Abstract
Breast cancer research is hampered by difficulties in obtaining and studying primary human breast tissue, and by the lack of in vivo preclinical models that reflect patient tumor biology accurately. To overcome these limitations, we propagated a cohort of human breast tumors grown in the epithelium-free mammary fat pad of SCID/Beige and NOD/SCID/IL2γ-receptor null (NSG) mice, under a series of transplant conditions. Both models yielded stably transplantable xenografts at comparably high rates (~21% and ~19%, respectively). Of the conditions tested, xenograft take rate was highest in the presence of a low-dose estradiol pellet. Overall, 32 stably transplantable xenograft lines were established, representing 25 unique patients. Most tumors yielding xenografts were “triple-negative” (ER-PR-HER2+) (n=19). However, we established lines from three ER-PR-HER2+ tumors, one ER+PR-HER2−, one ER+PR+HER2− and one “triple-positive” (ER+PR+HER2+) tumor. Serially passaged xenografts show biological consistency with the tumor of origin, are phenotypically stable across multiple transplant generations at the histologic, transcriptomic, proteomic, and genomic levels, and show comparable treatment responses as those observed clinically. Xenografts representing 12 patients, including two ER+ lines, showed metastasis to the mouse lung. These models thus serve as a renewable, quality-controlled tissue resource for preclinical studies investigating treatment response and metastasis.
Introduction
In translational breast cancer research, our ability to evaluate clinical responses of human tumors to new therapeutic agents is restricted experimentally. For example, we cannot evaluate the clinical response of a single treatment-naive tumor to multiple candidate therapeutics. Further, the number of in vivo preclinical human tumor models currently available remains limited, thus precluding conduct of xenograft-based “mouse clinical trials” reflecting the heterogeneity of human tumors using candidate therapeutic agents. These limitations severely compromise our ability to develop and test novel therapeutics, and to predict the best course of treatment for a given tumor subtype, and more importantly, an individual breast cancer patient.
Historically, in vivo experimental therapeutic research has relied on either genetically engineered mouse models, or “xenograft” transplantation models in which established human cancer cell lines are transplanted into immunocompromised host mice [1-3]. However, while mouse models mutant for TP53 do show a high degree of heterogeneity, genetically engineered animal models do not fully recapitulate the full spectrum of human breast cancers [4]. Similarly, a cell line represents only a single tumor type, and indeed only a single patient. Further, most available cell lines have been maintained in culture for years, or decades, and there has been debate whether these cell lines still accurately reflect the biological characteristics of the tumor of origin [5-7].
Early attempts to use primary breast cancer tissue xenografts (a.k.a. patient-derived xenografts (PDX) models, or “tumorgrafts”) as experimental models met with limited success [1, 2, 5, 6, 8-12], with typical rates of stable transplantation being 10% or less. Most of these attempts used athymic (“nude”) or NOD/SCID (non-obese diabetic/severe combined immunodeficiency disorder) mice, which lack B- and T-cell function but retain innate cellular immunity (natural killer (NK) cells, macrophages etc.) frequently leading to elimination of tumor cells over time [13, 14]. A vast majority of the stable xenografts produced have not expressed the estrogen receptor (ER-negative), but ER+ xenografts have recently begun to be reported [9, 11, 12, 15].
In a recent report, the efficiency of transplantation using normal human mammary epithelial cells (from reduction mammoplasty) was increased by “humanizing” the mammary fat pad of NOD/SCID mice via introducing an immortalized human fibroblast cell line, derived from a normal donor, into the mammary fat pad prior to transplantation [10]. The influence of these human fibroblasts on the growth of patient-derived breast cancer was not tested. In any case, because these fibroblasts were derived from a normal patient rather than from the patient-matched tumors, the presence of such fibroblasts may alter tumor biology significantly.
We sought to circumvent some of these limitations by propagating human tumors as xenografts in SCID/Beige immunocompromised mice, which were known to accept transplants of hematopoetic malignancies with higher efficiency than traditional immunocompromised models, and had not been used previously to establish breast cancer xenografts. SCID/Beige mice lack B-cell, T-cell, and NK cell function entirely, but show enhanced macrophage populations relative to wild type mice [13, 14]. Macrophages are required for mammary gland growth [16, 17], and immature myeloid cells of the macrophage lineage were recently shown to promote tumor invasion and metastasis [18]. Three different transplantation conditions were compared, and the optimal transplant condition also used to evaluate outgrowth rates in NOD/SCID/IL2γ-receptor null (NSG) immunocompromised mice. Resulting stably transplantable xenografts were characterized with respect to expression of clinically relevant biomarkers, and gene expression patterns (mRNA and protein), and a subset of patient/xenograft treatment responses, to lay the foundation for their use as preclinical models for breast cancer research.
Materials and Methods
Patient recruitment
Breast cancer patients were recruited from clinics in the Baylor College of Medicine Breast Center and Ben Taub General Hospital under IRB-approved protocols. Most patients received initial core needle biopsies at the time of diagnosis, and again either during or after treatment. Surgical samples were also obtained whenever possible.
Establishment of xenografts
The study design is outlined in Supplemental Figure 1. All mice were maintained and treated in accordance with the National Institutes of Health Guide for the Care and Use of Experimental Animals with approval from the Baylor College of Medicine Institutional Animal Care and Use Committee. A detailed surgical protocol was published elsewhere [19].
Pretreatment biopsy and post-treatment surgical specimens were received within an hour after excision. For fragment transplantation, samples were minced into ~1 mm3 fragments and transplanted directly into epithelium-free “cleared” fat pads [20] of recipient SCID/Beige (Charles River Laboratories, Wilmington, MA), or NSG mice (Jackson Laboratories, Bar Harbor, ME) (n=2 per patient). Transplants were performed under the following conditions: Condition 1: unmanipulated host mice, Condition 2: 17β-Estradiol supplementation (60-day release, 0.36 mg/pellet, Innovative Research of America, FL, Cat.#SE-121), or Condition 3: 17β-Estradiol supplementation with inclusion of 5 × 104 immortalized normal human fibroblasts (passages 35-41) (1:1 unirradiated:irradiated cells (4 Gy), as described previously [21] (fibroblasts generously provided by Dr. Charlotte Kuperwasser). Mice were palpated weekly, and tumor growth measured using calipers. Conditions found to be optimal for SCID/Bg mice were then tested using NSG mice (Condition 4).
We were also able to obtain a few samples from either pleural effusion or metastatic ascites. Fluid was centrifuged, and cells resuspended in a volume of 10-50ul, and injected into the cleared mammary fat pad using a Hamilton syringe. Xenografts derived from such samples were not included in the statistical analyses, but are included here for completeness of the collection.
Regardless of the source of tumor cells, when primary outgrowths reached 10mm in diameter, or if glands were suspected of carrying small primary outgrowths, fragments were re-transplanted into new hosts (n=3-4) as secondary xenografts. If no overt tumor formation was observed by 30 weeks, glands were harvested and processed for histological evaluation. Primary outgrowth take was defined as a surviving tissue fragment >1mm in diameter and demonstrated to be proliferative (Ki67 positivity). A xenograft line was defined as stable upon growth at transplant generation 3 (TG3).
Statistical analysis of clinical characteristics, biomarker expression, and outgrowth potential
The overall primary outgrowth and stable xenograft take rates were computed for each transplantation condition (Table 1) and compared using logistic regression. Clinical characteristics were summarized and compared across four different transplantation conditions using Fisher’s exact test. Within each transplantation condition, primary outgrowth as well as stable take rate were compared by clinical characteristics using Fisher’s exact test (Supplemental Table 1). There were a few patient samples appearing in both transplantation conditions 2 and 3. We performed a McNemar test to examine those overlapping samples and did not observe any significant difference. Thus, we analyzed the data as independent samples for different transplantation conditions.
Table 1.
Comparative xenograft take rate by transplant condition.
| Transplant Condition |
Host Strain | Estradiol Pellet |
Human Fibroblasts |
Number of Patients |
Primary Outgrowth Rate (%) |
Stable Xenograft Take Rate (%) |
|---|---|---|---|---|---|---|
| 1 | Scid/Beige | − | − | 38 | 18/38 (47.4) | 1/38 (2.6) |
| 2 | Scid/Beige | + | − | 70 | 28/70 (40) | 15/70 (21.4) |
| 3 | Scid/Beige | + | + | 29 | 13/29 (44.8) | 1/29 (3.4) |
| 4 | NSG | + | − | 32 | 10/32 (31.3) | 6/32 (18.8) |
Xenograft characterization
Xenografts were validated as unique by STR DNA fingerprinting using the AmpF_STR Identifiler kit according to manufacturer’s instructions (Applied Biosystems cat 4322288). The STR profiles were compared to known ATCC fingerprints (ATCC.org), and to the Cell Line Integrated Molecular Authentication database (CLIMA) version 0.1.200808 (http://bioinformatics.istge.it/clima/) [22]. The STR profiles of all xenograft lines were unique (Supplemental Table 2).
Histological evaluation of xenografts
Transplanted glands/tumors were fixed in 10% neutral buffered formalin, paraffin embedded, and hematoxylin-eosin (H&E) stained. Outgrowths were analyzed by immunohistochemistry for stability of expression of clinically-relevant biomarkers (ERα, PR, HER2, Ki67) (ERα, Novocastra Laboratories Ltd, United Kingdom, Cat. #NCL-ER-6F11; PR, DAKO, CA, Cat. #M3568; HER2, NeoMarkers, Lab Vision Corporation, CA, Cat.#RM-9103; Ki67, DAKO,CA, Cat.# M7240) with positive controls. Additional biomarkers included human cytokeratin 19 (NeoMarkers, Lab Vision Corporation, CA, Cat. #MS-198-P, 1:400), Cytokeratin 5/6 (CK5/6) (DAKO, CA, Cat.# M7237 at a 1:100 dilution); EGFR (pharmDx™ Kit, DAKO, CA, Cat.# K1492, ready to use) and TP53 (Vector, CA, Cat# VP-P958, 1:1000 dilutions). Biomarker expression patterns were compared with the tumor of origin whenever possible, or with the clinical pathology report (Table 2 and Supplemental Table 3).
Table 2.
Patient, Primary Tumor, and Xenograft Characteristics
| Patient | Xenograft Line(s) |
Xenograft PAM50 Intrinsic Subtype |
Transplant Condition |
Patient Ethnicity |
Tumor Source and Treatment Status |
Patient Tumor Type |
Estrogen Receptor Status |
Progest-erone Receptor Status |
HER2 Status |
BRCA Status |
Patient Nodal Status |
Patient Metastatic Site(s) |
Xenograft Metastasis Rate To Mouse Lung (%) |
Patient Clinical Treatment(s) |
Patient Clinical Response |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | BCM-2147 | Basal | 1 | AA | Pre, P.Br | rIDC | − | − | − | + | Brain | 0 | AC | Res | |
| BCM-2277 | Basal | 1 | Post, P.Br | − | − | − | 0 | ||||||||
| 2 | BCM-2665 | Basal | 3 | Hispanic | Post, P.Br | IDC, Mpap | - | − | − | − | 7.1 | AC, Doc | AC Sen, Doc. Res | ||
| 3 | BCM-3107 | Basal | 2 | Caucasian | Post, P.Br | IDC | − | − | − | − | 0 | Doc | Sen | ||
| 4 | BCM-3143 | Basal | 2 | Caucasian | Post wk6, P.Br | IDC | − | − | + | − | 0 | Lap −> Taxane + Trastuz | Lap Res; Taxane + Trastuz Res | ||
| BCM-3104 | Basal | 2 | Post wk4, P.Br | + | 0 | ||||||||||
| 5 | BCM-3204 | Basal | 2 | Caucasian | Pre, P.Br | IDC | 28.6 | AC | Res | ||||||
| 6 | BCM-3611 | Basal | 2 | AA | Pre, P.Br | IDC | − | − | − | + | 0 | AC + GSI −> Doc + GSI | AC + GSI Res; Doc + GSI Res | ||
| BCM-3824 | Basal | 2 | Post, P.Br | − | − | − | 0 | ||||||||
| 7 | BCM-3887 | Basal | 2 | AA | Pre, CWR | IDC | − | − | − | BRCA1 | − | Brain | 14.3 | Xeloda (5FU) | Res |
| 8 | BCM-3936 | Basal | 2 | Hispanic | Post, P.Br | IDC | − | − | − | + | 9.1 | AC | Sen | ||
| 9 | BCM-3807 | Basal | 2 | Hispanic | Pre, P.Br | IDC | − | − | − | − | 0 | AC −> Doc + GSI | AC Res; Doc + GSI Res | ||
| BCM-4400 | Basal | 2 | Post, P.Br | − | − | − | 0 | ||||||||
| 10 | BCM-3904 | Basal | 2 | Hispanic | Pre, P.Br | IDC | − | − | − | − | 18.2 | Doc | Res | ||
| 11 | BCM-4175 | HER2 | 4 | Hispanic | Post, P.Br | IDC | − | − | − | − | 0 | Das −> AC | Res | ||
| 12 | BCM-3963 | HER2 | 2 | Hispanic | Pre, P.Br | IDC | − | − | + | + | Brain | 13.6 | Lap + Trastuz | Sen | |
| BCM-4169 | HER2 | 4 | Post, P.Br | − | − | + | 8.3 | ||||||||
| 13 | BCM-4195 | Basal | 4 | Hispanic | Post, P.Br | IDC | − | − | − | − | 0 | Das −> Doc | Das Res; Doc Res | ||
| 14 | BCM-4013 | Basal | 4 | Caucasian | Pre, P.Br | IDC | − | − | − | + | 21.4 | Das + Doc | Res | ||
| 15 | BCM-4272 | Basal | 2 | Hispanic | Pre, P.Br | IDC | − | − | − | + | 28.6 | GSI + Doc | Sen | ||
| BCM-4849 | Basal | 2 | Post, P.Br | − | − | − | 0 | ||||||||
| 16 | BCM-4664 | Basal | 2 | AA | Pre, P.Br | IDC | − | − | − | + | 0 | Das + Doc | Res | ||
| 17 | BCM-4913 | Basal | 2 | AA | Post wk4, P.Br | IDC | − | − | − | BRCA1 | + | 0 | Doc | Res | |
| BCM-5438 | Basal | 2 | Post, P.Br | − | − | − | 0 | ||||||||
| 18 | BCM-4888 | HER2 | 4 | Hispanic | Post, P.Br | IDC, Mpap | + | + | + | + | 66.7 | AC −−> GSI + Doc | AC Res; GSI + Doc Sen | ||
| 19 | BCM-5097 | Basal | 2 | Caucasian | Post, P.Br | IDC | + | + | − | BRCA2 | + | 36.4 | Doc | Sen | |
| 20 | BCM-5156 | Basal | 2 | Hispanic | Post, P.Br | IDC | − | − | − | nr | 0 | GSI + Doc | Sen | ||
| 21 | BCM-5471 | Basal | 2 | Hispanic | Post, P.Br | IDC | − | − | − | + | 33.3 | GSI + Doc | Sen | ||
| 22 | BCM-5998 | Basal | 4 | Caucasian | Pre, CWR | IDC | − | − | − | nr | 0 | AC | Res | ||
| 23 | BCM-3613 | HER2 | 2 | Hispanic | Plural fluid | Met. | − | − | + | nr | Brain | 23.8 | AC, Pac, Traztuz, Lap, GSI + Doc, Others |
AC Res; Pac Res; Trastuz Res; Lap Res; GSI + Tax Res |
|
| 24 | BCM-3561 | HER2 | 2 | Caucasian | Ascites | Met. (ER+PR+ primary) |
− | − | − | 0 | Xeloda (5FU), Pac. Others | Xeloda Res; Pac Res | |||
| 25 | BCM-4189 | HER2 | 2 | Hispanic | Ascites | Met. (LCIS primary) |
+ | − | − | nr | 0 | AC, Pac, Xeloda, Arimidex, Faslodex |
AC Res; Pac Res; Xeloda Res; Arimidex Res; Faslodex Res |
Pre, pre-treatment; Post, post-treatment; P.Br, primary breast; CWR, chest wall recurrence; IDC, invasive ductal carcinoma; LCIS, lobular carcinoma in situ; Mpap, micropapillary; Met, metastatic disease; AC, doxorubicin (Adriamycin) and cyclophosphamide (Cytoxan) ; Doc, Docetaxel; Pac, Paclitaxel Lap, Lapatinib; Trastuz, Trastuzumab; GSI, gamma secretase inhibitor; Das, Dasatinib; Sen, ≥ 30% response; Res, < 30% response; AA, African American; nd, not determined; nr, not reported
Intrinsic subtype analysis
Xenografts were profiled as described previously using 244K human oligo microarrays (Agilent Technologies, Santa Clara, CA, USA) [23]. The probes or genes for all analyses were filtered by requiring the lowest normalized intensity values in both sample and control to be > 10. The normalized log2 ratios (Cy5 sample/Cy3 control) of probes mapping to the same gene (Entrez ID as defined by the manufacturer) were averaged to generate independent expression estimates. For Cy3-controls, we used Stratagene Human Universal Reference [24] enriched with equal amounts of RNA from the MCF7 and ME16C cell lines. Genes were median-centered and samples were standardized to zero mean and unit variance. To combine the BCM xenograft gene expression data with the UNC337 dataset, we estimated the technical bias from a subset of samples (n=35) of the UNC337 dataset that were also profiled on the 244K platform. Intrinsic subtype classification was performed in the combined BCM xenograft and UNC337 dataset using the PAM50 algorithm [25] and the 9-Cell Line Claudin-low Predictor [26] (Table 2 and Supplemental Table 3). All Agilent microarray data are available in the University of North Carolina (UNC) Microarray Database (https://genome.unc.edu/) and have been deposited in the Gene Expression Omnibus (GEO) under the accession number GEO: GSE34412.
Affymetrix Gene Expression Analysis
For most xenograft lines, Affymetrix gene expression arrays (U133 plus 2.0) were run on the first or second transplant generation (TG1 or TG2), and approximately every 5th transplant generation thereafter according to standard protocols recommended by Affymetrix (Affymetrix, Santa Clara, CA). Arrays were scanned on an Affymetrix GeneChip 3000 Scanner (Agilent, Palo Alto, CA). Raw data were then analyzed by ArrayAnalyzer (Insightful Corporation, Seattle, WA) for normalization and expression estimation. All Affymetrix microarray data have been deposited in GEO under the accession number GEO:GSE46106. An Affymetrix expression summary file is included as Supplemental Table 4. Sample information for Affymetrix data are shown in Supplemental Table 5 designated “affy.patient.info.final.color” (Supplemental Tables 4 and 5 are available by download only from www.bcxenograft.org).
Reverse Phase Protein Array Expression Analysis
Xenograft-derived tissue was harvested and frozen at −80 °C prior to use. Small pieces of tumor tissue were added to 2 ml tubes with ceramic beads together with ice-cold lysis buffer containing 1% Triton X-100, 50 mM Hepes pH 7.4, 150 mM NaCl, 1.5 mM MgCl2, 1 mM EGTA, 100 mM NaF, 10 mM NaPPi, 10% glycerol, 1 mM Na3VO4, Complete Protease Inhibitor Cocktail and Phosphatase Inhibitor cocktail (Roche Diagnostics). Protein supernatants were isolated as described previously [27], and protein concentration was determined by BCA assay (Pierce). Samples were diluted to a uniform protein concentration, and then denatured in 1% SDS sample buffer for 5 minutes at 95°C. Samples were stored at −80°C until use. RPPA analysis was performed as described previously [27, 28]. Data were obtained for 161 antibodies. A logarithmic value reflecting the relative amount of each protein in each sample was generated for analyses [29]. Similarity of proteomic gene expression was evaluated using cluster analysis as well as by Pearson distance correlation analysis. An RPPA data summary is shown in Supplemental Table 6. Sample information for the RPPA data is shown in Supplemental Table 7, designated “rppa.info.final.color”. Supplemental Tables 6 and 7 are available by download only from www.bcxenograft.org.
Sequenom Analysis
Polymerase chain reaction (PCR) and extension primers for each gene were designed using Sequenom, Inc. (San Diego, CA) Assay Design (Supplemental Table 8). PCR-amplified DNA was cleaned using EXO-SAP (Sequenom) primer extended by IPLEX chemistry, desalted using Clean Resin (Sequenom) and spotted onto Spectrochip matrix chips using a nanodispenser (Samsung). Chips were run in duplicate on a Sequenom MassArray MALDI-TOF MassArray system. Sequenom Typer Software and visual inspection were used to interpret mass spectra. Reactions that where ≥8% of the resultant mass run in the mutant site in both directions were scored as positive (Supplemental Table 9).
Metastasis rates
To evaluate metastatic behavior, lungs and liver were harvested from each host mouse at each transplant generation and evaluated grossly and histologically by H&E staining (3 sections per sample) (Table 2 and Supplemental Table 10).
Xenograft treatment response
Fresh xenograft tumor fragments of selected xenograft lines were transplanted into the cleared fat pad of recipient mice. When tumors reached a volume of ~200 mm3, mice were randomized and treated with either vehicle (9-10 mice), a single intraperitoneal injection of Docetaxel (20mg/kg) (3-9 mice), a single intraperitoneal injection of Doxorubicin (3mg/kg) (3-9 mice), or combined Trastuzumab and Lapatinib (10 mice) as described [30, 31], depending on the treatment the patient of origin received clinically. In some cases, patients were treated with chemotherapy in combination with an experimental targeted therapeutic (e.g. Dasatinib or a gamma secretase inhibitor). In such cases, resistance to both agents in the patient, and resistance to single agent in the xenograft were considered concordant. Tumor size was monitored twice weekly using calipers for a period of at least 2 weeks, and growth curves plotted. Sensitivity was defined as ≥30% regression (RECIST partial response or complete response); Resistance was defined as either <30% regression, stable disease, or continued growth (RECIST stable disease or progressive disease). Treatment responses in xenografts were compared to those of the primary tumor for concordance, and statistical significance of the difference between observed and expected concordance was evaluated by Fisher’s Exact Test. The degree of concordance above that expected by chance was evaluated using the kappa statistic.
Xenograft availability
Xenografts are available from the corresponding author for academic/nonprofit use on a cost recovery basis via a Material Transfer Agreement (mta@bcm.edu). Xenografts are maintained as viably frozen fragments (~1mm3 frozen slowly at −80°C and stored in liquid nitrogen; shipped on dry ice). Frozen fragments can be thawed rapidly and retransplanted. Fragments grow somewhat slower in the first transplantation after freezing, but revert to their characteristic growth pattern in subsequent transplant generations. All data presented in this report were derived from xenografts that had never been frozen.
Results
Establishment of xenografts using tumor fragments
Using SCID/Bg mice, three transplantation conditions were tested (Table 1 and Supplemental Table 1). Under transplantation condition 1 (unmanipulated hosts), primary tumor fragments representing 38 unique patients were implanted to a cleared fat pad to provide baseline take rates for comparison with experimental conditions. Upon harvest, 18 primary outgrowths were found after histological evaluation (~47% primary outgrowth rate). Only one stably transplantable xenograft line (BCM-2147) was established (2.6% stable take rate).
Under transplantation condition 2 (estradiol supplementation only), primary tumor fragments representing 70 unique patients were implanted with 28 primary outgrowths obtained (40% primary outgrowth rate) (Table 1). Fifteen patients yielded a stable xenograft (21.4% stable take rate), an approximately 8-fold increase above baseline, and consistent with recently published studies [1, 2, 5, 6, 8-12]. Stable lines represented 12 “triple negative” patients (Table 2), as well as two ER-PR-HER2+ patients (lines BCM-3143/3104 and BCM-3963/4169), and one ER+PR+HER2− patient (line BCM-5097). Notably, three xenograft lines established under this condition were generated from two BRCA1 mutation carriers (patient 7, line BCM-3887; and patient 17, lines BCM-4913 and BCM-5438), and one line was generated from a BRCA2 mutation carrier (patient 19, line BCM-5097) (Table 2).
In addition to these fragment transplant lines, under condition 2 we also established a stable line from ascites cells from one patient (BCM-3561). Ascites cells and the resulting xenograft were triple negative, whereas the primary tumor in this patient was ER+PR+. We also established a line from one ER+PR-HER2− patient (line BCM-4189) using ascites cells, as well as one line from an ER-PR-HER2+ patient (line BCM-3613), from cells derived from pleural effusion fluid (Table 2).
Given the success of estradiol supplementation, we hypothesized that addition of immortalized normal human fibroblasts would enhance the take rate further (as shown previously for normal human mammary epithelium [10]). Under these conditions (condition 3), primary and stable take rates were similar to those under condition 1 (Table 1), with 13 of 29 patients yielding primary outgrowths (44.8%), but only 1 of 29 patients (3.4%) yielding a stable xenograft line (line BCM-2665) (Table 2). Thus, rather than stimulating xenograft growth, the introduction of human fibroblasts derived from a normal patient was inhibitory to stable, but not primary, outgrowth. It is currently unclear why immortalized human fibroblasts would stimulate growth of normal human mammary tissue, but inhibit estradiol-enhanced growth of malignant tissue.
Overall, we found no significant differences in primary outgrowth rates between transplantation conditions using SCID/Bg host mice. However, there were significant differences for stable outgrowth rates, with condition 2 yielding more lines than condition 1 or 3 (OR: 11.0, 95% CI: 1.4-86.2, p=0.023; OR: 8.3, 95% CI:1.0-65.8, p=0.045, respectively).
Given that condition 2 (estradiol supplementation alone) provided our best stable take rate in SCID/Bg mice, these conditions were then tested in NOD/SCID/IL2γ-receptor null (NSG) immunocompromised mice (condition 4) (Table 1 and Supplemental table 1). Of 32 patients tested, ten (31.3%) yielded primary outgrowths, and six (18.8%) yielded stable xenograft lines. These included four triple negative lines (BCM-4175, BCM-4195, BCM-4013 and BCM-5998), one ER-PR-HER2+ line (BCM-4169), and one “triple positive” ER+PR+HER2+ line (BCM-4888) (Table 2). Primary outgrowth and stable take rate in NSG mice were not statistically different from those of SCID/Bg mice under condition 2 (logistic regression, p=0.40 and p=0.64, respectively).
In total, 32 stably transplantable xenograft lines were established representing 25 individual patients. Clinical features of the patient and primary tumor of origin for each stable xenograft line are shown in Table 2 and Supplemental Table 1. Of note, we established pre-treatment/post-treatment xenograft pairs (BCM-2147/2277; BCM-3611/3824; BCM 3807/4400; BCM 3963/4169; and BCM-4272/4849), from five patients. With respect to ethnicity, we established xenografts representing three major ethnic groups, including five African American (AA) patients, 13 Hispanic patients, and seven non-Hispanic Caucasian patients (Table 2). Finally, of the 22 patients used for fragment transplant, 11 patients were lymph node positive with 9 patients being node negative (two not reported). Also included were three definitively metastatic patients (patients 23-25).
Regardless of the host strain used initially, all ER-negative lines were propagated in the cleared fat pad of SCID/Beige mice without humanizing fibroblasts or estradiol after the third transplant generation. All ER+ lines were propagated in the presence of estradiol supplementation; their estrogen dependency and sensitivity to hormonal therapies is currently under investigation.
Xenograft take rate vs. patient clinical characteristics
To ensure that the difference in stable take rate across the transplantation conditions was not due to differences in patient characteristics we compared across four different transplantation conditions using Fisher’s exact test. Clinical characteristics of patients were similar in the four groups used under the different transplantation conditions (Supplemental Table 1).
To evaluate which clinical characteristics correlated with high tumor take rate within each transplantation condition, the primary and stable xenograft outgrowth rates were summarized and compared across or between levels of each clinical characteristic using Fisher’s exact test. Under condition 2, the stable xenograft take rate was significantly different between grades I or II vs grade III invasive carcinoma, with only grade III tumors yielding stably transplantable xenografts. The stable take rate of ER negative (~52%) and PR negative (~37%) tumors was significantly higher than that of ER positive (~2%) and PR positive (~3%) tumors, respectively. The rate of stable transplantation across ethnic groups was not statistically different (Supplemental Table 1).
Characteristics and stability of xenografts
To evaluate whether stable xenografts retained histological features and biomarker expression patterns consistent with the tumor of origin, we conducted comparative histological and immunohistochemical analyses of outgrowths using clinically relevant biomarkers (ER, PR, HER2), as well as TP53 (p53), epidermal growth factor receptor (EGFR, ErbB1), cytokeratin 19 (CK19), cytokeratin 5 (CK5) and Ki67, and compared biomarker status with the tumor of origin (Table 2 and Supplemental Table 3).
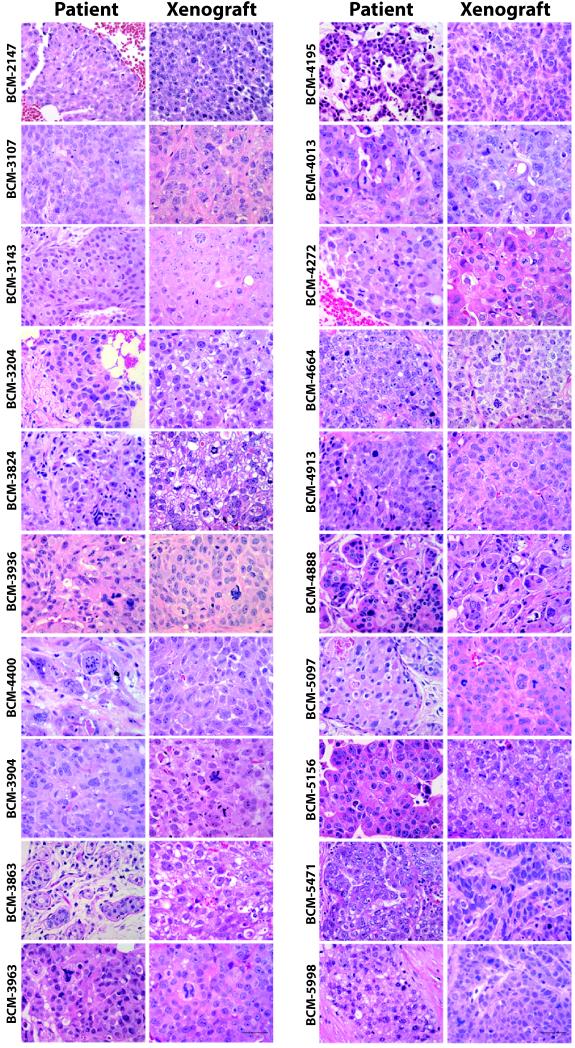
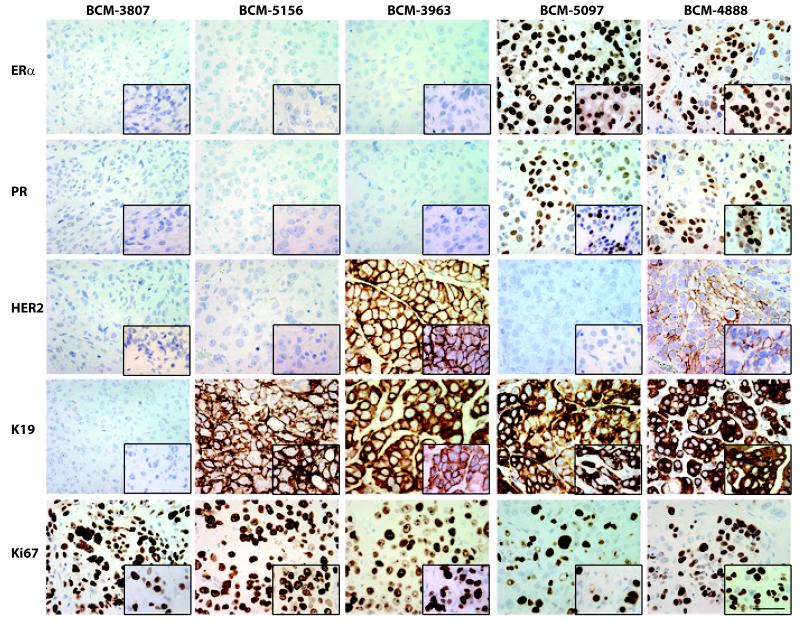
Figure 1 shows all xenograft lines for which matched primary patient samples were available for histological comparison (N=20) (unmatched xenograft lines are shown in Supplemental Figure 2). In all cases, patients retained their invasive histological phenotype as xenografts. Tumor cellularity was estimated to exceed 80% in all xenografts (Figure 1). Figure 2 shows five representative xenografts and their matched primary breast cancer with respect to the range of biomarker expression observed (biomarker expression fully summarized in Supplemental Table 3). Triple negative xenografts could be either CK19− (e.g. BCM-3807) or CK19+ (e.g. BCM-5156). All ER+ and/or HER2+ xenografts generated were CK19+. In all cases that could be compared, each xenograft retained the biomarker status observed in the patient. Further, all lines tested retained their histological phenotype and biomarker expression patterns of the tumor of origin over multiple passages (three representative xenografts are shown in Supplemental Figure 3). One xenograft (BCM-3807/4400) was found to grow as a fluid-filled mass at each transplant generation. The presence of this yellowish fluid was a clinical feature of the tumor of origin in the patient.
Figure 1. Comparisons of representative patient biopsies and their resulting xenografts.
Hematoxylin-eosin (H&E) stained sections. Patient biopsies are depicted in the left column; corresponding xenograft samples are depicted in the right column. Scale bar = 50 μm.
Figure 2. Biomarker expression in representative xenografts.
Five representative patient samples demonstrating retained biomarker status as xenografts. Xenograft line designations are shown above the column to which they apply. Biomarker designations are shown to the left of the row to which they apply. Inserts show the corresponding biomarker status in the tumor of origin. Scale bar = 50 μm.
Metastatic behavior of xenografts
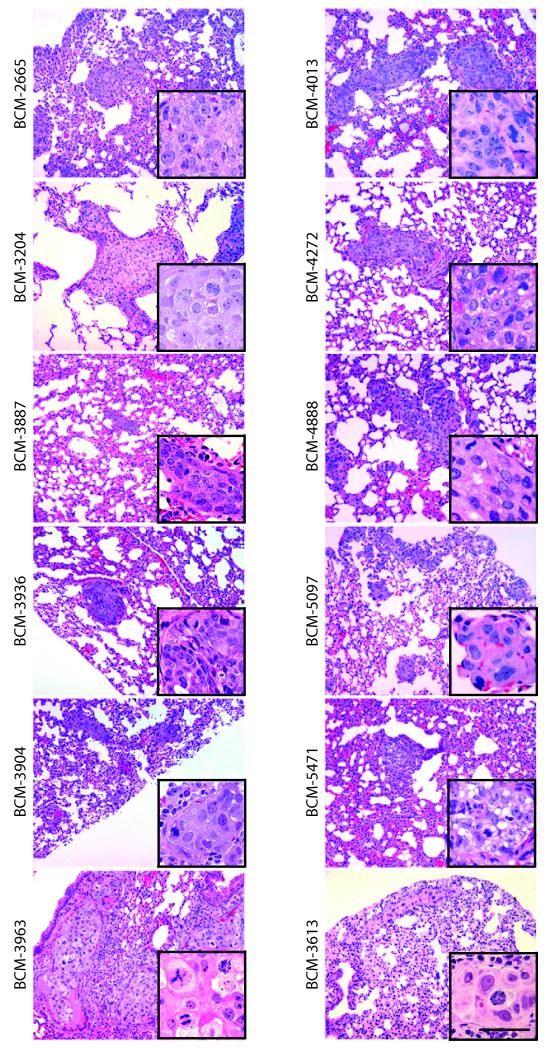
Because 18 of the 32 xenograft lines were derived from 14 patients that were either lymph node positive, or had metastatic breast cancer at other sites (brain, abdominal ascites, pleural cavity) (Table 2), we evaluated host mice for the presence of liver and lung metastases. Neither circulating tumor cells, nor lymph node, brain, or bone metastases, were examined in this study. Lungs and livers of xenograft-bearing mice were sampled at each transplant generation, and screened by histological examination. In xenograft lines representing 12 of 25 patients (48%), stable xenograft lines (including two ER+ xenograft lines) showed single or multiple foci of metastatic mammary adenocarcinoma as pulmonary metastases (Figure 3, Table 2 and Supplemental Table 10). No liver metastases were detected. The presence of lung metastasis in mice did not necessarily correlate with nodal/metastatic status in the patient of origin (Table 2 and Supplemental Table 10).
Figure 3. Histological analysis of lung metastases.
12 xenograft lines demonstrating lung metastases are shown. Insets show higher magnification images of the metastatic lesion. Scale bar = 50 μm.
Intrinsic subtype classification
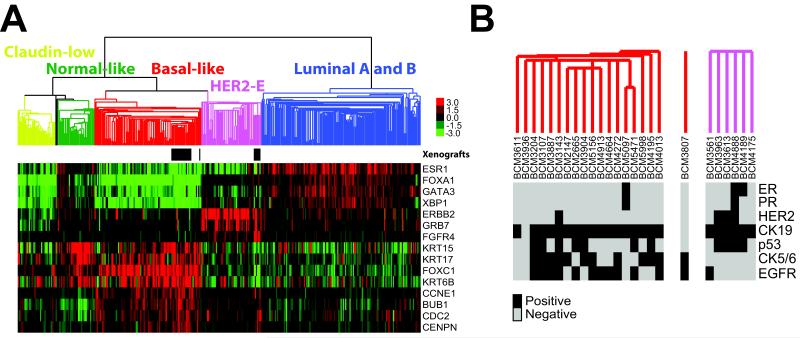
To determine the tumor subtype that these xenograft lines best resemble, we performed global gene expression analyses using 244K human oligo microarrays [23] (Agilent Technologies). Using the ~1900 intrinsic gene list [25], we co-clustered the xenografts samples with the UNC337 dataset which has an appropriate representation of all the intrinsic molecular subtypes (Luminal A, Luminal B, HER2-enriched, Basal-like and Claudin-low) [26]. Xenografts clustered with either the Basal-like or the HER2-enriched tumors and they did not cluster as a single group (Figure 4A-B). Interestingly, while most (3 of 4) HER2+ samples clustered with the HER2-enriched tumors, HER2+ BCM-3143/3104 line clustered with the Basal-like tumors. Conversely, three HER2− xenografts (BCM4175, BCM4189 and BCM3561) were identified as HER2-enriched. This observation is concordant with recent studies in human tumors showing that not all HER2+ tumors are HER2-enriched by gene expression, and that not all HER2-enriched tumors are HER2+ [32]. Regarding ER+/HER2-xenograft lines, BCM-4189 was identified as HER2-enriched, and BCM-5097 (also PR+) was identified as Basal-like. This observation is also concordant with a recent study showing that a subset of poor prognosis ER+ tumors can be identified as Basal-like [33]. Finally, identical intrinsic subtype calls were obtained when the PAM50 classifier was applied to the xenograft samples (Table 2).
Figure 4. Global gene expression analyses of the BCM xenografts combined with 337 breast samples of the UNC337 breast cancer dataset.
(A) Semi-unsupervised hierarchical clustering of 31 BCM xenografts and 337 breast samples using the 1900 intrinsic list [25, 26]. Localization of the BCM xenografts is shown by the black rectangles below the array tree. Expression of selected genes is shown in the heatmap. Each colored square represents the relative transcript abundance (in log2 space) for each sample with highest expression being red, average expression being black and lowest expression being green. (B) Detailed clusters of the BCM xenografts (one line per patient depicted) with the biomarker expression of each sample. Estrogen receptor (ER), progesterone receptor (PR), HER2, cytokeratin 19 (CK19), cytokeratin 5/6 (CK5/6) and EGFR.
Stability of xenografts with respect to their transcriptome, proteome and genome
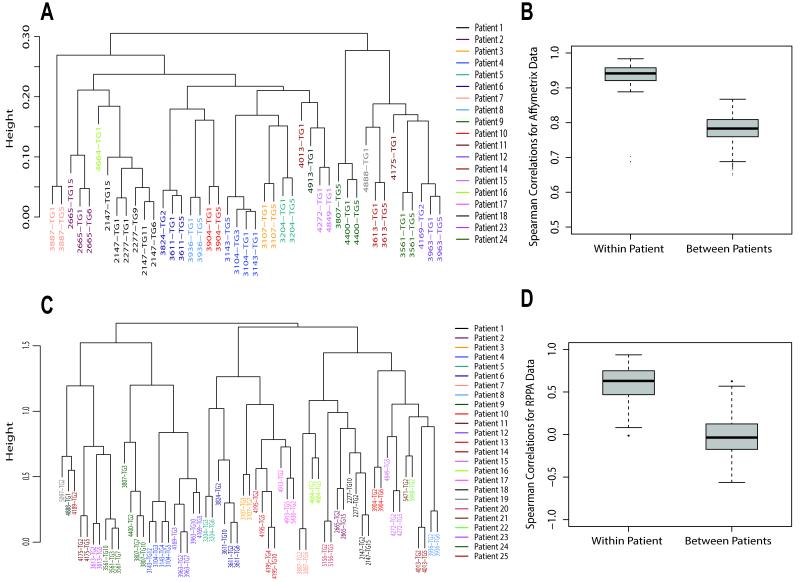
To demonstrate whether transplantable xenografts were stable at the transcriptome level, and to demonstrate whether xenografts established using pre-treatment biopsies were consistent with xenografts established with post-treatment biopsies, we conducted Affymetrix gene expression analysis using a majority of the xenograft lines, with arrays conducted roughly every 5th transplant generation whenever available. Within a given xenograft line, the transplant generations invariably clustered together thus demonstrating stability at the level of gene expression across at least five transplant generation (Figure 5A). In addition, lines established from both initial pre-treatment fragments (I) as well as post-treatment fragments (P) from the same patient also clustered together (Figure 5A). The within patient and between patients Pearson correlation coefficients are depicted in Figure 5B showing the within patient correlations to be consistently higher than the between patients correlations.
Figure 5. Transcriptome and Proteome Stability.
Xenograft lines are phenotypically stable over multiple transplant generations with respect to gene expression by Affymetrix microarray and Reverse Phase Proteomic Analysis (RPPA). A. Hierarchical cluster analysis using all probesets with detectable expression. Xenograft designation ([BCM-]####), and transplant generation number (TG#), are shown for each branch of the dendrogram. Patient of origin/xenograft association are designated by color. B. Box and whisker plot of Pearson correlation distances within patient and between patients. C. Hierarchical cluster analysis using 161 antibodies in RPPA. Xenograft designations are as shown for panel A. D. Box and whisker plot of Pearson correlation distances within patient and between patients.
To demonstrate whether stably transplantable xenografts were also stable at the proteome level, we conducted Reverse Phase Protein Array (RPPA) expression analysis on xenografts representing all 25 patients using 161 antibodies. For those xenografts having samples representing multiple transplant generations, the related samples invariably clustered together thus demonstrating stability at the level of the proteome using both hierarchical clustering and Pearson correlation coefficient comparison methods (Figures 5C and 5D). In addition, lines established from both initial pre-treatment fragments (I) as well as post-treatment fragments (P) from the same patient also invariably clustered together. As with the Affymetrix analysis, within patient correlations were consistently higher than between patients correlations.
To estimate whether xenografts were genomically stable, and to identify single nucleotide polymorphisms (SNPs) that may be relevant to xenograft biology, we conducted Sequenom analysis of SNPs for 155 known polymorphisms in 32 cancer-relevant genes (Supplemental Table 8) using each xenograft line, with each line represented at multiple transplant generations whenever possible. No tumor suppressor genes were evaluated in this study. A majority of the SNPs detected were in the PIK3CA gene (lines BCM-3613, BCM-3807/4400, and BCM-4888) (Supplemental Table 9) each showing a unique genetic alteration. However, a potentially activating SNP in c-Met (N375S_A1124G) was identified in both BCM-2147/2277, and BCM-3143/3104, with BCM-2277 showing an additional KRAS polymorphism (Q61LPR_A182TCG) relative to BCM-2147 that was either recovered as a function of sampling, or potentially selected for, or acquired, as a function of AC treatment. One activating AKT1 mutation (E17K_G49A) was identified in BCM-4175. Thus, these transplantable xenograft lines are genotypically stable across multiple transplant generations with respect to the genes represented in this assay.
Patient/Xenograft Treatment Response Concordance
To evaluate biological relevance and translational utility, we tested treatment responses of 11 xenograft lines derived from patients for whom we have a total of 13 observed clinical treatment responses to essentially the same drug. 12 of 13 xenograft responses matched the corresponding clinical responses (Supplemental Table 11 and Supplemental Figure 4). The sole discordant result was in a xenograft line derived from ascites. The ascites cells were resistant to Paclitaxel in the patient, but were modestly sensitive to Docetaxel when grown as a xenograft (33% decrease in tumor volume over the observation period). As most xenograft lines were resistant to the therapies used, one would expect about 60% concordance entirely by chance. However, our observed concordance of 92% was substantially higher (Kappa = 0.75, p=0.003), and there was a significant association between the xenograft and patient-derived results (Fisher’s exact test, p=0.04)
Discussion
The mouse mammary gland model derives its experimental power, in part, from the ability to transplant mammary epithelium from one animal to another. Transplantation allows one to expand normal, genetically modified, or neoplastic tissue into multiple hosts. With respect to therapeutic studies, tissue expansion by transplantation allows study of the in vivo effects of multiple agents on the behavior of the epithelium. In contrast, such studies are not possible in human patients, and experimental analysis of the human mammary gland and breast cancers has been limited by the lack of a suitable transplantation model in which malignant xenografts grow efficiently and tumor biology is, to the extent possible, faithful to the biology of the tumor of origin in the patient. In this paper, we demonstrate that the SCID/Beige and NSG immunocompromised mice are relatively permissive for growth of malignant tissue and allow the establishment of stably transplantable “triple negative”, HER2+, and ER+ xenograft lines from an ethnically diverse patient population. These xenografts are histologically and immunohistochemically indistinguishable from the tumor of origin, show comparable treatment responses, and are transcriptionally, translationally, post-translationally, and genomically, stable across multiple transplantation generations.
There have been numerous attempts to generate transplantable xenografts over the last three decades. Historically, stable take rates for xenografts were low (~10%, or less), but have improved in more recent studies [1, 2, 5, 6, 8-10, 34-41]. Our stable take rate under condition 2 (estradiol supplementation in SCID/Bg mice) was ~21%, with a statistically similar take rate observed in NSG mice (~19%). Thus, the rate we obtained in SCID/Bg and NSG mice is at the high end of the historical range. Similarly high per patient take rates (10 of 42 patients; ~24%) were shown recently by DeRose et al. [12] using tissue fragments coated in Matrigel and implanted into the epithelium-free fat pad of NOD/SCID mice supplemented with estradiol. An exceptionally high take rate was also obtained by Kabos et al. [15] (10 of 27 patients; 37%) using tissue fragments or metastatic cells (pleural fluid/ascites) coated in Matrigel and implanted into the intact fat pad of either NOD/SCID or NSG mice supplemented with estradiol. Elevated take rates in advanced tumors in our studies are consistent with the apparent elevated tumor-initiating cell (TIC) (a.k.a. cancer stem cell) frequency in high-grade tumors relative to lower grade tumors [42]. However, these data could also be explained by lack of one or more factors required for growth of low-grade tumors in the SCID/Bg and NSG mice rather than a decreased TIC frequency per se. Our data are also consistent with these recent studies showing that estradiol supplementation stimulates growth of breast cancer xenografts, including ER-negative xenografts [12, 15, 43, 44]. This stimulatory effect of estradiol supplementation on ER-negative tumor growth is, at least in part, due to an ERα-mediated effect on bone marrow-derived myeloid cells that promote angiogenesis and tumor growth [45].
Despite the observation that ~80% of all breast cancers in women are ER+, relatively few cell line or xenograft models are available. Recently, a few groups have succeeded in generating ER+ xenografts lines [12, 15]. In this work, we were also able to generate three new stably transplantable ER+ xenograft lines, two of which are metastatic to the mouse lung. Unfortunately, the rate of stable outgrowth was still low relative to triple negative tumors, and, perhaps due to the omission of Matrigel coating in our protocol, we did not recover stable lines representing the luminal subtypes. Thus, efficient establishment of ER+ xenografts will require further optimization. One potential method is via co-transplantation with mesenchymal stem cells, which were shown recently to enhance mammosphere formation in vitro [46], and to stimulate growth and metastasis of established xenografts in vivo [12, 47-49]. Alternatively, some mouse proteins (e.g. prolactin, hepatocyte growth factor, interleukin-6 (IL-6)) do not activate their human receptor counterparts [49-51]. Thus, tissue-appropriate expression of one or more of these human ligands as transgenes in an immunocompromised mouse background may be necessary to stimulate ER+ xenograft growth fully.
Two important features of a useful experimental model are the degree to which the experimental model faithfully recapitulates the tumor in the patient of origin, and the stability of the experimental model over time. In this study, all xenografts showed biological consistency with the tumor of origin both at the level of histology, and with respect to several clinically (ER, PR, HER2, Ki67) and biologically (TP53, EGFR, CK5/6, CK19) relevant biomarkers. With respect to the issue of stability over time, all xenografts for which Affymetrix RNA expression patterns or RPPA protein expression patterns were determined for multiple transplantation generations showed a high degree of stability with respect to gene expression at the mRNA and protein level, including post-translational modifications. Unfortunately, it was not possible to evaluate the degree to which the xenograft transcriptome or proteome were consistent with those of the tumor of origin because patients were enrolled in ongoing clinical trials and their array data could not yet be released, and remaining banked tissue was not sufficient to run RPPA without exhausting the sample entirely.
In addition to the molecular diversity demonstrated by this collection of xenografts, the lines established represent diversity with respect to the ethnic groups, as well as metastatic behavior. Of particular interest is the finding the two of the three ER+ breast cancer xenografts established were metastatic from the orthotopic site. Similar results were recently obtained by DeRose et al [12]. In contrast, we know of only one ER+ metastasis model in cell lines, a metastatic derivative of MCF7 developed recently [52]. Thus, these models should be useful in evaluating ethnic differences in tumor behavior, as well as evaluating therapeutic agents that might influence metastatic behavior, particularly of ER+ breast cancer.
Perhaps the most critical issue not addressed fully to date is the question of whether PDX models respond to a given treatment in a manner similar to the tumor of origin in the patient. This issue is important not only to establish relevance as experimental models, but particularly from a clinical/translational standpoint. If, in fact, the PDX models respond similarly to a given agent, it should be possible to identify predictive indicators of response that may ultimately be useful clinically. Further, there should be relevant biology underlying PDX treatment resistance that can be exploited to improve treatment clinically. A previous study evaluating 7 xenografts showed an observed concordance of 71% vs. an expected concordance of 47% [9]. However, statistical significance was not demonstrated using this sample size (Kappa=0.46, p=0.08). In our study, a majority of xenograft lines tested showed qualitatively identical treatment responses as the corresponding patient treated with a similar or identical agent, and statistical significance was achieved using this increased sample size. While the lack of a functional immune system may ultimately be shown to alter treatment responses to some agents, the observation that xenograft lines responded to two common chemotherapeutics in a qualitatively identical manner as the patient of origin strongly suggests that results obtained in “mouse clinical trials” will be generally relevant to patients.
Supplementary Material
Acknowledgements
The authors thank Dr. Charlotte Kuperwasser for generously providing the normal human fibroblast line used in this study. This work was supported in part by the Breast Cancer Research Foundation, the Emma Jacobs Clinical Breast Cancer Fund, the Helis Foundation, the Susan G. Komen Foundation, Cancer Fighters of Houston, BCM Cancer Center grant P30 CA125123, BCM Breast Cancer SPORE P50 CA50183, UNC Breast Cancer SPORE P50-CA58223, NIH/NCI grant R01 CA112305, NIH/NCI grant U54 CA149196, Cancer Prevention Research Institute of Texas Grant RP101251, Stand Up to Cancer Dream Team Translational Research Grant, a Program of the Entertainment Industry Foundation (SU2C-AACR-DT0209), CA16672 (RPPA core), and P01 CA30195 from the National Cancer Institute, K12-5611B through the University of Texas Health Sciences Center from the National Institute of Health, and US Army Medical Research and Materiel Command grants DAMD17-01-0132 and W81XWH-04-1-0468.
Footnotes
Author Contribution Statement
XZ conducted transplantations, and prepared samples for experiments, and carried out sample characterization and prepared the manuscript.
SC and GBM conducted RPPA, Sequenom, and STR analysis and prepared associated tables and text.
AP and CMP conducted Agilent oligonucleotide microarray with intrinsic subtype classification, and prepared associated figure and text.
LED, IP, QL, MDL, LW, RS, MG, and HW contributed to transplantation, and/or preparation and characterization of samples.
HW and SFW conducted Affymetrix microarray experiments.
AC, CG, and JH provided pathology support.
ACP and AMF collected, curated, and provided clinical information on patient samples
MFW, AT and SGH conducted statistical analyses, and prepared associated tables.
ESC, PZ, and CS constructed the www.bcxenograft.org database
CS conducted Affymetrix and RPPA statistical analysis and prepared associated figure.
MFR provided clinical material and information on patient samples.
JCC recruited patients to clinical trials, provided clinical material and information on patient samples, and supervised some Affymetrix microarray studies.
MTL conceived structure and content of the resource, designed experiments, coordinated and supervised the overall project, and prepared the manuscript.
Disclosures:
M.T.L. and J.C.C. are Founding Partners in StemMed Ltd.
C.M.P. is an equity stock holder in University Genomics and BioClassifier LLC, and is listed as an inventor on a patent application on the PAM50 assay.
Literature cited
- 1.Rae-Venter B, Reid LM. Growth of human breast carcinomas in nude mice and subsequent establishment in tissue culture. Cancer Res. 1980;40(1):95–100. [PubMed] [Google Scholar]
- 2.Sebesteny A, Taylor-Papadimitriou J, Ceriani R, Millis R, Schmitt C, Trevan D. Primary human breast carcinomas transplantable in the nude mouse. J Natl Cancer Inst. 1979;63(6):1331–1337. [PubMed] [Google Scholar]
- 3.Tentler JJ, Tan AC, Weekes CD, Jimeno A, Leong S, Pitts TM, Arcaroli JJ, Messersmith WA, Eckhardt SG. Patient-derived tumour xenografts as models for oncology drug development. Nature reviews Clinical oncology. 2012;9(6):338–350. doi: 10.1038/nrclinonc.2012.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Herschkowitz JI, Zhao W, Zhang M, Usary J, Murrow G, Edwards D, Knezevic J, Greene SB, Darr D, Troester MA, et al. Comparative oncogenomics identifies breast tumors enriched in functional tumor-initiating cells. Proc Natl Acad Sci U S A. 2011 doi: 10.1073/pnas.1018862108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Noel A, Borcy V, Bracke M, Gilles C, Bernard J, Birembaut P, Mareel M, Foidart JM. Heterotransplantation of primary and established human tumour cells in nude mice. Anticancer Res. 1995;15(1):1–7. [PubMed] [Google Scholar]
- 6.Murthy MS, Scanlon EF, Jelachich ML, Klipstein S, Goldschmidt RA. Growth and metastasis of human breast cancers in athymic nude mice. Clin Exp Metastasis. 1995;13(1):3–15. doi: 10.1007/BF00144013. [DOI] [PubMed] [Google Scholar]
- 7.Hampton OA, Den Hollander P, Miller CA, Delgado DA, Li J, Coarfa C, Harris RA, Richards S, Scherer SE, Muzny DM, et al. A sequence-level map of chromosomal breakpoints in the MCF-7 breast cancer cell line yields insights into the evolution of a cancer genome. Genome Res. 2009;19(2):167–177. doi: 10.1101/gr.080259.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Naundorf H, Fichtner I, Buttner B, Frege J. Establishment and characterization of a new human oestradiol- and progesterone-receptor-positive mammary carcinoma serially transplantable in nude mice. J Cancer Res Clin Oncol. 1992;119(1):35–40. doi: 10.1007/BF01209485. [DOI] [PubMed] [Google Scholar]
- 9.Marangoni E, Vincent-Salomon A, Auger N, Degeorges A, Assayag F, de Cremoux P, de Plater L, Guyader C, De Pinieux G, Judde JG, et al. A new model of patient tumor-derived breast cancer xenografts for preclinical assays. Clin Cancer Res. 2007;13(13):3989–3998. doi: 10.1158/1078-0432.CCR-07-0078. [DOI] [PubMed] [Google Scholar]
- 10.Kuperwasser C, Chavarria T, Wu M, Magrane G, Gray JW, Carey L, Richardson A, Weinberg RA. Reconstruction of functionally normal and malignant human breast tissues in mice. Proc Natl Acad Sci U S A. 2004;101(14):4966–4971. doi: 10.1073/pnas.0401064101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bergamaschi A, Hjortland GO, Triulzi T, Sorlie T, Johnsen H, Ree AH, Russnes HG, Tronnes S, Maelandsmo GM, Fodstad O, et al. Molecular profiling and characterization of luminal-like and basal-like in vivo breast cancer xenograft models. Mol Oncol. 2009;3(5-6):469–482. doi: 10.1016/j.molonc.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeRose YS, Wang G, Lin YC, Bernard PS, Buys SS, Ebbert TW, Factor R, Matsen C, Milash BA, Nelson E, et al. Patient-derived tumor grafts authentically reflect tumor pathology, growth, metastasis, and disease outcomes. Nature Medicine. 2011 doi: 10.1038/nm.2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xia Z, Taylor PR, Locklin RM, Gordon S, Cui Z, Triffitt JT. Innate immune response to human bone marrow fibroblastic cell implantation in CB17 scid/beige mice. J Cell Biochem. 2006;98(4):966–980. doi: 10.1002/jcb.20730. [DOI] [PubMed] [Google Scholar]
- 14.Clarke R. Human breast cancer cell line xenografts as models of breast cancer. The immunobiologies of recipient mice and the characteristics of several tumorigenic cell lines. Breast Cancer Res Treat. 1996;39(1):69–86. doi: 10.1007/BF01806079. [DOI] [PubMed] [Google Scholar]
- 15.Kabos P, Finlay-Schultz J, Li C, Kline E, Finlayson C, Wisell J, Manuel CA, Edgerton SM, Harrell JC, Elias A, et al. Patient-derived luminal breast cancer xenografts retain hormone receptor heterogeneity and help define unique estrogen-dependent gene signatures. Breast cancer research and treatment. 2012;135(2):415–432. doi: 10.1007/s10549-012-2164-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gouon-Evans V, Lin EY, Pollard JW. Requirement of macrophages and eosinophils and their cytokines/chemokines for mammary gland development. Breast Cancer Res. 2002;4(4):155–164. doi: 10.1186/bcr441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gouon-Evans V, Rothenberg ME, Pollard JW. Postnatal mammary gland development requires macrophages and eosinophils. Development. 2000;127(11):2269–2282. doi: 10.1242/dev.127.11.2269. [DOI] [PubMed] [Google Scholar]
- 18.Yang L, Huang J, Ren X, Gorska AE, Chytil A, Aakre M, Carbone DP, Matrisian LM, Richmond A, Lin PC, et al. Abrogation of TGF beta signaling in mammary carcinomas recruits Gr-1+CD11b+ myeloid cells that promote metastasis. Cancer Cell. 2008;13(1):23–35. doi: 10.1016/j.ccr.2007.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang X, Lewis MT. Establishment of Patient-Derived Xenograft (PDX) Models of Human Breast Cancer. Current Protocols in Mouse Biology. 2013 doi: 10.1002/9780470942390.mo120140. [DOI] [PubMed] [Google Scholar]
- 20.DeOme KB, Faulkin LJJ, Bern H. Development of mammary tumors from hyperplastic alveolar nodules transplanted into gland-free mammary fat pads of female C3H mice. Cancer Res. 1959;19:515–520. [PubMed] [Google Scholar]
- 21.Kuperwasser C, Chavarria T, Wu M, Magrane G, Gray JW, Carey L, Richardson A, Weinberg RA. Reconstruction of functionally normal and malignant human breast tissues in mice. Proc Natl Acad Sci U S A. 2004;101(14):4966–4971. doi: 10.1073/pnas.0401064101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Romano P, Manniello A, Aresu O, Armento M, Cesaro M, Parodi B. Cell Line Data Base: structure and recent improvements towards molecular authentication of human cell lines. Nucleic acids research. 2009;37:D925–932. doi: 10.1093/nar/gkn730. Database issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu Z, Fan C, Oh DS, Marron JS, He X, Qaqish BF, Livasy C, Carey LA, Reynolds E, Dressler L, et al. The molecular portraits of breast tumors are conserved across microarray platforms. BMC genomics. 2006;7:96. doi: 10.1186/1471-2164-7-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Novoradovskaya N, Whitfield ML, Basehore LS, Novoradovsky A, Pesich R, Usary J, Karaca M, Wong WK, Aprelikova O, Fero M, et al. Universal Reference RNA as a standard for microarray experiments. BMC genomics. 2004;5(1):20. doi: 10.1186/1471-2164-5-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parker JS, Mullins M, Cheang MC, Leung S, Voduc D, Vickery T, Davies S, Fauron C, He X, Hu Z, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2009;27(8):1160–1167. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prat A, Parker JS, Karginova O, Fan C, Livasy C, Herschkowitz JI, He X, Perou CM. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res. 2010;12(5):R68. doi: 10.1186/bcr2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vasudevan KM, Barbie DA, Davies MA, Rabinovsky R, McNear CJ, Kim JJ, Hennessy BT, Tseng H, Pochanard P, Kim SY, et al. AKT-independent signaling downstream of oncogenic PIK3CA mutations in human cancer. Cancer Cell. 2009;16(1):21–32. doi: 10.1016/j.ccr.2009.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tibes R, Qiu Y, Lu Y, Hennessy B, Andreeff M, Mills GB, Kornblau SM. Reverse phase protein array: validation of a novel proteomic technology and utility for analysis of primary leukemia specimens and hematopoietic stem cells. Mol Cancer Ther. 2006;5(10):2512–2521. doi: 10.1158/1535-7163.MCT-06-0334. [DOI] [PubMed] [Google Scholar]
- 29.Hennessy BT, Lu Y, Gonzalez-Angulo AM, Carey MS, Myhre S, Ju Z, Davies MA, Liu W, Coombes K, Meric-Bernstam F, et al. A Technical Assessment of the Utility of Reverse Phase Protein Arrays for the Study of the Functional Proteome in Non-microdissected Human Breast Cancers. Clinical Proteomics. 2010;6(4):129–151. doi: 10.1007/s12014-010-9055-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang YC, Morrison G, Gillihan R, Guo J, Ward RM, Fu X, Botero MF, Healy NA, Hilsenbeck SG, Phillips GL, et al. Different mechanisms for resistance to trastuzumab versus lapatinib in HER2-positive breast cancers--role of estrogen receptor and HER2 reactivation. Breast Cancer Res. 2011;13(6):R121. doi: 10.1186/bcr3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rimawi MF, Wiechmann LS, Wang YC, Huang C, Migliaccio I, Wu MF, Gutierrez C, Hilsenbeck SG, Arpino G, Massarweh S, et al. Reduced dose and intermittent treatment with lapatinib and trastuzumab for potent blockade of the HER pathway in HER2/neu-overexpressing breast tumor xenografts. Clin Cancer Res. 2011;17(6):1351–1361. doi: 10.1158/1078-0432.CCR-10-1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Prat A, Perou CM. Deconstructing the molecular portraits of breast cancer. Molecular oncology. 2011;5(1):5–23. doi: 10.1016/j.molonc.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Prat A, Parker JS, Fan C, Cheang MC, Miller LD, Bergh J, Chia SK, Bernard PS, Nielsen TO, Ellis MJ, et al. Concordance among gene expression-based predictors for ER-positive breast cancer treated with adjuvant tamoxifen. Annals of oncology: official journal of the European Society for Medical Oncology / ESMO. 2012 doi: 10.1093/annonc/mds080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beckhove P, Schutz F, Diel IJ, Solomayer EF, Bastert G, Foerster J, Feuerer M, Bai L, Sinn HP, Umansky V, et al. Efficient engraftment of human primary breast cancer transplants in nonconditioned NOD/Scid mice. Int J Cancer. 2003;105(4):444–453. doi: 10.1002/ijc.11125. [DOI] [PubMed] [Google Scholar]
- 35.Sakakibara T, Xu Y, Bumpers HL, Chen FA, Bankert RB, Arredondo MA, Edge SB, Repasky EA. Growth and metastasis of surgical specimens of human breast carcinomas in SCID mice. Cancer J Sci Am. 1996;2(5):291–300. [PubMed] [Google Scholar]
- 36.Outzen HC, Custer RP. Growth of human normal and neoplastic mammary tissues in the cleared mammary fat pad of the nude mouse. J Natl Cancer Inst. 1975;55(6):1461–1466. doi: 10.1093/jnci/55.6.1461. [DOI] [PubMed] [Google Scholar]
- 37.Sheffield LG, Welsch CW. Transplantation of human breast epithelia to mammary-gland-free fat-pads of athymic nude mice: influence of mammotrophic hormones on growth of breast epithelia. Int J Cancer. 1988;41(5):713–719. doi: 10.1002/ijc.2910410513. [DOI] [PubMed] [Google Scholar]
- 38.McManus MJ, Welsch CW. DNA synthesis of benign human breast tumors in the untreated athymic “nude” mouse. An in vivo model to study hormonal influences on growth of human breast tissues. Cancer. 1980;45(8):2160–2165. doi: 10.1002/1097-0142(19800415)45:8<2160::aid-cncr2820450825>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 39.Visonneau S, Cesano A, Torosian MH, Miller EJ, Santoli D. Growth characteristics and metastatic properties of human breast cancer xenografts in immunodeficient mice. Am J Pathol. 1998;152(5):1299–1311. [PMC free article] [PubMed] [Google Scholar]
- 40.Fichtner I, Becker M, Zeisig R, Sommer A. In vivo models for endocrine-dependent breast carcinomas: special considerations of clinical relevance. Eur J Cancer. 2004;40(6):845–851. doi: 10.1016/j.ejca.2003.11.030. [DOI] [PubMed] [Google Scholar]
- 41.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100(7):3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pece S, Tosoni D, Confalonieri S, Mazzarol G, Vecchi M, Ronzoni S, Bernard L, Viale G, Pelicci PG, Di Fiore PP. Biological and molecular heterogeneity of breast cancers correlates with their cancer stem cell content. Cell. 2010;140(1):62–73. doi: 10.1016/j.cell.2009.12.007. [DOI] [PubMed] [Google Scholar]
- 43.Gupta PB, Kuperwasser C. Contributions of estrogen to ER-negative breast tumor growth. J Steroid Biochem Mol Biol. 2006;102(1-5):71–78. doi: 10.1016/j.jsbmb.2006.09.025. [DOI] [PubMed] [Google Scholar]
- 44.Gupta PB, Proia D, Cingoz O, Weremowicz J, Naber SP, Weinberg RA, Kuperwasser C. Systemic stromal effects of estrogen promote the growth of estrogen receptor-negative cancers. Cancer Res. 2007;67(5):2062–2071. doi: 10.1158/0008-5472.CAN-06-3895. [DOI] [PubMed] [Google Scholar]
- 45.Iyer V, Klebba I, McCready J, Arendt LM, Betancur-Boissel M, Wu MF, Zhang X, Lewis MT, Kuperwasser C. Estrogen Promotes ER-Negative Tumor Growth and Angiogenesis through Mobilization of Bone Marrow-Derived Monocytes. Cancer Res. 2012 doi: 10.1158/0008-5472.CAN-11-3287. [DOI] [PubMed] [Google Scholar]
- 46.Klopp AH, Lacerda L, Gupta A, Debeb BG, Solley T, Li L, Spaeth E, Xu W, Zhang X, Lewis MT, et al. Mesenchymal stem cells promote mammosphere formation and decrease E-cadherin in normal and malignant breast cells. PLoS One. 2010;5(8):e12180. doi: 10.1371/journal.pone.0012180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu S, Ginestier C, Ou SJ, Clouthier SG, Patel SH, Monville F, Korkaya H, Heath A, Dutcher J, Kleer CG, et al. Breast cancer stem cells are regulated by mesenchymal stem cells through cytokine networks. Cancer Res. 2011;71(2):614–624. doi: 10.1158/0008-5472.CAN-10-0538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Karnoub AE, Dash AB, Vo AP, Sullivan A, Brooks MW, Bell GW, Richardson AL, Polyak K, Tubo R, Weinberg RA. Mesenchymal stem cells within tumour stroma promote breast cancer metastasis. Nature. 2007;449(7162):557–563. doi: 10.1038/nature06188. [DOI] [PubMed] [Google Scholar]
- 49.Utama FE, LeBaron MJ, Neilson LM, Sultan AS, Parlow AF, Wagner KU, Rui H. Human prolactin receptors are insensitive to mouse prolactin: implications for xenotransplant modeling of human breast cancer in mice. The Journal of endocrinology. 2006;188(3):589–601. doi: 10.1677/joe.1.06560. [DOI] [PubMed] [Google Scholar]
- 50.Rong S, Oskarsson M, Faletto D, Tsarfaty I, Resau JH, Nakamura T, Rosen E, Hopkins RF, 3rd, Vande Woude GF. Tumorigenesis induced by coexpression of human hepatocyte growth factor and the human met protooncogene leads to high levels of expression of the ligand and receptor. Cell growth & differentiation: the molecular biology journal of the American Association for Cancer Research. 1993;4(7):563–569. [PubMed] [Google Scholar]
- 51.Rong S, Bodescot M, Blair D, Dunn J, Nakamura T, Mizuno K, Park M, Chan A, Aaronson S, Vande Woude GF. Tumorigenicity of the met proto-oncogene and the gene for hepatocyte growth factor. Molecular and cellular biology. 1992;12(11):5152–5158. doi: 10.1128/mcb.12.11.5152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Barone I, Brusco L, Gu G, Selever J, Beyer A, Covington KR, Tsimelzon A, Wang T, Hilsenbeck SG, Chamness GC, et al. Loss of Rho GDIalpha and resistance to tamoxifen via effects on estrogen receptor alpha. J Natl Cancer Inst. 2011;103(7):538–552. doi: 10.1093/jnci/djr058. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.