Abstract
Progression of prostate cancer (CaP) relies on androgen receptor (AR) signaling, but AR-dependent events that underlie the lethal phenotype remain unknown. Recently, an indirect mechanism of androgen action in which effects of AR on CaP cells are mediated by Serum Response Factor (SRF) has been identified. This is the first mode of androgen action to be associated with aggressive CaP and disease recurrence. The manner in which androgen-responsive SRF activity controls aggressive CaP cell behavior is unknown. Here, the contribution of two representative SRF effector genes that are underexpressed, calponin 2 (CNN2), or overexpressed, sidekick-homolog 1 (SDK1), in clinical CaP specimens is studied. AR- and SRF- dependency of CNN2 and SDK1 expression was verified using synthetic and natural androgens, antiandrogens, and small interfering RNAs targeting AR or SRF, and evaluating the kinetics of androgen induction and SRF binding to endogenously and exogenously expressed regulatory gene regions in AR-positive CaP model systems that mimic the transition from androgen-stimulated to castration-recurrent disease. Small interfering RNA-mediated deregulation of CNN2 or SDK1 expression did not affect CaP cell proliferation or apoptosis but had marked effects on CaP cell morphology and actin cytoskeleton organization. Loss of CNN2 induced cellular protrusions and increased CaP cell migration, whereas silencing of SDK1 led to cell rounding and blunted CaP cell migration. Changes in cell migration did not involve epithelial-mesenchymal transition but correlated with altered β1-integrin expression. Taken together, individual androgen-responsive SRF target genes affect CaP cell behavior by modulating cell migration, which may have implications for therapeutic intervention downstream of AR and SRF.
Introduction
Androgen action is the main target for therapy in patients who suffer from non-organ-confined prostate cancer (CaP). First-line therapies targeted against androgen receptor (AR) inhibit the systemic production of androgens and/or interfere with the interaction between AR and androgens (1). Despite initial remission, the majority of CaPs reemerge during androgen-deprivation therapy (ADT), at which time the disease is referred to as castration-recurrent CaP (CR-CaP). AR remains essential for the growth of CR-CaP cells (2). Novel second-line ADT approaches that are based on insights into two of the mechanisms that underlie sustained AR action in CR-CaP, AR overexpression and intracrine androgen synthesis yield survival benefits in patients who have failed first-generation ADT (3–5). These beneficial effects, however, are again temporary, and the AR signaling axis is active in CaP that recurs (6–8). Although AR action remains critical during CaP progression, the AR-dependent events that drive the lethal phenotype are largely unknown. Identification of these events, their effector genes, and the manner in which they induce aggressive CaP behavior are of great interest as they may provide alternative means to target AR action in CaP cells.
Recently, our laboratory has identified a novel mechanism of androgen action in which the effects of androgens on CaP cells are mediated by Serum Response Factor (SRF) (9,10). SRF is a founding member of the MADS-box family of transcription factors and one of the best-characterized transactivating factors in the mammalian genome (11). SRF binds its consensus binding site, a 10bp cis element that consists of variations of a CC(A/T)6GG motif and is known as a CArG box, in regulatory regions of target genes in a constitutive manner. Stimulation of upstream signaling cascades or recruitment of one or more of the 60 SRF cofactors that have been identified to date activates SRF transcriptional activity (11–13). A set of 158 SRF target genes, which represents only 11% of the SRF-dependent transcriptome in CaP cells, was found to be androgen responsive (10). This mode of target gene activation is consistent with reports of gene specificity and context dependence for the manner in which SRF regulates expression of its target genes (14–17). At the same time, the androgen-responsive SRF target genes make up only a small fraction (5.5%) of androgen-regulated genes in CaP cells. Thus, the 158 SRF- and AR-dependent gene signature represents the transcriptional output of a discrete mechanism of androgen actions that controls select SRF effector genes. SRF, but not AR, is present at CArG boxes in regulatory regions of target genes under both androgen-deprived and androgen-supplemented conditions, and becomes activated, at least in part, via androgen control over the RhoA signaling axis, a well-known upstream mediator of SRF (9,18).
SRF was identified originally based on its involvement in the immediate early response (19). Since then, the CArGome, as the transcriptome under control of SRF is often referred to, has been shown to be enriched also in functions in organization of the cytoskeleton, cell adhesion, nucleic-acid binding and cell signaling (12). SRF is essential for the development and maintenance of almost every organism and organ system in which it has been studied (20). A role for SRF in the pathogenesis of human disease is emerging and involves among others, the cardiovascular, digestive and nervous systems (20). Observations of differential SRF expression in several human cancers and its requirement for metastatic spread of cancer cells (21–26) have generated interest in the relevance of SRF in human neoplasms. Remarkably, the small fraction of SRF activity that is androgen-regulated suffices to mediate clinically relevant AR action in CaP. Genes that rely on SRF for androgen-responsiveness are enriched in CaP, distinguish benign from malignant prostate, correlate with aggressive disease and are associated with disease recurrence after surgery. To our knowledge, this mode of SRF and AR action represents the first discrete mechanism of androgen action in human disease and the first AR-dependent signaling cascade that is relevant to CaP progression. How this select and androgen-dependent segment of SRF action conveys aggressive behavior to CaP cells remains unknown. Here, the impact of sidekick-homolog 1 (SDK1) and calponin 2 (CNN2), two androgen-responsive SRF target genes that are overexpressed or underexpressed, respectively, consistently between benign and malignant prostate specimens, on CaP cell behavior is examined.
Materials and methods
Cell culture
Lymph Node Carcinoma of the Prostate (LNCaP) and Vertebral Cancer of the Prostate (VCaP) cells were purchased from American Type Culture Collection (ATCC, Manassas, VA). C4-2 cells were obtained from UroCor (Oklahoma City, OK). LNCaP refractory (LN-Rf) cells were generated in the laboratory of Dr. Donald Tindall (27). LNCaP and C4-2 cells were maintained in phenol red-free RPMI1640 medium (Life Technologies, Grand Island, NY) that is supplemented with 9% fetal bovine serum (FBS) (Life Technologies) and 1% Anti-Anti (Life Technologies). VCaP cells were grown in phenol red-free Dulbecco’s modified Eagle’s medium (high glucose, Life Technologies) that is supplemented with 9% FBS and 1% Anti-Anti. LN-Rf cells were maintained in phenol red-free RPMI1640 medium containing 9% charcoal-stripped FBS (CSS) and 1% Anti-Anti. All cell lines were kept in a 37°C incubator at 5% CO2. For experiments that involved androgen treatment, cells were cultured in medium supplemented with CSS. Cells that were used in transfection studies were seeded in medium without antibiotics.
Reagents
Methyltrienolone (R1881) was purchased from DuPont (Boston, MA). Casodex (csdx) and 5-α-dihydrotestosterone was obtained from Sigma-Aldrich (St. Louis, MO). Antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA) (AR, SRF, β1-integrin), Cell Signaling (Beverly, MA; β-catenin, claudin, E-cadherin, ZO-1, poly (ADP ribose) polymerase (PARP), β-actin) and Sigma-Aldrich (SDK1). The antibody directed against CNN2 has been described before (28). Staurosporine was obtained from Enzo Life Sciences (Plymouth Meeting, PA). siGENOME SmartPools targeting AR, SRF, CNN2 or SDK1, and non-targeting control SmartPools were purchased from Thermo-Scientific (Lafayette, CO).
Western blotting
Cells were washed twice with ice-cold phosphate-buffered saline (PBS, Life Technologies) and harvested in whole-cell lysis buffer [110 mmol/l sodium dodecyl sulfate (SDS), 100 mmol/l dithiothreitol, 80 mmol/l Tris-HCl (pH 6.9), 10% glycerol]. Cell lysates were boiled for 5min and stored at −20°C until analysis. Equal amounts of protein were subjected to NuPAGE Novex gel electrophoresis (Life Technologies). CNN2 protein analysis was performed using 10% Bis-Tris NuPAGE gels, whereas SDK1 was studied using Tris-Acetate NuPAGE gels. Proteins were blotted onto nitrocellulose membranes (Bio-Rad, Hercules, CA). Chemiluminescence detection was performed using SuperSignal West Femto Chemiluminescent Substrate (ThermoFisher Scientific, Waltham, WA). Blots were stripped with Restore Plus Western blot Stripping buffer (ThermoFisher Scientific) and reprobed with an antibody against β-actin to control for protein loading differences.
RNA isolation
Cells were seeded into six-well plates at a density of 3 × 105 cells. At the appropriate time after treatment and/or transfection, cells were harvested. Cells were washed twice with ice-cold PBS, and 1 ml of TriZol (Life Technologies) was added to each well of a six-well plate. Cells were scraped and cell lysate was collected. To the lysate, 200 μl of chloroform was added. Samples were inverted vigorously for 15 s, and then incubated at room temperature for 15 min. Samples were centrifuged at 12 000g at 4°C for 15 min. The top aqueous phase was removed and placed into a new RNAse-free 1.5ml Eppendorf tube, and the bottom organic layer was discarded. Next, 500 μl isopropanol was added to the samples and samples were incubated for 10 min at room temperature. Samples were then centrifuged at 4°C for 10 min. The supernatant was removed, 1 ml of 75% ethanol was added and the samples were centrifuged again at 4°C at 7 500 g for 5 min. RNA pellets were allowed to air-dry, and RNA was resuspended in RNAse-free water and stored at −80°C until analysis.
Real-time reverse transcription–PCR
Complementary DNA (cDNA) was prepared from 3 μg total RNA using a SuperScript III first-strand synthesis system (Life Technologies). Real-time reverse transcription–PCR (RT–PCR) was done using SYBR Green PCR mastermix (Life Technologies) on a ABI 7300 RT–PCR system. Primers targeting human SDK1, prostate-specific antigen and glyceraldehyde 3-phosphate dehydrogenase have been described (9,18). Primer sequences used to analyze CNN2 expression were 5′-CCTGTTTGAGAGTGGGAACA-3′ (forward primer) and 5′-GTACTTGACGCCAATGTCCA-3′ (reverse primer).
Small interfering RNA transfection
Cells were seeded into six-well plates at a density of 3 × 105 cells in medium without antibiotics. One day later, cells were transfected with 60 pmol of siGENOME SmartPool siRNA (ThermoFisher Scientific) directed against CNN2, SDK1 or a non-targeting control SmartPool siRNA per well using Lipofectamine 2000 (Life Technologies). Medium was changed 16 h later. In experiments assessing the effects of androgens, 42 h after transfection, medium was removed and cells were washed once with medium containing 9% CSS. Fresh medium supplemented with 9% CSS was added to the cells and cells were treated with R1881 (5 nM) or ethanol vehicle for the indicated periods of time.
Generation of human CNN2 promoter-reporter constructs
Construct CNN2wt-luc was generated by cloning a 982bp human CNN2 promoter fragment into pGL3 vector (Promega, Madison, WI). The 982bp fragment was generated via PCR using genomic DNA from LNCaP cells and primers 5′-CTTAGGTACCCCACCTCAGCCTCCTCAGTAG-3′ (forward primer) and 5′-GTATGCTAGCGTTGAACTGCGTGGAGCTCATGG-3′ (reverse primer). The resulting amplicon was cloned into the KpnI and NheI restriction enzyme recognition sites within the multiple cloning site of the pGL3 vector. CNN2mut-luc was made by performing QuikChange site-directed mutagenesis (Agilent Technologies, Santa Clara, CA) using the mutagenic primer pair 5′-TCGGCGCCTCCAGGCCTTATAAAAACATTTGCGCTC-3′ (forward) and 5′-GAGCGCAAATGTTTTTATAAGGCCTGGAGGCGCCGT-3′ (reverse). All constructs were sequenced to verify sequence integrity.
Transient transfection
For experiments using promoter-reporter constructs, cells were seeded in six-well plates at a density of 3 × 105 per well in medium without antibiotics. The next day, transfection mixtures were prepared. For each well, 1 μg of promoter-reporter construct was added to 250 μl of Optimem-I medium (Life Technologies) (mixture A). For studies that included the use of siRNA, 20 pmol of siGENOME SmartPool directed against SRF or non-targeting control SmartPool was included in mixture A. In a second reaction mixture, 1 μl of Lipofectamine 2000 transfection reagent (Invitrogen) was added to 250 μl of Optimem-I medium (mixture B). Both reaction mixtures were incubated for 5 min at room temperature after which time mixture A was added to mixture B and the combined mixture was incubated for 20min at room temperature before it was added to the cells. The next day, medium was replaced. For experiments that assessed androgen effects, cells were washed once with medium supplemented with 9% CSS, fresh CSS-supplemented medium was added and cells were treated with 5nM R1881 or ethanol vehicle. After 2 days, cells were washed with PBS and lysed in 250 μl passive lysis buffer (Promega, Madison, WI). Aliquots (10 μl) of cleared lysates were analyzed for luciferase activity (Promega). Bio-Rad Protein Assays were performed on cell lysates to control for potential differences in cell numbers.
Rhodamine staining
LNCaP, C4-2 and LN-Rf cells were seeded on cover slips at a density of 2 × 105 cells per well in six-well plates and transfected using siRNA SmartPools as described above. Ninety-six hours after transfection, cells were washed two times with PBS at 37°C. Cells were incubated in fixing solution (4% formaldehyde in PBS) for 30 min at room temperature and washed two times for 5 min each with PBS at room temperature. Cells were incubated in permeabilization buffer (0.5% Triton X-100 in PBS) for 15 min at room temperature and washed three times for 5 min in PBS. Cells were stained with rhodamine phalloidin (Cytoskeleton, Denver, CO) for 30 min according to the manufacturer’s instructions. Cells were mounted using VectaShield DAPI mounting medium (Vector Laboratories, Burlingame, CA). Slides were kept in the dark at 4°C and imaged using a Nikon Eclipse TE 2000-E microscope (×40 magnification).
Chromatin immunoprecipitation assay
LNCaP cells were seeded at a density of 2 × 106 cells in 10cm dishes in medium containing CSS. Cells were stimulated with R1881 (5 nM) or vehicle 2 days later. After 16 h, cells were treated with 1% formaldehyde for 10 min at 37°C. Cells were then washed with ice-cold PBS two times, collected in PBS containing Complete Protease Inhibitor Cocktail (Roche Diagnostics, Mannheim, Germany) on ice and spun for 4 min at 2 000 r.p.m. at 4°C. A volume of 225 μl of SDS lysis buffer [1% SDS, 5 mM ethylenediaminetetraacetic acid (EDTA), 50 mM Tris-HCl, pH 8.1] was used to resuspend the pellet. The resuspension was incubated on ice for 15 min, followed by sonication (15 times at 30 s ON/30 s OFF cycle on high setting using a Diagenode Bioruptor, Diagenode, Liege, Belgium). Samples were centrifuged for 10 min at 13 000 r.p.m. at 4°C. Supernatants were divided into 100 μl aliquots and diluted 10-fold in chromatin immunoprecipitation (ChIP) dilution buffer (1% Triton X-100, 2 mM EDTA, 150 mM NaCl, 20 mM Tris-HCl pH8.1) containing protease inhibitors. Ten microliters of diluted sample was removed as input and stored at −20°C until reversal of protein-DNA cross-links. Sixty microliters of Dynal magnetic beads (Life Technologies) were washed once with 1ml of ChIP dilution buffer and added to the diluted supernatant samples. Immunoprecipitation was done using antibodies directed against SRF or non-specific IgGs. The samples were incubated overnight with nutation at 4°C. Beads were pelleted by 1 min incubation on a prechilled DynaMag (Life Technologies) on ice. Beads were washed once with 1ml of low-salt wash buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20mM Tris-HCl pH 8.1, 150mM NaCl), once with 1 ml of high-salt wash buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20mM Tris-HCl pH 8.1, 500 mM NaCl), once with 1 ml of LiCl wash buffer (0.25 M LiCl, 1% NP-40, 1% deoxycholate, 1 mM EDTA, 10mM Tris-HCl pH 8.1) and twice with 1ml of Tris-EDTA buffer. The complex was eluted twice with 100 μl elution buffer (1% SDS, 100 mM NaHCO3) and eluates were pooled. The formaldehyde cross-linking was reversed by adding 8 μl 5M NaCl and 5 h incubation at 65°C. Incubation was continued for another 30 min at 37°C of 1 μl of 10mg/ml RNase A. A subsequent incubation was done at 45°C after addition for 2 h following addition of 1 μl of 10mg/ml Proteinase K. DNA was recovered using a PCR purification kit (Qiagen, Valencia, CA). DNA was eluted in 50 μl Tris-EDTA. Input samples were diluted 1/10. Primers used to amplify the CArG box-containing CNN2 promoter fragment were 5′-AAGTCCGCGGCCGCAGGAATT-3′ (forward) and 5′-AACTGCGTGGAGCTCATGGCT-3′ (reverse).
Migration assays
Cells were seeded at a density of 2 × 105 cells per well of a six-well plate and transfected with siRNAs targeting CNN2 or SDK1, or non-targeting control small interfering RNAs (siRNAs). The next day, medium was changed. After 24 h, cells were trypsinized and seeded into culture inserts at a density of 5 × 104 cells per side of the insert. Culture inserts were removed the next day, and images were captured using Infinity Capture software (Lumenera, Ottawa, ON, Canada) (×2.5 magnification) daily for the next 2 days. Quantification was done using Optimas software. Images were uploaded in their original size and format. The distance among the wound edges (gap width) was measured by drawing five straight lines from one edge to the other across the length of the wound. The lines were converted into numerical values using a hexadecimal option in Optimas.
MTS assays
Cells were seeded in 96-well tissue culture plates at a density of 1 × 104 per well in their regular medium without added antibiotics. The next day, cells were transfected with siGENOME SmartPool directed against CNN2 or SDK1, or a control SmartPool as described. After 16 h, medium was changed. At the indicated time points, cell proliferation was assessed by means of a CellTiter 96 Aqueous One solution cell proliferation assay (Promega). Values from five wells were measured per treatment group for each time point.
Replication of results
Results shown are representative of two or more independent experiments. Student’s t-tests were done to determine statistical significance of data shown in Figure 3B.
Fig. 3.
CNN2 and SDK1 regulate CaP cell morphology, organization of the actin cytoskeleton and migration. (A) LNCaP cells were transfected with non-targeting control siRNA (top row), or siRNA targeting the expression of CNN2 (middle row) or SDK1 (bottom row). After 48 or 96 h, cells were fixed and stained with rhodamine phalloidin (red) and counterstained with DAPI (blue). (B) Quantification of morphological CaP cell features following siRNA-mediated silencing of CNN2 or SDK1. Each slide was divided into four quadrants and 12 or 13 random cells per quadrant (i.e. 50 cells in total) were evaluated for cell features such as the number of nuclei, presence of cell extensions, number of cell extensions, presence of stress fibers and number of fibers that are present. A more detailed description of individual cell data is provided in Supplementary Tables 2–4, available at Carcinogenesis Online. Student’s t-tests were done to determine statistical significance between control and CNN2 siRNA groups and between control and SDK1 siRNA groups. (C) LNCaP cells were transfected with non-targeting control siRNA, or siRNAs directed against CNN2 or SDK1 expression. The next day, cells were seeded into culture inserts. One day later, inserts were removed (day 0). Cell migration was evaluated at days 1, 2, and 3. (D) Quantification of wound closure. Black columns, cells transfected with non-targeting control siRNA; gray columns, cells transfected with siRNA targeting CNN2 (top panel) or SDK1 (bottom panel). Columns, means of values from five measurements; bars, standard error of the mean. Cell proliferation was assessed in parallel using MTS assays at day 1 as before (right panels).
Results
Expression of CNN2 and SDK1 is androgen-responsive
CNN2 and SDK1 belong to a 158-gene signature that was identified to be androgen-responsive in an SRF-dependent manner using an oligoarray screening approach (10). Both genes are part of a subset of genes that are consistently differentially expressed between malignant and benign prostate in CaP messenger RNA (mRNA) profiling studies, which used different methods of tissue procurement, RNA isolation and relied on diverse cDNA or oligoarray platforms (Supplementary Table 1, available at Carcinogenesis Online) (10,29–35). Although the manner of androgen regulation of a subset of these genes has been validated in detail (10), whether CNN2 and SDK1 represent bona fide examples of genes that rely on SRF to achieve full androgen-responsiveness remained to be determined. Treatment of the AR-positive CaP cell line LNCaP with the synthetic androgen R1881 led to marked increases in the expression of CNN2 and SDK1. These stimulatory effects were present at the mRNA and protein expression level (Figure 1A). Similar observations were done using the independent AR-positive cell line VCaP (Supplementary Figure 1, available at Carcinogenesis Online) or the natural ligand dihydrotestosterone (Supplementary Figure 2, available at Carcinogenesis Online). The specificity of the antibodies is shown in Supplementary Figure 3, available at Carcinogenesis Online. The AR dependency of these effects was verified by combining androgen treatment with siRNA directed against AR. As shown in Figure 1B, real-time RT–PCR and western blot analyses demonstrated that knockdown of AR prevents androgen induction of both CNN2 and SDK1. Pharmacological inhibition of AR using the antiandrogen bicalutamide (Casodex, csdx) also inhibited androgen-responsiveness of these SRF target genes (Figure 1C) at the mRNA and protein level. These data confirm that expression of CNN2 and SDK1 is subject to androgen regulation.
Fig. 1.
Expression of CNN2 and SDK1 is androgen regulated in an SRF-dependent manner. (A) LNCaP cells were treated with 5nM of the synthetic androgen R1881 (+) or ethanol vehicle (−) for 48 h. Cells were harvested and protein and RNA were isolated. RNA was converted into cDNA and real-time RT–PCR was done using primers that target CNN2 and SDK1 expression. Expression values were normalized to glyceraldehyde 3-phosphate dehydrogenase expression levels and expressed as relative expression, where the value obtained from one of the vehicle-treated samples was taken as 1. Black columns, vehicle-treated samples; gray columns, R1881-treated samples. Columns, means of values obtained from three independent biological replicates; bars, standard error of the mean values (top panel). Western blotting was performed using antibodies directed against CNN2 and SDK1. Blots were reprobed for β-actin (β-act) to control for loading differences (bottom panel). (B) LNCaP cells were transfected with siRNA directed against AR or a non-targeting control (c) siRNA. One day later, medium was replaced by CSS-supplemented medium. The next day, medium was replaced and cells were treated with vehicle or R1881 (5nM). After 48 h, cells were harvested for RNA and protein isolation. Real-time RT–PCR using primers directed against CNN2 and SDK1 was performed as described (left panel). Western blotting was performed with antibodies directed against CNN2, SDK1, AR and β-actin (β-act) (right panel). (C) LNCaP cells were treated with ethanol vehicle, R1881 (1nM), Casodex (10 μM, csdx) or R1881 and an excess of Casodex for 48 h. Cells were harvested for RNA, and cDNA synthesis followed by real-time RT–PCR was performed (left panel). LNCaP cells were treated with R1881 or Casodex at various concentrations either alone or in combination, and whole-cell protein extracts were subjected to western blotting using antibodies directed against CNN2 or SDK1 as above (right panel). (D) LNCaP cells were treated with 5nM R1881 or vehicle and harvested for RNA isolation 4 or 16 h later. Real-time RT–PCR was performed using primers directed against CNN2, SDK1 orprostate-specific antigen as above (top panel). LNCaP cells were treated with vehicle or R1881 (5 nM) and harvested after 0, 4, 8, 16, 24, 48 and 72 h. Western blot analysis was performed using antibodies directed against CNN2, SDK1 and β-actin (β-act) (bottom panel). (E) LNCaP cells were transfected with non-targeting siRNAs or siRNAs directed against SRF. One day later, medium was replaced by CSS-supplemented medium. The next day, medium was replaced and cells were treated with vehicle or R1881 (5nM). After 48 h, cells were harvested for protein and RNA isolation, and western blotting (right panel) and real-time RT–PCR (left panel) were done to evaluate CNN2 and SDK1 expression levels. (F) LNCaP cells were treated with either vehicle or R1881 (5nM) for 16 h. ChIP assays were performed as before using an antibody directed against SRF or non-targeting IgG. CNN2, CArG box containing CNN2 promoter fragment; control, similarly sized non-CArG box-containing exonic CNN2 gene fragment. (G) Graphical representation of the structure of the CNN2 promoter-reporter constructs. CNN2wt-luc, wild-type construct containing a 982 bp CNN2 promoter fragment that harbors a CArG box 133 base pairs upstream of the transcriptional start site; CNN2mut-luc, mutant construct in which the CArG box has been mutated. (H) LNCaP cells were transfected with CNN2wt-luc or CNN2mut-luc and treated with vehicle, R1881 and/or Casodex (csdx) for 48 h (left panel and right panel), with vehicle or R1881 in the presence of non-targeting siRNA or SRF-directed siRNA (middle panel). The next day, cells were treated with ethanol vehicle or R1881 (5nM). After 48 h, a luciferase assay was done. Columns, means of values obtained from three independent biological replicates; bars, standard error of the mean.
Androgen regulation of CNN2 and SDK1 is SRF-dependent
Androgen regulation of SRF activity constitutes an indirect mechanism of androgen action, which does not involve direct interaction of AR with its consensus binding site, an androgen response element (36,37). Indirect mechanisms of androgen action rely on an intermediate factor to convey androgen-responsiveness to target genes and are characterized by slower kinetics than direct mechanisms (38,39). Kinetics studies in which LNCaP cells were harvested for protein and RNA isolation at various time points after androgen stimulation were performed. These experiments showed that androgen induction of prostate-specific antigen, a direct androgen response element-driven AR target gene was evident already after 4 h. At that time point, expression of CNN2 and SDK1 was not yet affected; this required 16 h of androgen exposure, which is consistent with an indirect mechanism of action. These observations were done at both the mRNA and protein level (Figure 1D). Combining androgen treatment with an siRNA that specifically targets SRF led to a complete or severe inhibition of the androgen induction of CNN2 and SDK1, which corroborates the involvement of SRF in this indirect mechanism of androgen action (Figure 1E). The mechanism by which SRF regulates androgen-responsiveness of CNN2 and SDK1 was explored further by examining the promoter regions of the corresponding human genes for the presence of putative SRF-binding CArG boxes (12). In the case of CNN2, a motif that matched perfectly the consensus binding site for SRF (CCTTATAAGG) was identified 133bp upstream of the CNN2 transcriptional start site. ChIP experiments confirmed binding of SRF to this site but not to a non-specific region in the CNN2 coding region, both in the presence and absence of androgens (Figure 1F). Androgen treatment of LNCaP cells that were transfected with a reporter construct driven by a 982bp CNN2 promoter region that harbors the CArG box (Figure 1G, CNN2wt-luc) led to an increase in luciferase reporter activity. The level of androgen regulation achieved was similar to that observed for androgen induction of CNN2 mRNA levels. Addition of Casodex or siRNAs that are targeted specifically against SRF blunted androgen stimulation of reporter gene activity. A similar inhibition was seen following mutation of the CArG box in the CNN2 promoter in residues that are essential for SRF binding (Figure 1G and 1H). These findings confirmed that androgen regulation of CNN2 and SDK1 expression relies on SRF.
CNN2 and SDK1 do not affect CaP cell proliferation or apoptosis
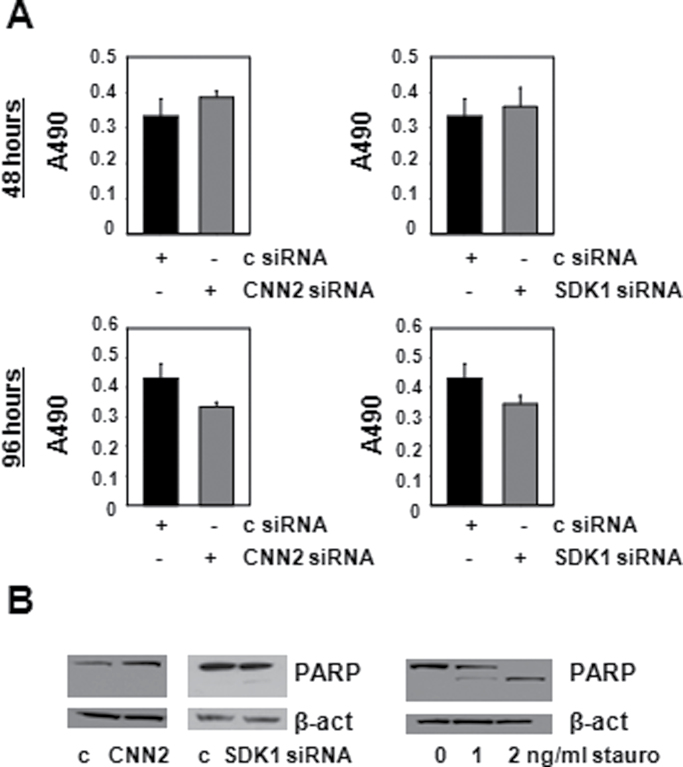
Our profiling studies were the first to implicate dysregulation of CNN2 and SDK1 expression in CaP (progression). The manner in which alterations in the expression of these genes affect CaP cell behavior is unknown. To address this question, the impact of CNN2 and SDK1 on CaP cell proliferation was investigated. LNCaP cells were transfected with siRNAs targeting CNN2 or SDK1, or non-targeting control siRNAs, and MTS assays were performed 48 and 96 h later. As shown in Figure 2A, silencing of CNN2 (left panel) or SDK1 (right panel) did not affect cell proliferation at either time points. These results were validated in independent cell-count experiments (data not shown). Loss of CNN2 or SDK1 did not notably affect cleavage of full-length (116kDa) poly (ADP ribose) polymerase (PARP), a widely recognized marker for cells undergoing apoptosis. This was not due to technical limitations in detecting PARP cleavage, as the same amount of cellular proteins obtained from cells induced to undergo apoptosis upon treatment with staurosporine yielded 89-kDa-cleaved PARP fragments that were readily detectable (Figure 2B). These findings rule out important roles for CNN2 and SDK1 in CaP cell proliferation or apoptosis.
Fig. 2.
CNN2 and SDK1 do not regulate CaP cell proliferation or apoptosis. (A) LNCaP cells were transfected with non-targeting control (c) siRNA (black columns) or siRNA directed against CNN2 and SDK1 (gray columns). Cell proliferation was assessed by performing MTS assays 48 h (top panels) and 96 h (bottom panels) after transfection under regular FBS-supplemented culture conditions. Absorbance at 490 nm (A490) was read. Columns, means of values from five biological replicates; bars, standard error of the mean values. (B) LNCaP cells were transfected with non-targeting control siRNAs (c) or siRNAs directed against CNN2 and SDK1. Ninety-six hours after transfection, cells were harvested for protein isolation and western blotting was performed using antibodies against PARP and β-actin (β-act) (left panel). Protein extracts from LNCaP cells that were treated with staurosporine (stauro) were subjected to western blotting using antibodies directed against PARP and β-actin (β-act) (right panel).
CNN2 and SDK1 induce changes in cell morphology and actin cytoskeleton
To explore further the role for CNN2 and SDK1 in CaP cell biology, their effects on cell morphology were studied. To this end, siRNA-mediated silencing of CNN2 or SDK1 was combined with rhodamine phalloidin staining, which is used to image the actin cytoskeleton. Figure 3A shows that loss of CNN2 led to distinct morphological changes: a marked increase in the number of cell protrusions was noted, as well as a general disorganization of the actin cytoskeleton (Figure 3A, middle row). These effects were present 48 h after transfection and were more pronounced at 96 h. Knockdown of SDK1, on the other hand, yielded a very different phenotype: cells were rounder and stress fibers became evident (Figure 3A, bottom row). These effects were most notable 96 h after transfection. Similar morphological changes were observed in VCaP cells following silencing of CNN2 and SDK1 (data not shown). Figure 3B summarizes and quantifies the effects of CNN2 or SDK1 silencing on features such as the percentage of cells with extensions, the average number of extensions per cell, the number of cells with above average number of extensions and the percentage of cells that display stress fibers, at the 96 time point. Upon quantification, loss of CNN2 was found to increase the average number of extensions per cell from 3 to 5, with 62% of cells that had lost CNN2 expression showing more than three extensions compared with 30% of control transfected cells. Similarly, upon silencing of SDK1 expression, 52% of cells showed pronounced intracellular actin filaments compared with 10% of control cells (Supplementary Tables 2–4, available at Carcinogenesis Online). Taken together, these findings point toward pronounced and differential effects of CNN2 and SDK1 on CaP cell morphology and actin cytoskeleton organization.
CNN2 and SDK1 affect CaP cell migration
In view of the importance of the actin cytoskeleton for cell movement and contractility, the possibility that CNN2 and SRF may play a role in CaP cell migration was explored. Following transfection with siRNA that is targeted against CNN2 or SDK1, or non-targeting siRNA, LNCaP cells were studied in wound-healing assays using tissue culture inserts. As shown in Figure 3C (top panel), upon loss of CNN2, wounds closed faster than under control conditions. By day 3 after culture insert removal, the gap was almost completely closed. In contrast, cells in which expression of SDK1 was silenced (bottom panel) displayed a slower gap closure than control cells. The impact of loss of CNN2 and SDK1 on cell migration was obvious already at day 1, when, consistent with observations described above, MTS assays done in parallel studies showed no effects on cell proliferation (Figure 3D, right panel). These data indicate that CNN2 limits CaP cell migration, whereas SDK1 induces CaP cell migration, and that these effects are not due to alterations in CaP cell proliferation.
Effects of CNN2 and SDK1 on actin cytoskeleton are associated with alterations in the expression levels of β1-integrin
Because CNN2 and SDK1 are involved in CaP cell migration, efforts were directed toward identifying the molecular determinants that underlie SRF target gene–dependent changes in cellular movements. Epithelial-mesenchymal transition (EMT) is a development program in which cells move through reduced cell adhesion and increased cell motility, and has been proposed to be involved also in the spread of malignant epithelial cells (40). EMT is assessed routinely through evaluation of molecular markers that mediate and/or control cell adhesion (e.g. E-cadherin, beta-catenin) or tight junctions (e.g. claudin-1, ZO-1). Protein extracts from LNCaP cells that had been transfected with siRNAs directed against CNN2 or SDK1, or non-targeting control siRNA, were analyzed in immunoblotting studies using antibodies against various markers of EMT, including β-catenin, claudin-1, E-cadherin, and ZO-1. As shown in Figure 4A, no changes in the expression of the EMT-related proteins were noted upon knockdown of CNN2 or SDK1, which indicates that neither SRF target gene is involved in EMT and that EMT does not mediate the observed changes in CaP cell migration.
Fig. 4.

Effect of CNN2 and SDK1 silencing on expression of EMT markers and β1-integrin. LNCaP cells were seeded and transfected with non-targeting control (c) siRNAs, or siRNAs directed against CNN2 or SDK1 expression. Four days after transfection, cells were harvested and western blot analysis was performed using antibodies directed against the EMT markers β-catenin, claudin-1, E-cadherin and ZO-1 (A), β1-integrin (B) and β-actin (β-act).
As EMT markers were not altered following CNN2- or SDK1-dependent changes in CaP cell migration, the involvement of other potential mediators of actin cytoskeleton organization or cell movement was explored. Integrins are a family of receptor proteins that function to regulate cell adhesion and cell migration (41,42). Following siRNA-mediated silencing of CNN2 or SDK1, LNCaP cell extracts were analyzed for the expression levels of β1-integrin, an integrin family member for which upregulated expression has been associated with increased CaP cell migration before (43). Following silencing of CNN2, an increase in β1-integrin protein expression was observed (Figure 4B). Interestingly, parallel with the minimal changes in cell movement, no alterations in β1-integrin expression were found upon loss of SDK1. These findings rule out EMT as a determinant of SRF target gene–dependent CaP migration but implicate β1-integrin in this process.
CNN2 and SDK1 action remains relevant in CaP throughout disease progression
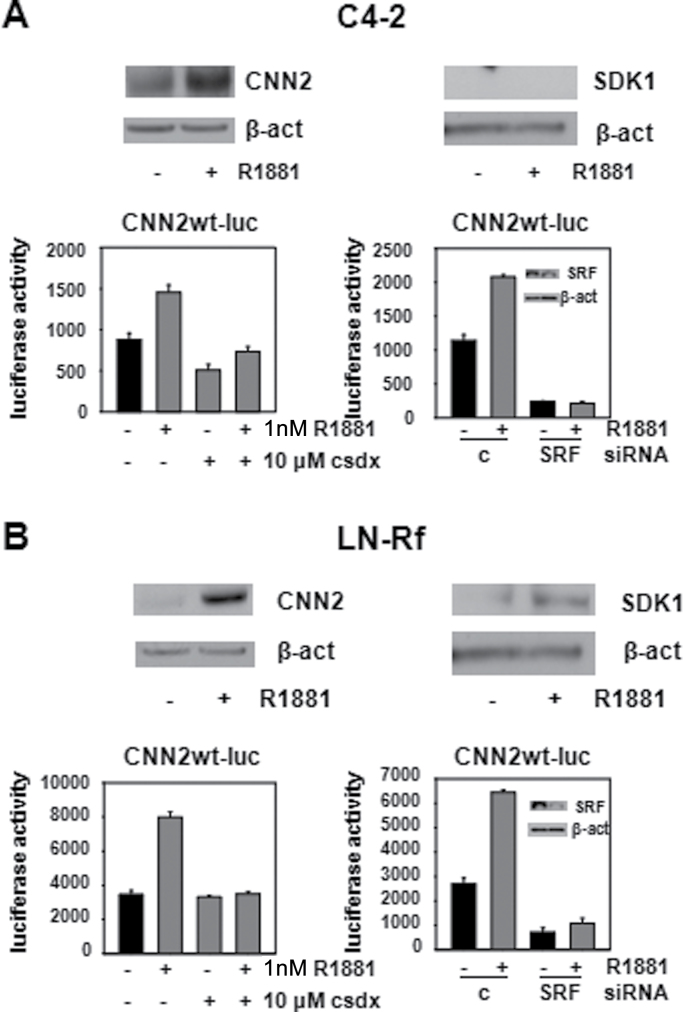
Androgen-dependent SRF action is associated with aggressive CaP disease and recurrence after surgical treatment (10). Therefore, the relevance of CNN2 and SDK1 to CR-CaP was explored. To this end, two isogenic cell line models that represent the progression from androgen-stimulated to CR-CaP and were derived following long-term androgen ablation of LNCaP cells in vivo (C4-2) (44) or in vitro (LN-Rf) (27), were used. Androgen treatment of both C4-2 and LN-Rf cells induced CNN2 expression, whereas androgen regulation of SDK1 was observed only in LN-Rf cells. In C4-2 cells cultured under androgen-deprived conditions, SDK1 fell below the level of detection (Figure 5A and B, top panels), which accounts for the lack of androgen regulation of SDK1 in C4-2 cells. Androgen-responsiveness of the human CNN2 promoter-reporter construct was maintained in C4-2 and LN-Rf cells, and use of the antiandrogen Casodex attenuated androgen regulation of reporter gene activity also in these CR-CaP cells. siRNA-mediated loss of SRF expression led to a decrease in basal reporter gene activity and prevented androgen regulation of luciferase activity (Figure 5A and B, bottom panels). Taken together, these findings demonstrated that androgen regulation of CNN2 and SDK1 is maintained in CR-CaP cells.
Fig. 5.
Expression of CNN2 and SDK1 is androgen regulated in an SRF-dependent manner in CR-CaP cell lines. (A) C4-2 cells were treated with vehicle or R1881 (5nM) for 96 h. Cells were harvested, total protein was isolated and subjected to western blotting using antibodies directed against CNN2, SDK1 and β-actin (β-act) (top panel). C4-2 cells were transfected with CNN2wt-luc and treated with vehicle, R1881 (1nM) and/or an excess of Casodex (csdx) (bottom panel, left), or with vehicle or R1881 (5nM) in the presence of non-targeting control siRNA or siRNA targeting SRF (bottom panel, right). After 2 days, cells were harvested and a luciferase activity was done. Columns, means of values obtained from three independent biological replicates; bars, standard error of the mean. Inset, western blotting control for the efficiency of siRNA-mediated SRF silencing. (B) The experiments described under (A) were performed using LN-Rf cells.
Rhodamine phalloidin staining was performed on C4-2 and LN-Rf cells in which expression of CNN2 or SDK1 had been silenced to determine if the effects on cell morphology and actin cytoskeleton that were observed in the androgen-stimulated LNCaP cell line carried over to its CR sublines. Figure 6 shows that, reminiscent of observations for LNCaP cells, in both C4-2 and LN-Rf cells, compared with control condition, knockdown of CNN2 induced cellular protrusions and disorganization of the cytoskeleton disorganization. Conversely, knockdown of SDK1 promoted a rounded-cell phenotype, as seen also in LNCaP cells. These observations confirm the androgen-responsiveness of CNN2 and SDK1 and their roles in regulation of CaP cell morphology in CR-CaP model systems.
Fig. 6.
CNN2 and SDK1 regulate organization of the actin cytoskeleton and cell migration in CR-CaP cells. C4-2 cells (A) and LN-Rf (B) cells were seeded on cover slips and transfected with non-targeting control siRNA (left image), or siRNAs targeted against CNN2 (middle image) and SDK1 (right image). After 96 h, cells were fixed and stained with rhodamine phalloidin as described (left panel). Protein extracts from C4-2 and LN-Rf cells that were transfected as above were analyzed via western blotting using antibodies directed against CNN2, SDK1 or β-actin (β-act) (right panel).
Discussion
SRF is one of the first isolated and best-characterized mammalian transcription factors. Although its role in development and physiology is well studied and understood increasingly, the relevance of SRF for human disease is largely unknown. Previous work from our laboratory identified SRF as a critical mediator of clinically relevant androgen action in CaP (10). Androgen activation of SRF represents the first discrete mode of SRF action in human neoplasms, and in CaP, it is the first mechanism of androgen action that is relevant to disease progression. Since the identification of SRF as a critical mediator of AR action, its importance to CaP development and progression has been validated by other research groups that have isolated SRF as a candidate driver of CaP development and implicated SRF in the emergence of disease recurrence after radical prostatectomy (26,45).
Although AR is the major target for treatment for patients with non-organ-confined CaP or CaP that recurs after initial surgery or radiation treatment with curative intent, the androgen-dependent events that drive CaP progression and insights into the molecular mechanisms by which androgens control these events remain largely elusive. Lack of this information contributes to the inevitable failure of ADT. CR-CaP that re-emerges during ADT accounts for the vast majority of CaP-related mortality, which is estimated at 28 170 deaths in the USA in 2012 (46), while continuing to rely on AR for growth (3,4,8,47). Identifying the upstream regulators that are responsible for conveying androgen-responsiveness to SRF and the manner in which effector genes regulate aggressive CaP cell behavior may identify novel means to interfere with the critical aspects of AR action that drives CaP progression, remain critical during progression from androgen-stimulated to CR-CaP and are not inhibited fully by current ADT.
Here, the contribution and relevance of two representative androgen-responsive SRF target genes, CNN2 and SDK1, to CaP behavior and progression were studied. Following an oligoarray profiling approach that combined information from CaP model systems and clinical specimens, CNN2 and SDK1 were identified as members of a 158 SRF- and AR-dependent gene signature and their expression was found to be decreased or increased, respectively, in CaP samples compared with benign prostate epithelium (10). Dysregulation of CNN2 and SDK1 expression in CaP was recognized originally in expression profiles that were derived from laser-captured microdissected prostate tissues and validated in multiple independent datasets that used diverse tissue dissection techniques, RNA procurement methods and different mRNA profiling platforms. Prior to this work, neither CNN2 nor SDK1 was known to be relevant to CaP cell biology or to be subject to androgen regulation. CNN2 is an actin-binding protein that inhibits actomyosin ATPase and stabilizes the actin cytoskeleton (48,49). Downregulation of the expression of the CNN2 homolog and calponin family member CNN1 contributes to the dysregulated expression of a 17 gene signature that mediates metastasis in adenocarcinoma, including CaP (50). SDK1 was characterized as an adhesion molecule with important roles in cell fate of retinal photoreceptors in D. melanogaster. In human disease, expression of SDK1 has been found to be upregulated in HIV-associated nephropathy and focal segmental glomerulosclerosis, and altered SDK1 expression has been associated with increased intercellular adhesion and loss of cytoskeletal integrity (51,52). The association of SDK1 with cancer is unknown.
Validating the oligoarray-based screening approach that was performed in LNCaP cells (10), this study confirms that expression of CNN2 and SDK1 is induced by androgens in an SRF-dependent manner in CaP model systems that mimic the progression from androgen-stimulated to CR disease. In the case of CNN2, androgen regulation was mediated through binding of SRF to a CArG box in its proximal promoter. Presence of a functional CArG box in the promoter region of the CNN2 gene has been described in other model systems derived from multiple species (14,17). In contrast, the promoter region of the gene encoding SDK1 did not contain a domain that resembles a consensus SRF binding site. Ongoing analysis of an SRF ChIP-chip dataset that was generated in our laboratory, however, has identified an SRF binding peak in an enhancer region more than 10 kb upstream of the SDK1 transcriptional start site (data not shown). The presence of a genomic SRF binding site at a location that allows for control over SDK1 transcription is in line with the SRF dependence and kinetics of androgen induction of SDK1 expression in CaP cells.
It should be noted that although expression of both CNN2 and SDK1 is induced following androgen exposure, in clinical specimens expression of CNN2 is decreased in CaP compared with benign prostate, whereas expression of SDK1 is increased. These expression patterns may be related to the specific molecular mechanisms by which SRF conveys androgen regulation to CNN2 and SDK1. SRF toggles among transcriptional programs and recruits different cofactors to modulate expression of individual SRF target genes (13,14). Differential recruitment of select cofactors could govern also androgen regulation of SRF target gene expression. Recent work from our laboratory has demonstrated that androgen regulation of most, but not all, SRF target genes is mediated by the RhoA signaling cascade, and RhoA-dependent recruitment of the SRF cofactor MAL to select CArG boxes (18). Androgen regulation of SDK1, but not CNN2, was under RhoA control (data not shown) (18). The manner in which activity of the RhoA signaling axis and other SRF cofactors or mediators of SRF action that control androgen regulation of SRF target genes is maintained or evolves during CaP progression may account for their expression levels in clinical specimens. Alternatively, additional SRF-independent regulation that contributes to SRF effector gene expression cannot be ruled out and may become more relevant for individual SRF target gene expression during CaP progression.
Individual silencing of the SRF effector genes CNN2 and SDK1 had pronounced and remarkable effects on CaP cell morphology, actin cytoskeleton organization and cell migration. In line with the expression patterns of CNN2 and SDK1 in clinical CaP specimens, loss of CNN2 promoted cell migration, whereas silencing of SDK1 blunted cell migration. Rearrangements in the actin cytoskeleton are vital for cancer cells to invade the surrounding stromal tissue, migrate and generate metastatic lesions. Involvement of SRF target genes such as CNN2 and SDK1 with CaP migratory behavior probably underlies the correlation of the SRF-dependent mechanism of androgen action with aggressive CaP. This correlation was defined as an increase in Gleason pattern number, which reflects increased divergence from the architecture of a benign prostate gland and takes into account shape and size of the gland, arrangement of cells within the gland and stromal invasion, and the presence of lymph node metastasis at the time of surgery (10). Lack of association between CNN2- and SDK1-induced changes in cell migration, alterations in molecular markers for EMT and the correlation with β1-integrin expression are consistent with previous literature reports. Experimental metastasis and underlying cytoskeletal changes depend on SRF in model systems for breast cancer and melanoma but are not accompanied by changes in EMT markers (21). The concept that mechanisms other than EMT can control CaP invasiveness is gaining momentum, as recently reviewed (53). Similarly, involvement of β1-integrin, for which upregulated expression in CaP has been associated with increases in CaP cell migration (43), has been reported downstream of SRF action (54).
The pronounced effects on CaP cell migration following alteration in expression levels of individual SRF target genes are remarkable and may have implications for therapeutic intervention. These observations suggest that, in addition to targeting the signaling upstream of SRF that mediates its androgen-responsiveness, interference with the activity of one or more SRF effectors may be effective also in blocking the androgen-dependent mechanism of SRF action in CaP cells. In this respect, ectopic expression of multiple copies of a short actin-binding module, known as a calponin-like repeat, that is found in calponin family members has been shown to reduce cell motility and colony formation in human adenocarcinoma cells, irrespective of major concomitant effects on cell proliferation (55). When considering targeting SRF target genes for CaP therapy, it should be noted that in addition to genes that function in organization of the actin cytoskeleton, this 158 gene signature also contains genes involved in lipid synthesis, transcription and protein synthesis, which have been shown to be important in CaP, as well as several genes involved in ion homeostasis, which may be viable pharmacological targets (10).
The preferential involvement of CNN2 and SDK1 in the regulation of CaP cell migration rather than proliferation may apply also to other SRF target genes. A small screen of other androgen-responsive SRF effector genes did not reveal notable effects on CaP cell proliferation, suggesting potential roles in other CaP-relevant processes such as cell migration and adhesion (18). These findings are consistent also within the emerging consensus that the relevance of SRF to development and physiology is due primarily to its roles in organization of actin cytoskeleton and cell contractility (14). Connecting the observations between androgen-responsiveness of CNN2 and SDK1 and their effects on CaP cell motility, ongoing studies in our laboratory corroborate a role for these genes also in androgen-dependent CaP cell migration. Consistent with literature reports, androgen exposure stimulated LNCaP cell migration in wound-healing assays. Loss of CNN2 and SDK1 has effects similar to those observed for cells cultured under regular FBS-supplemented conditions: CNN2 decreased androgen-dependent cell migration, whereas SDK1 functioned to increase. Silencing CNN2 or SDK1 did not affect androgen-dependent CaP cell proliferation (data not shown).
In conclusion, this study demonstrates that the association of the androgen-dependent fraction of SRF action with aggressive CaP is mediated by regulation of CaP cell migration. These findings indicate also that interference with the action of one or more androgen-responsive SRF target genes may be a viable approach to block clinically relevant androgen action downstream of SRF.
Supplementary material
Supplementary Tables 1–4 and Figures 1–3 can be found at http://carcin.oxfordjournals.org/
Funding
Department of Defense Prostate Cancer Research Program; the Roswell Park Alliance Foundation; the National Institutes of Health [CA 01605].
Conflict of Interest Statement: None declared.
Supplementary Material
Glossary
Abbreviations:
- ADT
androgen deprivation therapy
- AR
androgen receptor
- CaP
prostate cancer
- csdx
Casodex
- cDNA
complementary DNA
- ChIP
chromatin immunoprecipitation
- CNN2
calponin 2
- CR-CaP
castration-recurrent CaP
- CSS
charcoal-stripped FBS
- EDTA
ethylenediaminetetraacetic acid
- EMT
epithelial-mesenchymal transition
- FBS
fetal bovine serum
- LNCaP
Lymph Node Carcinoma of the Prostate
- LN-Rf
LNCaP refractory
- PARP
poly (ADP ribose) polymerase
- RT–PCR
reverse transcription–PCR
- SDS
sodium dodecyl sulfate
- SDK1
sidekick-homolog 1
- siRNAs
small interfering RNAs
- SRF
Serum Response Factor; VCaP, Vertebral Cancer of the Prostate.
References
- 1. Miyamoto H., et al. (2004). Androgen deprivation therapy for prostate cancer: current status and future prospects. Prostate, 61, 332–353 [DOI] [PubMed] [Google Scholar]
- 2. Ryan C.J., et al. (2011). Androgen receptor rediscovered: the new biology and targeting the androgen receptor therapeutically. J. Clin. Oncol., 29, 3651–3658 [DOI] [PubMed] [Google Scholar]
- 3. de Bono J.S., et al. ; COU-AA-301 Investigators (2011). Abiraterone and increased survival in metastatic prostate cancer. N. Engl. J. Med., 364, 1995–2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Scher H.I., et al. ; AFFIRM Investigators (2012). Increased survival with enzalutamide in prostate cancer after chemotherapy. N. Engl. J. Med., 367, 1187–1197 [DOI] [PubMed] [Google Scholar]
- 5. Ryan C.J., et al. ; COU-AA-302 Investigators (2013). Abiraterone in metastatic prostate cancer without previous chemotherapy. N. Engl. J. Med., 368, 138–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cai C., et al. (2011). Intratumoral de novo steroid synthesis activates androgen receptor in castration-resistant prostate cancer and is upregulated by treatment with CYP17A1 inhibitors. Cancer Res., 71, 6503–6513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mostaghel E.A., et al. (2011). Resistance to CYP17A1 inhibition with abiraterone in castration-resistant prostate cancer: induction of steroidogenesis and androgen receptor splice variants. Clin. Cancer Res., 17, 5913–5925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Richards J., et al. (2012). Interactions of abiraterone, eplerenone, and prednisolone with wild-type and mutant androgen receptor: a rationale for increasing abiraterone exposure or combining with MDV3100. Cancer Res., 72, 2176–2182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Heemers H.V., et al. (2007). Androgen induction of the androgen receptor coactivator four and a half LIM domain protein-2: evidence for a role for Serum Response Factor in prostate cancer. Cancer Res., 67, 10592–10599 [DOI] [PubMed] [Google Scholar]
- 10. Heemers H.V., et al. (2011). Identification of a clinically relevant androgen-dependent gene signature in prostate cancer. Cancer Res., 71, 1978–1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shore P., et al. (1995). The MADS-box family of transcription factors. Eur. J. Biochem., 229, 1–13 [DOI] [PubMed] [Google Scholar]
- 12. Sun Q., et al. (2006). Defining the mammalian CArGome. Genome Res., 16, 197–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Miano J.M. (2003). Serum Response Factor toggling between disparate programs of gene expression. J. Mol. Cell. Cardiol., 35, 577–593 [DOI] [PubMed] [Google Scholar]
- 14. Miano J.M., et al. (2007). Serum Response Factor master regulator of the actin cytoskeleton and contractile apparatus. Am. J. Physiol. Cell Physiol., 292, C70–C81 [DOI] [PubMed] [Google Scholar]
- 15. Cooper S.J., et al. (2007). Serum Response Factor binding sites differ in three human cell types. Genome Res., 17, 136–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sun Q., et al. (2009). Myocardin-dependent activation of the CArG box-rich smooth muscle gamma-actin gene: preferential utilization of a single CArG element through functional association with the NKX3.1 homeodomain protein. J. Biol. Chem., 284, 32582–32590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sullivan A.L., et al. (2011). Serum Response Factor utilizes distinct promoter- and enhancer-based mechanisms to regulate cytoskeletal gene expression in macrophages. Mol. Cell. Biol., 31, 861–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Schmidt L.J., et al. (2012). RhoA as a mediator of clinically relevant androgen action in prostate cancer cells. Mol. Endocrinol., 26, 716–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Treisman R. (1987). Identification and purification of a polypeptide that binds to the c-fos serum response element. EMBO J., 6, 2711–2717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Miano J.M. (2010). Role of Serum Response Factor in the pathogenesis of disease. Lab. Invest., 90, 1274–1284 [DOI] [PubMed] [Google Scholar]
- 21. Medjkane S., et al. (2009). Myocardin-related transcription factors and SRF are required for cytoskeletal dynamics and experimental metastasis. Nat. Cell Biol., 11, 257–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Park M.Y., et al. (2007). Expression of the Serum Response Factor in hepatocellular carcinoma: implications for epithelial-mesenchymal transition. Int. J. Oncol., 31, 1309–1315 [PubMed] [Google Scholar]
- 23. Kim H.J., et al. (2009). The expression and role of serum response factor in papillary carcinoma of the thyroid. Int. J. Oncol., 35, 49–55 [PubMed] [Google Scholar]
- 24. Choi H.N., et al. (2009). Serum response factor enhances liver metastasis of colorectal carcinoma via alteration of the E-cadherin/beta-catenin complex. Oncol. Rep., 21, 57–63 [PubMed] [Google Scholar]
- 25. Bai S., et al. (2009). MicroRNA-122 inhibits tumorigenic properties of hepatocellular carcinoma cells and sensitizes these cells to sorafenib. J. Biol. Chem., 284, 32015–32027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yu W., et al. (2011). FGFR-4 Arg388 enhances prostate cancer progression via extracellular signal-related kinase and Serum Response Factor signaling. Clin. Cancer Res., 17, 4355–4366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Murillo H., et al. (2001). Role of PI3K signaling in survival and progression of LNCaP prostate cancer cells to the androgen refractory state. Endocrinology, 142, 4795–4805 [DOI] [PubMed] [Google Scholar]
- 28. Hossain M.M., et al. (2003). Developmentally regulated expression of calponin isoforms and the effect of h2-calponin on cell proliferation. Am. J. Physiol. Cell Physiol., 284, C156–C167 [DOI] [PubMed] [Google Scholar]
- 29. Luo J., et al. (2001). Human prostate cancer and benign prostatic hyperplasia: molecular dissection by gene expression profiling. Cancer Res., 61, 4683–4688 [PubMed] [Google Scholar]
- 30. Vanaja D.K., et al. (2003). Transcriptional silencing of zinc finger protein 185 identified by expression profiling is associated with prostate cancer progression. Cancer Res., 63, 3877–3882 [PubMed] [Google Scholar]
- 31. Lapointe J., et al. (2004). Gene expression profiling identifies clinically relevant subtypes of prostate cancer. Proc. Natl. Acad. Sci. U. S. A., 101, 811–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dhanasekaran S.M., et al. (2005). Molecular profiling of human prostate tissues: insights into gene expression patterns of prostate development during puberty. FASEB J., 19, 243–245 [DOI] [PubMed] [Google Scholar]
- 33. Taylor B.S., et al. (2010). Integrative genomic profiling of human prostate cancer. Cancer Cell, 18, 11–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Varambally S., et al. (2005). Integrative genomic and proteomic analysis of prostate cancer reveals signatures of metastatic progression. Cancer Cell, 8, 393–406 [DOI] [PubMed] [Google Scholar]
- 35. Tomlins S.A., et al. (2007). Integrative molecular concept modeling of prostate cancer progression. Nat. Genet., 39, 41–51 [DOI] [PubMed] [Google Scholar]
- 36. Heinlein C.A., et al. (2004). Androgen receptor in prostate cancer. Endocr. Rev., 25, 276–308 [DOI] [PubMed] [Google Scholar]
- 37. Heemers H.V., et al. (2007). Androgen receptor (AR) coregulators: a diversity of functions converging on and regulating the AR transcriptional complex. Endocr. Rev., 28, 778–808 [DOI] [PubMed] [Google Scholar]
- 38. Verhoeven G., et al. (1999). Indirect mechanisms and cascades of androgen action. Mol. Cell. Endocrinol., 151, 205–212 [DOI] [PubMed] [Google Scholar]
- 39. Heemers H.V., et al. (2006). Androgen activation of the sterol regulatory element-binding protein pathway: Current insights. Mol. Endocrinol., 20, 2265–2277 [DOI] [PubMed] [Google Scholar]
- 40. Kalluri R., et al. (2009). The basics of epithelial-mesenchymal transition. J. Clin. Invest., 119, 1420–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rathinam R., et al. (2010). Important role of integrins in the cancer biology. Cancer Metastasis Rev., 29, 223–237 [DOI] [PubMed] [Google Scholar]
- 42. Scales T.M., et al. (2011). Spatial and temporal regulation of integrin signalling during cell migration. Curr. Opin. Cell Biol., 23, 562–568 [DOI] [PubMed] [Google Scholar]
- 43. Hu S., et al. (2010). Prosaposin down-modulation decreases metastatic prostate cancer cell adhesion, migration, and invasion. Mol. Cancer, 9, 30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wu H.C., et al. (1994). Derivation of androgen-independent human LNCaP prostatic cancer cell sublines: role of bone stromal cells. Int. J. Cancer, 57, 406–412 [DOI] [PubMed] [Google Scholar]
- 45. Gorlov I.P., et al. (2010). Prioritizing genes associated with prostate cancer development. BMC Cancer, 10, 599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Siegel R., et al. (2012). Cancer statistics, 2012. CA. Cancer J. Clin., 62, 10–29 [DOI] [PubMed] [Google Scholar]
- 47. Attard G., et al. (2011). Translating scientific advancement into clinical benefit for castration-resistant prostate cancer patients. Clin. Cancer Res., 17, 3867–3875 [DOI] [PubMed] [Google Scholar]
- 48. North A.J., et al. (1994). Calponin is localised in both the contractile apparatus and the cytoskeleton of smooth muscle cells. J. Cell Sci., 107 (Pt 3), 437–444 [DOI] [PubMed] [Google Scholar]
- 49. Wu K.C., et al. (2008). Calponin in non-muscle cells. Cell Biochem. Biophys., 52, 139–148 [DOI] [PubMed] [Google Scholar]
- 50. Ramaswamy S., et al. (2003). A molecular signature of metastasis in primary solid tumors. Nat. Genet., 33, 49–54 [DOI] [PubMed] [Google Scholar]
- 51. Kaufman L., et al. (2004). Sidekick-1 is upregulated in glomeruli in HIV-associated nephropathy. J. Am. Soc. Nephrol., 15, 1721–1730 [DOI] [PubMed] [Google Scholar]
- 52. Kaufman L., et al. (2010). Up-regulation of the homophilic adhesion molecule sidekick-1 in podocytes contributes to glomerulosclerosis. J. Biol. Chem., 285, 25677–25685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Nagle R.B., et al. (2011). Metastasis Update: Human Prostate Carcinoma Invasion via Tubulogenesis. Prostate Cancer, 2011, 249290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Brandt D.T., et al. (2009). SCAI acts as a suppressor of cancer cell invasion through the transcriptional control of beta1-integrin. Nat. Cell Biol., 11, 557–568 [DOI] [PubMed] [Google Scholar]
- 55. Lener T., et al. (2004). The role of calponin in the gene profile of metastatic cells: inhibition of metastatic cell motility by multiple calponin repeats. FEBS Lett., 556, 221–226 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.