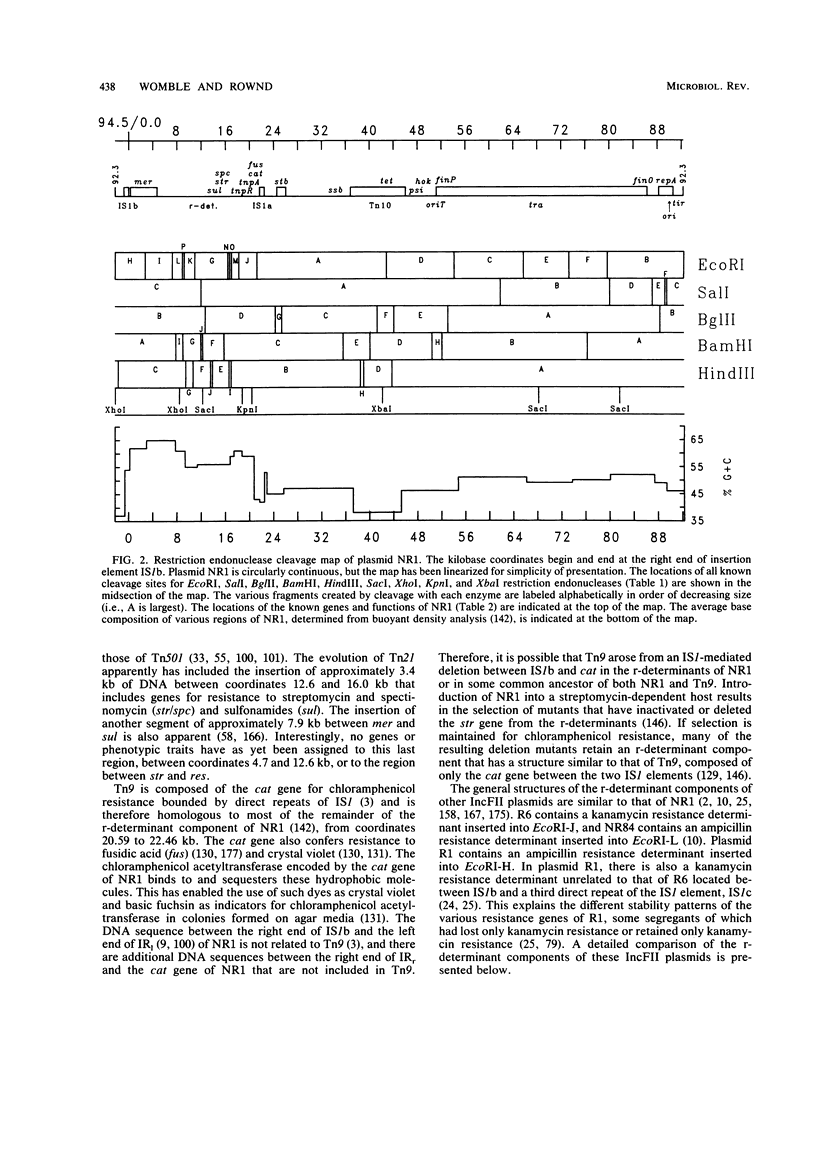
Full text
PDF

















Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alton N. K., Vapnek D. Molecular cloning of restriction fragments and construction of a physical and genetic map of the Escherichia coli plasmid R538-1. Plasmid. 1978 Jun;1(3):388–404. doi: 10.1016/0147-619x(78)90054-9. [DOI] [PubMed] [Google Scholar]
- Alton N. K., Vapnek D. Nucleotide sequence analysis of the chloramphenicol resistance transposon Tn9. Nature. 1979 Dec 20;282(5741):864–869. doi: 10.1038/282864a0. [DOI] [PubMed] [Google Scholar]
- Andrés I., Slocombe P. M., Cabello F., Timmis J. K., Lurz R., Burkardt H. J., Timmis K. N. Plasmid replication functions. II. Cloning analysis of the repA replication region of antibiotic resistance plasmid R6-5. Mol Gen Genet. 1979 Jan 5;168(1):1–25. doi: 10.1007/BF00267929. [DOI] [PubMed] [Google Scholar]
- Arber W., Sengstag C., Caspers P., Dalrymple B. Evolutionary relevance of genetic rearrangements involving plasmids. Basic Life Sci. 1985;30:21–31. doi: 10.1007/978-1-4613-2447-8_4. [DOI] [PubMed] [Google Scholar]
- Armstrong K. A., Ohtsubo H., Bauer W. R., Yoshioka Y., Miyazaki C., Maeda Y., Ohtsubo E. Characterization of the gene products produced in minicells by pSM1, a derivative of R100. Mol Gen Genet. 1986 Oct;205(1):56–65. doi: 10.1007/BF02428032. [DOI] [PubMed] [Google Scholar]
- Austin S., Abeles A. Partition of unit-copy miniplasmids to daughter cells. I. P1 and F miniplasmids contain discrete, interchangeable sequences sufficient to promote equipartition. J Mol Biol. 1983 Sep 15;169(2):353–372. doi: 10.1016/s0022-2836(83)80055-2. [DOI] [PubMed] [Google Scholar]
- Bagdasarian M., Bailone A., Bagdasarian M. M., Manning P. A., Lurz R., Timmis K. N., Devoret R. An inhibitor of SOS induction, specified by a plasmid locus in Escherichia coli. Proc Natl Acad Sci U S A. 1986 Aug;83(15):5723–5726. doi: 10.1073/pnas.83.15.5723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrineau P., Gilbert P., Jackson W. J., Jones C. S., Summers A. O., Wisdom S. The DNA sequence of the mercury resistance operon of the IncFII plasmid NR1. J Mol Appl Genet. 1984;2(6):601–619. [PubMed] [Google Scholar]
- Barton C. R., Warren R. L., Jezo P., Easton A. M., Rownd R. H. Sa/I restriction endonuclease maps of FII incompatibility group R plasmids. Plasmid. 1979 Jan;2(1):150–154. doi: 10.1016/0147-619x(79)90013-1. [DOI] [PubMed] [Google Scholar]
- Bennett P. M., Grinsted J., Choi C. L., Richmond M. H. Characterisation of Tn501, a transposon determining resistance to mercuric ions. Mol Gen Genet. 1978 Feb 7;159(1):101–106. doi: 10.1007/BF00401753. [DOI] [PubMed] [Google Scholar]
- Bergquist P. L., Saadi S., Maas W. K. Distribution of basic replicons having homology with RepFIA, RepFIB, and RepFIC among IncF group plasmids. Plasmid. 1986 Jan;15(1):19–34. doi: 10.1016/0147-619x(86)90011-9. [DOI] [PubMed] [Google Scholar]
- Bertrand K. P., Postle K., Wray L. V., Jr, Reznikoff W. S. Construction of a single-copy promoter vector and its use in analysis of regulation of the transposon Tn10 tetracycline resistance determinant. J Bacteriol. 1984 Jun;158(3):910–919. doi: 10.1128/jb.158.3.910-919.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand K. P., Postle K., Wray L. V., Jr, Reznikoff W. S. Overlapping divergent promoters control expression of Tn10 tetracycline resistance. Gene. 1983 Aug;23(2):149–156. doi: 10.1016/0378-1119(83)90046-x. [DOI] [PubMed] [Google Scholar]
- Brady G., Frey J., Danbara H., Timmis K. N. Replication control mutations of plasmid R6-5 and their effects on interactions of the RNA-I control element with its target. J Bacteriol. 1983 Apr;154(1):429–436. doi: 10.1128/jb.154.1.429-436.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo A., de Torrontegui G., Díaz R. Identification of components of a new stability system of plasmid R1, ParD, that is close to the origin of replication of this plasmid. Mol Gen Genet. 1987 Nov;210(1):101–110. doi: 10.1007/BF00337764. [DOI] [PubMed] [Google Scholar]
- Brown N. L., Misra T. K., Winnie J. N., Schmidt A., Seiff M., Silver S. The nucleotide sequence of the mercuric resistance operons of plasmid R100 and transposon Tn501: further evidence for mer genes which enhance the activity of the mercuric ion detoxification system. Mol Gen Genet. 1986 Jan;202(1):143–151. doi: 10.1007/BF00330531. [DOI] [PubMed] [Google Scholar]
- Brown N. L., Winnie J. N., Fritzinger D., Pridmore R. D. The nucleotide sequence of the tnpA gene completes the sequence of the Pseudomonas transposon Tn501. Nucleic Acids Res. 1985 Aug 12;13(15):5657–5669. doi: 10.1093/nar/13.15.5657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler M., Galas D. J. Studies on the transposition of IS1. Basic Life Sci. 1985;30:53–77. doi: 10.1007/978-1-4613-2447-8_7. [DOI] [PubMed] [Google Scholar]
- Chase J. W., Merrill B. M., Williams K. R. F sex factor encodes a single-stranded DNA binding protein (SSB) with extensive sequence homology to Escherichia coli SSB. Proc Natl Acad Sci U S A. 1983 Sep;80(18):5480–5484. doi: 10.1073/pnas.80.18.5480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheah K. C., Hirst R., Skurray R. finO sequences on conjugally repressed and derepressed F-like plasmids. Plasmid. 1987 May;17(3):233–239. doi: 10.1016/0147-619x(87)90031-x. [DOI] [PubMed] [Google Scholar]
- Cheah K. C., Ray A., Skurray R. Cloning and molecular analysis of the finO region from the antibiotic-resistance plasmid R6-5. Plasmid. 1984 Nov;12(3):222–226. doi: 10.1016/0147-619x(84)90050-7. [DOI] [PubMed] [Google Scholar]
- Clerget M., Chandler M., Caro L. Isolation of an IS1 flanked kanamycin resistance transposon from R1drd19. Mol Gen Genet. 1980;180(1):123–127. doi: 10.1007/BF00267360. [DOI] [PubMed] [Google Scholar]
- Clerget M., Chandler M., Caro L. The structure of R1drd19: a revised physical map of the plasmid. Mol Gen Genet. 1981;181(2):183–191. doi: 10.1007/BF00268425. [DOI] [PubMed] [Google Scholar]
- Clowes R. C. Molecular structure of bacterial plasmids. Bacteriol Rev. 1972 Sep;36(3):361–405. doi: 10.1128/br.36.3.361-405.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couturier M., Bex F., Bergquist P. L., Maas W. K. Identification and classification of bacterial plasmids. Microbiol Rev. 1988 Sep;52(3):375–395. doi: 10.1128/mr.52.3.375-395.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta N. Plasmids as organisms. Basic Life Sci. 1985;30:3–16. doi: 10.1007/978-1-4613-2447-8_2. [DOI] [PubMed] [Google Scholar]
- Davies J. E., Rownd R. Transmissible multiple drug resistance in Enterobacteriaceae. Science. 1972 May 19;176(4036):758–768. doi: 10.1126/science.176.4036.758. [DOI] [PubMed] [Google Scholar]
- Dempsey W. B., McIntire S. A. The finO gene of antibiotic resistance plasmid R100. Mol Gen Genet. 1983;190(3):444–451. doi: 10.1007/BF00331075. [DOI] [PubMed] [Google Scholar]
- Dempsey W. B., Willetts N. S. Plasmid co-integrates of prophage lambda and R factor R100. J Bacteriol. 1976 Apr;126(1):166–176. doi: 10.1128/jb.126.1.166-176.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diver W. P., Grinsted J., Fritzinger D. C., Brown N. L., Altenbuchner J., Rogowsky P., Schmitt R. DNA sequences of and complementation by the tnpR genes of Tn21, Tn501 and Tn1721. Mol Gen Genet. 1983;191(2):189–193. doi: 10.1007/BF00334812. [DOI] [PubMed] [Google Scholar]
- Dong X. N., Womble D. D., Rownd R. H. In-vivo studies on the cis-acting replication initiator protein of IncFII plasmid NR1. J Mol Biol. 1988 Aug 5;202(3):495–509. doi: 10.1016/0022-2836(88)90281-1. [DOI] [PubMed] [Google Scholar]
- Dong X. N., Womble D. D., Rownd R. H. Transcriptional pausing in a region important for plasmid NR1 replication control. J Bacteriol. 1987 Dec;169(12):5353–5363. doi: 10.1128/jb.169.12.5353-5363.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X., Womble D. D., Luckow V. A., Rownd R. H. Regulation of transcription of the repA1 gene in the replication control region of IncFII plasmid NR1 by gene dosage of the repA2 transcription repressor protein. J Bacteriol. 1985 Feb;161(2):544–551. doi: 10.1128/jb.161.2.544-551.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easton A. M., Rownd R. H. The incompatibility product of IncFII R plasmid NR1 controls gene expression in the plasmid replication region. J Bacteriol. 1982 Nov;152(2):829–839. doi: 10.1128/jb.152.2.829-839.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falkow S., Citarella R. V., Wohlhieter J. A. The molecular nature of R-factors. J Mol Biol. 1966 May;17(1):102–116. doi: 10.1016/s0022-2836(66)80097-9. [DOI] [PubMed] [Google Scholar]
- Fee B. E., Dempsey W. B. Cloning, mapping, and sequencing of plasmid R100 traM and finP genes. J Bacteriol. 1986 Jul;167(1):336–345. doi: 10.1128/jb.167.1.336-345.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlay B. B., Frost L. S., Paranchych W. Nucleotide sequences of the R1-19 plasmid transfer genes traM, finP, traJ, and traY and the traYZ promoter. J Bacteriol. 1986 May;166(2):368–374. doi: 10.1128/jb.166.2.368-374.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlay B. B., Frost L. S., Paranchych W. Origin of transfer of IncF plasmids and nucleotide sequences of the type II oriT, traM, and traY alleles from ColB4-K98 and the type IV traY allele from R100-1. J Bacteriol. 1986 Oct;168(1):132–139. doi: 10.1128/jb.168.1.132-139.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlay B. B., Frost L. S., Paranchych W., Willetts N. S. Nucleotide sequences of five IncF plasmid finP alleles. J Bacteriol. 1986 Aug;167(2):754–757. doi: 10.1128/jb.167.2.754-757.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster T. J. Plasmid-determined resistance to antimicrobial drugs and toxic metal ions in bacteria. Microbiol Rev. 1983 Sep;47(3):361–409. doi: 10.1128/mr.47.3.361-409.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost L. S., Finlay B. B., Opgenorth A., Paranchych W., Lee J. S. Characterization and sequence analysis of pilin from F-like plasmids. J Bacteriol. 1985 Dec;164(3):1238–1247. doi: 10.1128/jb.164.3.1238-1247.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes K., Bech F. W., Jørgensen S. T., Løbner-Olesen A., Rasmussen P. B., Atlung T., Boe L., Karlstrom O., Molin S., von Meyenburg K. Mechanism of postsegregational killing by the hok gene product of the parB system of plasmid R1 and its homology with the relF gene product of the E. coli relB operon. EMBO J. 1986 Aug;5(8):2023–2029. doi: 10.1002/j.1460-2075.1986.tb04459.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes K., Larsen J. E., Molin S. Stable inheritance of plasmid R1 requires two different loci. J Bacteriol. 1985 Jan;161(1):292–298. doi: 10.1128/jb.161.1.292-298.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes K., Molin S. Partitioning of plasmid R1. Structural and functional analysis of the parA locus. J Mol Biol. 1986 Aug 5;190(3):269–279. doi: 10.1016/0022-2836(86)90001-x. [DOI] [PubMed] [Google Scholar]
- Goebel W., Bonewald R. Class of small multicopy plasmids originating from the mutant antibiotic resistance factor R1 drd-19B2. J Bacteriol. 1975 Aug;123(2):658–665. doi: 10.1128/jb.123.2.658-665.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golub E. I., Low K. B. Unrelated conjugative plasmids have sequences which are homologous to the leading region of the F factor. J Bacteriol. 1986 May;166(2):670–672. doi: 10.1128/jb.166.2.670-672.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grindley N. D., Joyce C. M. Genetic and DNA sequence analysis of the kanamycin resistance transposon Tn903. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7176–7180. doi: 10.1073/pnas.77.12.7176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grinsted J., Brown N. L. A Tn21 terminal sequence within Tn501: complementation of tnpA gene function and transposon evolution. Mol Gen Genet. 1984;197(3):497–502. doi: 10.1007/BF00329949. [DOI] [PubMed] [Google Scholar]
- Grinsted J., de la Cruz F., Altenbuchner J., Schmitt R. Complementation of transposition of tnpA mutants of Tn3, Tn21, Tn501, and Tn1721. Plasmid. 1982 Nov;8(3):276–286. doi: 10.1016/0147-619x(82)90065-8. [DOI] [PubMed] [Google Scholar]
- Guerry P., Falkow S. Polynucleotide sequence relationships among some bacterial plasmids. J Bacteriol. 1971 Jul;107(1):372–374. doi: 10.1128/jb.107.1.372-374.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halling S. M., Simons R. W., Way J. C., Walsh R. B., Kleckner N. DNA sequence organization of IS10-right of Tn10 and comparison with IS10-left. Proc Natl Acad Sci U S A. 1982 Apr;79(8):2608–2612. doi: 10.1073/pnas.79.8.2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto H., Iyobe S., Mitsuhashi S. Unstable mutants of R factor. Jpn J Microbiol. 1969 Dec;13(4):343–349. doi: 10.1111/j.1348-0421.1969.tb00477.x. [DOI] [PubMed] [Google Scholar]
- Hashimoto H., Rownd R. H. Transition of the R factor NR1 and Proteus mirabilis: level of drug resistance of nontransitioned and transitioned cells. J Bacteriol. 1975 Jul;123(1):56–68. doi: 10.1128/jb.123.1.56-68.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillen W., Schollmeier K. Nucleotide sequence of the Tn10 encoded tetracycline resistance gene. Nucleic Acids Res. 1983 Jan 25;11(2):525–539. doi: 10.1093/nar/11.2.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota Y., Fujii T., Nishimua Y. Loss and repair of conjugal fertility and infectivity of the resistance factor and sex factor in Escherichia coli. J Bacteriol. 1966 Mar;91(3):1298–1304. doi: 10.1128/jb.91.3.1298-1304.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingshead S., Vapnek D. Nucleotide sequence analysis of a gene encoding a streptomycin/spectinomycin adenylyltransferase. Plasmid. 1985 Jan;13(1):17–30. doi: 10.1016/0147-619x(85)90052-6. [DOI] [PubMed] [Google Scholar]
- Hu S., Otsubo E., Davidson N., Saedler H. Electron microscope heteroduplex studies of sequence relations among bacterial plasmids: identification and mapping of the insertion sequences IS1 and IS2 in F and R plasmids. J Bacteriol. 1975 May;122(2):764–775. doi: 10.1128/jb.122.2.764-775.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huffman G. A., Rownd R. H. Transition of deletion mutants of the composite resistance plasmid NR1 in Escherichia coli and Salmonella typhimurium. J Bacteriol. 1984 Aug;159(2):488–498. doi: 10.1128/jb.159.2.488-498.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hänni C., Meyer J., Iida S., Arber W. Occurrence and properties of composite transposon Tn2672: evolution of multiple drug resistance transposons. J Bacteriol. 1982 Jun;150(3):1266–1273. doi: 10.1128/jb.150.3.1266-1273.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iida S., Hänni C., Echarti C., Arber W. Is the IS1-flanked r-determinant of the R plasmid NR1 a transposon? J Gen Microbiol. 1981 Oct;126(2):413–425. doi: 10.1099/00221287-126-2-413. [DOI] [PubMed] [Google Scholar]
- Inamoto S., Yoshioka Y., Ohtsubo E. Identification and characterization of the products from the traJ and traY genes of plasmid R100. J Bacteriol. 1988 Jun;170(6):2749–2757. doi: 10.1128/jb.170.6.2749-2757.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ippen-Ihler K. A., Minkley E. G., Jr The conjugation system of F, the fertility factor of Escherichia coli. Annu Rev Genet. 1986;20:593–624. doi: 10.1146/annurev.ge.20.120186.003113. [DOI] [PubMed] [Google Scholar]
- Johnson D. A., Willetts N. S. lambda-Transducing phages carrying transfer genes isolated from an abnormal prophage insertion into the traY gene of F. Plasmid. 1983 Jan;9(1):71–85. doi: 10.1016/0147-619x(83)90032-x. [DOI] [PubMed] [Google Scholar]
- Jorgensen R. A., Reznikoff W. S. Organization of structural and regulatory genes that mediate tetracycline resistance in transposon Tn10. J Bacteriol. 1979 Jun;138(3):705–714. doi: 10.1128/jb.138.3.705-714.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleckner N., Chan R. K., Tye B. K., Botstein D. Mutagenesis by insertion of a drug-resistance element carrying an inverted repetition. J Mol Biol. 1975 Oct 5;97(4):561–575. doi: 10.1016/s0022-2836(75)80059-3. [DOI] [PubMed] [Google Scholar]
- Kline B. C. A review of mini-F plasmid maintenance. Plasmid. 1985 Jul;14(1):1–16. doi: 10.1016/0147-619x(85)90027-7. [DOI] [PubMed] [Google Scholar]
- Komura I., Funaba T., Izaki K. Mechanism of mercuric chloride resistance in microorganisms. II. NADPH-dependent reduction of mercuric chloride and vaporization of mercury from mercuric chloride by a multiple drug resistant strain of Escherichia coli. J Biochem. 1971 Dec;70(6):895–901. doi: 10.1093/oxfordjournals.jbchem.a129719. [DOI] [PubMed] [Google Scholar]
- Komura I., Izaki K. Mechanism of mercuric chloride resistance in microorganisms. I. Vaporization of a mercury compound from mercuric chloride by multiple drug resistant strains of Escherichia coli. J Biochem. 1971 Dec;70(6):885–893. doi: 10.1093/oxfordjournals.jbchem.a129718. [DOI] [PubMed] [Google Scholar]
- Kopecko D. J., Brevet J., Cohen S. N. Involvement of multiple translocating DNA segments and recombinational hotspots in the structural evolution of bacterial plasmids. J Mol Biol. 1976 Dec;108(2):333–360. doi: 10.1016/s0022-2836(76)80124-6. [DOI] [PubMed] [Google Scholar]
- Lane D., Chandler M. Mapping of the drug resistance genes carried by the r-determinant of the R100.1 plasmid. Mol Gen Genet. 1977 Nov 29;157(1):17–23. doi: 10.1007/BF00268682. [DOI] [PubMed] [Google Scholar]
- Lane H. E. Replication and incompatibility of F and plasmids in the IncFI Group. Plasmid. 1981 Jan;5(1):100–126. doi: 10.1016/0147-619x(81)90079-2. [DOI] [PubMed] [Google Scholar]
- Light J., Molin S. Post-transcriptional control of expression of the repA gene of plasmid R1 mediated by a small RNA molecule. EMBO J. 1983;2(1):93–98. doi: 10.1002/j.1460-2075.1983.tb01387.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Light J., Molin S. The sites of action of the two copy number control functions of plasmid R1. Mol Gen Genet. 1982;187(3):486–493. doi: 10.1007/BF00332633. [DOI] [PubMed] [Google Scholar]
- Light J., Riise E., Molin S. Transcription and its regulation in the basic replicon region of plasmid R1. Mol Gen Genet. 1985;198(3):503–508. doi: 10.1007/BF00332947. [DOI] [PubMed] [Google Scholar]
- Liu C. P., Churchward G., Caro L. The repA2 gene of the plasmid R100.1 encodes a repressor of plasmid replication. Plasmid. 1983 Sep;10(2):148–155. doi: 10.1016/0147-619x(83)90067-7. [DOI] [PubMed] [Google Scholar]
- Loh S. M., Cram D. S., Skurray R. A. Nucleotide sequence and transcriptional analysis of a third function (Flm) involved in F-plasmid maintenance. Gene. 1988 Jun 30;66(2):259–268. doi: 10.1016/0378-1119(88)90362-9. [DOI] [PubMed] [Google Scholar]
- Lovett M. A., Helinski D. R. Method for the isolation of the replication region of a bacterial replicon: construction of a mini-F'kn plasmid. J Bacteriol. 1976 Aug;127(2):982–987. doi: 10.1128/jb.127.2.982-987.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MARMUR J., ROWND R., FALKOW S., BARON L. S., SCHILDKRAUT C., DOTY P. The nature of intergeneric episomal infection. Proc Natl Acad Sci U S A. 1961 Jul 15;47:972–979. doi: 10.1073/pnas.47.7.972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machida Y., Machida C., Ohtsubo E. Insertion element IS1 encodes two structural genes required for its transposition. J Mol Biol. 1984 Aug 5;177(2):229–245. doi: 10.1016/0022-2836(84)90454-6. [DOI] [PubMed] [Google Scholar]
- Manning P. A., Morelli G. DNA homology of the promoter-distal regions of the tra operons of sex factors F and R100 in Escherichia coli K-12. J Bacteriol. 1982 Apr;150(1):389–394. doi: 10.1128/jb.150.1.389-394.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masai H., Arai K. RepA and DnaA proteins are required for initiation of R1 plasmid replication in vitro and interact with the oriR sequence. Proc Natl Acad Sci U S A. 1987 Jul;84(14):4781–4785. doi: 10.1073/pnas.84.14.4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masai H., Kaziro Y., Arai K. Definition of oriR, the minimum DNA segment essential for initiation of R1 plasmid replication in vitro. Proc Natl Acad Sci U S A. 1983 Nov;80(22):6814–6818. doi: 10.1073/pnas.80.22.6814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntire S. A., Dempsey W. B. Fertility inhibition gene of plasmid R100. Nucleic Acids Res. 1987 Mar 11;15(5):2029–2042. doi: 10.1093/nar/15.5.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mickel S., Bauer W. Isolation, by tetracycline selection, of small plasmids derived from R-factor R12 in Escherichia coli K-12. J Bacteriol. 1976 Jul;127(1):644–655. doi: 10.1128/jb.127.1.644-655.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mickel S., Ohtsubo E., Bauer W. Heteroduplex mapping of small plasmids derived from R-factor R12: in vivo recombination occurs at IS1 insertion sequences. Gene. 1977;2(3-4):193–210. doi: 10.1016/0378-1119(77)90017-8. [DOI] [PubMed] [Google Scholar]
- Miki T., Easton A. M., Rownd R. H. Cloning of replication, incompatibility, and stability functions of R plasmid NR1. J Bacteriol. 1980 Jan;141(1):87–99. doi: 10.1128/jb.141.1.87-99.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki T., Easton A. M., Rownd R. H. Mapping of the resistance genes of the R plasmid NR1. Mol Gen Genet. 1978 Jan 17;158(3):217–224. doi: 10.1007/BF00267192. [DOI] [PubMed] [Google Scholar]
- Misra T. K., Brown N. L., Fritzinger D. C., Pridmore R. D., Barnes W. M., Haberstroh L., Silver S. Mercuric ion-resistance operons of plasmid R100 and transposon Tn501: the beginning of the operon including the regulatory region and the first two structural genes. Proc Natl Acad Sci U S A. 1984 Oct;81(19):5975–5979. doi: 10.1073/pnas.81.19.5975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra T. K., Brown N. L., Haberstroh L., Schmidt A., Goddette D., Silver S. Mercuric reductase structural genes from plasmid R100 and transposon Tn501: functional domains of the enzyme. Gene. 1985;34(2-3):253–262. doi: 10.1016/0378-1119(85)90134-9. [DOI] [PubMed] [Google Scholar]
- Morris C. F., Hashimoto H., Mickel S., Rownd R. Round of replication mutant of a drug resistance factor. J Bacteriol. 1974 Jun;118(3):855–866. doi: 10.1128/jb.118.3.855-866.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris C. F., Hershberger C. L., Rownd R. Strand-specific nick in open circular R-factor deoxyribonucleic acid: attachment of the linear strand to a proteinaceous cellular component. J Bacteriol. 1973 Apr;114(1):300–308. doi: 10.1128/jb.114.1.300-308.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NAKAYA R., NAKAMURA A., MURATA Y. Resistance transfer agents in Shigella. Biochem Biophys Res Commun. 1960 Dec;3:654–659. doi: 10.1016/0006-291x(60)90081-4. [DOI] [PubMed] [Google Scholar]
- Nakahara H., Kinscherf T. G., Silver S., Miki T., Easton A. M., Rownd R. H. Gene copy number effects in the mer operon of plasmid NR1. J Bacteriol. 1979 Apr;138(1):284–287. doi: 10.1128/jb.138.1.284-287.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahara H., Silver S., Miki T., Rownd R. H. Hypersensitivity to Hg2+ and hyperbinding activity associated with cloned fragments of the mercurial resistance operon of plasmid NR1. J Bacteriol. 1979 Oct;140(1):161–166. doi: 10.1128/jb.140.1.161-166.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen T. T., Postle K., Bertrand K. P. Sequence homology between the tetracycline-resistance determinants of Tn10 and pBR322. Gene. 1983 Nov;25(1):83–92. doi: 10.1016/0378-1119(83)90170-1. [DOI] [PubMed] [Google Scholar]
- Nikoletti S., Bird P., Praszkier J., Pittard J. Analysis of the incompatibility determinants of I-complex plasmids. J Bacteriol. 1988 Mar;170(3):1311–1318. doi: 10.1128/jb.170.3.1311-1318.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordström K., Ingram L. C., Lundbäck A. Mutations in R factors of Escherichia coli causing an increased number of R-factor copies per chromosome. J Bacteriol. 1972 May;110(2):562–569. doi: 10.1128/jb.110.2.562-569.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordström K., Molin S., Aagaard-Hansen H. Partitioning of plasmid R1 in Escherichia coli. I. Kinetics of loss of plasmid derivatives deleted of the par region. Plasmid. 1980 Sep;4(2):215–227. doi: 10.1016/0147-619x(80)90011-6. [DOI] [PubMed] [Google Scholar]
- Nordström K., Molin S., Light J. Control of replication of bacterial plasmids: genetics, molecular biology, and physiology of the plasmid R1 system. Plasmid. 1984 Sep;12(2):71–90. doi: 10.1016/0147-619x(84)90054-4. [DOI] [PubMed] [Google Scholar]
- Nordström M., Nordström K. Control of replication of FII plasmids: comparison of the basic replicons and of the copB systems of plasmids R100 and R1. Plasmid. 1985 Mar;13(2):81–87. doi: 10.1016/0147-619x(85)90060-5. [DOI] [PubMed] [Google Scholar]
- Novick R. P. Plasmid incompatibility. Microbiol Rev. 1987 Dec;51(4):381–395. doi: 10.1128/mr.51.4.381-395.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata R. T., Winters C., Levine R. P. Nucleotide sequence analysis of the complement resistance gene from plasmid R100. J Bacteriol. 1982 Aug;151(2):819–827. doi: 10.1128/jb.151.2.819-827.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura T., Hiraga S. Mini-F plasmid genes that couple host cell division to plasmid proliferation. Proc Natl Acad Sci U S A. 1983 Aug;80(15):4784–4788. doi: 10.1073/pnas.80.15.4784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura T., Hiraga S. Partition mechanism of F plasmid: two plasmid gene-encoded products and a cis-acting region are involved in partition. Cell. 1983 Feb;32(2):351–360. doi: 10.1016/0092-8674(83)90454-3. [DOI] [PubMed] [Google Scholar]
- Ohtsubo E., Feingold J., Ohtsubo H., Mickel S., Bauer W. Unidirectional replication in Escherichia coli of three small plasmids derived from R factor R12. Plasmid. 1977 Nov;1(1):8–18. doi: 10.1016/0147-619x(77)90004-x. [DOI] [PubMed] [Google Scholar]
- Ohtsubo H., Ohtsubo E. Nucleotide sequence of an insertion element, IS1. Proc Natl Acad Sci U S A. 1978 Feb;75(2):615–619. doi: 10.1073/pnas.75.2.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsubo H., Ryder T. B., Maeda Y., Armstrong K., Ohtsubo E. DNA replication of the resistance plasmid R100 and its control. Adv Biophys. 1986;21:115–133. doi: 10.1016/0065-227x(86)90018-3. [DOI] [PubMed] [Google Scholar]
- Otsubo E., Nishimura Y., Hirota Y. Transfer-defective mutants of sex factors in Escherichia coli. I. Defective mutants and complementation analysis. Genetics. 1970 Feb;64(2):173–188. doi: 10.1093/genetics/64.2.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsubo E. Transfer-defective mutants of sex factors in Escherichia coli. II. Deletion mutants of an F-prime and deletion mapping of cistrons involved in genetic transfer. Genetics. 1970 Feb;64(2):189–197. doi: 10.1093/genetics/64.2.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlman D., Rownd R. H. Transition of R factor NR1 in Proteus mirabilis: molecular structure and replication of NR1 deoxyribonucleic acid. J Bacteriol. 1975 Sep;123(3):1013–1034. doi: 10.1128/jb.123.3.1013-1034.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlman D., Rownd R. H. Two origins of replication in composite R plasmid DNA. Nature. 1976 Jan 29;259(5541):281–284. doi: 10.1038/259281a0. [DOI] [PubMed] [Google Scholar]
- Perlman D., Twose T. M., Holland M. J., Rownd R. H. Denaturation mapping of R factor deoxyribonucleic acid. J Bacteriol. 1975 Sep;123(3):1035–1042. doi: 10.1128/jb.123.3.1035-1042.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson B. C., Rownd R. H. Drug resistance gene amplification of plasmid NR1 derivatives with various amounts of resistance determinant DNA. J Bacteriol. 1985 Mar;161(3):1042–1048. doi: 10.1128/jb.161.3.1042-1048.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson B. C., Rownd R. H. Homologous sequences other than insertion elements can serve as recombination sites in plasmid drug resistance gene amplification. J Bacteriol. 1983 Oct;156(1):177–185. doi: 10.1128/jb.156.1.177-185.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson B. C., Rownd R. H. Recombination sites in plasmid drug resistance gene amplification. J Bacteriol. 1985 Dec;164(3):1359–1361. doi: 10.1128/jb.164.3.1359-1361.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proctor G. N., McKell J., Rownd R. H. Chloramphenicol acetyltransferase may confer resistance to fusidic acid by sequestering the drug. J Bacteriol. 1983 Aug;155(2):937–939. doi: 10.1128/jb.155.2.937-939.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proctor G. N., Rownd R. H. Rosanilins: indicator dyes for chloramphenicol-resistant enterobacteria containing chloramphenicol acetyltransferase. J Bacteriol. 1982 Jun;150(3):1375–1382. doi: 10.1128/jb.150.3.1375-1382.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptashne K., Cohen S. N. Occurrence of insertion sequence (IS) regions on plasmid deoxyribonucleic acid as direct and inverted nucleotide sequence duplications. J Bacteriol. 1975 May;122(2):776–781. doi: 10.1128/jb.122.2.776-781.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray A., Skurray R. Stabilization of the cloning vector pACYC184 by insertion of F plasmid leading region sequences. Plasmid. 1984 May;11(3):272–275. doi: 10.1016/0147-619x(84)90036-2. [DOI] [PubMed] [Google Scholar]
- Riise E., Molin S. Purification and characterization of the CopB replication control protein, and precise mapping of its target site in the R1 plasmid. Plasmid. 1986 May;15(3):163–171. doi: 10.1016/0147-619x(86)90034-x. [DOI] [PubMed] [Google Scholar]
- Rosen J., Ohtsubo H., Ohtsubo E. The nucleotide sequence of the region surrounding the replication origin of an R100 resistance factor derivative. Mol Gen Genet. 1979 Mar 27;171(3):287–293. doi: 10.1007/BF00267583. [DOI] [PubMed] [Google Scholar]
- Rosen J., Ryder T., Inokuchi H., Ohtsubo H., Ohtsubo E. Genes and sites involved in replication and incompatibility of an R100 plasmid derivative based on nucleotide sequence analysis. Mol Gen Genet. 1980;179(3):527–537. doi: 10.1007/BF00271742. [DOI] [PubMed] [Google Scholar]
- Rosen J., Ryder T., Ohtsubo H., Ohtsubo E. Role of RNA transcripts in replication incompatibility and copy number control in antibiotic resistance plasmid derivatives. Nature. 1981 Apr 30;290(5809):794–797. doi: 10.1038/290794a0. [DOI] [PubMed] [Google Scholar]
- Rownd R. H., Womble D. D., Dong X. N., Luckow V. A., Wu R. P. Incompatibility and IncFII plasmid replication control. Basic Life Sci. 1985;30:335–354. doi: 10.1007/978-1-4613-2447-8_26. [DOI] [PubMed] [Google Scholar]
- Rownd R., Mickel S. Dissociation and reassociation of RTF and r-determinants of the R-factor NR1 in Proteus mirabilis. Nat New Biol. 1971 Nov 10;234(45):40–43. doi: 10.1038/newbio234040a0. [DOI] [PubMed] [Google Scholar]
- Rownd R., Nakaya R., Nakamura A. Molecular nature of the drug-resistance factors of the Enterobacteriaceae. J Mol Biol. 1966 Jun;17(2):376–393. doi: 10.1016/s0022-2836(66)80149-3. [DOI] [PubMed] [Google Scholar]
- Rownd R. Replication of a bacterial episome under relaxed control. J Mol Biol. 1969 Sep 28;44(3):387–402. doi: 10.1016/0022-2836(69)90368-4. [DOI] [PubMed] [Google Scholar]
- Ryder T. B., Davidson D. B., Rosen J. I., Ohtsubo E., Ohtsubo H. Analysis of plasmid genome evolution based on nucleotide-sequence comparison of two related plasmids of Escherichia coli. Gene. 1982 Mar;17(3):299–310. doi: 10.1016/0378-1119(82)90146-9. [DOI] [PubMed] [Google Scholar]
- SUGINO Y., HIROTA Y. Conjugal fertility associated with resistance factor R in Escherichia coli. J Bacteriol. 1962 Nov;84:902–910. doi: 10.1128/jb.84.5.902-910.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saadi S., Maas W. K., Hill D. F., Bergquist P. L. Nucleotide sequence analysis of RepFIC, a basic replicon present in IncFI plasmids P307 and F, and its relation to the RepA replicon of IncFII plasmids. J Bacteriol. 1987 May;169(5):1836–1846. doi: 10.1128/jb.169.5.1836-1846.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R., Hong G. F., Hill D. F., Petersen G. B. Nucleotide sequence of bacteriophage lambda DNA. J Mol Biol. 1982 Dec 25;162(4):729–773. doi: 10.1016/0022-2836(82)90546-0. [DOI] [PubMed] [Google Scholar]
- Schimke R. T. Gene amplification in cultured cells. J Biol Chem. 1988 May 5;263(13):5989–5992. [PubMed] [Google Scholar]
- Scott J. R. Regulation of plasmid replication. Microbiol Rev. 1984 Mar;48(1):1–23. doi: 10.1016/b978-0-12-048850-6.50006-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P. A., Cohen S. N., Davidson N. Electron microscope heteroduplex studies of sequence relations among plasmids of Escherichia coli. II. Structure of drug resistance (R) factors and F factors. J Mol Biol. 1973 Apr 5;75(2):235–255. doi: 10.1016/0022-2836(73)90018-1. [DOI] [PubMed] [Google Scholar]
- Silver L., Chandler M., de la Tour E. B., Caro L. Origin and direction of replication of the drug resistance plasmid R100.1 and of a resistance transfer factor derivative in synchronized cultures. J Bacteriol. 1977 Sep;131(3):929–942. doi: 10.1128/jb.131.3.929-942.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skurray R. A., Guyer M. S., Timmis K., Cabello F., Cohen S. N., Davidson N., Clark A. J. Replication region fragments cloned from Flac+ are identical to EcoRI fragment f5 of F. J Bacteriol. 1976 Sep;127(3):1571–1575. doi: 10.1128/jb.127.3.1571-1575.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D. H. R factors mediate resistance to mercury, nickel, and cobalt. Science. 1967 May 26;156(3778):1114–1116. doi: 10.1126/science.156.3778.1114. [DOI] [PubMed] [Google Scholar]
- Summers A. O. Organization, expression, and evolution of genes for mercury resistance. Annu Rev Microbiol. 1986;40:607–634. doi: 10.1146/annurev.mi.40.100186.003135. [DOI] [PubMed] [Google Scholar]
- Swedberg G., Sköld O. Plasmid-borne sulfonamide resistance determinants studied by restriction enzyme analysis. J Bacteriol. 1983 Mar;153(3):1228–1237. doi: 10.1128/jb.153.3.1228-1237.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabuchi A., Min Y. N., Kim C. K., Fan Y. L., Womble D. D., Rownd R. H. Genetic organization and nucleotide sequence of the stability locus of IncFII plasmid NR1. J Mol Biol. 1988 Aug 5;202(3):511–525. doi: 10.1016/0022-2836(88)90282-3. [DOI] [PubMed] [Google Scholar]
- Tanak N., Cramer J. H., Rownd R. H. EcoRI restriction endonuclease map of the composite R plasmid NR1. J Bacteriol. 1976 Jul;127(1):619–636. doi: 10.1128/jb.127.1.619-636.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M., Yamamoto T., Sawai T. Evolution of complex resistance transposons from an ancestral mercury transposon. J Bacteriol. 1983 Mar;153(3):1432–1438. doi: 10.1128/jb.153.3.1432-1438.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanimoto K., Iino T., Ohtsubo H., Ohtsubo E. Identification of a gene, tir of R100, functionally homologous to the F3 gene of F in the inhibition of RP4 transfer. Mol Gen Genet. 1985;198(2):356–357. doi: 10.1007/BF00383019. [DOI] [PubMed] [Google Scholar]
- Taylor D. P., Cohen S. N. Structural and functional analysis of cloned DNA segments containing the replication and incompatibility regions of a miniplasmid derived from a copy number mutant of NR1. J Bacteriol. 1979 Jan;137(1):92–104. doi: 10.1128/jb.137.1.92-104.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor D. P., Greenberg J., Rownd R. H. Generation of miniplasmids from copy number mutants of the R plasmid NR1. J Bacteriol. 1977 Dec;132(3):986–995. doi: 10.1128/jb.132.3.986-995.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmerman K. P., Tu C. P. Complete sequence of IS3. Nucleic Acids Res. 1985 Mar 25;13(6):2127–2139. doi: 10.1093/nar/13.6.2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmis K. N., Andrés I., Achtman M. Fertility repression of F-like conjugative plasmids: physical mapping of the R6--5 finO and finP cistrons and identification of the finO protein. Proc Natl Acad Sci U S A. 1978 Dec;75(12):5836–5840. doi: 10.1073/pnas.75.12.5836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmis K. N., Andrés I., Slocombe P. M. Plasmid incompatibility: cloning analysis of an incFII determinant of R6-5. Nature. 1978 May 4;273(5657):27–32. doi: 10.1038/273027a0. [DOI] [PubMed] [Google Scholar]
- Timmis K. N., Cabello F., Cohen S. N. Cloning and characterization of EcoRI and HindIII restriction endonuclease-generated fragments of antibiotic resistance plasmids R6-5 and R6. Mol Gen Genet. 1978 Jun 14;162(2):121–137. doi: 10.1007/BF00267869. [DOI] [PubMed] [Google Scholar]
- Timmis K., Cabello F., Cohen S. N. Cloning, isolation, and characterization of replication regions of complex plasmid genomes. Proc Natl Acad Sci U S A. 1975 Jun;72(6):2242–2246. doi: 10.1073/pnas.72.6.2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchimoto S., Ohtsubo H., Ohtsubo E. Two genes, pemK and pemI, responsible for stable maintenance of resistance plasmid R100. J Bacteriol. 1988 Apr;170(4):1461–1466. doi: 10.1128/jb.170.4.1461-1466.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Völker T. A., Iida S., Bickle T. A. A single gene coding for resistance to both fusidic acid and chloramphenicol. J Mol Biol. 1982 Jan 25;154(3):417–425. doi: 10.1016/s0022-2836(82)80004-1. [DOI] [PubMed] [Google Scholar]
- WATANABE T., FUKASAWA T. Episome-mediated transfer of drug resistance in Enterobacteriaceae. I. Transfer of resistance factors by conjugation. J Bacteriol. 1961 May;81:669–678. doi: 10.1128/jb.81.5.669-678.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WATANABE T. Infective heredity of multiple drug resistance in bacteria. Bacteriol Rev. 1963 Mar;27:87–115. doi: 10.1128/br.27.1.87-115.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber P. C., Palchaudhuri S. Incompatibility repressor in a RepA-like replicon of the IncFI plasmid ColV2-K94. J Bacteriol. 1986 Jun;166(3):1106–1112. doi: 10.1128/jb.166.3.1106-1112.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willetts N., Skurray R. The conjugation system of F-like plasmids. Annu Rev Genet. 1980;14:41–76. doi: 10.1146/annurev.ge.14.120180.000353. [DOI] [PubMed] [Google Scholar]
- Willetts N., Wilkins B. Processing of plasmid DNA during bacterial conjugation. Microbiol Rev. 1984 Mar;48(1):24–41. doi: 10.1128/mr.48.1.24-41.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Womble D. D., Dong X., Luckow V. A., Wu R. P., Rownd R. H. Analysis of the individual regulatory components of the IncFII plasmid replication control system. J Bacteriol. 1985 Feb;161(2):534–543. doi: 10.1128/jb.161.2.534-543.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Womble D. D., Dong X., Wu R. P., Luckow V. A., Martinez A. F., Rownd R. H. IncFII plasmid incompatibility product and its target are both RNA transcripts. J Bacteriol. 1984 Oct;160(1):28–35. doi: 10.1128/jb.160.1.28-35.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Womble D. D., Sampathkumar P., Easton A. M., Luckow V. A., Rownd R. H. Transcription of the replication control region of the IncFII R-plasmid NR1 in vitro and in vivo. J Mol Biol. 1985 Feb 5;181(3):395–410. doi: 10.1016/0022-2836(85)90228-1. [DOI] [PubMed] [Google Scholar]
- Womble D. D., Taylor D. P., Rownd R. H. Method for obtaining more-accurate covalently closed circular plasmid-to-chromosome ratios from bacterial lysates by dye-buoyant density centrifugation. J Bacteriol. 1977 Apr;130(1):148–153. doi: 10.1128/jb.130.1.148-153.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu R. P., Womble D. D., Rownd R. H. Incompatibility mutants of IncFII plasmid NR1 and their effect on replication control. J Bacteriol. 1985 Sep;163(3):973–982. doi: 10.1128/jb.163.3.973-982.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa M. Identification and mapping of the replication genes of an R factor, R100-1, integrated into the chromosome of Escherichia coli K-12. J Bacteriol. 1974 Jun;118(3):1123–1131. doi: 10.1128/jb.118.3.1123-1131.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshioka Y., Ohtsubo H., Ohtsubo E. Repressor gene finO in plasmids R100 and F: constitutive transfer of plasmid F is caused by insertion of IS3 into F finO. J Bacteriol. 1987 Feb;169(2):619–623. doi: 10.1128/jb.169.2.619-623.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z. X., Chandler M., Hipskind R., Clerget M., Caro L. Dissection of the r-determinant of the plasmid R100.1: the sequence at the extremities of Tn21. Nucleic Acids Res. 1981 Dec 11;9(23):6265–6278. doi: 10.1093/nar/9.23.6265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Cruz F., Grinsted J. Genetic and molecular characterization of Tn21, a multiple resistance transposon from R100.1. J Bacteriol. 1982 Jul;151(1):222–228. doi: 10.1128/jb.151.1.222-228.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]


