantification purposes. Rather, in-silico libraries can be created directly from compound structures and can be used to annotate mass spectra using scoring algorithms. We have shown that LipidBlast can be successfully applied to tandem mass spectral data from more than 40 different mass spectrometer types. The current array of plant, animal, virus and bacterial lipid tandem mass spectra in LipidBlast can be easily extended to many other important lipid classes.
ONLINE METHODS
Creation of molecular structure templates
Compound structures were generated with three different combinatorial chemistry software tools. For commonly known lipids the publicly available LIPID MAPS Tools9, 16 (v1.0, http://www.lipidmaps.org/) were used to create a starting set of 45,000 glycerophospholipid and 444,080 glycerolipid structures using the Perl scripts provided by LIPID MAPS. The number of carbons and position of double bonds was based on LIPID MAPS nomenclature17. File sizes of around 5.7 MByte per 10,000 compounds were generated; hence, the structure library file of 45,000 glycerophospholipid species yielded a 256 MByte file. For lipid classes generating even larger structure files, structure generation was performed sequentially class-by-class in order to manage computational time and memory size. For example, using the LIPID MAPS Tools we initially generated a cardiolipin library of a total of 32 million structures which would have required larger computational resources than were available to us. For this reason, we have employed a different way to generate individual structures for cardiolipins, triacylglycerides and all bacterial lipoglycans and plant glycolipids (MGDG, DGDG, SQDG, Ac2PIM1, Ac2PIM2, Ac3PIM2, Ac4PIM2, LipidA-PP). In order to avoid combinatorial explosion of the number of structures generated, constraints were applied. For example, the cardiolipin library was limited to only 25,426 structures by constraining the lengths of acyl carbon chains to C14-C22 and the number of double bonds in a single acyl chain to 0-6 and by removing stereo- and regioisomeric structures. The triacylglycerol library was reduced from over one million compounds to only 2,640 relevant structures by limiting carbon numbers from C12-C22 and allowing 0-6 double bonds in each individual acyl chain. Not all of the computationally generated structures may actually exist in nature, while other potentially existing structures may have been missed due the constraints applied here. Mass spectral libraries, including the most prominent NIST library as well as the LipidBlast library presented here, will therefore continue to grow in breadth and volume over time.
We have used the ChemAxon Reactor software11, the ChemAxon Markush structure generator and the SMILIB (v2.0) virtual synthesis software13 for building these structure libraries. A scaffold of the core structure and fifteen fatty acid building blocks were entered. Only the fifteen most important fatty acid residues known from the literature were taken into account and stereochemistry of the double bonds was removed. Only the total carbon chain length and double bond number were considered. Due to the molecular symmetry of the cardiolipins a canonization (creation of a unique hash code) was performed with the original InChI and InChIKey software to remove duplicate structures (http://www.iupac.org/inchi/download/). The obtained SDF files for each class were loaded into Instant-JChem desktop structure database (Instant-JChem v2.4, 2008, ChemAxon, http://www.chemaxon.com/) and additional calculations were performed including the exact isotopic mass and the octanol/water partition coefficient (logP). The resulting libraries were exported to separate Microsoft Excel worksheets for each lipid class. Additional data such as exact isotopic masses and molecular formulas were calculated with Instant-JChem. The lipid name, short name, side chain length, number of side chain double bonds, accurate masses for possible adducts and possible and observed side chain losses were included. The LIPID MAPS nomenclature name was included when available.
Modeling fragment and ion abundances and spectra creation
Accurate masses of ten different electrospray adducts (e.g. [M+H]+, [M+Na]+, [M+NH4]+, [M+Li]+, [M-H]−) were obtained for positive and negative electrospray conditions by summing up the accurate masses of the adduct ions, head groups and their alkyl and acyl side chains (sn1 and sn2). For lipid reference standards available to us, ion trap mass spectra were obtained at six different voltages (see below). For compound classes that yielded spectra that differed from published mass spectra, additional product ions were included such as specific losses for phosphatidylcholine [M-18]+, [M-59]+ and [M-183]+ in order to correctly reflect experimental ion trap spectra in the virtual MS/MS library. Fragmentation rules for all 26 lipid classes were obtained from at least two standard compounds with different degree of unsaturation, either by investigating in-house obtained data or fragmentation experiments from the literature.
Creating the virtual MS/MS LipidBlast library from the structures involved several steps. First, all lipid structure files were imported into Instant-JChem, and exact isotopic masses and molecular formulas were calculated within the database. Tandem mass spectra for each lipid class had to be modeled specifically. MS/MS fragments were investigated by associating the experimental MS/MS spectrum with the structure and performing a mass spectral fragmentation reaction for each class. Lipid classes were not only determined by the head groups but also by the chemistry of the side chains. For example, mass spectra of alkyl and alkenyl ethers are very distinct from MS/MS spectra of acyl ester species, and hence, such lipid classes had to be modeled separately and are counted as unique classes in Table 1. Product ion fragmentations and ion abundances were modeled in LipidBlast by a three step method: (i) obtaining specific fragments from commercially available standards; (ii) querying key structures in LIPID MAPS or using the LIPID MAPS MS tools; (iii) validating known fragmentations with literature data. Each MS/MS spectrum was modeled with major chemical adducts. For all interpretable and characteristic product ions we calculated molecular formulas, exact isotopic masses and added short textual descriptions of neutral losses and specific product ions. Such peak annotations were incorporated for each in-silico spectrum and can guide practitioners during manual inspection of MS/MS spectra. Unlike other approaches, the virtual MS/MS LipidBlast library not only contains fragment ions, but also includes heuristically modeled ion abundances. Overall, the accurate modeling of fragment ion abundances is an unresolved problem and clearly depends on the fragmentation parameters and instruments design. Recently machine learning algorithms were used to train and predict mass spectral peak intensities using peak intensities from experimentally measured spectra18. However currently no validated thermo-chemical or quantum-mechanical ab-initio model exists for the calculation of mass spectral peak abundances given a compound structure only1. We observed that low abundant fragment ions detected in one instrument (e.g., an ion trap MS/MS experiment) were usually also low abundant in a different mass spectrometer (e.g. a QTOF MS/MS experiment) because intensity of product ions depends on the internal energy and chemical structure of the precursor ions, and much less on the way the collision energy was technically applied. Hence, for similar collision energies, we found lipid mass spectra, including ion abundances, to be comparable across certain instrument types. Therefore we chose to heuristically model all MS/MS peak abundances to yield a lipid spectral library that can be used across platforms, for all other cases we decided to perform custom modeling of spectra. The modeling of ion abundances further helps in annotation of lipids by mathematical scoring. Product ion abundances were coded in a static manner based on our observation of experimental mass spectra under ion trap MS/MS conditions. For special cases (such as very high collision energies or TOF instrument settings) we customized the modeled abundances by including multiple tables with ion abundances for each product ion. Regiospecific analysis of the specific position of alky or acyl side chain on the glycerol backbone would require MS3 or adduct experiments and therefore could not be correctly modeled by our LipidBlast MS/MS library. All modeled mass spectra were compiled in a Microsoft Excel sheet and subsequently exported to MS formats such as MSP files containing accurate masses and fragment information. The Excel sheet contained a Visual Basic macro program of around 6,000 lines of code that automatically created all MS/MS libraries in mass spectral export format (NIST MSP ASCII). The creation of all 212,516 MS/MS spectra took around 90 seconds. MSP files are text based and can be imported into any vendor specific mass spectral library search application. MSP files can also be converted into other library formats with existing software tools when necessary. The MSP format contains the following information: name of the compound, accurate precursor mass, positive or negative mode, comment with short name, long name, lipid class name, formula, the number of peaks in the spectrum, m/z and intensity pairs, annotation and explanation of all m/z peaks. For fast pre-screening of accurate masses, a lookup table of all ions in LipidBlast is provided as a separate LipidBlast EXCEL macro-enabled worksheet (LipidBlast-mz-lookup). Such a lookup table can also be used for accurate mass instruments without MS/MS or MSE capability and can provide lipid-class, carbon and double bond numbers depending on mass accuracy settings in an automated way.
Custom modeling of mass spectral abundances and fragments
It is well established that mass spectra of lipids can be largely different when comparing tandem mass spectra across mass spectrometry platforms and fragmentation energies. Older linear ion trap instruments suffer from low mass cut-off in CID mode and cannot record product ions at less than 1/3 of the mass of the precursor ions. Hence, certain fragment ions are missing from ion trap spectra, for example, the ion m/z=184.07 referring to the phosphocholine head group (C5H15NO4P) of phosphatidylcholines. On the other hand, these ions can be very abundant on QTOF instruments, QTRAP hybrid instruments or Orbitrap analyzers with HCD activation. We have custom modeled such well-established fragment ions into the LipidBlast library and further characteristic fragments and ion abundance can be added via customized templates. Misidentification of lipids can thus be avoided using such customized templates, as shown for QTOF MS/MS spectra of PC 36:2 as [M+H]+ and as [M+HCOO]− adduct species (see Supplementary Figure 3). The LipidBlast software can be easily extended, e.g. for adducts not yet listed in the library such as potassium adducts. The software can also be used for fragment generation of completely new lipid classes, once standard compounds are available or consistent mass spectral fragmentation patterns were reported in literature. LipidBlast scoring works best with fragmentation-rich product ion spectra. Such voltage optimizations should be performed for each instrument type and each lipid class. LipidBlast currently contains common mammalian and plant fatty side chains as defined in LIPID MAPS. For less common side chains such as highly unsaturated and branched carbon chains synthesized by plants and bacteria, customized libraries need to be constructed through the combinatorial chemistry and structure-space approach implemented in the LipidBlast software. As an example, we included tuberculostearic acid (10-Methyloctadecanoic acid) into specific glycolipid structures. These bacterial acids are observed in patients with tuberculosis and are important biomarkers from mycobacterial cells19, 20. The stereochemistry of lipid species including tetrahedral (R/S) and double bonds (Z/E) cannot be detected with the current version of LipidBlast. This step would either require the complete chromatographic resolution and multi-stage tandem mass spectrometry (MSn). Selective annotation of regiospecific isomers such as the different position of double bonds as well as the correct determination of sn1, sn2, sn3 acyl chain positions in triacylglycerols are not yet feasible based on existing experimental fragmentation rules. We kept all phospholipid species downloaded from LIPID MAPS in the LipidBlast library even for stereo- and regioisomers to enumerate the correct number of compounds which can be expected using a chromatographic separation. In principle, the versatility of lipid scoring in LipidBlast can be extended by using further constraints. For example, molecular descriptors such as octanol/water partition coefficients (logP and logD) can be calculated directly from the molecular structures and may serve in multipredictor models to predict retention times in liquid chromatography. Such constraints can be used to exclude false positive annotations by retention time modeling. Moreover, the structure centric approach in LipidBlast enables the use of the database for other purposes, for example to integrate the library with other fragmentation prediction software such as MassFrontier21 or for use in cheminformatics software for systematic naming and comparisons of structure similarities.
MS/MS library search with precursor ion filtering and product ion matching
Tandem mass spectra are generally searched in two steps. The appropriate LipidBlast library is selected according to positive or negative ionization mode. First, a precursor ion filter removes all spectra that are outside a specific precursor m/z window. For low resolution instruments (such as ion trap mass spectrometers), a precursor search window of ± 0.4 Da can be applied, whereas for mass spectrometers with high resolving power and high mass accuracy, a precursor search window of ± 0.005 Da should be selected. A search for a negative ion mode electrospray MS/MS spectrum of a lipid with a precursor ion of m/z=750.540 Da will result in only three hits out of 134,204 possible LipidBlast hits, whereas a search of the same ion from a low resolution instrument will result in 153 candidates. Hence, a precursor ion filter can remove up to 99.99% of all false positive hits for high resolution instruments. However, high resolving power does not suffice for lipid annotation: a search of m/z=760.500 Da in negative electrospray mode will yield 201 hits with a precursor search window of ± 0.005 Da. In addition, the identity of side chains cannot be easily determined without MS/MS fragmentation (however it is possible to use in-source fragmentation). Because LipidBlast also covers different acyl chain lengths and double bond counts in the product ion spectra even isobaric species can be annotated. For example the triacylglycerol TG(56:6) as ammonium ion [M+NH4]+, at m/z=924.8015 can cover species TG(16:0/20:2/20:4) and TG(18:1/18:1/20:4) and 22 other isomers. The accurate mass precursor matching and the stringent matching of abundant product ion peaks will exclude all other unlikely species based on the scoring threshold. In case of very few product ions, the matching algorithm is still functional on the precursor level, but less specific due to the missing product ion peaks. In such cases the scoring algorithm detects the correct lipid class, carbon number and double bonds, but information on specific acyl chains is limited.
Use of LipidBlast with mass spectral search programs
The freely available NIST MS Search GUI program (version 2.0f, build April 2010, http://chemdata.nist.gov/mass-spc/ms-search/) was used for mass spectral library searches. The program uses a very fast indexing method with search results in a 200,000 entry library usually represented within milliseconds. The program is capable of MS/MS mass spectral search and requires precursor and product ion m/z tolerances to be set. The program presents multiple search scores, including dot-product, probability matched and reverse-dot-product as the result of a library search. A perfect match obtains a search score of 999 and lower confidence matches result in lower match scores22. The GUI is valuable for manual inspection (see Fig. 2) of MS/MS spectra by comparing head to tail view and inspecting LipidBlast peak fragment annotations (see Supplementary Figure 4). A faster command line version of the search program (NIST MSPepSearch mass spectral library search program ver. 0.9 build 04/22/2010, http://peptide.nist.gov) was used for batch searches across multiple MS/MS spectra. The search speed was up to 1000 spectra/seconds and depends on the library size. A LC-MS/MS MGF file with 10,000 precursors is searched against LipidBlast within ten seconds. Parallel searches allow for even higher annotation rates, by starting multiple instances of the MSPepSearch program. The tool directly presents a spreadsheet with compound names and hit scores for each tandem mass spectrum.
The NIST MS/MS library was created from the LipidBlast MSP files using the Lib2NIST converter tool. The LipidBlast library in NIST format consisting of 212,516 MS/MS spectra has a size of only 150 Mbytes. Due to the large size of structure files, these were not included in the NIST MS/MS library although in principle, the NIST MS program can handle associated structures. The library search is used with the MS/MS search option by setting a precursor and product ion m/z tolerance. In case of low-resolution ion trap mass spectra, the precursor accuracy was set to ±0.4 Da and the product ion tolerance to ±0.8 Da. For high mass resolution data, the windows can be narrowed down to ±0.005 Da, depending on the mass accuracy of the instrument. The peptide scoring options are all turned off; however the QTOF search option and the score threshold setting have an influence on the result scores and were set to low or turned off.
All calculations were performed on a Monarch Computer Dual Opteron 254 (2.8 GHz) with an ARECA-1120 Raid-6 array using WD Raptor hard disks (max hard disk burst read/write transfer rate 500 MByte/s) equipped with 2.8 GByte RAM running a 32-bit Windows XP. An additional RamDisk (QSoft Ramdisk Enterprise) was used for file based operations allowing burst read-write rates of 1,000 MByte/s.
Validation settings and procedures
The validation was needed to determine thresholds for mass spectral library scoring and to determine the figures of merit for database search. We therefore counted as true positive identification if both the lipid class and the number of carbon and double bonds of the side chains were correctly identified. At present, there is no large MS/MS database of lipid species available for downloads and validation purposes. Therefore, non-equal distribution had to be assumed for some of the performed steps.
LipidBlast self-search settings: For positive ionization mode the algorithm detected the correct lipid class in 99.99% for all 78,314 positive mode MS/MS spectra. The lipid class and the associated correct side chains (acyl, plasmenyl and alkyl ethers) including the carbon number and double bond number were found in 99.54% of the cases. For negative ionization mode, all 134,202 spectra yielded the correct lipid class and correct side chains in 100% of the cases.
Decoy search settings: Using the Peptide Atlas consensus library of 337 human Albumin peptides MS/MS spectra against all LipidBlast MS/MS spectra in positive ionization mode and a precursor ion filter of ±0.4 Da, not a single peptide yielded hit scores of more than 277 with a median score of only 29. Only 5 peptides (1.5%) had hit scores of larger than 200 and 16 peptides (4.7%) yielded reverse scores larger than 300, defining potential lower thresholds for MS/MS identification in LipidBlast scoring.
NIST08 settings: This library contains 14,802 tandem mass spectra of 5,308 precursor ions. It contains 131 MS/MS spectra from 47 unique lipid species. These spectra were not used during the development of the LipidBlast libraries. A search of all NIST08 spectra against LipidBlast using a simple found/not-found strategy and precursor ion filter of ±0.4 Dalton without scoring revealed a sensitivity (true positive rate) of 65%, specificity of 74% and a false positive rate of 26%. Of these false positive spectra, some lipid classes were found more often than other classes such as phosphatidic acids, monoacylglycerols, lysoPC, MGDG and lysoPE. Annotation of spectra for these lipid classes should be validated by visual inspection of spectra or constrained retention time filtering. For better hit rates, we advise to use accurate mass precursor selections. We then enabled the commonly used MS/MS scoring algorithm23 and the sensitivity (true positive rate) increased to 89%, the specificity increased to 96% and the false positive rate dropped to 4%.
NIST11 settings: We performed an additional independent validation with 104 negative ion mode and 220 positive ion mode ESI tandem mass spectra measured with different ionization voltages on an Agilent 6530 QTOF instrument. These spectra were obtained from the NIST11 database and were not available during the time of development. In negative mode 94% and in positive mode 84% of all spectra were correctly annotated. Reasons for such false annotations, which occurred mostly in positive ionization mode, included missing product ions that reflect an acyl chain loss or product ion spectra with very few peaks. Overall 87% of all 325 validation MS/MS spectra were correctly annotated. For 76% of all combined cases, each individual acyl chain was correctly assigned.
Settings for literature spectra: We found several MS/MS spectra that were published in the literature, but in fact associated wrong structures to the published spectra, or that contained MS/MS spectra of compound mixtures. After cleaning spectra of mixtures and wrong annotations a total of 117 spectra remained. For very few lipid classes, the literature-based validation could not be performed completely independent from the LipidBlast library construction, for example for phosphatidylinositol mannosides and ceramide phosphates, for which only two tandem mass spectra were found in literature and for which no commercial authentic reference standards were available.
Experimental settings for MS/MS infusion and LC-MS/MS
Experimental spectra were obtained on a LTQ linear ion trap mass spectrometer, a hybrid LTQ-FT-ICR mass spectrometer (Thermo Fisher Scientific) and a 6530 QTOF mass spectrometer (Agilent). All lipid standards were obtained from Sigma/Aldrich and Avanti Polar Lipids. The infusion of lipid standards and extracted lipid samples was performed using a chip based nano-electrospray infusion (Advion Nanomate). Plasma lipids were extracted using methyl-tert-butyl ether (MTBE)24. In brief, methanol (225 μL) was added to 30 μL blood plasma and shaken with an additional 750 μL of methyl-tert-butyl ether solvent. Phase separation of this extract was induced by adding 187.5 μL of water, vortexing and centrifuging the mixture at 14,000 g for 2 min. The upper organic phase was collected and dried in a vacuum centrifuge. After adding 10 μL of 100 mM ammonium acetate to 90 μL of the supernatant, lipid extracts were infused into the mass spectrometers using an Advion Nanomate chip-based infusion system (nanoESI). Ion trap mass spectra were collected in low resolution mode (1,500 resolving power) on the linear ion trap. The data collection method performed a full scan and a data dependent MS/MS scan of the most abundant ions. Different CID voltages in the range from 0V to 100V were used for evaluation of spectra. For abundance calculations standard spectra were scanned in low-resolution mode with 15V, 20V, 25V, 35V, 45V and 55V CID voltage to obtain specific MS/MS fragmentations. All spectra were recorded with the Thermo Xcalibur software. An infusion time of 30 seconds was set up in full scan mode with 0V CID with an additional 30 seconds of data dependent MS/MS scans to obtain tandem mass spectra for the largest peaks. For each sample, around 50 MS/MS scans were averaged. NIST SRM 1950 blood plasma samples were infused for around 10 minutes to allow the acquisition of a higher number of MS/MS scans.
The 6530 QTOF mass spectrometer for measurement of reference compounds was operated with the following parameters. An Agilent JetStream electrospray source was used in infusion mode at a flow rate of 0.25 ml/min for acquiring QTOF MS and MS/MS spectra. Data were collected with a 0.25 s scan rate in both profile and centroid modes, and mass calibration was maintained by constant infusion of reference ions at 121.0509 and 922.0098 m/z. MS/MS data was generated utilizing data-dependent MS/MS triggering with dynamic exclusion. Precursor ions, with a minimum 1 k signal intensity were isolated with a 4 m/z isolation width (medium setting), and a variable collision energy was applied based on precursor ion m/z (10 eV + 0.03 eV × ion m/z). Data were exported into the open exchange format mzXML. Samples were measured in negative and positive mode. For lipid profiling with liquid chromatography/quadrupole time-of-flight mass spectrometry (LC-MS/MS) we used settings from an external reference25, except we choose a scan rate of 4-8 spectra per scan event and collision energies ranging from 20-40eV.
Use of LipidBlast for LC-MS/MS and direct-infusion MS/MS experiments
Experimental mass spectra were exported as MGF files using DeconMSn26 and for AB SCIEX, Agilent, Bruker, Thermo Fisher Scientific and Waters files the freely available Proteowizard27 tools can be used. The MGF files are simple container files holding multiple data dependent MS/MS scans. Prior merging data into MGF formats and in order to reduce the number of similar tandem spectra, a spectral clustering based on the precursor ion selection was performed for direct-infusion data using MSCluster28. Such a clustering algorithm computes consensus spectra from multiple MS/MS scans. The MGF files were directly imported into the NIST MS Peptide Search program to either perform manual search or create batch lists of results. To perform batch searches, the NIST MS Search program can either be started in batch mode (command line par=4 which creates NISTLOG.TXT) or the freely available NIST MSPepSearch can be used for high-throughput batch annotations. The NIST MS search reports hit scores from 0-999 in addition to dot product scores from 0-999 and probability match scores ranging from 0-99%. For each category, higher scores mean more confident lipid annotations. During our validation tests, we found that hit scores > 950 were generally true positive hits and hit scores > 750 were potential true positive hits but required manual investigations. We recommend using further criteria for correct lipid annotations; for example, fractionation schemas or retention time information that will improve probability of correct annotation of lipid species. For determining false positive annotation rates and lower thresholds for MS queries, we used a peptide database as decoy database from PeptideAtlas (http://www.peptideatlas.org/speclib/) with a mass cut off of 1100 Dalton. The source was the NIST consensus library of peptide ion fragmentation spectra (Human Serum Albumin, HSA, http://peptide.nist.gov/) with fully assigned peptide names.
For the direct-infusion experiment we collected data dependent MS/MS scans during fifteen minute infusion time. 1,332 MS/MS spectra from unique precursor ions were extracted in positive mode and 1,060 MS/MS spectra were identified in negative mode. All spectra were searched against the LipidBlast libraries using a 0.4 Da precursor search window and obtaining reverse search scores ranging from 0 to 999 (0: no result, 999: highest confidence). In order to rank the results, we defined sub-scores based on the prior scores from the validation of the library. Reverse dot product scores 999-600 were acceptable; scores in the range 300-600 were manually confirmed. All scores lower than 300 were considered as false positives as given by the validation thresholds.
The results of lipid identifications by defined (and static) MS/MS transition experiments in triple-quadrupole mass spectrometry may have a high false positive discovery rate29, unless lipids are clearly pre-separated into the different lipid classes by liquid chromatography or fractionation methods. Unfortunately, false positive discovery rates were rarely published in the past for shotgun lipidomics approaches. The diversity of lipid structures and lipid mass spectra render it highly likely that there are false-positive annotations in shotgun lipidomics reports due to unexpected product ions and lack of full MS/MS spectral validation. On hybrid triple quadrupole instruments systems an enhanced product ion scan (EPI) can be performed to obtain MS/MS spectra for validation. Nevertheless, the inclusion of analytical figures of merit for compound identification such as sensitivity, specificity and error rates should be included for all methodologies and approaches.
The LipidBlast software, 212,516 accurate mass and fully annotated tandem mass spectra (MS/MS) from 119,200 lipid structures, as well all development Microsoft Excel templates and validation materials are freely available for commercial and non-commercial use under a Creative Commons License (By Attribution, CC-BY) at the authors website at (http://fiehnlab.ucdavis.edu/projects/LipidBlast/).
Supplementary Material
ACKNOWLEDGMENTS
We thank the LIPID MAPS consortium and the National Institute of General Medical Sciences (NIGMS/US) for providing extensive lipid identification and database services. We thank the NIST Mass Spectrometry (US) group for providing the freely available NIST MS Search GUI program and help with the Lib2NIST converter. We thank ModLab (University Frankfurt am Main) for providing the free SMILIB enumeration tool. We thank ChemAxon for a free research license for Marvin and Instant-JChem cheminformatics tools. Funding for K.H.L was supported by the National Research Foundation of Korea, Ministry of Education, Science and Technology (Grants 2010-0021368), the Korea Healthcare Technology R&D Project, Ministry of Health & Welfare (Grant No.: A103017), and the Cooperative Research Program for Agriculture Science & Technology Development (Project No. PJ00948604), Rural Development Administration, Republic of Korea. Funding for T.K. and O.F. was supported by NSF MCB 1139644, NIH P20 HL113452 and U24 DK097154.
Footnotes
AUTHOR CONTRIBUTIONS
T.K., K.H.L, D.Y.L. and O.F. designed the experiments. T.K., K.H.L, B.D., J.K.M. and D.Y.L. performed mass spectrometric experiments. T.K. and K.H.L performed mass spectral fragmentation analysis and compound annotations. T.K. created the compound structures and developed the in-silico MS/MS libraries and wrote and validated the algorithm. T.K. and O.F. wrote the manuscript in interaction with all contributing authors.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Note: Supplementary information is available on the Nature Methods website.
References
- 1.Kind T, Fiehn O. Bioanalytical Reviews. 2010;2:23–60. doi: 10.1007/s12566-010-0015-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Song H, Hsu FF, Ladenson J, Turk J. Journal of the American Society for Mass Spectrometry. 2007;18:1848–1858. doi: 10.1016/j.jasms.2007.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yetukuri L, et al. BMC Syst Biol. 2007;1:12. doi: 10.1186/1752-0509-1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Forrester JS, Milne SB, Ivanova PT, Brown HA. Mol Pharmacol. 2004;65:813–821. doi: 10.1124/mol.65.4.813. [DOI] [PubMed] [Google Scholar]
- 5.Yang K, Cheng H, Gross RW, Han X. Analytical chemistry. 2009;81:4356–4368. doi: 10.1021/ac900241u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bou Khalil M, et al. Mass spectrometry reviews. 2010;29:877–929. doi: 10.1002/mas.20294. [DOI] [PubMed] [Google Scholar]
- 7.Taguchi R, Ishikawa M. Journal of Chromatography A. 2010;1217:4229–4239. doi: 10.1016/j.chroma.2010.04.034. [DOI] [PubMed] [Google Scholar]
- 8.Herzog R, et al. PLoS One. 2012;7:e29851. doi: 10.1371/journal.pone.0029851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fahy E, Sud M, Cotter D, Subramaniam S. Nucleic acids research. 2007;35:W606–W612. doi: 10.1093/nar/gkm324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sud M, et al. Nucl. Acids Res. 2007;35:D527–532. doi: 10.1093/nar/gkl838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pirok G, et al. Journal of Chemical Information and Modeling. 2006;46:563–568. doi: 10.1021/ci050373p. [DOI] [PubMed] [Google Scholar]
- 12.Instant-JChem, Marvin . Reaktor v5 - software for database management, 2010-2012. ChemAxon; http://www.chemaxon.com. [Google Scholar]
- 13.Schuller A, Hahnke V, Schneider G. QSAR & Combinatorial Science. 2007;26 [Google Scholar]
- 14.Quehenberger O, et al. Journal of lipid research. 2010;51:3299–3305. doi: 10.1194/jlr.M009449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao X, et al. Analytical and Bioanalytical Chemistry. 2012;402:2923–2933. doi: 10.1007/s00216-012-5773-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sud M, Fahy E, Subramaniam S. Journal of Cheminformatics. 2012;4:23. doi: 10.1186/1758-2946-4-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Subramaniam S, et al. Chemical reviews. 2012;111:6452. doi: 10.1021/cr200295k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kangas LJ, et al. Bioinformatics. 2012;28:1705–1713. doi: 10.1093/bioinformatics/bts194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sartain MJ, Dick DL, Rithner CD, Crick DC, Belisle JT. Journal of lipid research. 2011;52:861–872. doi: 10.1194/jlr.M010363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Layre E, et al. Chemistry & biology. 2011;18:1537–1549. doi: 10.1016/j.chembiol.2011.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sheldon MT, Mistrik R, Croley TR. Journal of the American Society for Mass Spectrometry. 2009;20:370–376. doi: 10.1016/j.jasms.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 22.Stein SE, Scott DR. Journal of the American Society for Mass Spectrometry. 1994;5:859–866. doi: 10.1016/1044-0305(94)87009-8. [DOI] [PubMed] [Google Scholar]
- 23.Stein S. Analytical Chemistry. 2012;84:7274–7282. doi: 10.1021/ac301205z. [DOI] [PubMed] [Google Scholar]
- 24.Matyash V, Liebisch G, Kurzchalia TV, Shevchenko A, Schwudke D. The Journal of Lipid Research. 2008;49:1137. doi: 10.1194/jlr.D700041-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sandra K, Pereira A.d.S., Vanhoenacker G, David F, Sandra P. Journal of Chromatography A. 2010;1217:4087–4099. doi: 10.1016/j.chroma.2010.02.039. [DOI] [PubMed] [Google Scholar]
- 26.Mayampurath AM, et al. Bioinformatics. 2008;24:1021–1023. doi: 10.1093/bioinformatics/btn063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chambers MC, et al. Nature Biotechnology. 2012;30:918–920. doi: 10.1038/nbt.2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frank AM, et al. J. Proteome Res. 2008;7:113–122. doi: 10.1021/pr070361e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stein SE, Heller DN. Journal of the American Society for Mass Spectrometry. 2006;17:823–835. doi: 10.1016/j.jasms.2006.02.021. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.