Abstract
The human microbiota is a complex assemblage of the microbes inhabiting many sites in the human body. Recent advances in technology have enabled deep sequencing and analysis of the members and structures of these communities. Two sites, the vagina and gastrointestinal tract, are highlighted to exemplify how technological advances have enhanced our knowledge of the host–microbiota system. These examples represent low- and high-complexity communities, respectively. In each example, certain community structures are identified that can be extrapolated to larger collections representing multiple individuals and potential disease or health states. One common feature is the unexpected diversity of the microbiota at any of these locations, which poses a challenge for relating the microbiota to health and disease. However, we anticipate microbiota compositional measurements could become standard clinical practice in the future and may become diagnostic for certain diseases or increased susceptibility to certain disorders. The microbiota of a number of disease states are currently being examined to identify potential correlations. In line with these predictions, it is possible that existing conditions may be resolved by altering the microbiota in a positive way.
Keywords: microbiota, bacterial community structure, host-pathogen interaction, gastrointestinal tract, vagina
Introduction
Despite our generally anthropocentric view of the world, it is the microbial population that dominates life on this planet in global diversity and in numbers. The human body itself serves as a scaffold for a multitude of bacteria, archaea, viruses, and eukaryotic microbes that inhabit discrete anatomical niches and outnumber our own somatic and germ cells by an order of magnitude (1). Until recently, the complex and dynamic nature of our microbiota was not fully recognized, owing to the technological limitations of in vitro microbiological cultivation techniques and limited throughput of sequencing technologies. Large-scale endeavors such as the Human Microbiome Project (HMP) (2), hosted by the US National Institutes of Health, and the Metagenomics of the Human Intestinal Tract (MetaHIT) project (3), through the European Commission, have initiated extensive programs aimed at surveying the repertoire of microbial genes and genomes collectively termed the microbiome. The efforts of the HMP have produced >70 million 16S ribosomal gene sequences characterizing the microbial composition across 15 body sites, and >3.5 tera–base pairs (Tbp) of whole-genome shotgun metagenomic data encoding >60 million predicted genes (4, 5).
The role of human-associated microbiota in health and disease has received newfound appreciation owing to our ability to quantify and qualify the types and the metabolic and functional capabilities of the microbial consortia associated with our bodies (6). For example, a critical link has recently been established for the resident gastrointestinal microbiota in the promotion of atherosclerosis (7). Wang and colleagues (7) delineated a two-step metabolic pathway involving the microbially mediated metabolism of dietary phosphatidylcholine, resulting in the production of the metabolite trimethylamine N-oxide (TMAO), a predictor of cardiovascular disease (CVD) risk. Additionally, a recent study by Sellitto et al. (8) described the development of the microbiota among individuals with a genetic predisposition for celiac disease. This study also highlighted the utility of incorporating studies of the metabolic capabilities of the microbiota, resulting in the potential identification of biomarkers for the development of celiac disease (8). Development of a mechanistic understanding of the human microbiota in relation to human health and disease and incorporation of the microbiome as a key component of the entire human genomic framework are fundamental for the advancement of personalized medicine.
Concomitant with technological advances, the application of ecological theory to the host– microbiota system has generated profound insight into human health. The human microbiota performs essential functions that define and contribute to the physiology of the host, sharing a unique biological relationship termed a symbiosis. According to the botanist Heinrich Anton de Bary, symbiosis is a broad term to describe “different organisms living together” and encompasses the gamut of biological interactions: mutualism, commensalism, and parasitism (9). In the human microbiome literature, the definition of symbiosis ranges from a commensalistic relationship, wherein the interaction is decidedly beneficial for one of the partners (the host), to mutualistic, involving beneficial outcomes for all organisms involved. In this review, we discuss the variety of symbiotic interactions of the human host and microbiota in the context of maintaining homeostasis, focusing on the host–microbiota systems of the vagina and gastrointestinal tract, and how perturbations of these interactions lead to dysbiosis.
Tools and Technologies to Access our Microbiota
In many complex communities, the majority of the microbial members identified using molecular techniques have resisted cultivation efforts (10). The advent of community genomics and high-throughput cultivation-independent molecular techniques has yielded remarkable insight into the complex diversity of “our microbial selves” (11), previously obscured from researchers focused on readily cultivated microorganisms from human samples. Sequencing technologies have advanced the investigation of these complex communities by increasing the throughput. During the past decade, sequencing strategies have changed from using Sanger sequencing to interrogate a full-length 16S rRNA gene (∼1,200 bp) to only examining hypervariable portions of the gene with second and third generation sequencing technology. These changes in sequencing strategy are indicative of the length of sequencing reads, from the long (800–1,000 bp) Sanger reads to the short sequencing reads of the Roche 454 and Illumina platforms (∼400 and 100 bp, respectively). Additionally, the high throughput of the new sequencing technologies has removed biases associated with cloning and allowed the simultaneous interrogation of hundreds, if not thousands, of samples. This level of throughput has altered the experimental design, as it is now economically and technically feasible to examine large numbers of samples from large patient or subject cohorts. However, the increase in sequencing throughput has resulted in challenges that have driven the development of novel tools for analysis, examined below.
Taxonomic Characterization
The overwhelming majority of human-associated microbiota studies have focused on characterizing the microbial community composition based on comparative molecular sequence analyses of the highly conserved small-subunit ribosomal gene from bacteria (16S rRNA). The bacterial 16S rRNA gene is amenable as a microbial genetic marker owing to highly conserved (or “universal”) regions for PCR primer design, fast-evolving (or “hypervariable”) regions for phylogenetic resolution, and extensive databases with millions of reference sequences collected from numerous habitats globally (see sidebar, The Pitfalls of Sequencing Technologies). Moreover, these high-throughput sequencing biodiversity surveys have established an extensive catalogue of the microbial phylogenetic groups, or phylotypes, associated with a given body site, such as the gastrointestinal tract, skin, or vagina (12–14). Strikingly, the principal determinant of microbial community composition is anatomical site, a feature that can in fact be used to identify mislabeled samples (15). Bioinformatic analyses and integration of ecological measurements, such as community similarity, richness, and structure, have contributed much to our understanding of the microbial consortia.
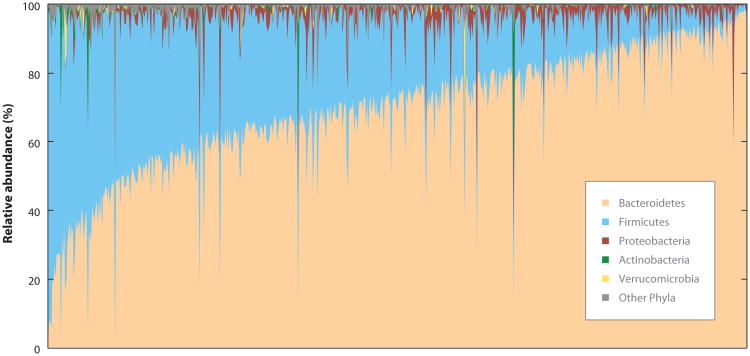
The level of community variation within the same anatomical site across individuals is considerable (Figure 1). This interindividual variation becomes a confounding issue for comparative cohort studies and challenges our ability to generalize about what constitutes a “normal” healthy or “disease-associated” microbial community. Substantial effort has focused on identifying the “core” constituents of the microbiota, particularly for the gastrointestinal tract (1, 16). Definitions of a common core microbiota encompass both highly abundant members and those that are prevalent across multiple human hosts. Although initial hypotheses have centered on the concept that a deviation from the microbial core results in dysbiosis or an unhealthy state, there is growing appreciation for the alternative concept of an individual baseline composition and variation within an individual's own range. Two pivotal studies investigating the bacterial taxonomic composition within the gastrointestinal tract (17) and vagina (14) have revealed discrete community types, providing, for the first time, a comparative framework for large cohort studies. We explore these findings in subsequent sections.
Figure 1.
Interindividual variation of the gastrointestinal microbiota. Distribution of bacterial phyla across 648 samples collected as part of the NIH Human Microbiome Project (HMP). Samples are arranged on the x-axis by abundance of dominant organism. The V3–V5 region of the bacterial 16S rRNA gene was amplified from total genomic DNA extracted from fecal samples and sequenced using Roche 454 pyrosequencing. Protocols and sample collection details are available at the Data Analysis and Coordination Center (DACC, http://www.hmpdacc.org/).
Functional Characterization
Metagenomic analyses have begun to capture the breadth of microbial functional and metabolic potential, in some cases revealing significant metabolic discrepancies between diseased and healthy individuals (see sidebar, Microbiome: The Importance of Terminology). Despite considerable variation in the microbiota composition across individuals, the functional repertoire appears to be more stable (18). Whole-genome shotgun metagenomic data can be assigned functional properties on the basis of comparisons to databases such as the Clusters of Orthologous Groups (COG) from the National Center for Biotechnology Information (NCBI) and the Kyoto Encyclopedia of Genes and Genomes (KEGG). Improvement of these databases has resulted from the increased genome representation from human-associated microbial isolates (16). With improved and expanded reference databases, exciting integrative systems biology methods are now being applied to decipher changes in host–microbial networks associated with obesity and inflammatory bowel disease (IBD) (19).
Model Animal Systems
In model animal systems, the influences of host genetics and environmental factors (such as diet) can be highly controlled. The use of animal models, particularly gnotobiotic and humanized mice, has provided crucial information regarding host-microbiota interactions (20). For example, using a mouse model, Vaishnava and colleagues established that epithelial MyD88 and RegIIIγ are key host components regulating the microbiota at the intestinal surface (21). This work provided significant insight into a previously unspecified immune mechanism that promotes host–microbial spatial interactions. However, findings from animal models do not always translate to humans, both in terms of reproducing clinical outcomes for a given disease (22) and in some of the observed experimental features. For instance, early studies evaluating microbiomes in lean and obese mice indicated that changes in the relative abundances of the two main phyla, Bacteroidetes and Firmicutes, were associated with obesity (23). These initial observations in animal models have been challenged by more recent studies demonstrating that the relationship is more complicated than simply the ratio of Bacteroidetes to Firmicutes in associating gastrointestinal microbiota with obesity in humans (24, 25).
Spatial and Temporal Considerations
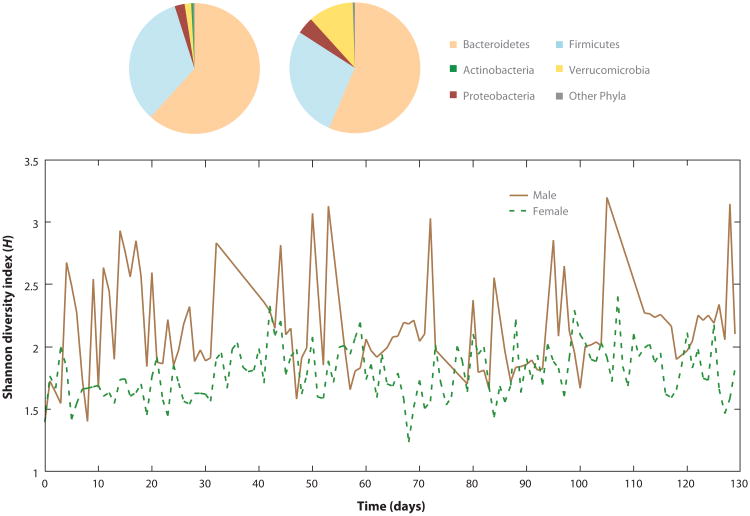
Studies incorporating temporal (longitudinal) experimental designs afford more observations than a single “snapshot” of the microbial community (26). The dynamics of the host– microbiota system can be appreciated at daily, monthly, and even yearly resolution. Fluctuations of the microbiota composition can indicate a highly individual temporal profile (Figure 2). Similarly, spatial resolution is significant in the context of the biochemical interactions and activities occurring on the micrometer scale. For example, the oral microbiota is remarkably unique, restricted to microniches associated with supra- and subgingival plaques, saliva and tongue surfaces within an individual's mouth (27–29). As we describe below, an eloquent study of the vaginal microbial composition of 32 reproductive-age women over a 16-week period has provided profound insight into the temporal dynamics that microbiota studies need to take into account in order to better quantify and attempt to define microbial “normality” for human health.
Figure 2.
Intraindividual variation over time. Depending on the temporal scale, an individual's gastrointestinal microbiota can vary as measured using the overall community diversity measure, Shannon diversity (H). Data reconstructed from two individuals, male and female, left and right pie charts respectively. Size of pie sector indicates proportion of community; color represents dominant phylum; from Reference 78.
The Vaginal Ecosystem
It is thought that the bacterial communities present in the vagina of reproductive-aged women are the cornerstone of a multifaceted antimicrobial defense system, but the mechanisms by which the microbiota is protective are poorly understood. The vaginal microbiota appears to play a significant role in preventing bacterial vaginosis, yeast infections, sexually transmitted infections, urinary tract infections, and HIV infection (30–34). The prevailing hypothesis is that the lactic acid–producing bacteria (mainly Lactobacillus sp.), common colonizing bacteria in the human vagina, are the key players in maintaining homeostasis of the microbiota (26, 35). These species are hypothesized to facilitate the protective response by lowering the environmental pH through lactic acid production (36, 37), by producing various bacteriostatic and bacteriocidal compounds, or through competitive exclusion (38, 39). A cross-sectional study by Ravel et al. (26) categorized the microbiota of reproductive-aged women into five distinct community types: four that were dominated by Lactobacillus species (L. iners, L. crispatus, L. gasseri, or L. jensenii) and one that was not dominated by a single species but consisted of mainly anaerobic bacteria. This study was the first to speciate the Lactobacilli, providing a greater understanding of the specific microbial species that constitute the microbial community in the vagina. These findings are in direct contrast to the extreme species-level diversity that is observed in the gastrointestinal tract, discussed later in this review.
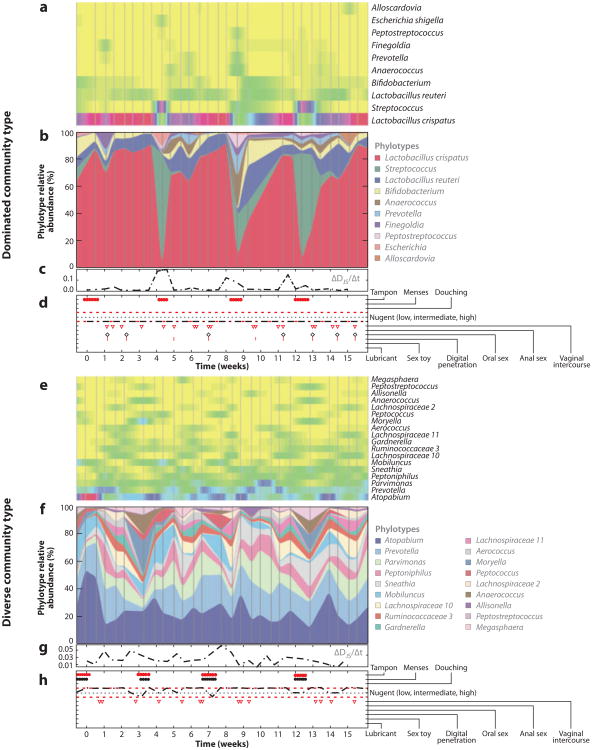
The presence of an established symbiotic microbial community in the host may prevent potential pathogens from colonizing and disrupting the resident microbiota, but environmental factors, in the form of sexual activity as well as monthly menstrual and hormonal cycles, can substantially impact the stability of the community. These environmental factors play a significant role in the dynamics of the vaginal microbiota (Figure 3). A recent longitudinal study of the vaginal microbiota of 32 women, examined daily over a 16-week period, confirmed the five major community types previously identified in the cross-sectional study, but, importantly, was also able to examine the temporal dynamics of the microbiota (35). It is clear from the measures of the microbiota composition and the association with sexual activity, menstrual cycle, and other environmental factors that there are periods of community-wide stability as well as periods of extreme variability. The data presented in Figure 3 are the longitudinal profiles from two individuals. The first contains a community that is dominated by L. crispatus, where, other than during menses, the community structure is extremely stable (panels a–d ); the other individual exhibits a much more diverse microbiota over the sample acquisition period (panels e–h). These two individuals exhibit distinct microbiota community profiles, and although one community is more diverse in composition, it appears to be more stable over the length of the study period. Ecological theory suggests that the less stable a community is, the more susceptible it is to invasion by incoming organisms (40). At this point, we can draw an important conclusion: a diverse community composition does not reflect the health status of the environment it was obtained from, and only through long-term longitudinal studies performed using large cohorts will we be able to identify microbiota signatures that are useful from the standpoint of health and human disease.
Figure 3.
Measurements of the vaginal microbiota of two individuals representing a Lactobacillus-dominated community type and a diverse community type. Panels a and e are heatmaps that represent the presence or absence of the species at each sampled time point (represented on the X axis in panels d and h). Colors indicate the relative abundance, with yellow being absent and red being dominant. Panels b and f display the relative abundance of the phylotype at each time point. Panels c and g are measures of the diversity of microbiota, and panels d and h indicate environmental factors such as menses, sexual activity, and hygienic processes. The data clearly demonstrate a dynamic microbiota within the human vagina and highlight that diversity or stability cannot be determined by cross-sectional studies but only through frequent sampling of large cohorts. Additionally, diversity does not correlate with disease or impairment of function. These are representative data generated from the studies within the Ravel laboratory; further details of the study can be found in Reference 35.
Case Study: Longitudinal Samples Matter
The studies by Ravel and colleagues (26, 35) are instructional on two fronts. The first is that there is significant diversity among samples collected from the same individual that does not correlate with a disease state. Second, frequent sampling can provide insight into the dynamics of the microbiota. As mentioned above, microbiota studies in the past have been hampered by the inability to process and sequence large numbers of samples. Advances in sequencing technologies have made it financially and programmatically feasible to examine large numbers of samples from large patient cohorts, enabling the interrogation of the microbiota at unprecedented levels. If the sampling and data generated for Figure 3 are examined only on the days of menses compared to days prior or subsequent to menses, an entirely different clinical picture appears. Overwhelmingly, the majority of previous microbiota studies utilized a cross-sectional study design, providing only a snapshot of the diversity of the microbiota with little insight into its dynamics. Understanding both diversity and dynamics is essential for developing therapeutics or diagnostics based on the microbiota. By way of analogy to chemical therapeutics, examining the microbiota in a cross-sectional study is like measuring the effective dose of a new drug only at 2 hours post administration and trying to determine its efficacy at 2 days post administration. It is impossible to determine an outcome or impact by examining a single time point in a rapidly evolving and developing system.
As mentioned above, the study of the vaginal microbiota has identified five community types, four of which are dominated by single species. As we describe in the following section, the microbial community of the gastrointestinal tract contrasts with the vaginal community in high species-level diversity and separate community dynamics.
The Complexities of the Gastrointestinal Microbiota
The assemblage of microorganisms present in the human gastrointestinal tract presumably plays a central role in health and disease, yet the fundamental mechanisms through which these host-associated microbiota function remain elusive. Collectively, the gastrointestinal microbial consortia possess metabolic activity equivalent to a “virtual organ within an organ” (41), and humans have been termed a “supraorganism” (1). Our indigenous microbes reside at the interface of the mucosal epithelial barrier, mediating host defense, immune development, and nutritional state. The vast majority of studies to date have investigated the composition, structure, and functional repertoire of the microbial community inhabiting the human gastrointestinal tract through the use of fecal samples (12, 17, 19, 42, 43, 44). Methodologies utilizing fecal material have proven effective and noninvasive yet remain only a proxy for the autochthonous members adherent to the epithelial mucosa at different sites throughout the gastrointestinal tract (45). Additionally, few studies have addressed the importance of archaeal, fungal, and viral components to the overall functioning of the gastrointestinal microbial system. Recent findings have shed light on the viral component as a reservoir of genetic heterogeneity (46), as well as the interaction of the resident microbiota in promoting viral replication and transmission (47, 48). Future efforts to integrate information regarding all microbial and viral players will be crucial in our deeper understanding of the host–microbiota system.
Humans and their gastrointestinal microbiota maintain an intimate relationship that begins at birth and develops in a concerted and coordinated fashion throughout the human lifespan. The gastrointestinal community appears to be seeded by mother-derived microbes during either vaginal delivery or Caesarean section and initially features low complexity and species richness (43, 49, 50). By the first year of life, the microbiota has dynamically progressed and converged toward a stable, phylogenetically diverse, adult-like profile according to distinct “environmental” events, e.g., the shift from breast milk to solid foods (43, 50). Age-related changes in community composition are apparent throughout the lifespan of an individual; for example, Faecalibacterium prausnitzii, the presumptive sentinel of health and anti-inflammation (51), is found at conspicuously low abundances in the elderly and centenarians (52, 53). However, the specific trajectory of age-related shifts has yet to be fully characterized. For instance, relatively little is known regarding the gastrointestinal microbiota in pubescent children, which is a time of significant growth, dramatic hormonal changes, and bodily maturation (54). Recently, Flores et al. delineated a trend toward increased alpha diversity with age using a bivariate linear regression model (55). A highly attractive hypothesis derived from ecological theory is that a succession of rapid changes after birth leads to an equilibrium microbial composition or “climax community” of gastrointestinal microbes in adulthood (11). Despite considerable taxonomic variation among adult individuals, a functional climax equilibrium is a readily testable concept in need of rigorous experimentation. Further study of the temporal dynamics over the lifespan is certainly a priority in an effort to fully describe developmental changes in the microbiome and their relation to global human health.
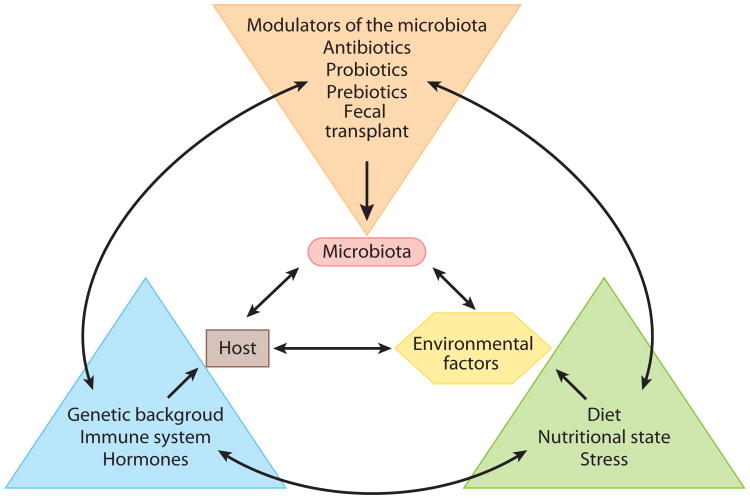
The basic principles determining how environmental factors (i.e., diet or antibiotic usage) and host genetics shape the complexity of the symbiotic gastrointestinal microbiota are largely unknown (Figure 4). To date, no comprehensive microbiota study has incorporated human host genetic background. Recently, Benson and colleagues (56) performed a systematic study of the factors affecting gastrointestinal microbiota composition in a mouse model of known genetic background. Remarkably, the authors identified a subset of host genetic loci controlling individual microbial species, related taxa, and even putative pleiotropic effects on groups of distantly related organisms. Their results suggest that gastrointestinal microbial individuality is a complex polygenic trait, influenced and shaped both by the host genotype and by external, and sometimes stochastic, triggers. Key to our understanding of the gastrointestinal microbiota will be to identify the timing, specific influence, and extent of contributions by the host and environment to the composition and function of the community (Figure 4).
Figure 4.
Framework of interactions among the host, microbiota, and environment. Although these three features are discussed in the context of human homeostasis in this review, there are factors that interact independently of these primary features. All of these features are interacting in the environments (vagina and gastrointestinal tract) discussed in the text, each providing some perturbation in the system. This figure is not meant as an exhaustive list of features but only to illustrate the complexity of interactions that may affect measurements of the microbiota.
The “microflora hypothesis” suggests that reduced microbial exposure at an early age inhibits normal maturation of the intestinal microbiota, altering immune development in a way that enhances symptoms of “allergic hyper-sensitivity” (57). A recent study examined the development of the microbiota among children genetically predisposed to develop celiac disease (8). Among these children, infants exposed to gluten early in life mounted an immune response and developed celiac disease more frequently than did infants with delayed gluten exposure. The microbiota was characterized by an overall lack of bacteria from the phylum Bacteroidetes along with a high abundance of Firmicutes. Interestingly, the microbiota does not resemble that of adults even at two years of age (43). The data from this study highlight the interplay between host features (genetic predisposition), environmental factors (exposure to gluten), and microbiota as contributors to the health and development of infants. This interrelatedness is highlighted in Figure 4, and one can most likely identify other factors on each arm of the figure that could play a role in the development and maintenance of the microbiota.
The pivotal work of Arumugam and colleagues identified three main community types, termed enterotypes, from human fecal samples by using both bacterial 16S ribosomal gene and metagenomic sequencing (17, 58). Importantly, this work provides a framework for gastrointestinal microbiota studies that has thus far been lacking because of the interindividual nature and intraindividual variability of the community, as well as the complexity of the system. However, the implications for the stratified nature of the enterotypes remain unclear. Identifying the drivers of the the development of enterotypes in the gastrointestinal tract or the five dominant community types within the vaginal environment, whether host-mediated or negotiated at the microbial level, will be fundamental in dissecting the ecological basis for these groupings.
As one thinks about the environmental and physical processes that occur in the human gastrointestinal tract, it is not surprising that there is a great deal of microbial diversity and variability. Each of us eats a distinct diet, containing its own microbial communities including potential food-borne pathogens, which are then introduced into the distinct regions of the gastrointestinal tract, which in turn produce enzymes and mount immune responses that are dictated by host genetics and previous bacterial exposures. These factors are all also influenced by the constant environmental onslaught of factors that could potentially influence the microbiota, including chemical and biological entities.
We now highlight two examples of how our symbiotic gastrointestinal microbiota can be exploited by invasive pathogens, underscoring the subtle microbial homeostatic mechanisms we are only beginning to uncover. In many cases it is unclear how this homeostasis is altered, as the commensal species are highly similar to some of the pathogens. Many theories have been suggested from the immunological side (59), where it has been proposed that the host can sense the potential danger of pathogens, as well as from the microbial side, where Joel Doré at a recent International Human Microbiome Consortium conference suggested, “Pathogens shout, commensals whisper” (94). A great deal remains to be elucidated in the host-commensal-pathogen dynamic.
Case Study: An Invasive Pathogen Defies Host Defense
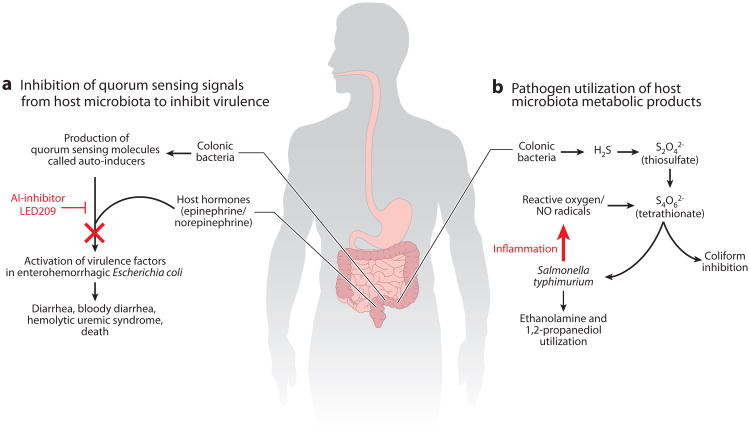
The gastrointestinal microbiota is thought to be one of the first lines of protection against incoming pathogens, hosting an arsenal of defense mechanisms to counter a potential pathogenic invasion. This protection against colonization by enteric pathogens, termed colonization resistance, acts through three distinct mechanisms: (a) the direct inhibition of pathogen growth by microbiota-derived substances, (b) nutrient depletion by microbiota growth, and (c) microbiota-induced stimulation of innate and adaptive immune responses (60). The invasive enteric pathogen Salmonella enterica serotype Typhimurium (S. Typhimurium) causes acute intestinal inflammation through invasion of the intestinal epithelium and survival in mucosal macrophages (61). Recently, an eloquent set of studies revealed the ability of S. Typhimurium to outcompete resident members of the microbiota through utilization of the respiratory chain electron acceptor tetrathionate, produced by reactive oxygen species during inflammation (61). The intestinal inflammation subsequently confers a growth advantage for S. Typhimurium to metabolize ethanolamine as a carbon source, which cannot be utilized by the resident microbiota (62). Remarkably, these subtle changes in the mode of respiration for S. Typhimurium (anaerobic respiration) appear to be a fundamental mechanism governing the dynamics of the host-associated microbiota (Figure 5, right).
Figure 5.
Two examples of the interaction of the microbiota, host, and pathogen. (a) Development of an antivirulence-signaling therapeutic (71, 80). The quorum sensing signal from the host microbiota and the hormone signals from the host are inhibited by a compound known as LED209. The inhibition of these signals prevents the activation of virulence factors, and the bacteria are removed through physical processes and/or the immune system. (b) Interaction of an incoming pathogen, Salmonella, and the host microbiota production of H2S to inhibit other pathogens and provide a terminal electron acceptor for Salmonella. The production of these molecules provides an opening for the pathogen to colonize and cause disease (61).
Case Study: Therapeutic Development Inhibits Microbiota Signaling
The communication of bacterial members of the microbiota has been studied for years in the context of bacterial quorum sensing (63–65). Quorum sensing is the ability to regulate gene expression in response to variations in cell density. Usually the term refers to self-communication, but in the context of the microbiota, there are potentially endless variations in terms of signal producers and responders. Cell-cell signaling between bacterial species is known as intrakingdom signaling. Additionally, it has been demonstrated that bacterial pathogens can respond to human hormones such as epinephrine and norepinephrine to increase the expression of virulence factors (66–68). This type of signaling is known as interkingdom signaling (69, 70). It has been proposed that if a therapeutic could be developed that would inhibit the bacterial signaling for virulence then the bacteria would not activate virulence mechanisms and would either pass through the human body or be easily eliminated by the immune system. A study by Rasko et al. (71) identified a compound known as LED209 that appeared to inhibit both the intra- and interkingdom signals in the gastrointestinal tract for the enterohemorrhagic Escherichia coli isolates, demonstrated to cause hemolytic uremic syndrome and death (72). This compound was demonstrated to inhibit the signal from the host microbiota, known as auto-inducer 3 (AI-3), as well as the bacterial sensing of the host hormones, epinephrine and norepinephrine (Figure 5, left). Additionally, the bacterial protein that was the sensor of the signals was conserved in other pathogens, and it was demonstrated that LED209 decreased the virulence of both Salmonella species and Francisella tularensis in animal models (71). Other examples of this strategy have been described for Vibrio cholerae (73, 74). It is thought that in contrast to traditional antibiotics, which seek to eliminate the bacterium, signaling inhibitors will be much better tolerated and the decreased negative pressure will prevent the rapid development of resistance to these compounds. These therapies still require a great deal of study before they will be available for human treatment but may signal a shift in the ideology of therapeutic antimicrobial design.
Elucidating Transitions from Homeostasis to Dysbiosis
Ultimately, we seek to develop a fundamental understanding of how our microbiota contributes to our overall health and well-being, and in turn, that knowledge can be used to manipulate the microbiota to restore a healthy state from a diseased state (Figure 4). However, features or biological markers within the microbiome that are indicative of diseases such as atherosclerosis, obesity and the metabolic syndrome, and IBD have yet to be identified. There is mounting evidence against a one-to-one causal relationship in which a single pathogen (or virulence-related gene) is the etiological agent of a complex disease.
Compositional shifts in relevant members of the microbiota have been shown to be highly associated with certain disease states; the prime example is IBD, which includes Crohn's disease and ulcerative colitis (75). Marked changes in the composition of the gut microbiota and a decrease in overall community diversity have been readily observed in IBD patients compared to healthy individuals (16). The functional implications of these shifts in the microbiota remain unexamined, and a source of potential research in the future. Table 1 highlights further examples of associations between microbiota and certain “disease states.” However, these studies include small cohorts, and we cannot yet determine causation or diagnostic capabilities from these limited studies.
Table 1. Examples of putative associations between microbiota and “disease states”.
| Clinical condition | Observed differences in microbiota compared to a “healthy state” | References |
|---|---|---|
|
| ||
| Bacterial vaginosis (BV) | Greater bacterial diversity observed in women with BV; BV-associated bacteria found to correlate with diagnostic criteria (Amsel's clinical criteria) | 82, 83 |
|
| ||
| Inflammatory bowel disease (IBD; colitis) | Enterobacteriaceae found to correlate with colitis; global microbiota profiles of phylotype and/or gene content distinguish IBD individuals | 75, 84, 85 |
|
| ||
| Type 1 diabetes | Bacterial diversity decreased over time in children; functionally aberrant gene content | 87, 88 |
|
| ||
| Type 2 diabetes | Relative proportions of Clostridia (phylum Firmicutes) significantly reduced in diabetic group | 89 |
|
| ||
| Rheumatoid arthritis | Segmented filamentous bacteria or Lactobacillus sp. can activate TH17 cells, resulting in inflammation | Reviewed in 90 |
|
| ||
| Colorectal cancer | Increased abundances of Fusobacterium sp.; colitis can promote tumorigenesis by altering microbial composition | 91–93 |
For the gastrointestinal microbiota, a recurring theme is the concept of homeostasis and avoidance of an inflammatory state leading to dysbiosis. Members of the genus Akkermansia, mucus degraders within the deeply rooted phylum Verrucomicrobia, have been implicated recently as a potential biomarker for a healthy intestine owing to their ability to produce oligosaccharides and short-chain fatty acids that stimulate the growth of resident mucus-colonizing gastrointestinal microbiota (76). Furthermore, investigation of specific compounds, in addition to microbial groups, has identified protective properties; for example, Bacteroides fragilis capsular polysaccharide A is protective against experimental autoimmune encephalomyelitis in mice, the most widely used animal model for multiple sclerosis (77). Diagnostic and prognostic applications based on an understanding of our microbial partners, particularly to identify precursor markers of disease that might be treated, hold remarkable promise as “microbiome therapeutics.” However, we will require a much greater understanding of our microbial communities before we can accurately shape the microbiota in the ways that are advantageous to human health. A major shortcoming in all microbiota research is that we currently lack the ability to differentiate cause and effect in studies that link changes in the microbiota with disease. The ability to harness genomic and microbiome data for use in clinical practice is still in its infancy.
Future Directions
Although a wealth of knowledge has been gained from the predominantly DNA-based community characterizations, these methods only provide information about functional potential. Whether identified metabolic pathways are functional, to what extent, and under what conditions remain to be quantified. Similarly, whole-community DNA-based surveys rely heavily on computational methods to relate a molecular signature back to the cell, yet this molecular signature may be from a living, active microbial cell or from a dormant or even dead cell. Advances in other “-omics” technologies, including transcriptomics, proteomics, and metabolomics, will provide fundamental information about the active constituents of the microbial consortia and what metabolic products are in flux at a given time (Table 2). Additionally, computational capacity and novel statistical methodology are advancing toward a systems biology approach to integrate clinical outcome, host metadata, and the microbiome. The availability of new tools, such as the maximal information coefficient as a measure of dependence for two-variable relationships (9, 86), will provide crucial mechanisms to associate health and the microbiome. In years past, the bottleneck to analysis was the feasibility of large-scale studies, limited by sequencing and financial concerns. Now, the new technologies can generate large amounts of data, rapidly, at relatively low cost; however, the methodologies to properly analyze and compare these data are still in development.
Table 2. Culture-independent methods to interrogate the human microbiome.
| Technology | Target molecule | Methodological platform | Analytical tools | Biological questions |
|---|---|---|---|---|
|
| ||||
Metagenomics
|
DNA | Sanger (ABI 3730)
|
QIIME, mothur, MG-RAST, CAMERA, VAMPS, IMG/M, SmashCommunity, MEGAN |
|
|
| ||||
| Meta-transcriptomics | RNA |
|
TopHat, Bowtie, HUMAnN |
|
|
| ||||
| Meta-proteomics | Proteins | High-throughput mass spectroscopy | Developing | What proteins are being produced and are potentially interacting |
Concluding Remarks
It appears that microbiome variability far exceeds human genetic variation. The evident plasticity of the microbiome presents the possibility of modulating the microbiota to promote health. We have examined the concepts of the microbiota and human health within the two body sites that have garnered the most attention and thus have the most complete datasets and advanced hypotheses. The human vagina appears to contain a limited number of community types, the majority of which are dominated by one or more Lactobacilli species. In contrast, the microbiota of the human gastrointestinal tract displays a great deal of variability, but large cohorts of frequently sampled individuals are not yet available. Further investigation of the dynamics of the gastrointestinal enterotypes over time (78) will continue to reveal the range of community variation and provide context for the impact of microbial modulators, such as antibiotics and probiotics. In particular, antibiotic use brings about a regime shift in the resident community that fails to fully return to baseline (79).
The further study of host–microbiota interaction cannot be undertaken by microbiologists alone. It will require an integrative, systems biology approach that includes input from immunologists, cell biologists, ecologists, and physicians to understand these processes.
The Pitfalls of Sequencing Technologies.
DNA sequencing technologies have revolutionized the field of microbiology, spawning the field of metagenomics and generating a deluge of high-complexity data. All human microbiome studies, to date, utilize three main sequencing platforms: capillary-based sequencing (Sanger, such as Applied Biosystems 3730xl), pyrosequencing (including Roche 454 GS, FLX, and FLX Titanium), and Illumina clonal arrays (Illumina GAIIx, HiSeq 2000, and MiSeq). Each technology has distinct characteristics, including read length, coverage depth, accuracy, scalability, relative cost, and time to generate data. We refer the reader to an extensive review of sequencing methodologies for the study of the human microbiome (81). Surprisingly, there is no “standard” sequencing platform, methodology, or computational tool uniformly used by researchers in the field, rendering cross-study comparisons difficult. The choices of technology and bioinformatic tools to mine the data are critically important considerations for study design and subsequent interpretation of the results. For example, Sanger sequencing produces long read lengths (approximately 800–1,000 bp) compared to other technologies (<500 bp), enabling better phylogenetic resolution for ribosomal-based classification, but it yields four orders of magnitude fewer reads than the Illumina sequencing platform. Understanding the strengths and weaknesses of a particular sequencing platform, as well as the computational and analytical limitations, is crucial if we are to integrate knowledge of the microbiome as a diagnostic tool for human health and disease.
Microbiome: The Importance of Terminology.
Although the term microbiome is widely used to describe studies ranging from bacterial amplicon sequencing to metagenomic surveys, there is an important distinction. The human microbiome is the collection of genes and genomes within the microbial community associated with a distinct body region. Characterizing the microbial phylogenetic composition from a given body site using 16S ribosomal gene sequence data describes only a portion of the microbiome. The term metagenome refers to the totality of genomes from an environment, including bacteria, archaea, viruses, and eukaryotic microbes. The terminology is significant because ribosomal gene sequencing and metagenomics are different measures of the microbial components, and each contributes a distinct microbial readout for interpretation and application to human health.
Summary Points.
New technologies have allowed the interrogation of the human microbiome at multiple body sites at unprecedented levels.
Frequent longitudinal sampling is required to identify meaningful differences in microbiota studies.
Identification of community types, particularly for the vaginal and gastrointestinal tract ecosystems, has provided an important interpretive framework for future studies.
Despite great strides in understanding the microbiota and its relation to human health, much remains unknown.
The subtle mechanisms through which pathogens are able to exploit our symbiotic microbiota to cause disease are only beginning to be uncovered.
Acknowledgments
We thank the investigators who have produced microbiota data and released them into the public domain. The data utilized in Figures 1–3 were obtained from the HMP datasets stored at the NIH Human Microbiome Project Data Analysis and Coordination Center (http://www.hmpdacc.org/). Additionally, we thank Jonathan Crabtree for assistance with HMP datasets, Drs. Emmanuel F. Mongodin and Garry S.A. Myers for helpful comments during the preparation of this manuscript, and Dr. Jacques Ravel for the data represented in Figure 3. E.A.E.F. is funded by NIH U19 AI082655 NIAID, NIH U01 AT002388 and U01 AT002952. D.A.R. is funded by NIH U19 AI082655 and RC4 AI092828A and receives funds from the State of Maryland.
Glossary
- Microbiota
the collection of microbial organisms inhabiting a defined environment, such as a specific body site
- Microbiome
the collection of genes and genomes within the microbiota
- Metagenomics
the study of the collective genomic content from an environment
- Dysbiosis
alterations in the relative abundance of microbial groups or functions that cause an imbalance compared to a healthy state, generally leading to a detrimental change in health
- Biodiversity
the variety of organisms present in an ecosystem
- Phylotype
taxon-neutral description of microorganisms based on phylogenetic relationship
- Bioinformatics
application of computational methods and algorithms to decipher biological sequence data
- IBD
inflammatory bowel disease
- Gnotobiotic mice
germ-free mice
- Humanized mice
germ-free mice into which fresh or frozen human fecal microbial communities have been transplanted
- Autochthonous
indigenous or resident to a given body region
- Alpha diversity
measure of diversity describing the biological components of a single ecosystem
Footnotes
Disclosure Statement: The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
Literature Cited
- 1.Turnbaugh PJ, Ley RE, Hamady M, et al. The Human Microbiome Project. Nature. 2007;449:804–10. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peterson J, Garges S, Giovanni M, et al. The NIH Human Microbiome Project. Genome Res. 2009;19:2317–23. doi: 10.1101/gr.096651.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ehrlich SD . METAHIT Consortium. MetaHIT: the European Union Project on Metagenomics of the Human Intestinal Tract. In: Nelson KE, editor. Metagenomics of the Human Body. New York: Springer; 2011. pp. 307–16. [Google Scholar]
- 4.The Human Microbiome Project Consortium. A framework for human microbiome research. Nature. 2012;486:215–21. doi: 10.1038/nature11209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.The Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–14. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hess M, Sczyrba A, Egan R, et al. Metagenomic discovery of biomass-degrading genes and genomes from cow rumen. Science. 2011;331:463–67. doi: 10.1126/science.1200387. [DOI] [PubMed] [Google Scholar]
- 7.Wang ZN, Klipfell E, Bennett BJ, et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. 2011;472:57–82. doi: 10.1038/nature09922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sellitto M, Bai G, Serena G, et al. Proof of concept of microbiome-metabolome analysis and delayed gluten exposure on celiac disease autoimmunity in genetically at-risk infants. PLoS ONE. 2012;7:e33387. doi: 10.1371/journal.pone.0033387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paracer S, Ahmadijian V. Symbiosis: An Introduction to Biological Associations. Oxford, UK: Oxford Univ. Press; 2000. [Google Scholar]
- 10.Rappé MS, Giovannoni SJ. The uncultured microbial majority. Annu Rev Microbiol. 2003;57:369–94. doi: 10.1146/annurev.micro.57.030502.090759. [DOI] [PubMed] [Google Scholar]
- 11.Gonzalez A, Clemente JC, Shade A, et al. Our microbial selves: what ecology can teach us. EMBO Rep. 2011;12:775–84. doi: 10.1038/embor.2011.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eckburg PB, Bik EM, Bernstein CN, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–38. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grice EA, Kong HH, Conlan S, et al. Topographical and temporal diversity of the human skin microbiome. Science. 2009;324:1190–92. doi: 10.1126/science.1171700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ravel J, Gajer P, Abdo Z, et al. Vaginal microbiome of reproductive-age women. Proc Natl Acad Sci USA. 2011;108:4680–87. doi: 10.1073/pnas.1002611107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Knights D, Kuczynski J, Charlson ES, et al. Bayesian community-wide culture-independent microbial source tracking. Nat Methods. 2011;8:761–63. doi: 10.1038/nmeth.1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qin J, Li R, Raes J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arumugam M, Raes J, Pelletier E, et al. Enterotypes of the human gut microbiome. Nature. 2011;473:174–80. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Turnbaugh PJ, Hamady M, Yatsunenko T, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–84. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Greenblum S, Turnbaugh PJ, Borenstein E. Metagenomic systems biology of the human gut microbiome reveals topological shifts associated with obesity and inflammatory bowel disease. Proc Natl Acad Sci USA. 2012;109:594–99. doi: 10.1073/pnas.1116053109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Samuel BS, Gordon JI. A humanized gnotobiotic mouse model of host-archaeal-bacterial mutualism. Proc Natl Acad Sci USA. 2006;103:10011–16. doi: 10.1073/pnas.0602187103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vaishnava S, Yamamoto M, Severson KM, et al. The antibacterial lectin RegIIIγ promotes the spatial segregation of microbiota and host in the intestine. Science. 2011;334:255–58. doi: 10.1126/science.1209791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kau AL, Ahern PP, Griffin NW, et al. Human nutrition, the gut microbiome and the immune system. Nature. 2011;474:327–36. doi: 10.1038/nature10213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Turnbaugh PJ, Ley RE, Mahowald MA, et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–131. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 24.Jumpertz R, Le DS, Turnbaugh PJ, et al. Energy-balance studies reveal associations between gut microbes, caloric load, and nutrient absorption in humans. Am J Clin Nutr. 2011;94:58–65. doi: 10.3945/ajcn.110.010132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Turnbaugh PJ, Gordon JI. The core gut microbiome, energy balance and obesity. J Physiol. 2009;587:4153–58. doi: 10.1113/jphysiol.2009.174136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ravel J, Gajer P, Abdo Z, et al. Vaginal microbiome of reproductive-age women. Proc Natl Acad Sci USA. 2011;108(Suppl. 1):4680–87. doi: 10.1073/pnas.1002611107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.von Ohle C, Gieseke A, Nistico L, et al. Real-time microsensor measurement of local metabolic activities in ex vivo dental biofilms exposed to sucrose and treated with chlorhexidine. Appl Environ Microbiol. 2010;76:2326–34. doi: 10.1128/AEM.02090-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Diaz PI. Microbial diversity and interactions in subgingival biofilm communities. Front Oral Biol. 2012;15:17–40. doi: 10.1159/000329669. [DOI] [PubMed] [Google Scholar]
- 29.Aas JA, Paster BJ, Stokes LN, et al. Defining the normal bacterial flora of the oral cavity. J Clin Microbiol. 2005;43:5721–32. doi: 10.1128/JCM.43.11.5721-5732.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Donders GG, Bosmans E, Dekeersmaecker A, et al. Pathogenesis of abnormal vaginal bacterial flora. Am J Obstet Gynecol. 2000;182:872–78. doi: 10.1016/s0002-9378(00)70338-3. [DOI] [PubMed] [Google Scholar]
- 31.Cherpes TL, Meyn LA, Krohn MA, et al. Association between acquisition of herpes simplex virus type 2 in women and bacterial vaginosis. Clin Infect Dis. 2003;37:319–25. doi: 10.1086/375819. [DOI] [PubMed] [Google Scholar]
- 32.Martin HL, Richardson BA, Nyange PM, et al. Vaginal lactobacilli, microbial flora, and risk of human immunodeficiency virus type 1and sexually transmitted disease acquisition. J Infect Dis. 1999;180:1863–68. doi: 10.1086/315127. [DOI] [PubMed] [Google Scholar]
- 33.Sobel JD. Is there a protective role for vaginal flora? Curr Infect Dis Rep. 1999;1:379–83. doi: 10.1007/s11908-999-0045-z. [DOI] [PubMed] [Google Scholar]
- 34.Watts DH, Fazzari M, Minkoff H, et al. Effects of bacterial vaginosis and other genital infections on the natural history of human papillomavirus infection in HIV-1-infected and high-risk HIV-1-uninfected women. J Infect Dis. 2005;191:1129–39. doi: 10.1086/427777. [DOI] [PubMed] [Google Scholar]
- 35.Gajer P, Brotman RM, Bai G, et al. Temporal dynamics of the human vaginal microbiota. Sci Transl Med. 2012;4:132ra52. doi: 10.1126/scitranslmed.3003605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boskey ER, Telsch KM, Whaley KJ, et al. Acid production by vaginal flora in vitro is consistent with the rate and extent of vaginal acidification. Infect Immun. 1999;67:5170–75. doi: 10.1128/iai.67.10.5170-5175.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boskey ER, Cone RA, Whaley KJ, et al. Origins of vaginal acidity: high D/L lactate ratio is consistent with bacteria being the primary source. Hum Reprod. 2001;16:1809–13. doi: 10.1093/humrep/16.9.1809. [DOI] [PubMed] [Google Scholar]
- 38.Klebanoff SJ, Hillier SL, Eschenbach DA, et al. Control of the microbial flora of the vagina by H2O2-generating lactobacilli. J Infect Dis. 1991;164:94–100. doi: 10.1093/infdis/164.1.94. [DOI] [PubMed] [Google Scholar]
- 39.Kaewsrichan J, Peeyananjarassri K, Kongprasertkit J. Selection and identification of anaerobic lactobacilli producing inhibitory compounds against vaginal pathogens. FEMS Immunol Med Microbiol. 2006;48:75–83. doi: 10.1111/j.1574-695X.2006.00124.x. [DOI] [PubMed] [Google Scholar]
- 40.Dunstan PK, Johnson CR. Linking richness, community variability, and invasion resistance with patch size. Ecology. 2006;87:2842–50. doi: 10.1890/0012-9658(2006)87[2842:lrcvai]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 41.O'Hara AM, Shanahan F. The gut flora as a forgotten organ. EMBO Rep. 2006;7:688–93. doi: 10.1038/sj.embor.7400731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gill SR, Pop M, Deboy RT, et al. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312:1355–59. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Palmer C, Bik EM, DiGiulio DB, et al. Development of the human infant intestinal microbiota. PLoS Biol. 2007;5:e177. doi: 10.1371/journal.pbio.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yatsunenko T, Rey FE, Manary MJ, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–27. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aguirre de Carcer D, Cuiv PO, Wang T, et al. Numerical ecology validates a biogeographical distribution and gender-based effect on mucosa-associated bacteria along the human colon. ISME J. 2011;5:801–9. doi: 10.1038/ismej.2010.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Minot S, Sinha R, Chen J, et al. The human gut virome: inter-individual variation and dynamic response to diet. Genome Res. 2011;21:1616–25. doi: 10.1101/gr.122705.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kuss SK, Best GT, Etheredge CA, et al. Intestinal microbiota promote enteric virus replication and systemic pathogenesis. Science. 2011;334:249–52. doi: 10.1126/science.1211057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kane M, Case LK, Kopaskie K, et al. Successful transmission of a retrovirus depends on the commensal microbiota. Science. 2011;334:245–49. doi: 10.1126/science.1210718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dominguez-Bello MG, Costello EK, Contreras M, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci USA. 2010;107:11971–75. doi: 10.1073/pnas.1002601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Koenig JE, Spor A, Scalfone N, et al. Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci USA. 2011;108:4578–85. doi: 10.1073/pnas.1000081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sokol H, Pigneur B, Watterlot L, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci USA. 2008;105:16731–36. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mueller S, Saunier K, Hanisch C, et al. Differences in fecal microbiota in different European study populations in relation to age, gender, and country: a cross-sectional study. Appl Environ Microbiol. 2006;72:1027–33. doi: 10.1128/AEM.72.2.1027-1033.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Biagi E, Nylund L, Candela M, et al. Through ageing, and beyond: gut microbiota and inflammatory status in seniors and centenarians. PLoS ONE. 2010;5:e10667. doi: 10.1371/journal.pone.0010667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Agans R, Rigsbee L, Kenche H, et al. Distal gut microbiota of adolescent children is different from that of adults. FEMS Microbiol Ecol. 2011;77:404–12. doi: 10.1111/j.1574-6941.2011.01120.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Flores R, Shi J, Gail MH, et al. Assessment of the human faecal microbiota: II Reproducibility and associations of 16S rRNA pyrosequences. Eur J Clin Invest. 2012;42:855–63. doi: 10.1111/j.1365-2362.2012.02659.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Benson AK, Kelly SA, Legge R, et al. Individuality in gut microbiota composition is a complex polygenic trait shaped by multiple environmental and host genetic factors. Proc Natl Acad Sci USA. 2010;107:18933–38. doi: 10.1073/pnas.1007028107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shreiner A, Huffnagle GB, Noverr MC. The “microflora hypothesis” of allergic disease. GI Microbiota Regul Immune Syst. 2008;635:113–34. doi: 10.1007/978-0-387-09550-9_10. [DOI] [PubMed] [Google Scholar]
- 58.Wu GD, Chen J, Hoffmann C, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;333:105–8. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Matzinger P. The danger model: a renewed sense of self. Science. 2002;296:301–5. doi: 10.1126/science.1071059. [DOI] [PubMed] [Google Scholar]
- 60.Stecher B, Hardt WD. Mechanisms controlling pathogen colonization of the gut. Curr Opin Microbiol. 2011;14:82–91. doi: 10.1016/j.mib.2010.10.003. [DOI] [PubMed] [Google Scholar]
- 61.Winter SE, Thiennimitr P, Winter MG, et al. Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature. 2010;467:426–29. doi: 10.1038/nature09415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thiennimitr P, Winter SE, Winter MG, et al. Intestinal inflammation allows Salmonella to use ethanolamine to compete with the microbiota. Proc Natl Acad Sci USA. 2011;108:17480–85. doi: 10.1073/pnas.1107857108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fuqua C, Greenberg EP. Self perception in bacteria: quorum sensing with acylated homoserine lactones. Curr Opin Microbiol. 1998;1:183–89. doi: 10.1016/s1369-5274(98)80009-x. [DOI] [PubMed] [Google Scholar]
- 64.Fuqua C, Parsek MR, Greenberg EP. Regulation of gene expression by cell-to-cell communication: acyl-homoserine lactone quorum sensing. Annu Rev Genet. 2001;35:439–68. doi: 10.1146/annurev.genet.35.102401.090913. [DOI] [PubMed] [Google Scholar]
- 65.Miller MB, Bassler BL. Quorum sensing in bacteria. Annu Rev Microbiol. 2001;55:165–99. doi: 10.1146/annurev.micro.55.1.165. [DOI] [PubMed] [Google Scholar]
- 66.Sperandio V, Torres AG, Jarvis B, et al. Bacteria-host communication: the language of hormones. Proc Natl Acad Sci USA. 2003;100:8951–56. doi: 10.1073/pnas.1537100100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kaper JB, Sperandio V. Bacterial cell-to-cell signaling in the gastrointestinal tract. Infect Immun. 2005;73:3197–209. doi: 10.1128/IAI.73.6.3197-3209.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Waldor MK, Sperandio V. Adrenergic regulation of bacterial virulence. J Infect Dis. 2007;195:1248–49. doi: 10.1086/513281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pacheco AR, Sperandio V. Inter-kingdom signaling: chemical language between bacteria and host. Curr Opin Microbiol. 2009;12:192–98. doi: 10.1016/j.mib.2009.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hughes DT, Sperandio V. Inter-kingdom signalling: communication between bacteria and their hosts. Nat Rev Microbiol. 2008;6:111–20. doi: 10.1038/nrmicro1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rasko DA, Moreira CG, Li de R, et al. Targeting QseC signaling and virulence for antibiotic development. Science. 2008;321:1078–80. doi: 10.1126/science.1160354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rasko DA, Webster DR, Sahl JW, et al. Origins of the E. coli strain causingan outbreak of hemolyticuremic syndrome in Germany. N Engl J Med. 2011;365:709–17. doi: 10.1056/NEJMoa1106920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hung DT, Shakhnovich EA, Pierson E, et al. Small-molecule inhibitor of Vibrio cholerae virulence and intestinal colonization. Science. 2005;310:670–74. doi: 10.1126/science.1116739. [DOI] [PubMed] [Google Scholar]
- 74.Hung DT, Rubin EJ. Chemical biology and bacteria: not simply a matter of life or death. Curr Opin Chem Biol. 2006;10:321–26. doi: 10.1016/j.cbpa.2006.06.016. [DOI] [PubMed] [Google Scholar]
- 75.Frank DN, St Amand AL, Feldman RA, et al. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci USA. 2007;104:13780–85. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Belzer C, de Vos WM. Microbes inside—from diversity to function: the case of Akkermansia. ISME J. 2012;6:1449–58. doi: 10.1038/ismej.2012.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ochoa-Reparaz J, Mielcarz DW, Wang Y, et al. A polysaccharide from the human commensal Bacteroides fragilis protects against CNS demyelinating disease. Mucosal Immunol. 2010;3:487–95. doi: 10.1038/mi.2010.29. [DOI] [PubMed] [Google Scholar]
- 78.Caporaso JG, Lauber CL, Costello EK, et al. Moving pictures of the human microbiome. Genome Biol. 2011;12:R50. doi: 10.1186/gb-2011-12-5-r50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dethlefsen L, Relman DA. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc Natl Acad Sci USA. 2011;108:4554–61. doi: 10.1073/pnas.1000087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rasko DA, Sperandio V. Anti-virulence strategies to combat bacteria-mediated disease. Nat Rev Drug Discov. 2010;9:117–28. doi: 10.1038/nrd3013. [DOI] [PubMed] [Google Scholar]
- 81.Kuczynski J, Lauber CL, Walters WA, et al. Experimental and analytical tools for studying the human microbiome. Nat Rev Genet. 2012;13:47–58. doi: 10.1038/nrg3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Fredricks DN, Fiedler TL, Marrazzo JM. Molecular identification of bacteria associated with bacterial vaginosis. N Engl J Med. 2005;353:1899–911. doi: 10.1056/NEJMoa043802. [DOI] [PubMed] [Google Scholar]
- 83.Srinivasan S, Hoffman NG, Morgan MT, et al. Bacterial communities in women with bacterial vaginosis: High resolution phylogenetic analyses reveal relationships of microbiota to clinical criteria. PLoS ONE. 2012;7(6):e37818. doi: 10.1371/journal.pone.0037818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Garrett WS, Gallini CA, Yatsunenko T, et al. Enterobacteriaceae act in concert with the gut microbiota to induce spontaneous and maternally transmitted colitis. Cell Host Microbe. 2010;8:292–300. doi: 10.1016/j.chom.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Qin J, Li R, Raes J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Reshef DN, Reshef YA, Finucane HK, et al. Detecting novel associations in large data sets. Science. 2011;334:1518–24. doi: 10.1126/science.1205438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Giongo A, Gano KA, Crabb DB, et al. Toward defining the autoimmune microbiome for type 1 diabetes. ISME J. 2011;5:82–89. doi: 10.1038/ismej.2010.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Brown CT, Davis-Richardson AG, Giongo A, et al. Gut microbiome metagenomics analysis suggests a functional model for the development of autoimmunity for type 1 diabetes. PLoS ONE. 2011;6(10):e25792. doi: 10.1371/journal.pone.0025792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Larsen N, Vogensen FK, van den Berg FWJ, et al. Gut microbiota in human adults with type 2 diabetes differs from non-diabetic adults. PLoS ONE. 2010;5(2):e9085. doi: 10.1371/journal.pone.0009085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Scher JU, Abramson SB. The microbiome and rheumatoid arthritis. Nat Rev Rheumatol. 2011;7(10):569–78. doi: 10.1038/nrrheum.2011.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Castellarin M, Warren RL, Freeman JD, et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2012;22:299–306. doi: 10.1101/gr.126516.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kostic AD, Gevers D, Pedamallu CS, et al. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res. 2012;22:292–298. doi: 10.1101/gr.126573.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Arthur JC, Perez-Chanona E, Mühlbauer M, et al. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science. 2012;338:120–23. doi: 10.1126/science.1224820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Doré J. Screening of metagenomic clones; Presented at Int. Hum. Microbiome Consort; Vancouver. Mar. 9–11.2012. [Google Scholar]