Abstract
The ginkgolides, acting as anti-platelet-activating factors, have been studied for many years. The biosynthetic pathway of ginkgolides is still far away from unveiling at the level of molecular genetics and biochemistry. There are at least 11 kinds of enzymes having been cloned from Ginkgo biloba L., which catalyze the formation of ginkgolides via a series of reactions. Some researchers have indicated that the addition of precursors and elicitors can influence the accumulation of ginkgolides in the suspension cell cultures of G. biloba. There are also other factors that can influence the production of ginkgolides. This review focuses on the aforementioned aspects to discuss the biosynthetic pathways of the ginkgolides.
Keywords: Biosynthetic pathways, cell culture, enzymes, genes, Ginkgo biloba, ginkgolides
INTRODUCTION
Ginkgo biloba L. (ginkgo), the only remaining species of the order Ginkgoales, is a living fossil plant as it has been existing on the earth for more than 200 million years. The leaf extract of ginkgo has been employed for treating cerebrovascular and cardiovascular diseases for centuries. It has been well studied in recent years as it contains many bioactive constituents, such as flavanoid compounds, diterpene lactone compounds, polysaccharides, etc.[1,2,3] The diterpene lactones contain ginkgolides A, B, C, J, and M and bilobalide compounds [Figure 1].[1] Recently, two new diterpenoid compounds, ginkgolides P and Q, have been isolated from the leaves of ginkgo.[2] Bilobalide exhibits neuroprotective effects by decreasing the release of excitotoxic amino acids, particularly glutamate and aspartate.[3] Ginkgolides have many pharmacological activities such as acting as specific platelet-activating factor antagonists,[4] selective antagonists of glycine receptors,[5] etc., The commercial ginkgolides are produced merely from G. biloba plants, especially from the ginkgo leaves. But the contents of ginkgolides in the native ginkgo plants are very low. Furthermore, the native ginkgo plant materials are limited. Some studies have been made on ginkgo cell and tissue cultures with the aim of producing ginkgolides, but the concentrations of ginkgolide and bilobalide in ginkgo cells and tissues were even lower than that of natural plants.[6] The chemical synthesis of ginkgolides has been accomplished, but the procedures were too complicated to fulfill commercial-scale production of ginkgolides.[7] Therefore, detailed understanding of the biosynthetic pathway and the involved enzymes would contribute to improve the biological production yield. On the basis of molecular genetics of ginkgolide biosynthesis, ginkgolides might be obtained alternatively through metabolic engineering, including breaking the committed step, blocking the branch ways of ginkgolide biosynthesis, and fluxing the secondary metabolite pools to ginkgolide biosynthesis.[8]
Figure 1.
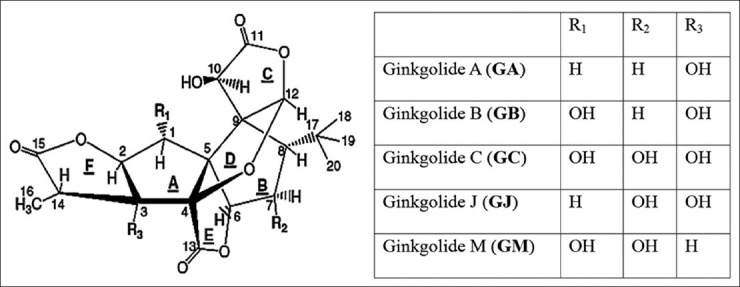
Ginkgolide structures from Ginkgo biloba[1]
This review focuses on the biosynthetic pathways of ginkgolides, the enzymes related to the biosynthesis of the ginkgolides, the genes encoding the enzymes cloned from the ginkgo, and the factors influencing the production of ginkgolides in cell cultures.
BIOSYNTHETIC PATHWAYS OF GINKGOLIDES
Terpenoids such as ginkgolides are biosynthesized from a universal 5-carbon building block: Isopentenyl diphosphate (IPP).[9] IPP can be derived from two pathways: One is the classical cytosolic mevalonic acid (MVA) pathway and the other is the plastidial methylerythritol 4-phosphate (MEP) pathway, which is mevalonate independent. The MVA pathway in the cytosol, starting from 3 acetyl-CoA to finally yield IPP, is responsible for synthesizing sesquiterpenoids and sterols. The MEP pathway producing IPP and dimethylallyl diphosphate (DMAPP) from pyruvate and D-glyceraldehyde 3-phosphate (GAP) is mainly responsible for forming monoterpenoids, diterpenoids constituents.
The ginkgolides biosynthetic pathway can be described in three major stages: (1) The plastid MEP pathway providing universal isoprenoid precursors; (2) the condensing steps producing direct precursors; and (3) the ensuing modifying procedures yielding ginkgolides.[10]
There are seven enzymes involved in the MEP pathway to biosynthesize the GAP into IPP and DMAPP.[11] Among the seven enzymes in ginkgo MEP pathway, condensation of pyruvate and GAP into 1-deoxy-D-xylulose 5-phosphate (DXP) is catalyzed by 1-deoxy-D-xylulose 5-phosphate synthase (GbDXS), which initiates the biosynthesis of ginkgolides.[12] 1-deoxy-D-xylulose 5-phosphate reductoisomerase (DXR) is the second enzyme of the MEP pathway for ginkgolide biosynthesis, which catalyzes the conversion of DXP into MEP.[13] 2-C-methyl-D-erythritol 4-phosphate cytidyltransferase (MECT), the third enzyme, catalyzes the formation of 4-(cytidine 5’- diphospho)-2-C-methyl-D-erythritol from MEP.[14] The following two steps are then catalyzed by 4-(cytidine 5’- diphospho)-2-C-methyl-D-erythritol kinase (CMEK)[15] and 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase (MECS), respectively.[16] Then, 2-C-methyl-D-erythritol 2,4-cyclodiphosphate is catalyzed by 1-hydroxy-2-methyl-2-(E)-butenyl-4-diphosphate synthase (HDS), forming 1-hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate (HMBPP).[17] In the final step of the MEP pathway, HMBPP is reduced by HMBPP reductase (HDR) to produce both IPP and its isomer DMAPP.[10,18,19]
In plastids, the key universal diterpene precursor geranylgeranyl diphosphate (GGDP) is biosynthesized from IPP and DMAPP by catalysis of geranylgeranyl diphosphate synthase (GGPPS).[8] Levopimaradiene is biosynthesized from GGDP via a series of reactions, and levopimaradiene synthase (LPS) performs the first committed step of cyclization of GGPP in ginkgolide biosynthesis.[20] C ring is further oxidized and rearranged to produce the ginkgolides. The plastid MEP biosynthetic pathway of ginkgolide is shown in Figure 2.
Figure 2.

Methylerythritol 4-phosphate pathway in the isoprenoid biosynthesis of Ginkgo biloba. DXS = 1-deoxy-D-xylulose 5-phosphate synthase, DXR = 1-deoxy-D-xylulose 5-phosphate reductoisomerase, MECT = 2-C-methyl-D-erythritol 4-phosphate cytidyltransferase, CMEK = 4-(cytidine 5’-diphospho)-2-C-methyl-D-erythritol kinase, MECS = 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase, HDS = 1-hydroxy-2-methyl-2-(E)- butenyl 4-diphosphate synthase, HDR = 1-hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate reductase, IDI = Isopentenyl diphosphate isomerase, GGPPS = Geranylgeranyl diphosphate synthase, and LPS = Levopimaradiene synthase
GINKGOLIDES BIOSYNTHETIC-RELATED ENZYMES AND GENES
Many genes encoded enzymes have been isolated and cloned from the G. biloba, including 1-Deoxy-D-xylulose 5-phosphate synthase (DXS), DXR, MECT, CMEK, MECS, HDS, HDR, 3-Hydroxy-3-methylglutaryl coenzyme A reductase (HMGR), GGPPS, LPS, and mevalonate diphosphate decarboxylase (MVD), which are involved in the biosynthesis of ginkgolides.
1-Deoxy-D-xylulose 5-phosphate synthase
The initial step of the MEP pathway is the formation of DXP catalyzed by DXS, which may be considered the first committed step of the MEP pathway for ginkgolides biosynthesis. The full-length cDNA of DXS was isolated and characterized from the G. biloba.[12] The full-length cDNA sequence of GbDXS with 2,795 bp was deduced and subsequently confirmed by sequencing, containing a 2,154 bp open reading frame (ORF) encoding a protein of 717 amino acids. Comparative and bioinformatic analyses indicated that GbDXS had high homology with DXSs sequences from other plant species, containing a conserved transit peptide for plastid import, histidine residue, a putative thiamine diphosphate-binding site, and a transketolase motif. GbDXS could be expressed in roots, stems, leaves, and other tested tissues.
The accumulation of ginkgolide B was increased after induction of methyl jasmonate, arachidonic acid, acetylsalicylic acid, and ceric ammonium sulfate in tissue culture of G. biloba. RT-PCR analyses indicated that the expression of GbDXS was also strongly increased with elicitor treatments. The increase in the DXS mRNA accumulation correlates with ginkgolide accumulation, which suggested that DXS might influence the biosynthesis of ginkgolides.
1-Deoxy-D-xylulose 5-phosphate reductoisomerase
DXR, catalyzed the formation of 2-C-methyl-D-erythritol 4-phosphate from DXP in the presence of NADPH,[21] may be considered the second step of MEP pathway, catalyzing in the earlier step of ginkgolide biosynthesis. The full-length cDNA sequence of GbDXR was cloned and characterized.[13] The full-length cDNA was 1720 bp containing a 1431 bp ORF encoding a peptide of 477 amino acids with a calculated molecular mass (Mr) of 52 kDa and a isoelectric point (pI) of 6.58. GbDXR had high homology with DXRs from other plant species and can be expressed in all tissues including roots, stems, leaves, pericarps, and seeds.
2-C-methyl-D-erythritol 4-phosphate cytidyltransferase
MECT, the third enzyme of the MEP pathway, catalyzes the formation of 4-(cytidine 5’- diphospho)-2-C-methyl-D-erythritol from MEP.[14] GbMECT was cloned and characterized from G. biloba embryonic roots presumably, which is involved in ginkgolide biosynthesis. The full-length cDNA of GbMECT was 1,411 bp, consisting of a 984-bp ORF, which was obtained by combining the sequence information of 3’-and 5’- RACE fragments. The deduced amino acid sequence of GbMECT consisted of 327 residues, with theoretical Mr and pI values of 36.3 kDa and 9.08, respectively. Transcription levels of GbMECT remained generally constant in embryonic roots and leaves for 1 month. As shown by protein-targeting analysis with GFP as a reporter protein in Arabidopsis thaliana protoplasts, the full 88 N-terminal residues were necessary to deliver the protein into the chloroplast.
4-(cytidine 5’- diphospho)-2-C-methyl-D-erythritol kinase
CMEK, the fourth enzyme of the 2-C-methyl-D-erythritol 4-phosphate pathway, phosphorylates the 2-hydroxyl group of 4-(cytidine 5’- diphospho)-2-C-methyl-D-erythritol in the presence of ATP.[15] The classes of genes encoding CMEK (GbCMEK1 and GbCMEK2) were cloned and characterized from G. biloba. GbCMEK1 and GbCMEK2 were comprised of 433 and 428 amino acid residues, and their theoretical Mr and pI values were 39.2 kDa and 5.12, and 39.9 kDa and 5.19, respectively. They showed high amino acid sequence identities with other CMEKs. ChloroP program predicted that GbCMEKs had the chloroplast transit peptides.
2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase
MECS from G. biloba (GbMECS), the fifth enzyme in the MEP pathway sequence, transforming 2-phospho-4-(cytidine 5’-diphospho)-2-C-methyl-D-erythritol into 2-C-methyl-D-erythritol 2,4-cyclo-diphosphate, had been cloned and characterized.[16] The full-length GbMECS cDNA was 935 bp, consisting of 717 bp long ORF and 3’-and 5’- untranslated regions (UTR). The deduced protein contained 238 amino acid residues. The N-terminal chloroplast transit peptide in the deduced GbMECS, which consisted of 59 residues, was predicted by the ChloroP program. The theoretical Mr and pI of GbMECS were 19.3 kDa and 6.47, respectively. Furthermore, the quantification of GbMECS transcript indicated that, the transcription level in the embryo roots was at least 2 times higher than that in the embryo leaves throughout the 1-month period of embryo culture.
1-hydroxy-2-methyl-2-(E)-butenyl-4-diphosphate synthase
HDS converts 2-C-methyl-D-erythritol-2,4-cyclodiphosphate (MEcPP) into HMBPP, which is the sixth enzyme of the MEP pathway, supplying building blocks for plant isoprenoids of ginkgolides. The full-length cDNA encoding HDS (GbHDS) had been isolated from G. biloba.[17] The full-length cDNA of GbHDS was 2,763 bp containing 164 bp 5’- UTR and 193 bp 3’-UTR. This cDNA contained an ORF of 2,226 bp encoding a protein consisting of 741 amino acids. The deduced amino acid sequence of GbHDS contained 679 residues, with theoretical Mr of 75.6 kDa and pI of 5.5. The copy number of GbHDS transcript in the 1-week-old embryo roots was slightly lower than that of the leaves. But from the week 2 onward, the level in the roots was always higher than that in the leaves, and at week 3, it reached 2 times higher level. The ChloroP program predicted that GbHDS had a chloroplast transit peptide consisting of 62 amino acid residues, suggesting its putative localization in the plastids.
1-Hydroxy-2-methyl-2-(E)-butenyl-4-diphosphate reductase
HDR is proved to be the terminal-acting enzyme in the plastid MEP pathway which provides isoprenoid precursors for the biosynthesis of ginkgolides. The full-length cDNA encoding HDR (GbHDR) was isolated from G. biloba and characterized.[10] The full-length cDNA of GbHDR was 1,827 bp containing 1,422-bp ORF, with a 3’- UTR of 374 bp downstream from the stop codon and a 5’- UTR of 28 bp upstream of the start codon. There were 474 amino acid residues for the deduced GbHDR, with a calculated Mr of 53.2 kDa and pI of 5.76. The deduced GbHDR protein showed high identity to HDRs of other plant species, and four conserved cysteine residues were found in all plant HDRs. It was predicted by the targetP algorithm to possess a chloroplast transit peptide. Transcription pattern analysis revealed that GbHDR had high transcription in roots, and low in leaves and stems.
Geranylgeranyl diphosphate synthase
GGPPS catalyzed the biosynthesis of GGPP, which was a key precursor for ginkgolides biosynthesis. The full-length cDNA encoding GGPPS from G. biloba (designed as GbGGPPS) had been cloned.[8] The full-length cDNA of GbGGPPS was 1657 bp long. It contained 1176 bp ORF encoding a 391 amino acid residues, with Mr of 42.5 kDa and theoretical pI of 5.98. GbGGPPS had high homology with other plant GGPPSs and the PSI-BLAST results indicated that GbGGPPS belonged to the GGPPS family. Moreover, the TargetP had shown that there was a 79-amino acid transit peptide at the N-terminal of GbGGPPS, for targeting to the plastids, in which ginkgolides were biosynthesized.
Levopimaradiene synthase
LPS, which catalyzes the initial cyclization step in ginkgolide biosynthesis, was cloned and characterized.[20] The cDNA library from G. biloba was prepared from cultivated seedling roots and degenerate primers were designed based on conserved sequence regions in gymnosperm terpene synthases. The sequencing revealed a similar 2,681 bp cDNA similar to other diterpene synthases, but LPS lacked the initiation codon. The full-length cDNA encoded an 873 amino acid ORF with a predicted Mr of 100,289 Da. Compared to other terpenoid synthases, the predicted LPS polypeptide sequence had high sequence identity to A. grandis abietadiene synthase. It maintained most amino acid residues that were highly conserved in mono-, sesqui-, and diterpene synthases.
3-Hydroxy-3-methylglutaryl coenzyme a reductase
HMGR catalyzes the NADP-dependent synthesis of MVA from 3-hydroxy-3-methylglutaryl-CoA, which is the first committed step in MVA pathway for biosynthesis of isoprenoids.[22] The full-length cDNA encoding GMGR (GbHMGR) had been isolated from G. biloba. The full-length GbHMGR cDNA was 2,237 bp with partial poly (A), containing 1716 bp ORF encoding a protein with 571 amino acid. There was a 5’-UTR of 120 bp upstream from the start codon with G as the putative transcript start site. The coding region of GbHMGR was followed by 3’-UTR (401 bp) downstream from the stop codon. The Mr and pI of the deduced GbHMGR protein were predicted to be 60.87 kDa and 6.43, respectively. There was a high homology between the GbHMGR and other plant HMGRs, while GbHMGR diverged earlier than other plant species. The Southern blot and RT-PCR assay results indicated that GbHMGR belonged to a small gene family, and expressed in a tissue-specific manner with a low level expression being only found in root.
Mevalonate diphosphate decarboxylase
MVD catalyzes the conversion of mevalonate diphosphate to IPP, which is important for the biosynthesis of secondary metabolites. A full-length cDNA of MVD from G. biloba (GbMVD) was isolated and characterized.[23] The full-length cDNA of GbMVD was 1,958 bp with a poly (A) tailing, containing an ORF of 1,290 bp encoding 430 amino acids. There was a 5’-UTR of 291 bp upstream from the start codon with G as putative transcription start, and the coding region was followed by 3’-UTR that was 345 bp-long downstream from the stop codon. It was predicted that the calculated Mr and pI of the deduced GbMVD protein were 48 kDa and 5.83, respectively. It also showed a high homology with other MVDs from some species. Transcript accumulation analysis revealed that GbMVD was transcribed in root, stem, and leaf tissues.
Up to now, the genes encoded whole enzymes in the MEP pathway have been cloned from G. biloba, but the enzymes and genes involved in the MVA pathway still need further research. Furthermore, how levopimaradiene transforms into ginkgolides is still under investigation.
INFLUENCING FACTORS IN ACCUMULATION OF GINKGOLIDES
Some elicitors, including fungal and bacterial elicitors, have been studied and investigated the influences on the accumulation of ginkgolides in G. biloba cell cultures. Kang found that native Staphylococcus aureus KCTC 1916 and Candida albicans KCTC 7121 as biotic elicitors produced more of ginkgolide A (GA) and ginkgolide B (GB) than the control groups.[24] Effect of mycelium extract of Rhizopus japonicus on the production of GB was the most significant in the studied 10 kinds of fungi.[25]
The precursors involved in the terpenoids biosynthesis had also been investigated. Kang et al. found that treatments with HMG-CoA, GPP, and IPP, which were precursors related to MVA and MEP pathways, had different promoting effects on GA and GB productions, with 2.7 folds to 4.25 folds compared to the control groups.[26] Dai had investigated the effects of isoprene and geraniol on the GB production in suspension cultured cells of G. biloba.[25] Compared with the control, the total GB yields were enhanced by 69%, 13.8%, and 11.4% when adding 100 mg/L of isoprene, 10 mg/L isoprene, and 50 mg/L isoprene in the media, respectively.
Furthermore, strains in the fermentation liquid of endophytic fungi influenced the ginkgolide accumulation.[27] The production patterns in the leaf, stem bark, and stem of female and male trees were also different.[28]
FUTURE PERSPECTIVES
As demonstrated above, many researches about G. biloba have been made involved extraction, purification, structural elucidation, and bioactive determination. The investigations of ginkgolide biosynthetic pathways have also made in some progresses, for example, 11 kinds of enzymes having been cloned from G. biloba, which catalyze the formation of ginkgolides via a series of reactions. However, the biosynthetic pathway of ginkgolides is still far away from unveiling at the level of molecular genetics and biochemistry: The biosynthesis of ginkgolides was mainly regulated by MEP pathway or MVA pathway, or both the two pathways, and the pathways of the GGPP is transformed into ginkgolides, etc., are still under investigation. Those need scientists to find out the enzymes and genes included in the pathways which are still not clear. Moreover, it provides us an opportunity to find out a way to improve the accumulation of ginkgolides in the cultured cells which would be hard to determine them in natural G. biloba.
Footnotes
Source of Support: Nil
Conflict of Interest: None declared.
REFERENCES
- 1.Strømgaard K, Nakanishi K. Chemistry and biology of terpene trilactones from Ginkgo biloba. Angew Chem Int Ed Engl. 2004;43:1640–58. doi: 10.1002/anie.200300601. [DOI] [PubMed] [Google Scholar]
- 2.Liao HJ, Zheng YF, Li HY, Peng GP. Two new ginkgolides from the leaves of Ginkgo biloba. Planta Med. 2011;77:1818–21. doi: 10.1055/s-0030-1271153. [DOI] [PubMed] [Google Scholar]
- 3.Huang SH, Duke RK, Chebib M, Sasaki K, Wada K, Johnston GA. Bilobalide, a sesquiterpene trilactone from Ginkgo biloba, is an antagonist at recombinant alpha1 beta2 gamma2 L GABA(A) receptors. Eur J Pharmacol. 2003;464:1–8. doi: 10.1016/s0014-2999(03)01344-x. [DOI] [PubMed] [Google Scholar]
- 4.Etienne A, Hecquet F, Soulard C, Spinnewyn B, Clostre F, Braquet P. In vivo inhibition of plasma protein leakage and Salmonella enteritidis-induced mortality in the rat by a specific paf-acether antagonist: BN 52021. Agents Actions. 1986;17:368–70. doi: 10.1007/BF01982649. [DOI] [PubMed] [Google Scholar]
- 5.van Beek TA. Ginkgolides and bilobalide: Their physical, chromatographic and spectroscopic properties. Bioorg Med Chem. 2005;13:5001–12. doi: 10.1016/j.bmc.2005.05.056. [DOI] [PubMed] [Google Scholar]
- 6.Laurain D, Tremouillaux-Guillerm J, Chenieux JC, van Beek TA. Production of ginkgolide and bilobalide in transformed and gametophyte derived cell cultures of Ginkgo biloba. Phytochem. 1997;46:127–30. [Google Scholar]
- 7.Crimmins MT, Pace JM, Nantermet PG, Kim-Meade AS, Thomas JB, Watterson SH, et al. The total synthesis of (t/2)-ginkgolide B”. J Am Chem Soc. 2000;122:8453–63. [Google Scholar]
- 8.Liao Z, Chen M, Gong Y, Guo L, Tan Q, Feng X, et al. A new geranylgeranyl diphosphate synthase gene from Ginkgo biloba, which intermediates the biosynthesis of the key precursor for ginkgolides. DNA Seq. 2004;15:153–8. doi: 10.1080/10425170410001667348. [DOI] [PubMed] [Google Scholar]
- 9.Han YS, Roytrakul S, Verberne MC, van der Heijden R, Linthorst HJ, Verpoorte R. Cloning of a cDNA encoding 1-deoxy-D-xylulose 5-phosphate synthase from Morinda citrifolia and analysis of its expression in relation to anthraquinone accumulation. Plant Sci. 2003;164:911–7. [Google Scholar]
- 10.Lu J, Wu W, Cao S, Zhao H, Zeng H, Lin L, et al. Molecular cloning and characterization of 1-hydroxy-2-methyl-2-(E)-butenyl-4-diphosphate reductase gene from Ginkgo biloba. Mol Biol Rep. 2008;35:413–20. doi: 10.1007/s11033-007-9101-7. [DOI] [PubMed] [Google Scholar]
- 11.Eisenreich W, Bacher A, Arigoni D, Rohdich F. Biosynthesis of isoprenoids via the non-mevalonate pathway. Cell Mol Life Sci. 2004;61:1401–26. doi: 10.1007/s00018-004-3381-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gong YF, Liao ZH, Guo BH, Sun XF, Tang KX. Molecular cloning and expression profile analysis of Ginkgo biloba DXS gene encoding 1-deoxy-D-xylulose 5-phosphate synthase, the first committed enzyme of the 2-C-methyl-D-erythritol 4-phosphate pathway. Planta Med. 2006;72:329–35. doi: 10.1055/s-2005-916234. [DOI] [PubMed] [Google Scholar]
- 13.Gong Y, Liao Z, Chen M, Zuo K, Guo L, Tan Q, et al. Molecular cloning and characterization of a 1-deoxy-D-xylulose 5-phosphate reductoisomerase gene from Ginkgo biloba. DNA Seq. 2005;16:111–20. doi: 10.1080/10425170500058869. [DOI] [PubMed] [Google Scholar]
- 14.Kim SM, Kuzuyama T, Chang YJ, Kwon HJ, Kim SU. Cloning and functional characterization of 2-C-methyl-D-erythritol 4-phosphate cytidyltransferase (GbMECT) gene from Ginkgo biloba. Phytochemistry. 2006;67:1435–41. doi: 10.1016/j.phytochem.2006.05.034. [DOI] [PubMed] [Google Scholar]
- 15.Kim SM, Kim YB, Kuzuyama T, Kim SU. Two copies of 4-(cytidine 5’-diphospho)-2-C-methyl-D-erythritol kinase (CMK) gene in Ginkgo biloba: Molecular cloning and functional characterization. Planta. 2008;228:941–50. doi: 10.1007/s00425-008-0794-1. [DOI] [PubMed] [Google Scholar]
- 16.Kim SM, Kuzuyama T, Chang YJ, Kim SU. Cloning and characterization of 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase (MECS) gene from Ginkgo biloba. Plant Cell Rep. 2006;25:829–35. doi: 10.1007/s00299-006-0136-3. [DOI] [PubMed] [Google Scholar]
- 17.Kim SM, Kim SU. Characterization of 1-hydroxy-2-methyl-2-(E)-butenyl-4-diphosphate synthase (HDS) gene from Ginkgo biloba. Mol Biol Rep. 2010;37:973–9. doi: 10.1007/s11033-009-9771-4. [DOI] [PubMed] [Google Scholar]
- 18.Altincicek B, Kollas A, Eberl M, Wiesner J, Sanderbrand S, Hintz M, et al. LytB, a novel gene of the 2-C-methyl-D-erythritol 4-phosphate pathway of isoprenoid biosynthesis in Escherichia coli. FEBS Lett. 2001;499:37–40. doi: 10.1016/s0014-5793(01)02516-9. [DOI] [PubMed] [Google Scholar]
- 19.Hecht S, Eisenreich W, Adam P, Amslinger S, Kis K, Bacher A, et al. Studies on the nonmevalonate pathway to terpenes: The role of the GcpE (IspG) protein. Proc Natl Acad Sci U S A. 2001;98:14837–42. doi: 10.1073/pnas.201399298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schepmann HG, Pang J, Matsuda SP. Cloning and characterization of Ginkgo biloba levopimaradiene synthase which catalyzes the first committed step in ginkgolide biosynthesis. Arch Biochem Biophys. 2001;392:263–9. doi: 10.1006/abbi.2001.2438. [DOI] [PubMed] [Google Scholar]
- 21.Takahashi S, Kuzuyama T, Watanabe H, Seto H. A 1-deoxy-D-xylulose 5-phosphate reductoisomerase catalyzing the formation of 2-C-methyl-D-erythritol 4-phosphate in an alternative nonmevalonate pathway for terpenoid biosynthesis. Proc Natl Acad Sci U S A. 1998;95:9879–84. doi: 10.1073/pnas.95.17.9879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shen G, Pang Y, Wu W, Liao Z, Zhao L, Sun X, et al. Cloning and characterization of a root-specific expressing gene encoding 3-hydroxy-3-methylglutaryl coenzyme A reductase from Ginkgo biloba. Mol Biol Rep. 2006;33:117–27. doi: 10.1007/s11033-006-0014-7. [DOI] [PubMed] [Google Scholar]
- 23.Pang Y, Shen G, Bergesc T. Molecular cloning, characterization and heterologous exp ression in saccharomyces cerevisiae of a mevalonate diphosphate decarboxylase cDNA from Ginkgo biloba. Physiol Plant. 2006;127:19–27. [Google Scholar]
- 24.Kang SM, Min JY, Kim YD, Karigar CS, Kim SW, Goo GH, et al. Effect of biotic elicitors on the accumulation of bilobalide and ginkgolides in Ginkgo biloba cell cultures. J Biotechnol. 2009;139:84–8. doi: 10.1016/j.jbiotec.2008.09.007. [DOI] [PubMed] [Google Scholar]
- 25.Dai JG, Zhu WH, Wu YQ, Hu WQ, Zhang DY. Effects of precursors and fungal elicitors on GKB production in suspension cultured cells of Ginkgo biloba L. Acta Pharmaceut Sin. 2000;35:151–5. [Google Scholar]
- 26.Kang SM, Min JY, Kim YD, Park DJ, Jung HN, Karigar CS, et al. Effect of supplementing terpenoid biosynthetic precursors on the accumulation of bilobalide and ginkgolides in Ginkgo biloba cell cultures. J Biotechnol. 2006;123:85–92. doi: 10.1016/j.jbiotec.2005.10.021. [DOI] [PubMed] [Google Scholar]
- 27.Yan ZY, Luo J, Guo XH, Zeng QQ. Screening of ginkgolides-producing endophytic fungi and optimal study on culture condition. Nat Prod Res Dev. 2007;19:554–8. [Google Scholar]
- 28.Park YG, Kim SJ, Jung HY, Kang YM, Kang SM, Prasad DT, et al. Variation of ginkgolides and bilobalide contents in leaves and cell cultures of Ginkgo biloba. Biotechnol Bioproc Eng. 2004;9:35–40. [Google Scholar]


