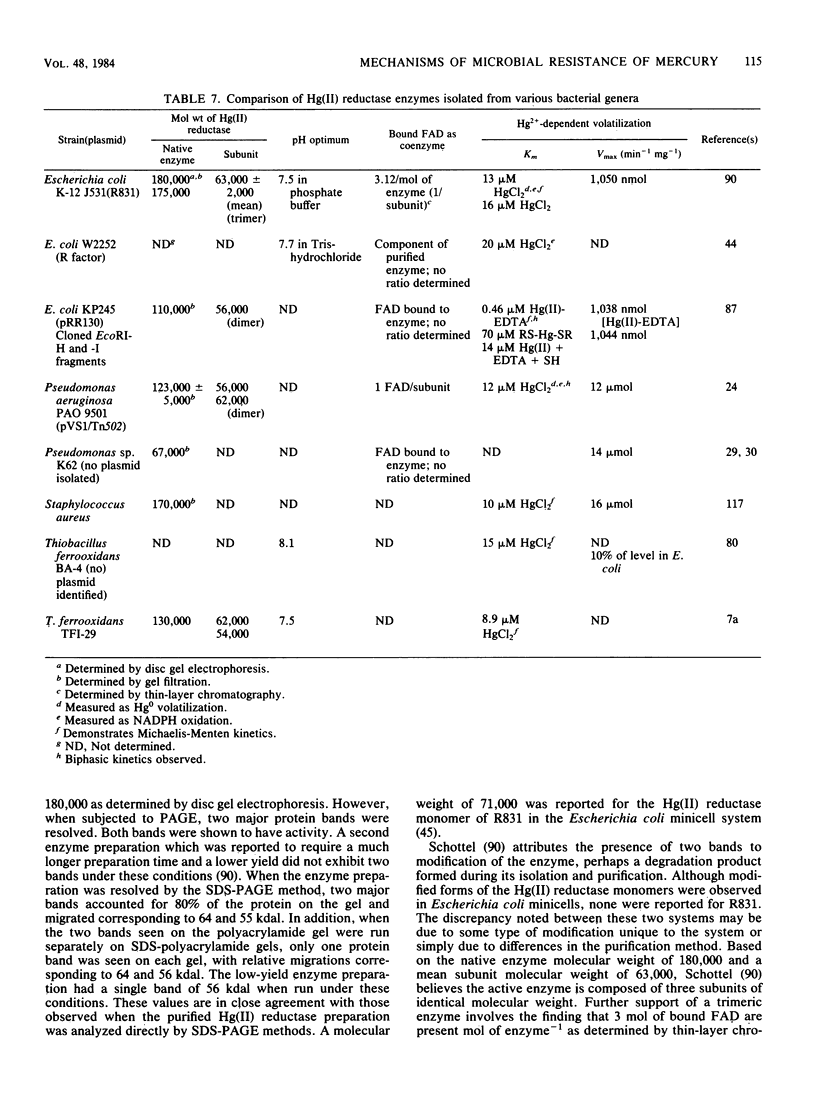
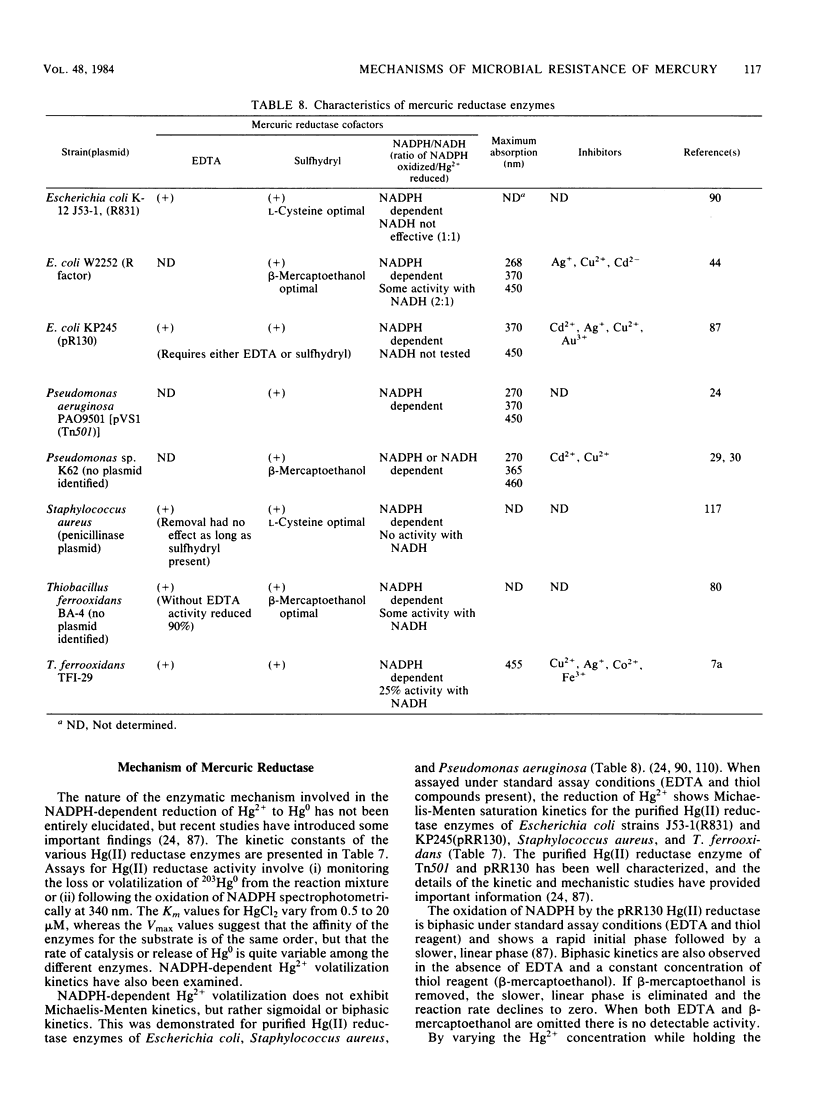
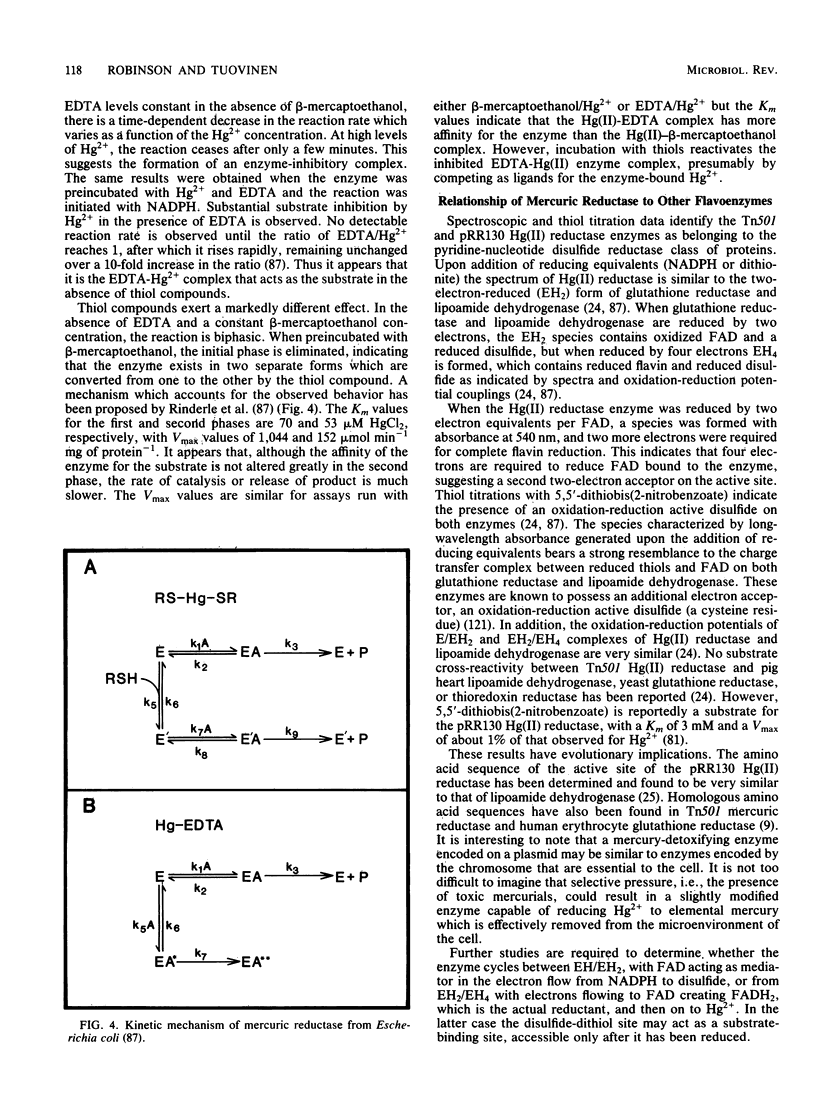
Full text
PDF






















Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bennett P. M., Grinsted J., Choi C. L., Richmond M. H. Characterisation of Tn501, a transposon determining resistance to mercuric ions. Mol Gen Genet. 1978 Feb 7;159(1):101–106. doi: 10.1007/BF00401753. [DOI] [PubMed] [Google Scholar]
- Bertilsson L., Neujahr H. Y. Methylation of mercury compounds by methylcobalamin. Biochemistry. 1971 Jul 6;10(14):2805–2808. doi: 10.1021/bi00790a024. [DOI] [PubMed] [Google Scholar]
- Bohlander F. A., Summers A. O., Meagher R. B. Cloning a promoter that puts the expression of tetracycline resistance under the control of the regulatory elements of the mer operon. Gene. 1981 Dec;15(4):395–403. doi: 10.1016/0378-1119(81)90182-7. [DOI] [PubMed] [Google Scholar]
- Brown N. L., Choi C. L., Grinsted J., Richmond M. H., Whitehead P. R. Nucleotide sequences at the ends of the mercury resistance transposon, Tn501. Nucleic Acids Res. 1980 May 10;8(9):1933–1945. doi: 10.1093/nar/8.9.1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown N. L., Ford S. J., Pridmore R. D., Fritzinger D. C. Nucleotide sequence of a gene from the Pseudomonas transposon Tn501 encoding mercuric reductase. Biochemistry. 1983 Aug 16;22(17):4089–4095. doi: 10.1021/bi00286a015. [DOI] [PubMed] [Google Scholar]
- Clark D. L., Weiss A. A., Silver S. Mercury and organomercurial resistances determined by plasmids in Pseudomonas. J Bacteriol. 1977 Oct;132(1):186–196. doi: 10.1128/jb.132.1.186-196.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSimone R. E., Penley M. W., Charbonneau L., Smith S. G., Wood J. M., Hill H. A., Pratt J. M., Ridsdale S., Williams R. J. The kinetics and mechanism of cobalamin-dependent methyl and ethyl transfer to mercuric ion. Biochim Biophys Acta. 1973 May 28;304(3):851–863. doi: 10.1016/0304-4165(73)90232-8. [DOI] [PubMed] [Google Scholar]
- Dempsey W. B., McIntire S. A. Lambda transducing phages derived from a FinO- R100::lambda cointegrate plasmid: proteins encoded by the R100 replication/incompatibility region and the antibiotic resistance determinant. Mol Gen Genet. 1979 Nov;176(3):319–334. doi: 10.1007/BF00333094. [DOI] [PubMed] [Google Scholar]
- Dempsey W. B., McIntire S. A., Willetts N., Schottel J., Kinscherf T. G., Silver S., Shannon W. A., Jr Properties of lambda transducing bacteriophages carrying R100 plasmid DNA: mercury resistance genes. J Bacteriol. 1978 Dec;136(3):1084–1093. doi: 10.1128/jb.136.3.1084-1093.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dempsey W. B., Willetts N. S. Plasmid co-integrates of prophage lambda and R factor R100. J Bacteriol. 1976 Apr;126(1):166–176. doi: 10.1128/jb.126.1.166-176.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster T. J., Nakahara H. Deletions in the r-determinant mer region of plasmid R100-1 selected for loss of mercury hypersensitivy. J Bacteriol. 1979 Oct;140(1):301–305. doi: 10.1128/jb.140.1.301-305.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster T. J., Nakahara H., Weiss A. A., Silver S. Transposon A-generated mutations in the mercuric resistance genes of plasmid R100-1. J Bacteriol. 1979 Oct;140(1):167–181. doi: 10.1128/jb.140.1.167-181.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox B. S., Walsh C. T. Mercuric reductase: homology to glutathione reductase and lipoamide dehydrogenase. Iodoacetamide alkylation and sequence of the active site peptide. Biochemistry. 1983 Aug 16;22(17):4082–4088. doi: 10.1021/bi00286a014. [DOI] [PubMed] [Google Scholar]
- Fox B., Walsh C. T. Mercuric reductase. Purification and characterization of a transposon-encoded flavoprotein containing an oxidation-reduction-active disulfide. J Biol Chem. 1982 Mar 10;257(5):2498–2503. [PubMed] [Google Scholar]
- Grinsted J., Bennett P. M., Higginson S., Richmond M. H. Regional preference of insertion of Tn501 and Tn802 into RP1 and its derivatives. Mol Gen Genet. 1978 Nov 9;166(3):313–320. doi: 10.1007/BF00267624. [DOI] [PubMed] [Google Scholar]
- Groves D. J., Short H., Thewaini A. J., Young F. E. Epidemiology of antibiotic and heavy metal resistance in bacteria: resistance patterns in staphylococci isolated from populations in Iraq exposed and not exposed to heavy metals or antibiotics. Antimicrob Agents Chemother. 1975 May;7(5):622–628. doi: 10.1128/aac.7.5.622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groves D. J., Young F. E. Epidemiology of antibiotic and heavy metal resistance in bacteria: resistance patterns in staphylococci isolated from populations not known to be exposed to heavy metals. Antimicrob Agents Chemother. 1975 May;7(5):614–621. doi: 10.1128/aac.7.5.614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamdy M. K., Noyes O. R. Formation of methyl mercury by bacteria. Appl Microbiol. 1975 Sep;30(3):424–432. doi: 10.1128/am.30.3.424-432.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm H. W., Cox M. F. Transformation of elemental mercury by bacteria. Appl Microbiol. 1975 Apr;29(4):491–494. doi: 10.1128/am.29.4.491-494.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S., Otsubo E., Davidson N., Saedler H. Electron microscope heteroduplex studies of sequence relations among bacterial plasmids: identification and mapping of the insertion sequences IS1 and IS2 in F and R plasmids. J Bacteriol. 1975 May;122(2):764–775. doi: 10.1128/jb.122.2.764-775.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hänni C., Meyer J., Iida S., Arber W. Occurrence and properties of composite transposon Tn2672: evolution of multiple drug resistance transposons. J Bacteriol. 1982 Jun;150(3):1266–1273. doi: 10.1128/jb.150.3.1266-1273.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iida S., Hänni C., Echarti C., Arber W. Is the IS1-flanked r-determinant of the R plasmid NR1 a transposon? J Gen Microbiol. 1981 Oct;126(2):413–425. doi: 10.1099/00221287-126-2-413. [DOI] [PubMed] [Google Scholar]
- Imura N., Sukegawa E., Pan S. K., Nagao K., Kim J. Y., Kwan T., Ukita T. Chemical methylation of inorganic mercury with methylcobalamin, a vitamin B12 analog. Science. 1971 Jun 18;172(3989):1248–1249. doi: 10.1126/science.172.3989.1248. [DOI] [PubMed] [Google Scholar]
- Izaki K. Enzymatic reduction of mercurous ions in Escherichia coli bearing R factor. J Bacteriol. 1977 Aug;131(2):696–698. doi: 10.1128/jb.131.2.696-698.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izaki K., Tashiro Y., Funaba T. Mechanism of mercuric chloride resistance in microorganisms. 3. Purification and properties of a mercuric ion reducing enzyme from Escherichia coli bearing R factor. J Biochem. 1974 Mar;75(3):591–599. doi: 10.1093/oxfordjournals.jbchem.a130427. [DOI] [PubMed] [Google Scholar]
- Jackson W. J., Summers A. O. Biochemical characterization of HgCl2-inducible polypeptides encoded by the mer operon of plasmid R100. J Bacteriol. 1982 Aug;151(2):962–970. doi: 10.1128/jb.151.2.962-970.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson W. J., Summers A. O. Polypeptides encoded by the mer operon. J Bacteriol. 1982 Feb;149(2):479–487. doi: 10.1128/jb.149.2.479-487.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen S., Jernelöv A. Biological methylation of mercury in aquatic organisms. Nature. 1969 Aug 16;223(5207):753–754. doi: 10.1038/223753a0. [DOI] [PubMed] [Google Scholar]
- Jernelöv A., Martin A. L. Ecological implications of metal metabolism by microorganisms. Annu Rev Microbiol. 1975;29:61–77. doi: 10.1146/annurev.mi.29.100175.000425. [DOI] [PubMed] [Google Scholar]
- Jernelöv A., Wallin T. Air-borne mercury fall-out on snow around five Swedish chlor-alkali plants. Atmos Environ. 1973 Feb;7(2):209–214. doi: 10.1016/0004-6981(73)90170-4. [DOI] [PubMed] [Google Scholar]
- Joensuu O. I. Fossil fuels as a source of mercury pollution. Science. 1971 Jun 4;172(3987):1027–1028. doi: 10.1126/science.172.3987.1027. [DOI] [PubMed] [Google Scholar]
- Kamps L. R., Carr R., Miller H. Total mercury-monomethylmercury content of several species of fish. Bull Environ Contam Toxicol. 1972 Nov;8(5):273–279. doi: 10.1007/BF01684556. [DOI] [PubMed] [Google Scholar]
- Kleckner N. Translocatable elements in procaryotes. Cell. 1977 May;11(1):11–23. doi: 10.1016/0092-8674(77)90313-0. [DOI] [PubMed] [Google Scholar]
- Kleckner N. Transposable elements in prokaryotes. Annu Rev Genet. 1981;15:341–404. doi: 10.1146/annurev.ge.15.120181.002013. [DOI] [PubMed] [Google Scholar]
- Komura I., Funaba T., Izaki K. Mechanism of mercuric chloride resistance in microorganisms. II. NADPH-dependent reduction of mercuric chloride and vaporization of mercury from mercuric chloride by a multiple drug resistant strain of Escherichia coli. J Biochem. 1971 Dec;70(6):895–901. doi: 10.1093/oxfordjournals.jbchem.a129719. [DOI] [PubMed] [Google Scholar]
- Komura I., Izaki K. Mechanism of mercuric chloride resistance in microorganisms. I. Vaporization of a mercury compound from mercuric chloride by multiple drug resistant strains of Escherichia coli. J Biochem. 1971 Dec;70(6):885–893. doi: 10.1093/oxfordjournals.jbchem.a129718. [DOI] [PubMed] [Google Scholar]
- Kondo I., Ishikawa T., Nakahara H. Mercury and cadmium resistances mediated by the penicillinase plasmid in Staphylococcus aureus. J Bacteriol. 1974 Jan;117(1):1–7. doi: 10.1128/jb.117.1.1-7.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopecko D. J., Brevet J., Cohen S. N. Involvement of multiple translocating DNA segments and recombinational hotspots in the structural evolution of bacterial plasmids. J Mol Biol. 1976 Dec;108(2):333–360. doi: 10.1016/s0022-2836(76)80124-6. [DOI] [PubMed] [Google Scholar]
- Landner L. Biochemical model for the biological methylation of mercury suggested from methylation studies in vivo with Neurospora crassa. Nature. 1971 Apr 16;230(5294):452–454. doi: 10.1038/230452a0. [DOI] [PubMed] [Google Scholar]
- Lane D., Chandler M. Mapping of the drug resistance genes carried by the r-determinant of the R100.1 plasmid. Mol Gen Genet. 1977 Nov 29;157(1):17–23. doi: 10.1007/BF00268682. [DOI] [PubMed] [Google Scholar]
- Matsumura F., Gotoh Y., Boush G. M. Phenylmercuric acetate: metabolic conversion by microorganisms. Science. 1971 Jul 2;173(3991):49–51. doi: 10.1126/science.173.3991.49. [DOI] [PubMed] [Google Scholar]
- Meissner P. S., Falkinham J. O., 3rd Plasmid-encoded mercuric reductase in Mycobacterium scrofulaceum. J Bacteriol. 1984 Feb;157(2):669–672. doi: 10.1128/jb.157.2.669-672.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki T., Easton A. M., Rownd R. H. Mapping of the resistance genes of the R plasmid NR1. Mol Gen Genet. 1978 Jan 17;158(3):217–224. doi: 10.1007/BF00267192. [DOI] [PubMed] [Google Scholar]
- Murphy E., Novick R. P. Physical mapping of Staphylococcus aureus penicillinase plasmid pI524: characterization of an invertible region. Mol Gen Genet. 1979 Aug;175(1):19–30. doi: 10.1007/BF00267851. [DOI] [PubMed] [Google Scholar]
- Nakahara H., Ishikawa T., Sarai Y., Kondo I. Frequency of heavy-metal resistance in bacteria from inpatients in Japan. Nature. 1977 Mar 10;266(5598):165–167. doi: 10.1038/266165a0. [DOI] [PubMed] [Google Scholar]
- Nakahara H., Ishikawa T., Sarai Y., Kondo I., Kozukue H. Mercury resistance and R plasmids in Escherichia coli isolated from clinical lesions in Japan. Antimicrob Agents Chemother. 1977 Jun;11(6):999–1003. doi: 10.1128/aac.11.6.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahara H., Ishikawa T., Sarai Y., Kondo I., Kozukue H., Silver S. Linkage of mercury, cadmium, and arsenate and drug resistance in clinical isolates of Pseudomonas aeruginosa. Appl Environ Microbiol. 1977 Apr;33(4):975–976. doi: 10.1128/aem.33.4.975-976.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahara H., Kinscherf T. G., Silver S., Miki T., Easton A. M., Rownd R. H. Gene copy number effects in the mer operon of plasmid NR1. J Bacteriol. 1979 Apr;138(1):284–287. doi: 10.1128/jb.138.1.284-287.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahara H., Silver S., Miki T., Rownd R. H. Hypersensitivity to Hg2+ and hyperbinding activity associated with cloned fragments of the mercurial resistance operon of plasmid NR1. J Bacteriol. 1979 Oct;140(1):161–166. doi: 10.1128/jb.140.1.161-166.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neet K. E., Ainslie G. R., Jr Hysteretic enzymes. Methods Enzymol. 1980;64:192–226. doi: 10.1016/s0076-6879(80)64010-5. [DOI] [PubMed] [Google Scholar]
- Nelson J. D., Blair W., Brinckman F. E., Colwell R. R., Iverson W. P. Biodegradation of phenylmercuric acetate by mercury-resistant bacteria. Appl Microbiol. 1973 Sep;26(3):321–326. doi: 10.1128/am.26.3.321-326.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni'Bhriain N. N., Silver S., Foster T. J. Tn5 insertion mutations in the mercuric ion resistance genes derived from plasmid R100. J Bacteriol. 1983 Aug;155(2):690–703. doi: 10.1128/jb.155.2.690-703.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick R. P., Edelman I., Schwesinger M. D., Gruss A. D., Swanson E. C., Pattee P. A. Genetic translocation in Staphylococcus aureus. Proc Natl Acad Sci U S A. 1979 Jan;76(1):400–404. doi: 10.1073/pnas.76.1.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick R. P., Roth C. Plasmid-linked resistance to inorganic salts in Staphylococcus aureus. J Bacteriol. 1968 Apr;95(4):1335–1342. doi: 10.1128/jb.95.4.1335-1342.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson G. J., Porter F. D., Rubinstein J., Silver S. Mercuric reductase enzyme from a mercury-volatilizing strain of Thiobacillus ferrooxidans. J Bacteriol. 1982 Sep;151(3):1230–1236. doi: 10.1128/jb.151.3.1230-1236.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinney R. J. Survival of plasmid-containing strains of Escherichia coli, Pseudomonas aeruginosa and Staphylococcus aureus in phenylmercuric nitrate and thiomersal. J Pharm Pharmacol. 1978 Apr;30(4):228–232. doi: 10.1111/j.2042-7158.1978.tb13210.x. [DOI] [PubMed] [Google Scholar]
- Porter F. D., Silver S., Ong C., Nakahara H. Selection for mercurial resistance in hospital settings. Antimicrob Agents Chemother. 1982 Nov;22(5):852–858. doi: 10.1128/aac.22.5.852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramamoorthy S., Cheng T. C., Kushner D. J. Effect of microbial life stages on the fate of methylmercury in natural waters. Bull Environ Contam Toxicol. 1982 Aug;29(2):167–173. doi: 10.1007/BF01606145. [DOI] [PubMed] [Google Scholar]
- Ridley W. P., Dizikes L. J., Wood J. M. Biomethylation of toxic elements in the environment. Science. 1977 Jul 22;197(4301):329–332. doi: 10.1126/science.877556. [DOI] [PubMed] [Google Scholar]
- Rinderle S. J., Booth J. E., Williams J. W. Mercuric reductase from R-plasmid NR1: characterization and mechanistic study. Biochemistry. 1983 Feb 15;22(4):869–876. doi: 10.1021/bi00273a025. [DOI] [PubMed] [Google Scholar]
- Rowland I. R., Davies M. J., Grasso P. Volatilisation of methylmercuric chloride by hydrogen sulphide. Nature. 1977 Feb 24;265(5596):718–719. doi: 10.1038/265718a0. [DOI] [PubMed] [Google Scholar]
- Rowland I. R., Grasso P., Davies M. J. The methylation of mercuric chloride by human intestinal bacteria. Experientia. 1975 Sep 15;31(9):1064–1065. doi: 10.1007/BF02326961. [DOI] [PubMed] [Google Scholar]
- Schottel J. L. The mercuric and organomercurial detoxifying enzymes from a plasmid-bearing strain of Escherichia coli. J Biol Chem. 1978 Jun 25;253(12):4341–4349. [PubMed] [Google Scholar]
- Schottel J., Mandal A., Clark D., Silver S., Hedges R. W. Volatilisation of mercury and organomercurials determined by inducible R-factor systems in enteric bacteria. Nature. 1974 Sep 27;251(5473):335–337. doi: 10.1038/251335a0. [DOI] [PubMed] [Google Scholar]
- Spangler W. J., Spigarelli J. L., Rose J. M., Flippin R. S., Miller H. H. Degradation of methylmercury by bacteria isolated from environmental samples. Appl Microbiol. 1973 Apr;25(4):488–493. doi: 10.1128/am.25.4.488-493.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spangler W. J., Spigarelli J. L., Rose J. M., Miller H. M. Methylmercury: bacterial degradation in lake sediments. Science. 1973 Apr 13;180(4082):192–193. doi: 10.1126/science.180.4082.192. [DOI] [PubMed] [Google Scholar]
- Stanisich V. A., Bennett P. M., Richmond M. H. Characterization of a translocation unit encoding resistance to mercuric ions that occurs on a nonconjugative plasmid in Pseudomonas aeruginosa. J Bacteriol. 1977 Mar;129(3):1227–1233. doi: 10.1128/jb.129.3.1227-1233.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers A. O., Kight-Olliff L. Tn1 generated mutants in the mercuric ion reductase of the Inc P plasmid, R702. Mol Gen Genet. 1980;180(1):91–97. doi: 10.1007/BF00267356. [DOI] [PubMed] [Google Scholar]
- Summers A. O., Knight-Olliff L., Slater C. Effect of catabolite repression on the mer operon. J Bacteriol. 1982 Jan;149(1):191–197. doi: 10.1128/jb.149.1.191-197.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers A. O., Lewis E. Volatilization of mercuric chloride by mercury-resistant plasmid-bearing strains of Escherichia coli, Staphylococcus aureus, and Pseudomonas aeruginosa. J Bacteriol. 1973 Feb;113(2):1070–1072. doi: 10.1128/jb.113.2.1070-1072.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers A. O., Silver S. Mercury resistance in a plasmid-bearing strain of Escherichia coli. J Bacteriol. 1972 Dec;112(3):1228–1236. doi: 10.1128/jb.112.3.1228-1236.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers A. O., Silver S. Microbial transformations of metals. Annu Rev Microbiol. 1978;32:637–672. doi: 10.1146/annurev.mi.32.100178.003225. [DOI] [PubMed] [Google Scholar]
- Summers A. O., Sugarman L. I. Cell-free mercury(II)-reducing activity in a plasmid-bearing strain of Escherichia coli. J Bacteriol. 1974 Jul;119(1):242–249. doi: 10.1128/jb.119.1.242-249.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers A. O., Weiss R. B., Jacoby G. A. Transposition of mercury resistance from a transferable R plasmic of Escherichia coli. Plasmid. 1980 Jan;3(1):35–47. doi: 10.1016/s0147-619x(80)90032-3. [DOI] [PubMed] [Google Scholar]
- Tanak N., Cramer J. H., Rownd R. H. EcoRI restriction endonuclease map of the composite R plasmid NR1. J Bacteriol. 1976 Jul;127(1):619–636. doi: 10.1128/jb.127.1.619-636.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M., Harafuji H., Yamamoto T. A gene and its product required for transposition of resistance transposon Tn2603. J Bacteriol. 1982 Aug;151(2):723–728. doi: 10.1128/jb.151.2.723-728.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M., Yamamoto T., Sawai T. Evolution of complex resistance transposons from an ancestral mercury transposon. J Bacteriol. 1983 Mar;153(3):1432–1438. doi: 10.1128/jb.153.3.1432-1438.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tezuka T., Tonomura K. Purification and properties of a second enzyme catalyzing the splitting of carbon-mercury linkages from mercury-resistant Pseudomonas K-62. J Bacteriol. 1978 Jul;135(1):138–143. doi: 10.1128/jb.135.1.138-143.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tezuka T., Tonomura K. Purification and properties of an enzyme catalyzing the splitting of carbon-mercury linkages from mercury-resistant Pseudomonas K-62 strain. I. Splitting enzyme 1. J Biochem. 1976 Jul;80(1):79–87. doi: 10.1093/oxfordjournals.jbchem.a131261. [DOI] [PubMed] [Google Scholar]
- Timoney J. F., Port J., Giles J., Spanier J. Heavy-metal and antibiotic resistance in the bacterial flora of sediments of New York Bight. Appl Environ Microbiol. 1978 Sep;36(3):465–472. doi: 10.1128/aem.36.3.465-472.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonomura K., Kanzaki F. The reductive decomposition of organic mercurials by cell-free extract of a mercury-resistant pseudomonad. Biochim Biophys Acta. 1969 Jun 17;184(1):227–229. doi: 10.1016/0304-4165(69)90124-x. [DOI] [PubMed] [Google Scholar]
- Vonk J. W., Sijpesteijn A. K. Studies on the methylation of mercuric chloride by pure cultures of bacteria and fungi. Antonie Van Leeuwenhoek. 1973;39(3):505–513. doi: 10.1007/BF02578894. [DOI] [PubMed] [Google Scholar]
- Walker J. D., Colwell R. R. Mercury-resistant bacteria and petroleum degradation. Appl Microbiol. 1974 Jan;27(1):285–287. doi: 10.1128/am.27.1.285-287.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss A. A., Murphy S. D., Silver S. Mercury and organomercurial resistances determined by plasmids in Staphylococcus aureus. J Bacteriol. 1977 Oct;132(1):197–208. doi: 10.1128/jb.132.1.197-208.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss H. V., Koide M., Goldberg E. D. Mercury in a Greenland ice sheet: evidence of recent input by man. Science. 1971 Nov 12;174(4010):692–694. doi: 10.1126/science.174.4010.692. [DOI] [PubMed] [Google Scholar]
- Westö G. Methylmercury as percentage of total mercury in flesh and viscera of salmon and sea trout of various ages. Science. 1973 Aug 10;181(4099):567–568. doi: 10.1126/science.181.4099.567. [DOI] [PubMed] [Google Scholar]
- Williams J. W., Morrison J. F. The kinetics of reversible tight-binding inhibition. Methods Enzymol. 1979;63:437–467. doi: 10.1016/0076-6879(79)63019-7. [DOI] [PubMed] [Google Scholar]
- Wood J. M. Biological cycles for toxic elements in the environment. Science. 1974 Mar 15;183(4129):1049–1052. doi: 10.1126/science.183.4129.1049. [DOI] [PubMed] [Google Scholar]
- Wood J. M., Cheh A., Dizikes L. J., Ridley W. P., Rakow S., Lakowicz J. R. Mechanisms for the biomethylation of metals and metalloids. Fed Proc. 1978 Jan;37(1):16–21. [PubMed] [Google Scholar]
- Wood J. M., Kennedy F. S., Rosen C. G. Synthesis of methyl-mercury compounds by extracts of a methanogenic bacterium. Nature. 1968 Oct 12;220(5163):173–174. doi: 10.1038/220173a0. [DOI] [PubMed] [Google Scholar]
- de la Cruz F., Grinsted J. Genetic and molecular characterization of Tn21, a multiple resistance transposon from R100.1. J Bacteriol. 1982 Jul;151(1):222–228. doi: 10.1128/jb.151.1.222-228.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]


