Abstract
Background:
DNA barcode-based molecular characterization is in practice for plants, but yet lacks total agreement considering the selection of marker. Plant species of subfamily Rauvolfioideae have long been used as herbal medicine by the majority of tribal people in Northeast (NE) India and at present holds mass effect on the society. Hence, there is an urgent need of correct taxonomic inventorization vis-à-vis species level molecular characterization of important medicinal plants.
Objective:
To test the efficiency of matK in species delineation like DNA barcoding in Rauvolfiadae (Apocynaceae).
Materials and Methods:
In this study, the core DNA barcode matK and trnH-psbA sequences are examined for differentiation of selected ethnomedicinal plants of Apocynaceae. DNA from young leaves of selected species was isolated, and matK gene (~800 bp) and trnH-psbA spacer (~450 bp) of Chloroplast DNA was amplified for species level identification.
Results:
The ~758 bp matK sequence in comparison to the trnH-psbA showed easy amplification, alignment, and high level of discrimination value among the medicinal Rauvolfioidae species. Intergenic spacer trnH-psbA is also exhibited persistent problem in obtaining constant bidirectional sequences. Partial matK sequences exhibited 3 indels in multiple of 3 at 5 end. Evidently, generated matK sequences are clustered cohesively, with their conspecific Genbank sequences. However, repeat structures with AT-rich regions, possessing indels in multiple of 3, could be utilized as qualitative molecular markers in further studies both at the intra-specific and shallow inter-specific levels like the intergenic spacers of CpDNA.
Conclusion:
matK sequence information could help in correct species identification for medicinal plants of Rauvolfioideae.
Keywords: Apocynaceae, DNA barcoding, ethnomedicinal, indels, matK
INTRODUCTION
DNA barcoding is emerged as powerful technique of species identification and exemplified with its wide application in monitoring and documentation of bio-resource.[1,2,3,4] The technique utilizes ~650 bp region of mitochondrial COI in animals[5] and various chloroplast regions (matK, rbcL, and trnH-psbA) in plants.[6,7,8] The application of the technique emphasizes some thrust areas, like documentation of the important and vulnerable ethnomedicinal plant bio-resources, dealing with which is recently defined as the subject “Ethnobotany Genomics.”[9] The principal issues in ethnobotany emphasized the importance of correct species identification and deciphering of indigenous and conventional knowledge of restorative plant usage and their transfer for the promotion of bio-prospect in human health care. Apocynaceae is one of the 10 largest angiosperm families (including Asclepiadaceae) and comprises of several prominent medicinal plants, like Rauvolfioideae subfamily of Apocynaceae is known for the rich source of typical laticiferous tissues, which produce various alkaloids and cardenolides being used in traditional medicines for stomach ulcer, fever, asthma, whooping cough, etc. Similarly, Catharanthus roseus is the source of very important drug viz. vinblastine, vincristine used in cancer chemotherapy.[10] Calotropis gigantea is also a potential candidate source for anti-cancer drugs,[11] and Allamanda cathartica possess a remarkable wound healing function.[12]
Indian saga has a long heritage of using numerous medicinal and aromatic plants (MAPs) for human health care, and the nation is bestowed with rich resources of plant bio-diversity distributed in various ecological conditions. It is the home of about 17,000 of global plant species and expected to be fully explored. It is reported that above 2000 species of ethnobotany plants have been utilizing by various medicines in Northeast India.[13] Amidst, the galaxy of rich traditional knowledge of herbal medicine in use by the majority of tribal people in NE India, there is an urgent need of correct taxonomic inventorization vis-à-vis species level molecular characterization of medicinal plants from this region in the globe. The conventional morphological techniques involve difficulties in species identification from any unstructured plant part. Thus, the ethnomedicinal resources of NE India seem least explored and found fragmentary. It entails the need of intervention of modern tools to characterize the molecular marker of important and vulnerable medicinal plants for correct species-level identification as well as their inventorization.
The DNA barcoding is rapidly evolving, but yet provides full agreement on which region(s) of DNA should be universally used for plants. In the current study, we have explored the effectiveness of matK and trnH-psbA spacer in differentiation of selected ethnomedicinal plants (Catharanthus roseus (L.) G. Don, Alstonia scholaris (L.) R. Br., Thevetia Peruviana (Pers.) Merrill, Allamanda cathartica L. Allamanda, Tabernaemontana divaricata (L.) Alston, Calotropis gigantea L. R. Br. Ex Ait) belonging to the family Apocynaceae inhabiting in NE India. The matK is located in the large single-copy region of chloroplast genome, nested between the 5´ and 3´ exons of trnK, t-RNA–lysin. In matK, rates of substitution among all the 3 codon positions are reported almost equal,[14] leading to the high rate of substitution, which results from non-synonymous mutations, but amino acid replacements occur as chemically-conserved, preserving its structural and biochemical properties.[15] The trnH-psbA spacer is among the most variable plastid regions in angiosperms. It is a popular tool for plant population genetic and species-level authentication.[16,17] The study shows the efficiency of matK in species delineation like DNA barcoding in Rauvolfiadae, and bears insights of effective utilization of matK indels in multiple of 3 for studies both at the intra-specific and shallow inter-specific levels in the entire family Apocynaceae.
MATERIALS AND METHODS
Sample collection, DNA Isolation, and PCR amplification
Young leaves of selected ethnomedicinal plants of Rauvolfioideae were collected aseptically from different sources in Southern Assam, India. All the species examined in the study were carefully identified by expert. About 40 mg, wet young leaves were homogenized in the DNA extraction buffer (50 mM Tris HCl pH 8.0, 25 mM EDTA pH 8.0, 150 mM NaCl, and 2 μL/mL β- mercaptoethanol). Genomic DNA was extracted through successive steps using 5 M Potassium acetate (pH 9.0), Phenol:Chloroform:Isoamylalchol (25:24:1), Chloroform:Isoamylalchol (24:1). To obtain high-quality DNA, free from polysaccharides and other metabolites that might interfere during PCR amplification, purified DNA concentration of each sample was estimated both fluorometrically and by comparison of ethidium bromide-stained band intensities against standard λ DNA. PCR was performed using primers pair, matK-F 5´-TAATTTACGATCAATTCATTC-3´, matK-R 5´-GTTCTAGCACAAGAAAGTCG-3´ and trnH-F 5´-CGCGCATGGTGGATTCACAATCC-3´ and psbA-R 5´-GTTATGCATGAACGTAATGCTC-3´ for matK and trnH-psbA, respectively.[18] The PCR reaction of 30 μl mixture contained 20 ng genomic DNA, 20 pmole each primer, 0.2 mM of each dNTPs, 0.5 units of high fidelity Taq polymerase enzyme (Applied Biosystem), 1Xbuffer, and 1.5 mM MgCl2. PCR thermal conditions were 94°C for 3 minutes, 30 cycles at 94°C for 1 minute, 48°C for 45 seconds, 72°C for 45 seconds in the case of matK, and 94°C for 3 minutes, 30 cycles at 94°C for 1 minute; 51°C for 45 seconds, 72°C for 45 seconds for trnH-psbA and a final extension at 72°C for 10 minutes for both the cases. The PCR products were checked by 1.5% agarose gel electrophoresis.
Purification of PCR Products and DNA sequencing
The PCR products of presumed size were extracted using QIA quick PCR purification kit (QIAGEN, Cat. No.28704). The purified PCR products were sequenced both bi-directionally using automated DNA sequencer (ABI 3700).
Sequence analysis
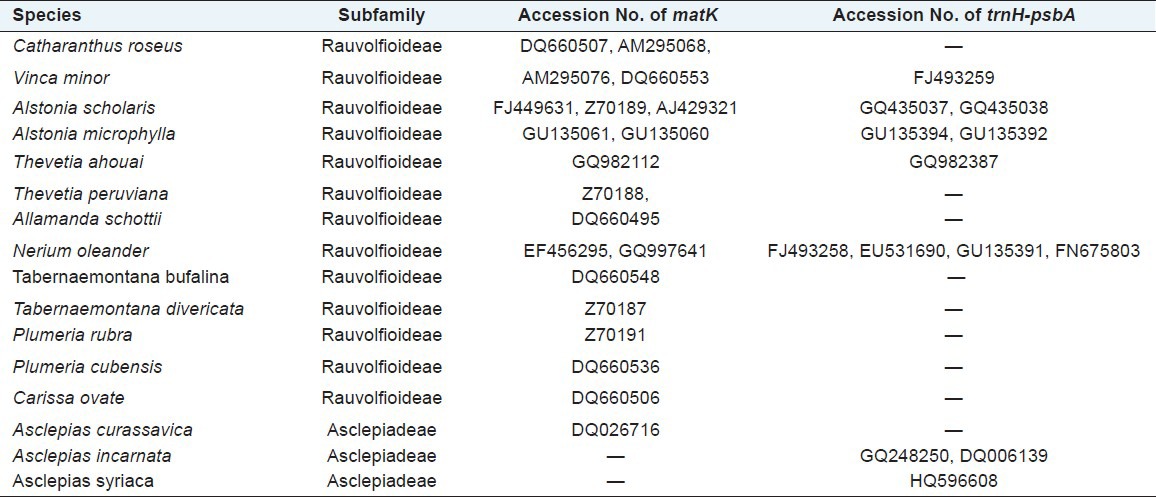
Raw traces were manually edited, and both forward and reverse sequences were subsequently aligned to generate targeted sequences. The 3´ and 5´ terminals were clipped to generate consensus sequences for each taxon for sequence length of ~ 758 bp (Nt. 520-1278) for matK and ~450 bp for trnH-psbA. The Open Reading Frame (ORF) for matK was checked, and correct amino acid sequences were determined by online software ORF prediction (http://www.ncbi.nlm.nih.gov/gorf/gorf.html). These matK and trnH-psbA sequences were aligned individually for combined data set using the ClustalX program.[19] The aligned sequences were corrected manually, and nucleotide compositions were calculated using BioEdit program.[20] Neighbor-joining (NJ) method was used for calculating intra- and interspecies divergence. In addition, 20 sequences of matK and 13 sequences of trnH-psbA intergenic spacer for same or related taxa of the studied specimen were obtained from GenBank (http://www.ncbi.nlm.nih.gov/genbank/) [Table 1]. The generated sequences of both matK and trnH-psbA for the studied species of Apocynaceae were subsequently submitted to NCBI.
Table 1.
matK and trnH-psbA sequences from NCBI with their Accesion No also given
Phylogenetic analysis
Pair-wise nucleotide sequence divergences were calculated using the Kimura-2-parameter (K2P) model to generate the distance matrices, and the neighbor-joining (NJ) analysis was done in MEGA 4.2[21] to examine phylogenetic relationship between 14 taxa from a subfamily Rauvolfioideae, and two taxa from the subfamily Asclepiadaceae of Apocynaceae. K2P distances were used following the guidelines of the Consortium for the Barcoding of Life (CBOL) to evaluate performance barcoding locus (http://www.barcoding.si.edu/protocols.html). A total of 1000 bootstraps replicates were calculated for the NJ tree construction.
RESULTS
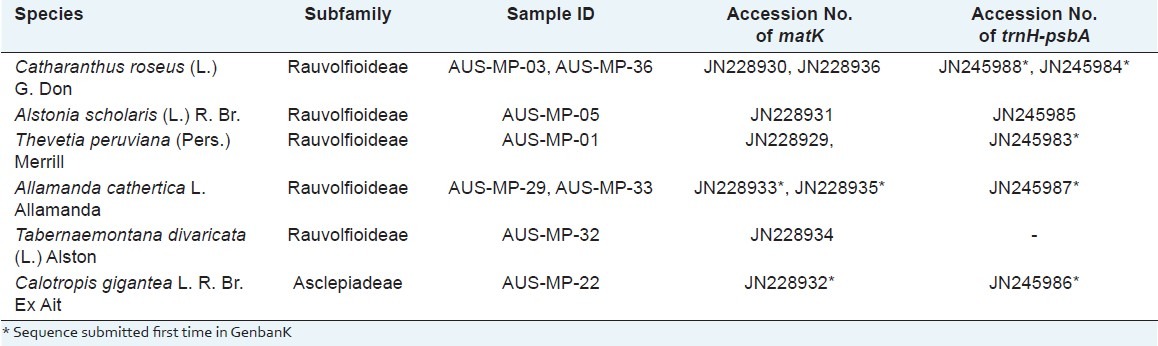
In this study, we uncovered 8 sequences of the matK region and 6 sequences of trnH-psbA spacer from the studied specimens, which include the few sequences that have been determined for the first time. The matK sequences of Allamanda cathartica (JN228933, JN228935) and Calotropis gigantea (JN228932), and trnH-psbA spacer of Catharanthus roseus (JN245984 and JN245989), A. cathartica (JN245987), C. gigantea (JN245986) T. peruviana (JN245983) are the novel sequences contributed from the study [Table 2].
Table 2.
List of Plant sample of Apocynaceae examined in this study scientific name, subfamily, Voucher, Accession Number of sequences of matK and trnH-psbA also given
Due to length variations in the matK sequences, only 758 aligned nucleotide positions were used in sequence analysis, of which a total of 189 variable and 157 parsimony-informative positions were found. However, the trnH-psbA sequences were not included in the subsequent analysis because the alignment was impossible across the Apocynaceae family [Figure 1]. Also, very few sequences are available from Rauvolfioideae in GenBank. Nucleotide composition of matK sequences of Apocynaceae is strong A+T bias (average 65.6% for all codon) where a percentage of T (36.5%) is higher than A (29.1%). The rates of substitution among the 3 codon position were almost equal [Figure 2].
Figure 1.
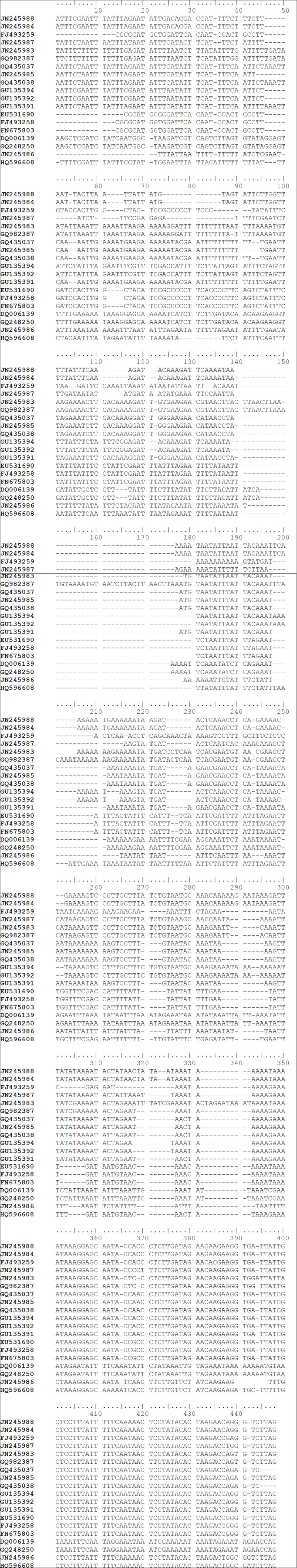
Showing Alignment of 19 sequences of trnH-psbA of Apocynaceae, containing indels of different regions
Figure 2.
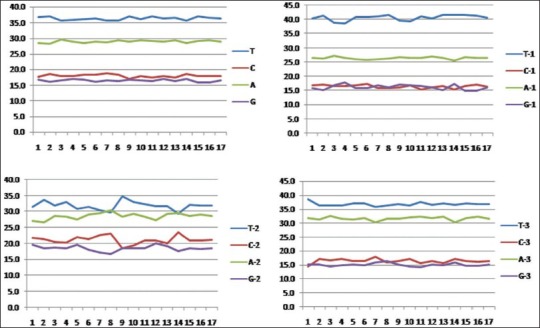
Nucleotide compositions of ~758 bp partial matK for the different species of Apocynaceae plants. The frequencies of nucleotide in sequences are present as the total average value for the all the codon positions and for each codon position separately with the accuracy to tenths of a percent. (A, T, G, C shown average value for all codon positions. A-1, T-1, G-1, C-1 shown average value for first codon position. A-2, T-2, G-2, C-2 shown average value for second codon position. A-3, T-3, G-3, C-3 shown average value for third codon position. A+T, A1+T1, A2+T2, A3+A3 represent the average value of A+T bias of total and each codon position.)
matK sequences exhibited indels in multiple of 3 at 5´ end where a 12 bp insertion (641-652 region) was found in Tabernaemontana divaricata, Tabernaemontana bufalina, Calotropis gigantea, and Asclepias curassavica; next 12 bp insertion (677-688 region) was found in Tabernaemontana divaricata and Tabernaemontana bufalina, while the other 6 bp insertion (1124-1129 region) was found only in Calotropis gigantea and Asclepias curassavica of a subfamily Asclepiadaceae [Figure 3].
Figure 3.
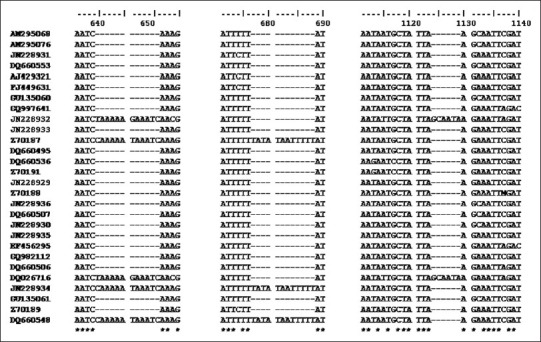
Showing Alignment of 28 sequences of matK of Apocynaceae, containing indels of 3 different regions. A 12 bp insertion found in Tabernaemontana divaricata (Z70187, JN228934), Tabernaemontana bufalina (DQ660548), Calotropis gigantea (JN228932), Asclepias curassavica (DQ026716) (641-652 region) and in Tabernaemontana divaricata (Z70187, JN228934), Tabernaemontana bufalina (DQ660548) (677-688 region) and 6 bp insertion in Calotropis gigantea (JN228932), Asclepias curassavica (DQ026716) (1124-1129 region).
* indicate conserve nucleotide
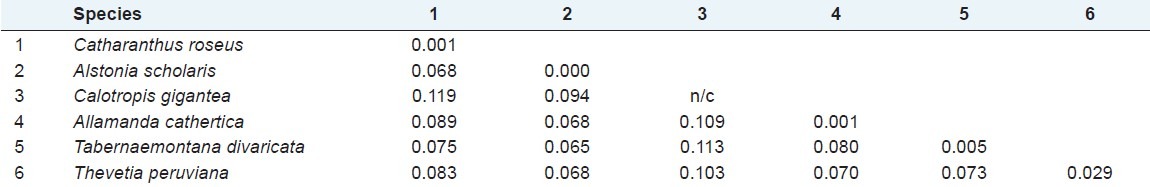
To evaluate the degree of DNA polymorphism, sequence divergence between and within species were calculated by Kimura 2-parameter (K2P) that revealed high average inter-specific and low intra-specific distances. Highest inter specific distance was 0.119 between Catharanthus roseus and Calotropis gigantea. Thevetia peruviana, Tabernaemontana divaricata, Allamanda cathartica, and Alstonia scholaris also showed high distance with Calotropis gigantea. Minimum inter-specific (0.065) was found between Catharanthus roseus and Tabernaemontana divaricata; maximum mean divergence within species (0.029) was found in Thevetia peruviana, and minimum mean divergence (0.00) was found in Alstonia scholaris [Table 3]. The accuracy of barcoding depends on the barcode gap between intra-specific and inter-specific variation. Sequence variation between species has to be high enough to tell them apart, while the distance within species must be low for them to cluster together.
Table 3.
Mean divergence (K2P) within (bold number on diagonal) and among (below diagonal) the 6 species of Apocynaceae from southern Assam. (n/c indicates comparable due to only one accession number)
The different species of Apocynaceae have formed distinctive clusters. Evidently, all the database sequences and the conspecific generated sequences of Catharanthus roseus, Thevetia peruviana, Tabernaemontana divaricata, Allamanda cathartica, and Alstonia scholaris with Genbank accession numbers are clustered cohesively. However, the members of Asclepiadaceae subfamily, Calotropis gigantea and Asclepias curassavica, were located at the basal position, hence used as an out group of the phylogenetic tree [Figure 4].
Figure 4.
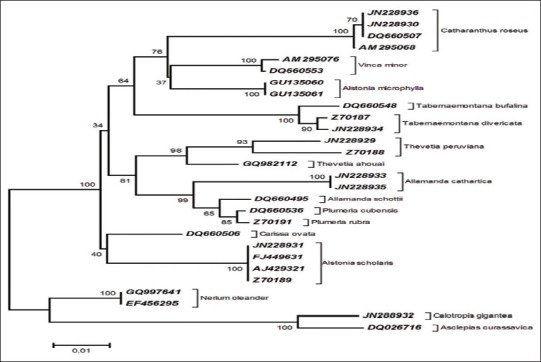
Neighbor-Joining analysis of Kimura2-parameter (K2P) distance of matK sequences of Apocynaceae ~758 aligned nucleotide positions of matK (Nt. 520-1278) were used in phylogenetic analysis. A total of 1000 bootstrap replicates were calculated for the NJ tree construction.
DISCUSSION
Analyzes of the targeted single loci matK (~ 750 bp, Nt. 520-1278) sequences depicted repeat structures with AT-rich regions possessing indels in multiple of 3, and high rate of substitution contributed a considerable number of characters for resolving the phylogeny of the ethnomedicinal plants of Apocynaceae. Occurrences of indels in matK sequences have also been explored to the extent of their applicability as qualitative molecular markers depending upon the size, position, and influence of open reading frame.[22] Several molecular processes are known to create indels, viz., polymerase slippages during DNA replication so called slipped-strand mispairing,[23] and due to addition or subtraction of short repeat sequences, which are primarily AT rich.[22] In general, microstructural changes in DNA, such as, insertions and deletions (indels), and inversions in introns and intergenic spacers, have been importantly used both for resolving phylogenetic relationships among the angiosperms[24,25] and for inferring relationships among more closely related taxa.[26] Imperatively, these changes in protein coding gene are very rare phenomenon, because these changes would lead into non-synonymous mutation. But, the observed indels in the presumed barcode region of matK happened in multiple of 3 nucleotides, thereby reduced the chances of frameshift mutation and did not interrupt the site of maturase activity in X domain. So, matK indels could be utilized as qualitative molecular marker for studies both at the intra-specific and shallow inter-specific levels like the intergenic spacers of CpDNA.
The sequence divergence among the studied ethnomedicinal plants of Apocynaceae revealed the highest divergence (0.119) between Catharanthus roseus and Calotropis gigantea [Table 3]. Moreover, Calotropis gigantea being a member of subfamily Asclepiadoideae always consistently high rate of divergences with other 5 studied members of subfamily Rauvolfioideae. Thus, following the notional DNA barcode concept, it can be justifiably infer that use of the partial matK sequence having reliable barcode gap as characterized in the study would be appreciably applicable to the species level discrimination of the important ethnomedicinal plants belonging to the family Apocynaceae.
Furthermore, NJ tree showed that the member of Rauvolfioideae subfamily Apocynaceae formed one clade where different species clustered into different subclade. The generated sequences of Allamanda cathartica is found closely related to Allamanda schottii. It is also close to genera Thevetia and Plumeria. Although Alstonia microphylla and Alstonia scholaris are the congeners but placed in different clades, which may be due to polyphyletic nature of Alstonieae.[27] Two sequences from Neruim oleander of subfamily Apocynoideae, and two members, viz. Calotropis gigantea and Asclepias curassavica of subfamily Asclepiadoideae, formed two distinct clades at the basal position of phylogenetic tree. Large sample sizes are required to increase the power of the test in Asclepiadaceae subfamily members, but the poor number of matK sequences of Asclepiadaceae in the database remained a limitation of the study, which entail the study using large sample sizes from different geographical location.
Recently, the CBOL Plant Working Group (2009) confirmed and suggested the combination of matK with rbcL as a universal plant DNA barcode[28] though the low discriminating power of rbcL gene is severally reported.[6,8] On the contrary, insertions, deletions, and short sequence repeats were common and often more numerous than single base pair substitution that has been the limitation on the part of trnH-psbA, hence remained unable to fulfill the criteria of plant DNA barcoding.[7] Nevertheless, in the present study, intergenic spacer trnH-psbA also exhibited persistent problem in obtaining constant bidirectional sequences. Our study showed that species identification of Rauvolfioideae subfamily is possible using phylogenetic analyzes constructed from partial matK sequences (Nt. 520-1278), which is comparable to that of the full-length sequences, also had species discrimination power. The observed divergences among the studied species using the partial matK sequences maintained a reliable gap, which holds good to the concept of species discrimination through DNA barcoding.[29] Furthermore, the NJ phylogenic tree, based on K2P model, also efficiently distinguished the species under study using the partial matK sequence. This gene has been identified as a universal DNA barcode for flowering plants.[30]
CONCLUSION
The matK sequences within and among the Rauvolfioideae sub-family have shown indels in multiple of 3, particularly N-terminal regions. The matK indels could be utilized as studies both at the intra-specific and shallow inter-specific levels like intergenic spacers of CpDNA. To evaluate the indel containing regions, a more powerful algorithm is needed to calculate the intra- and inter-species comparisons. Our result suggests that matK sequence information could help in correct species identification for medicinal plants of Rauvolfioideae and in providing diagnostics for rapid and easier identification of mal species forensics in herbal formulation, which bear insights of similar application in family Apocynaceae.
Footnotes
Source of Support: Infrastructural support from Department of Biotechnology, Govt. of India
Conflict of Interest: No.
REFERENCES
- 1.Hollingsworth PM, Graham SW, Little DP. Choosing and using a plant DNA barcode. PLoS One. 2011;6:e19254. doi: 10.1371/journal.pone.0019254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kress WJ, Wurdack KJ, Zimmer EA, Weigt LA, Janzen DH. Use of DNA barcodes to identify flowering plants. Proc Natl Acad Sci U S A. 2005;102:8369–74. doi: 10.1073/pnas.0503123102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu Z, Chen SL, Song JY, Zhang SJ, Chen KL. Application of deoxyribonucleic acid barcoding in Lauraceae plants. Pharmacogn Mag. 2012;8:4–11. doi: 10.4103/0973-1296.93301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stoeckle MY, Gamble CC, Kirpekar R, Young G, Ahmed S, Little DP. Commercial teas highlight plant DNA barcode identification successes and obstacles. Sci Rep. 2011;1:42. doi: 10.1038/srep00042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hebert PD, Cywinska A, Ball SL, deWaard JR. Biological identifications through DNA barcodes. Proc Biol Sci. 2003;270:313–21. doi: 10.1098/rspb.2002.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Asahina H, Shinozaki J, Masuda K, Morimitsu Y, Satake M. Identification of medicinal Dendrobium species by phylogenetic analyses using matK and rbcL sequences. J Nat Med. 2010;64:133–8. doi: 10.1007/s11418-009-0379-8. [DOI] [PubMed] [Google Scholar]
- 7.Bruni I, De Mattia F, Galimberti A, Galasso G, Banfi E, Casiraghi M, et al. Identification of poisonous plants by DNA barcoding approach. Int J Legal Med. 2010;124:595–603. doi: 10.1007/s00414-010-0447-3. [DOI] [PubMed] [Google Scholar]
- 8.Sun XQ, Zhu YJ, Guo JL, Peng B, Bai MM, Hang YY. DNA barcoding the Dioscorea in China, a vital group in the evolution of monocotyledon: Use of matK gene for species discrimination. PLoS One. 2012;7:e32057. doi: 10.1371/journal.pone.0032057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Newmaster SG, Ragupathy S. Ethnobotany genomics - discovery and innovation in a new era of exploratory research. J Ethnobiol Ethnomed. 2010;6:2. doi: 10.1186/1746-4269-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Van Der Heijden R, Jacobs DI, Snoeijer W, Hallard D, Verpoorte R. The Catharanthus alkaloids: Pharmacognosy and biotechnology. Curr Med Chem. 2004;11:607–28. doi: 10.2174/0929867043455846. [DOI] [PubMed] [Google Scholar]
- 11.Wong SK, Lim YY, Abdullah NR, Nordin FJ. Antiproliferative and phytochemical analyses of leaf extracts of ten Apocynaceae species. Pharmacogn Res. 2011;3:100–6. doi: 10.4103/0974-8490.81957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nayak S, Nalabothu P, Sandiford S, Bhogadi V, Adogwa A. Evaluation of wound healing activity of Allamanda cathartica. L. and Laurus nobilis. L. extracts on rats. BMC Complement Altern Med. 2006;6:12. doi: 10.1186/1472-6882-6-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kala CP, Dhyani PP, Sajwan BS. Developing the medicinal plants sector in northern India: Challenges and opportunities. J Ethnobiol Ethnomed. 2006;2:32. [Google Scholar]
- 14.Hilu KW, Borsch T, Muller K, Soltis DE, Soltis PS, Savolainen V, et al. Angiosperm phylogeny based on matK sequence information. Am J Bot. 2003;90:1758–76. doi: 10.3732/ajb.90.12.1758. [DOI] [PubMed] [Google Scholar]
- 15.Barthet MM, Hilu KW. Evaluating evolutionary constraint on the rapidly evolving gene matK using protein composition. J Mol Evol. 2008;66:85–97. doi: 10.1007/s00239-007-9060-6. [DOI] [PubMed] [Google Scholar]
- 16.Liu Y, Zhang L, Liu Z, Luo K, Chen S, Chen K. Species identification of Rhododendron (Ericaceae) using the chloroplast deoxyribonucleic acid PsbA-trnH genetic marker. Pharmacogn Mag. 2012;8:29–36. doi: 10.4103/0973-1296.93311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Strochova H, Olson MS. The architechture of the chloroplast psbA-trnH non coding region in angiosperms. Plant Syst Evol. 2007;268:235–56. [Google Scholar]
- 18.Ragupathy S, Newmaster SG, Murugesan M, Balasubramaniam V. DNA barcoding discriminates a new cryptic grass species revealed in an ethnobotany study by the hill tribes of the Western Ghats in southern India. Mol Ecol Resour. 2009;9(Suppl s1):S164–71. doi: 10.1111/j.1755-0998.2009.02641.x. [DOI] [PubMed] [Google Scholar]
- 19.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–82. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hall TA. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–8. [Google Scholar]
- 21.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–9. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 22.Hilu KW, Alice LA. Evolutionary implications of matK indels in Poaceae. Am J Bot. 1999;86:1735–41. [PubMed] [Google Scholar]
- 23.Kelchner SA. The evolution of non-coding chloroplast DNA and its application in plant systematic. Ann Mo Bot Gard. 2000;87:499–527. [Google Scholar]
- 24.Graham SW, Reeves PA, Burns AC, Olmstead RG. Microstructural changes in non-coding DNA: Interpretation, evolution and utility of indels and inversions in basal angiosperm phylogenetic inference. Int J Plant Sci. 2000;161:S83–96. [Google Scholar]
- 25.Ingvarsson PK, Ribstein S, Taylor DR. Molecular evolution of insertions and deletion in the chloroplast genome of silene. Mol Biol Evol. 2003;20:1737–40. doi: 10.1093/molbev/msg163. [DOI] [PubMed] [Google Scholar]
- 26.Golenberg EM, Clegg MT, Durbin ML, Doebley J, Ma DP. Evolution of a noncoding region of the chloroplast genome. Mol Phylogenet Evol. 1993;2:52–64. doi: 10.1006/mpev.1993.1006. [DOI] [PubMed] [Google Scholar]
- 27.Simoes AO, Livshultz T, Conti E, Endress ME. Phylogeny and systematic of the Rauvolfioideae (Apocynaceae) based on molecular and morphological evidence. Ann Mo Bot Gard. 2007;94:268–97. [Google Scholar]
- 28.Hollingsworth MP, Forrest LL, Spouge LJ, Hajibabaei M, Ratnasingham S, van der Bank M, et al. A DNA barcode for land plants. Proc Natl Acad Sci U S A. 2009;106:12794–7. doi: 10.1073/pnas.0905845106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gao T, Sun Z, Yao H, Song J, Zhu Y, Ma X, et al. Identification of Fabaceae plants using the DNA barcode matK. Planta Med. 2011;77:92–4. doi: 10.1055/s-0030-1250050. [DOI] [PubMed] [Google Scholar]
- 30.Lahaye R, van der Bank M, Bogarin D, Warner J, Pupulin F, Gigot G, et al. DNA barcoding the floras of biodiversity hotspots. Proc Natl Acad Sci U S A. 2008;105:2923–8. doi: 10.1073/pnas.0709936105. [DOI] [PMC free article] [PubMed] [Google Scholar]


