Abstract
An RNA-dependent association of Ku antigen with nuclear DNA helicase II (NDH II), alternatively named RNA helicase A (RHA), was found in nuclear extracts of HeLa cells by immunoprecipitation and by gel filtration chromatography. Both Ku antigen and NDH II were associated with hnRNP complexes. Two-dimensional gel electrophoresis showed that Ku antigen was most abundantly associated with hnRNP C, K, J, H and F, but apparently not with others, such as hnRNP A1. Unexpectedly, DNA-dependent protein kinase (DNA-PK), which comprises Ku antigen as the DNA binding subunit, phosphorylated hnRNP proteins in an RNA-dependent manner. DNA-PK also phosphorylated recombinant NDH II in the presence of RNA. RNA binding assays displayed a preference of DNA-PK for poly(rG), but not for poly(rA), poly(rC) or poly(rU). This RNA binding affinity of DNA-PK can be ascribed to its Ku86 subunit. Consistently, poly(rG) most strongly stimulated the DNA-PK-catalyzed phosphorylation of NDH II. RNA interference studies revealed that a suppressed expression of NDH II altered the nuclear distribution of hnRNP C, while silencing DNA-PK changed the subnuclear distribution of NDH II and hnRNP C. These results support the view that DNA-PK can also function as an RNA-dependent protein kinase to regulate some aspects of RNA metabolism, such as RNA processing and transport.
INTRODUCTION
Ku antigen is a heterodimeric protein consisting of 86 and 70 kDa subunits that was originally identified in patients with the autoimmune diseases systemic lupus erythematosus (SLE), scleroderma, polymyositis and Sjogren’s syndrome (1). Earlier studies demonstrated that Ku antigen is a chromosomal DNA binding protein (2). Its preferred binding to dsDNA termini (3) is necessary for the suggested in vivo functions in DNA double-strand break repair and V(D)J recombination (4). In addition to dsDNA ends, Ku antigen also displays affinity to other DNA structures such as nicks, single-strand gaps or any type of the single- to double-stranded DNA transitions (5,6). Moreover, some data show that Ku antigen can also bind to specific DNA sequences, e.g. transcription regulatory elements and origins of DNA replication (7). Furthermore, Ku antigen possesses a dsDNA unwinding activity that has been alternatively named human DNA helicase II (HDH II) (8). It is noteworthy to point out that HDH II is different from the here described helicase NDH II. Most importantly, Ku antigen is the DNA binding subunit of DNA-dependent protein kinase (DNA-PK) that, as a holoenzyme, consists of the Ku heterodimer and an ∼470 kDa catalytic subunit (DNA-PKcs), which belongs to the PI-3 kinase family (9). DNA-PK phosphorylates many DNA-binding proteins to regulate their functions in DNA repair, recombination, replication and transcription (9).
Nuclear DNA helicase II (NDH II) was initially isolated from bovine tissue as a monomeric 140 kDa protein (10). The human ortholog unwound RNA and was therefore named RNA helicase A (RHA) (11). Subsequently it was shown that NDH II, or RHA, is involved in transcription (12) and RNA transport (13). Recent results revealed that NDH II is also a component of hnRNPs and mediates the attachment of hnRNP complexes to actin filaments in the nucleus (14). Moreover, NDH II localizes to specific transcriptionally active loci, such as the nucleolus in mouse cells (15). The maleless protein (MLE) of Drosophila, the dipterian ortholog of NDH II and RHA (16), is associated with the single X chromosome in males (17). MLE helicase doubles the expression of this chromosome and thereby compensates for the double gene dosis of females. Based on this dosage compensation effect, MLE is considered as a transcriptional activator.
To identify further binding partners of human NDH II immunoprecipitation experiments were performed. Un expectedly, one of these partners turned out to be Ku antigen that bound to NDH II in an RNA-dependent manner. We also show that Ku antigen is an RNA-binding protein that associates with hnRNP complexes. These results confirm earlier reports suggesting an RNA binding propensity of Ku antigen (18–20). Moreover, DNA-PK consisting of Ku antigen and a kinase subunit (DNA-PKcs) was able to perform an RNA-dependent phosphorylation of hnRNP proteins and of NDH II. Thus, in addition to its functions in DNA double-strand break repair and V(D)J recombination, DNA-PK may also be involved in RNA metabolism.
MATERIALS AND METHODS
Antibodies
Rabbit antiserum against NDH II was described before (21). Mouse monoclonal antibodies against Ku86 (B-1) and Ku70 (A-9) and goat polyclonal antibodies against hnRNP K (P-20) and hnRNPF (N-15) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). An antibody against DNA-PKcs (25–4) was obtained from NeoMarkers (Fremont, CA). Mouse monoclonal antibodies against hnRNP C (4F4) and hnRNP A1 (4B10) were provided by G. Dreyfuss (Howard Hughes Medical Institute, University of Pennsylvania). In western blot experiments a dilution of 1:1000 was used for rabbit anti-NDH II, mouse anti-Ku86 (B-1) and Ku70 (A-9), a 1:5000 dilution was used for mouse anti-hnRNP C (4F4) and anti-hnRNP A1 (4B10), and a 1:200 dilution was used for goat anti-hnRNP K (P-20), anti-hnRNP-F (N-15) and mouse anti-DNA-PKcs (25–4).
Immunoprecipitations
The HeLa cell pellet (0.5 ml) was suspended in 1 ml RSB-100 (10 mM Tris–HCl pH 7.2, 10 mM NaCl, 1.5 mM MgCl2) plus proteinase inhibitors (5 µg/ml each of aprotinin, leupeptin and pepstatin). The cells were disrupted with a Teflon homogenizer and centrifuged at 3000 g for 5 min at 4°C. The nuclear pellet was suspended in 1 ml 50 mM Tris–HCl pH 8.0, 0.35 M NaCl and 0.1 mM EDTA plus proteinase inhibitors (see above) and rocked for 45 min at 4°C, followed by centrifugation at 12 000 g for 5 min at 4°C. The resulting supernatant was used as nuclear extract. The extraction was repeated one more time and both nuclear extracts were combined for carrying out the immunoprecipitations. Nuclear extract was equally divided and antibodies against NDH II (10 µl rabbit serum), Ku86 (20 µg) and hnRNP C (4F4, 5 µl) were added. The same amounts of preimmune IgG from rabbit or mouse served as controls. The nuclear extracts were incubated on a rocker table with the respective antibodies for 2 h at 4°C. Then protein A–agarose beads (50 µl each) were added and the suspensions were incubated for another hour. Washing and recovery of proteins for western blotting was as described before (14).
Immunoprecipitation of hnRNP complexes from the nucleoplasm of HeLa cells was performed by following a previously described protocol (22). HeLa cells were placed on a culture dish (95 cm diameter) to ≈80% density and incubated for 20 h with Dulbecco’s modified Eagle’s medium containing [35S]methionine (20 µCi/ml) and one-tenth of the normal methionine. HeLa cells were then collected and opened by four passages through a 25 gauge needle in a buffer (10 mM Tris–HCl pH 7.4, 100 mM NaCl, 2.5 mM MgCl2 plus proteinase inhibitors as above) containing 0.5% Triton X-100, followed by centrifugation at 3000 g for 5 min at 4°C. The nuclei pellet was suspended in the above buffer (500 µl) and treated three times with a Branson sonifier at an output of 50, each for 10 s, interrupted by intervals on ice for cooling down the sample. The sonicated nuclear contents were centrifuged through a 30% sucrose cushion in the above buffer at 4000 g for 15 min at 4°C and the supernatant was collected as nucleoplasm. Immunoprecipitations were performed with this nucleoplasmic extract using the mouse monoclonal antibody against Ku86 (B-1) or hnRNP C (4F4). The immunoprecipitated proteins were analyzed by two-dimensional gel electrophoresis using a customized system from Amersham for isoelectric focusing (IEF). The proteins from protein A–agarose beads were eluted with the same buffer as used for rehydrating the IEF strip (7 cm, pH 3–10). The IEF was run in a recommended program of 500 V for 250 Vh, 1000 V for 500 Vh and finally 8000 V for 8000 Vh. The second dimension of electrophoresis was achieved on a 10% SDS–polyacrylamide gel. Separated 35S-labeled protein bands were detected by fluorography. Alternatively, western blots with the antibody dilutions given above were performed to confirm the hnRNP proteins derived from immunoprecipitation with the non-radioactively labeled cells.
Gel filtration chromatography
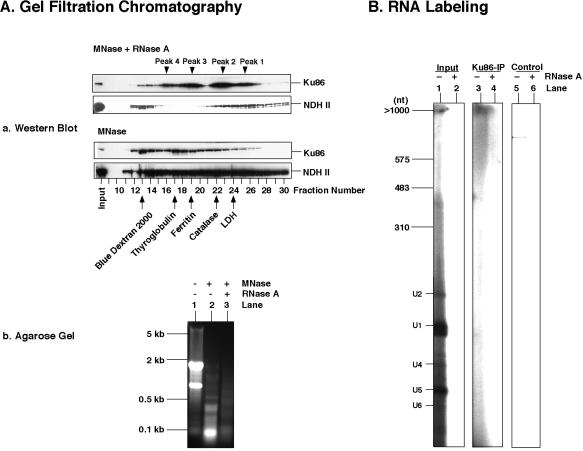
First, 0.35 M NaCl nuclear extracts from HeLa cells were prepared as described and loaded onto a Sepharose 6B column (0.8 × 12 cm), pre-equilibrated in 50 mM Tris–HCl pH 8.0, 150 mM NaCl, 1 mM EDTA, 10% glycerol and 7 mM β-mercaptoethanol. One milliliter fractions were collected and the eluted protein was analyzed by western-blotting. The column was calibrated with blue dextran (≈2000 kDa) for the void volume and the molecular weight standards shown in Figure 2.
Figure 2.
RNA binding of Ku antigen in nuclear extract. (A) Gel filtration chromatography of HeLa nuclear extracts (2 ml) on Sepharose 6B. Elution of Ku antigen and NDH II after digestion of nuclear extracts by MNase (150 U/ml) plus RNase A (0.1 mg/ml) or by MNase (150 U) only for 15 min at room temperature was shown by western blotting (Aa). The multiple elution peaks of Ku86 from MNase plus RNase A treated nuclear extracts are indicated. The Sepharose 6B column was calibrated with the indicated molecular weight standards. Nucleic acids from the nuclease-treated extracts are shown by agarose gel electrophoresis (Ab). (B) RNAs were immunoprecipitated from 0.35 M NaCl nuclear extracts by the antibody against Ku86 and labeled with T4 RNA ligase and [5′-32P]pCp after protein extraction with phenol–chloroform and RNA precipitation with ethanol. After labeling, the solutions were divided for digestion with RNase A as indicated. DNase I (0.1 mg/ml) was always present to remove possible contaminating DNA fragments. The labeling mixtures were then separated by electrophoresis through a 7 M urea–8% polyacrylamide gel. The RNA length standard was obtained from Roche; the positions of small nuclear RNAs (U1, U2, U4, U5 and U6) are indicated.
Phosphorylation of hnRNP proteins by DNA-PK
For examining a DNA-PK catalyzed phosphorylation, hnRNP complexes were immunoprecipitated from HeLa nucleoplasmic extract as described. Protein A–agarose pellets were washed and subsequently treated with the 10-fold bead volume of the DNA-PK reaction buffer [50 mM HEPES pH 7.5, 100 mM KCl, 10 mM MgCl2, 0.2 mM EGTA, 0.1 mM EDTA, 1 mM dithiothreitol (DTT), 0.2 mM ATP and 80 µg/ml bovine serum albumin (BSA)]. Immunoprecipitates (10 µl agarose beads each) were mixed with 10 µl DNA-PK reaction buffer, followed by the addition of 2 µCi [γ-32P]ATP (5000 Ci/mmol) and 25 U of DNA-PK (Promega). After 10 min incubation at 30°C the reactions were stopped by the addition of SDS–PAGE sample buffer (5 µl each) and heating to 95°C for 5 min. Proteins were separated through a 10% SDS polyacrylamide gel, stained with Coomassie brilliant blue, dried and exposed to an X-ray film.
Filter binding assays
For filter binding assays poly(rA), poly(rC), poly(rG) and poly(rU) were radiolabeled by [γ-32P]ATP and T4 polynucleotide kinase, followed by purification through a nick column (Sephadex G-50, Amersham) to remove unincorporated nucleotides. Increasing amounts of DNA-PK were incubated with 32P-labeled RNA polynucleotides in 50 µl binding buffer (20 mM HEPES pH 8.0, 50 mM NaCl, 1 mM DTT, 1 mM EDTA and 100 µg/ml BSA) for 15 min at room temperature. Then the mixtures were applied to a nitrocellulose membrane (Protran B85, 0.45 µM, Schleicher and Schuell, Dassel, Germany) mounted onto a 96-well vacuum blotter. Prior to use the nitrocellulose membrane had been pretreated with 0.3 M NaOH for 10 min, washed twice with distilled water, each for 5 min, and equilibrated in binding buffer for ∼16 h. After sample loading the nitrocellulose membrane was washed three times with 500 µl binding buffer for each well by applying a vacuum. Then the membrane was removed from the blotter, dried and cut. The retained radioactivity was measured by scintillation counting.
Northwestern blot assays
For northwestern blot assays Ku antigen was immunoprecipitated from HeLa nuclear extracts and eluted from protein A–agarose after a treatment by RNase A (0.1 mg/ml) for 15 min at room temperature to remove unspecifically RNA-bound proteins. Then the obtained Ku86/70 complex was separated on a 10% SDS–polyacrylamide gel and transferred to a Hybond-C nitrocellulose membrane (Amersham), where RNA binding was examined with 32P-labeled synthetic polymers. After electrotransfer the protein was renatured by incubating the membrane with 8 M urea in Tris-buffered saline (25 mM Tris, pH 7.4, 140 mM NaCl and 3 mM KCl) and then washing out the urea by 10 steps of 2- to 3-fold dilutions with Tris-buffered saline, each for 10 min. Then the membrane was treated with 5% (w/v) blocking reagent (Roche) in binding buffer (25 mM NaCl, 10 mM MgCl2, 10 mM HEPES pH 8.0, 0.1 mM EDTA and 1 mM DTT) for 1 h at room temperature. 32P-labeled RNA probes were added to the membrane in binding buffer and incubation was continued for 30 min at room temperature. The membrane was washed three times with binding buffer, each for 5 min, and exposed to an X-ray film.
RNA interference
siRNAs, designed from nucleotides 236–256 of human NDH II and nucleotides 122–142 of DNA-PKcs, were synthesized (Dharmacon) and transfected into HeLa cells with oligofectamine (Invitrogene). The effect of RNA interference was examined by western blotting. Proteins were obtained from cells lysed with SDS–PAGE sample buffer 48 h after transfection. For immunofluorescence the transfected cells were fixed in 4% paraformaldehyde as described before (15).
RESULTS
NDH II co-immunoprecipitated with Ku antigen
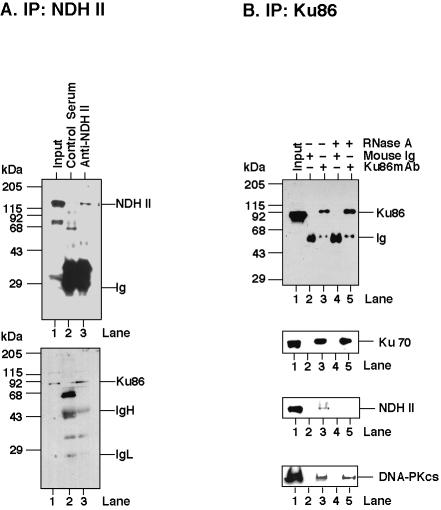
A systematic search for proteins that associate with NDH II of HeLa nuclear extracts revealed that Ku86 protein co-immunoprecipitated with NDH II to a significant extent (Fig. 1A). This association was verified by performing the immunoprecipitations with an antibody against Ku86 (Fig. 1B). The observed binding between NDH II and Ku depended on the presence of RNA since treatment of the Ku immunoprecipitates with RNase decreased the associated NDH II signals to nearly background levels (Fig. 1B). On the other hand, RNA degradation did not significantly affect the association between Ku and DNA-PKcs, which was expected since these subunits interact directly with each other (Fig. 1B).
Figure 1.
RNA-dependent co-immunoprecipitation of NDH II with Ku antigen from HeLa nuclear extracts. (A) Co-immunoprecipitation of NDH II with Ku86 by a rabbit polyclonal antibody against NDH II. (B) Co-immunoprecipitation of Ku 86/70 with NDH II and DNA-PKcs by a mouse monoclonal antibody against Ku86. To digest RNA the thrice washed immunoprecipitates were incubated with RNase A (0.1 mg/ml) for 15 min at room temperature. After this treatment the immunoprecipitates were washed one more time and processed for western blotting. The immunoglobulin heavy chain (IgH) and light chain (IgL) are indicated.
RNA association of Ku antigen in HeLa nuclear extracts
Ku antigen binding to RNA was measured by loading HeLa nuclear extract onto a Sepharose 6B gel filtration column. Ku antigen from undigested extract eluted as a single peak close to the void volume of the column (data not shown), whereas in MNase plus RNase A treated extract Ku86 eluted in multiple peaks corresponding to lower molecular weights (Fig. 2Aa). In contrast, digestion by MNase alone released very little amounts of the smaller complexes of Ku86 (Fig. 2Aa). NDH II behaved somewhat differently from Ku antigen; after treatment by both RNase A and MNase NDH II eluted as a completely released monomeric protein or as part of a nuclease-resistant macromolecular complex in the void volume of the gel filtration column (Fig. 2Aa). However treatment by MNase alone led to the elution of NDH II in many different fractions that probably contained the remaining RNA-containing complexes of different sizes (Fig. 2Aa). The effects of MNase, alone or together with RNase A, were analyzed by agarose gel electrophoresis of the nucleic acids prepared from nuclear extract. A complete RNA degradation was achieved when both nucleases were administered (Fig. 2Ab). Further evidence for RNA binding of Ku antigen was obtained from immunoprecipitations with the Ku antibody and subsequent detection of the coprecipitated RNA. For this the immunoprecipitate was deproteinized by phenol–chloroform; the remaining nucleic acids were enriched by ethanol precipitation and identified by isotopic labeling of the dissolved precipitates with T4 RNA ligase and [5′-32P]pCp. This revealed that Ku antigen was abundantly associated with nuclear RNAs in the size range from 500 to >1000 nt (Fig. 2B, lanes 3 and 4). In contrast, there were no significant amounts of RNAs that coprecipitated with the mouse control IgG antibody (Fig. 2B, lanes 5 and 6). The efficiency of RNA labeling can be assessed from the detection of RNAs of the total nuclear extract that contained large amounts of uridine-rich small nuclear RNA species (U1, U2, U4, U5 and U6) (Fig. 2B, lanes 1 and 2).
Association of Ku antigen with hnRNP complexes
We expected that most of the identified RNA-binding Ku antigen was present in hnRNP complexes. This was further examined by immunoprecipitation of HeLa nuclear extract with an antibody against hnRNP C. This revealed that Ku antigen was indeed a component of hnRNP complexes, although a co-immunoprecipitation of Ku with the anti-hnRNP C antibody required the presence of intact RNA (Fig. 3). NDH II, as shown before (14), was also tightly associated with hnRNP C. But in contrast with the situation with Ku antigen, degradation of the immunoprecipitates by RNase A hardly reduced the amount of the hnRNP C co-immunoprecipitated with NDH II (Fig. 3).
Figure 3.
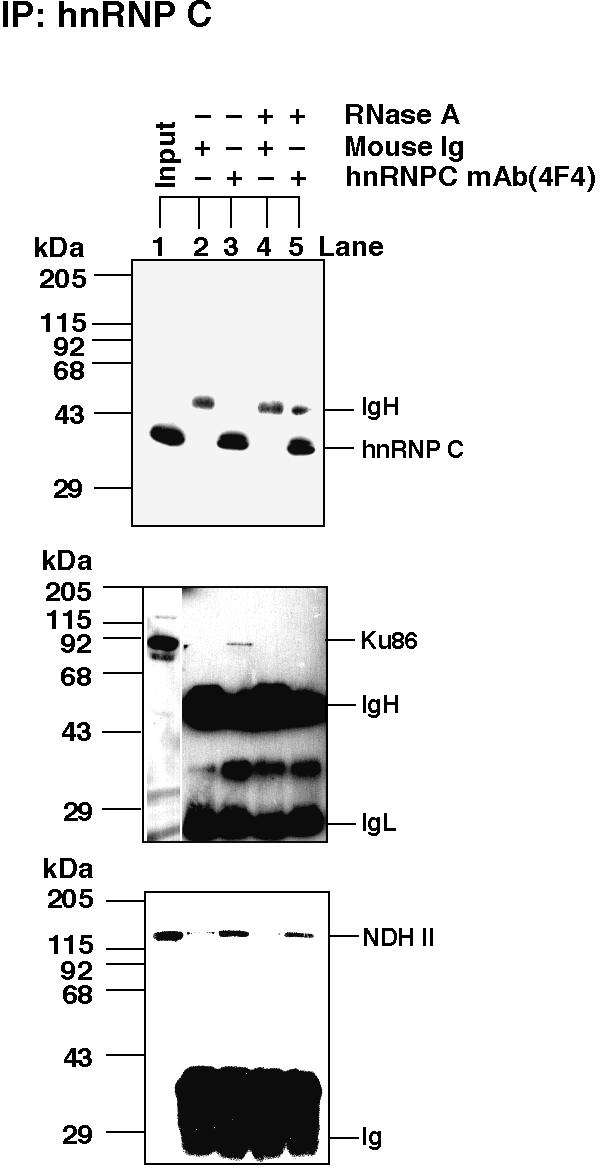
Co-immunoprecipitation of hnRNP C, NDH II and Ku antigen. Immunoprecipitation was performed with a mouse monoclonal antibody against hnRNP C protein (4F4) from 0.35 M NaCl nuclear extracts, followed by western blotting to visualize hnRNP C, NDH II and Ku86. The conditions were as described in Figure 1. The immunoglobulin heavy chain (IgH) and light chain (IgL) are indicated.
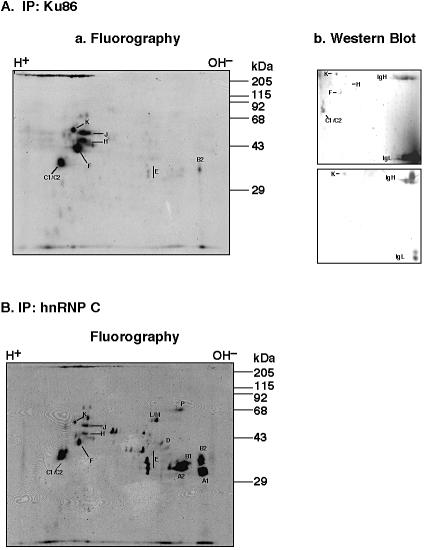
The presence of Ku antigen in hnRNP complexes was also examined by a previously established method for revealing hnRNP complexes by two-dimensional gel electrophoresis (22). In these experiments immunoprecipitations were performed with an operationally defined nucleoplasmic extract that was obtained as the supernatant from centrifugation of the sonicated nuclear extract through a sucrose cushion, again with the antibody against Ku antigen (Fig. 4Aa) or hnRNP C (Fig. 4B). A comparison of the immunoprecipitates of Ku antigen and that of hnRNP C revealed several [35S]methionine-labeled proteins after two-dimensional electrophoresis that were in common. These proteins could also be identified by western blots with a mixture of antibodies against hnRNP C, hnRNP K and hnRNP F (Fig. 4Ab, upper panel), or with the antibody against hnRNP K alone (Fig. 4Ab, lower panel). It turned out that co-immunoprecipitated Ku/hnRNP complexes contained abundant amounts of hnRNP K, J, H, F and C1/C2, whereas hnRNP E and B2 were present at much lower levels. No detectable amounts of hnRNP A1, A2 or B1 were seen in these complexes, although A1, A2 and B1 are usually associated with hnRNP complexes that co-immunoprecipitated with hnRNP C (Fig. 4b).
Figure 4.
Ku86 co-immunoprecipitated with hnRNP complexes from the nucleoplasm of HeLa cells. Nucleoplasmic extracts of HeLa cells were prepared after metabolic labeling with [35S]methionine, followed by immunoprecipitations with a mouse antibody against (A) Ku86 or (B) hnRNP C. Proteins were separated by two-dimensional electrophoresis, followed by fluorography (Aa and B) or western blotting (Ab) with a mixture of antibodies against hnRNP K (P-20), hnRNP-F (N-15) that also cross-react with hnRNP H and hnRNP C (4F4) (Ab, upper panel) or only an antibody against hnRNP K (p20) (Ab, lower panel). Letters denote the different hnRNP proteins.
RNA-dependent phosphorylation of hnRNP proteins and NDH II
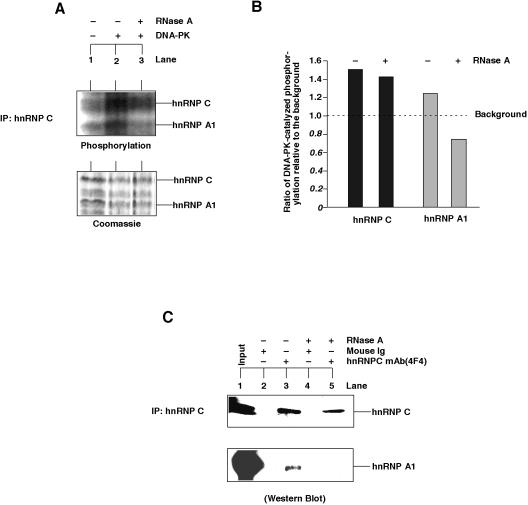
Ku antigen is the DNA binding subunit of DNA-PK that recruits the catalytic subunit (DNA-PKcs) to DNA repairosomes and recombination complexes and thereby induces their phosphorylation (9). Therefore it was of interest to learn whether the hnRNP-bound Ku antigen was able to induce protein phosphorylation. To this end the 4F4 antibody against hnRNP C was used to immunoprecipitate hnRNP complexes that contained both hnRNP C and hnRNP A1 (Fig. 5A). After addition of DNA-PK to the resulting immunoprecipitates a phosphorylation of hnRNP C and A1 was detected that was clearly above the relative low level of phosphorylation without exogenously added DNA-PK (Fig. 5A). The background level was probably due to the presence of endogenous protein kinases. Interestingly, the phosphorylation of hnRNP A1 was suppressed to a level lower than background when the hnRNP complexes were digested by RNase prior to the addition of DNA-PK, while the phosphorylation of hnRNP C was not so much affected (Fig. 5B). The observed RNase-resistant phosphorylation of hnRNP C by DNA-PK indicates a stronger protection of RNA packed by hnRNP C than by hnRNP A1. This explanation is in accordance with the finding that RNase A treatment of hnRNP complexes preferentially released hnRNP A1 (Fig. 5C). An RNase-sensitive association of hnRNP A1 with hnRNP complexes could also be shown by centrifugation of the nucleoplasmic extracts through a 10–40% sucrose gradient. Here, after the same RNase treatment nearly all hnRNP A1 molecules were at the top of the gradient, whereas sedimentation of hnRNP C was much less affected (data not shown).
Figure 5.
RNA-dependent phosphorylation of hnRNP proteins by DNA-PK. hnRNP proteins were obtained from HeLa nucleoplasmic extracts by immunoprecipitations with the antibody against hnRNP C. (A) DNA-PK catalyzed an RNA-dependent phosphorylation of hnRNP C and A1. Where indicated the samples were digested with RNase A (0.1 mg/ml) for 15 min at room temperature prior to the addition of DNA-PK. (B) The phosphorylation levels of hnRNP C and A1 were measured by scintillation counting of the excised protein bands and expressed as values relative to the background (no addition of DNA-PK). (C) Western blot of hnRNP C and A1 from hnRNP C immunoprecipitates eluted from protein-A agarose beads. RNase treatment was as described above.
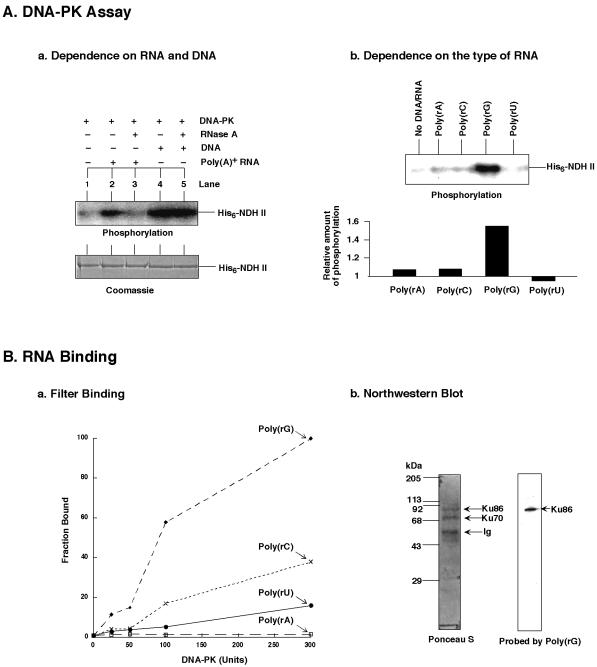
Since NDH II was also a component of hnRNP complexes (Fig. 3) we also wanted to know whether DNA-PK could catalyze an RNA-dependent phosphorylation of this enzyme. Unlike the Coomassie-stainable proteins hnRNP C and A1 (Fig. 5), NDH II is much less abundant in hnRNP complexes and must be detected by more sensitive methods such as immunoblotting. To detect phosphorylation of NDH II by DNA-PK we had to add purified recombinant enzyme (23). In such an assay NDH II was phosphorylated by DNA-PK, and this modification reaction was strikingly stimulated by poly(A)-containing RNA from mouse spleen (Fig. 6Aa). The phosphorylation of NDH II could be abolished by the addition of RNase A (lane 3), which confirmed the RNA-dependent activity of DNA-PK. DNA-PK also catalyzed the DNA-dependent phosphorylation of NDH II that was unaffected by RNase (lanes 4 and 5). An RNA- and DNA-stimulated phosphorylation of NDH II by DNA-PK is consistent with the notion that this protein functions in both RNA and DNA metabolism (24).
Figure 6.
RNA-dependent phosphorylation of NDH II by DNA-PK. (A) (Aa) DNA-PK phosphorylated NDH II in the presence of native RNA. Purified His6-tagged NDH II (≈1 µg each) (23) was used as protein substrate for the DNA-PK assay. Poly(A)+-RNA was from mouse spleen (Clontech) and added to ≈0.1 mg/ml. DNA was from calf thymus and used at 0.05 mg/ml. RNase A treatment was as described in Figure 5. (Ab) Phosphorylation of NDH II in the presence of synthetic RNAs. DNA-PK was incubated with His6-tagged NDH II (≈1 µg each) in the presence of poly(rA), poly(rC), poly(rG) and poly(rU). Phosphorylation levels relative to the background (i.e. in the absence of nucleic acids) are also presented. The signals were quantified with a PhosphorImager. (B) RNA binding assay of DNA-PK. (Ba) Filter binding of DNA-PK with the indicated 32P-labeled ribopolymers poly(rA), poly(rC), poly(rG) and poly(rU). (Bb) Northwestern blot of the Ku heterodimer. Ku86/70 was obtained from HeLa nuclear extract by immunoprecipitation as described in Figure 1. After separation by SDS–PAGE the proteins were transferred to a Hybond-C nitrocellulose membrane (Amersham) and probed by the same RNAs as used for filter binding. Only the Ponceau S protein staining and the membrane probed with poly(rG) are shown. Ig, immunoglobulin.
The observed RNA-dependent protein phosphorylation by DNA-PK suggests that Ku antigen acts as the RNA binding entity that brings the catalytic subunit of DNA-PK to an RNA-bound protein substrate. To measure the hypothesized RNA binding affinity of Ku antigen, filter binding assays were performed with DNA-PK and the 32P-labeled RNAs poly(rA), poly(rC,) poly(rG) and poly(rU). Among all the RNA polynucleotides examined, DNA-PK preferred binding to poly(rG) (Fig. 6Ba). To examine which of the subunits of Ku binds RNA, northwestern blots were performed. To this end the Ku 86/70 heterodimer obtained by immunoprecipitation (see above) was separated by SDS–PAGE and subsequently blotted onto a nitrocellulose membrane. After this treatment the Ku86 and Ku70 were visible by Ponceau S staining (Fig. 6Bb). Incubation of the immobilized proteins with the 32P-labeled RNA probes poly(rA), poly(rC), poly(rG) and poly(rU) revealed a poly(rG)-induced binding signal for the Ku86 subunit, while none of the Ku subunits bound poly(rA), poly(rC) or poly(rU) (data not shown). The coincidence between the preference of Ku86 and DNA-PK for poly(rG) supports the idea that Ku86 is in fact the RNA-binding subunit of DNA-PK. This was further tested with NDH II as the phosphorylation substrate for DNA-PK and the synthetic RNAs already used. Consistently, DNA-PK phosphorylated NDH II most extensively in the presence of poly(rG) (Fig. 6Ab). This further supports the view that DNA-PK can act as an RNA-dependent protein kinase and that its binding to RNA is mediated by the Ku86 subunit.
Subnuclear distributions of NDH II and hnRNP C after silencing NDH II and DNA-PKcs by siRNAs
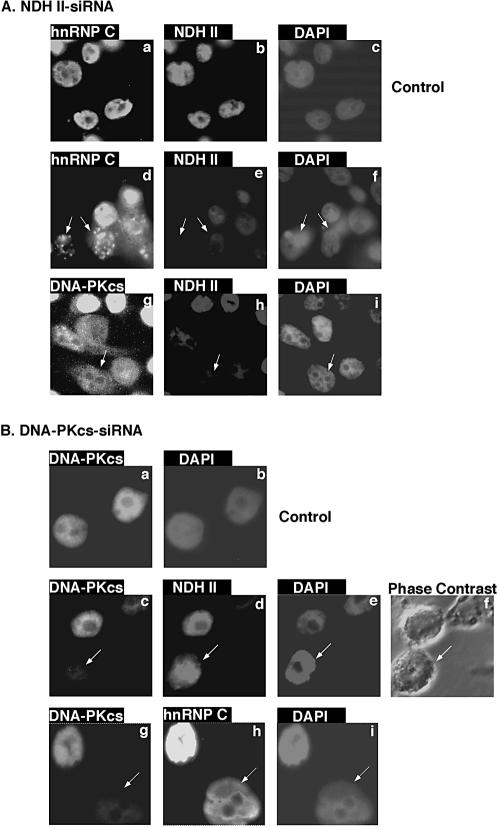
Until now the RNA-dependent phophorylation of several proteins by DNA-PK was only shown in vitro. To get first indications for a functional relationship between NDH II, hnRNP proteins and DNA-PK in the living cell, protein silencing experiments were performed. siRNAs targeting NDH II or DNA-PK were transfected into HeLa cells and the resulting shut-off of the corresponding proteins was examined by western blotting (data not shown) and immunofluorescence. Compared with mock-transfected cells (Fig. 7Aa–c), silencing of NDH II (Fig. 7Ae) seriously disturbed the nuclear distribution of the hnRNP C protein (Fig. 7Ad) that lost its otherwise homogeneous staining pattern (Fig. 7Aa) and aggregated strikingly into small dots over the entire nucleoplasm (Fig. 7Ad). The subnuclear redistribution of hnRNP C as a result of a reduced NDH II expression points to an intimate functional relationship, which was already suggested by the tight association between these two proteins during immunoprecipitation (Fig. 3). On the other hand, silencing NDH II by siRNA (Fig. 7Ah) did not change the cellular localization of DNA-PKcs (Fig. 7Ag). siRNA silencing of DNA-PKcs was also performed and a mock transfection was used as a control (Fig. 7Ba-b). When DNA-PKcs was silenced (Fig. 7Bc), a different subnuclear distribution of NDH II was found (Fig. 7Bd), which now diffused throughout the whole nuclear area and lost its characteristic exclusion from the nucleoli (see Fig. 7Ab). DNA-PKcs silencing (Fig. 7Bg) also led to an accumulation of hnRNP C at the nuclear boundary and increased signals of hnRNP C at the nucleolar periphery (Fig. 7Bh). These results provide some evidence that DNA-PK is necessary for the maintenance of the nucleoplasmic localization of NDH II and hnRNP C. Therefore DNA-PK may also be involved in some regulatory aspects of RNA metabolism.
Figure 7.
Immunofluorescence of HeLa cells after transfection of siRNAs against NDH II and DNA-PK. siRNAs against the expression of (A) NDH II and (B) DNA-PKcs were transfected into HeLa cells that were attached to cover slips. Mock transfected controls were also carried out (Aa–c and Ba and b). Forty-eight hours after transfection immunofluorescence was performed with a rabbit antibody against NDH II (Ab, e and h; Bd), a mouse monoclonal antibody against hnRNP C (4F4) (Aa and d; Bh) or an antibody against the catalytic subunit of DNA-PK (DNA-PKcs) (Ag; Ba, c and g). (Bf) Phase contrast image of HeLa cells. DNA is shown by DAPI stainings in (Ac, f and i) and (Bb, e and i). Arrows indicate HeLa cells that exhibit siRNA silencing of NDH II- or DNA-PKcs expression.
DISCUSSION
Until now Ku antigen was mainly considered as the DNA binding subunit of DNA-PKcs, a protein kinase that is involved in the DNA damage response and sites of DNA recombination. The results from this work demonstrate that Ku antigen is also associated with hnRNP complexes. It was already known that Ku antigen induces the formation of DNA-loop structures that bear similarities to transcriptionally active chromatin loops (25). An association of DNA-PK with sites of RNA synthesis was also supported by the finding that DNA-PK could be extracted from nuclei by lower salt (26). Ku antigen may preferentially target transcriptionally active chromatin by direct interactions with RNA polymerase II (27,28) or with promoter DNA that adopts an open conformation for the entry of Ku (29). Moreover, physical interactions between Ku antigen and several transcription factors have been described (30–32). The physical links between Ku antigen and transcription domains point to a role in transcription-coupled DNA double-strand break repair or recombination (33). Also, DNA-PK has been shown to exert either negative or positive regulatory effects on transcription. DNA-PK, for example, represses the transcriptional initiation of RNA polymerase I (34,35), but is a nuclear factor indispensable for the re-initiation of RNA synthesis catalyzed by RNA polymerase II (36).
The RNA-dependent association between Ku antigen and NDH II observed here suggests a cooperation in vivo. This could be achieved, for example, by the DNA unwinding activity of NDH II that in turn may facilitate the entry of Ku into some chromosomal binding sites, since DNA unwinding is required for the loading of Ku antigen to specific sequences of transcriptional regulation (37). A functional cooperation can also be concluded from the observation that NDH II (like Ku) is an autoantigen in patients suffering from systemic lupus erythematosus (SLE) (38). Moreover, some autosera against Ku antigen cross-react with RNA-binding proteins and components of snRNPs (2), which support the view that all these proteins may form a functional complex in vivo. Taken together, we suggest that Ku antigen is first bound to transcriptionally active chromatin loops and subsequently loaded onto hnRNAs, where it might recognize specific sequences or structures for which it has a high affinity (18–20). Here Ku may accompany the hnRNP proteins and NDH II, for which a similar loading mechanism is assumed.
Immunoprecipitations with antibodies against the abundant components of hnRNP complexes, i.e. hnRNP C or hnRNP A1 (39), revealed that Ku antigen was not associated with all the hnRNP proteins. In particular, hnRNP A1, which is an RNA transport protein accompanying mature mRNPs from the nucleus to the cytosol (40), was not present in two-dimensional gels of Ku-directed immunoprecipitates. On the other hand, the hnRNP proteins C1/2, F, H, J and K were abundantly found in Ku immunoprecipitates. hnRNP C prefers binding to (U)-rich sequences, while the hnRNPs F, H and E prefer (G)-rich RNAs, and hnRNP K/J prefer (C)-rich RNAs (41). In this respect Ku antigen behaved like hnRNP F/H/E, since all these proteins bound poly(rG) with considerable affinity (41). G-rich sequences are found in hnRNAs at the 5′ end of intronic elements that facilitate splice site selection; they are also found at exon enhancers that promote splicing of upstream introns (42). However, to establish a functional relationship between Ku antigen and the G-rich RNA binding proteins beyond the coincidence reported here requires further investigations.
Previously it has been shown that the nucleic acid binding properties and the cellular localization of hnRNP proteins are regulated by phosphorylation (43,44). Most recently phosphorylation of hnRNP C protein was induced by administering low concentrations of H2O2, where casein kinase II seemed to be one of the phosphorylating kinases (45). An RNA-dependent protein kinase that phosphorylated hnRNP C and was different from both casein kinase II and dsRNA activated protein kinase (DAI) had been found in nuclear extract (46). This RNA-dependent protein kinase was not further characterized but could be identical to DNA-PK. Indeed, a specific binding of Ku antigen to RNA derived from the SELEX procedure (Systematic Evolution of Ligands by Exponential Enrichment) has been previously demonstrated (18). Unfortunately, the DNA-PK/SELEX-RNA complex failed to promote protein phosphorylation, probably due to the lack of an appropriate substrate (that also has to bind to SELEX-RNA) or to the lack of RNA sequences that stimulate the kinase activity. The positive outcome of our study is most likely due to the use of natural RNA-binding proteins (NDH II and hnRNP proteins) and natural RNAs with G-rich sequences that are preferred binding partners of DNA-PK. We expect that DNA-PK can catalyze an RNA-dependent phosphorylation in a similar manner and to the same extent as it can phosphorylate DNA-bound proteins. In both cases Ku antigen brings DNA-PK into the proximity of its protein targets, either on DNA or on RNA. By analogy with what has been observed in DNA metabolism, a phosphorylation by DNA-PK may signal the dissociation of distinct RNA binding proteins from hnRNA and/or the modulation of protein–protein interactions within the hnRNA–protein complexes in order to regulate RNA processing and/or transport. Further investigations are certainly required to understand the precise functions of DNA-PK in the regulation of the intracellular RNA metabolism.
Acknowledgments
ACKNOWLEDGEMENTS
The IMB is a Gottfried-Wilhelm-Leibniz Institute and is financially supported by the Federal Government of Germany and the State of Thuringia.
REFERENCES
- 1.Reeves W.H. (1992) Antibodies to the p70/p80 (Ku) antigens in systemic lupus erythematosus. Rheum. Dis. Clin. North Am., 18, 391–414. [PubMed] [Google Scholar]
- 2.Mimori T., Hardin,J.A. and Steitz,J.A. (1986) Characterization of the DNA binding protein antigen Ku recognized by autoantibodies from patients with rheumatic disorders. J. Biol. Chem., 261, 2274–2278. [PubMed] [Google Scholar]
- 3.Mimori T. and Hardin,J.A. (1986) Mechanism of interaction between Ku protein and DNA. J. Biol. Chem., 261, 10375–10379. [PubMed] [Google Scholar]
- 4.Taccioli G.E., Gottlieb,T.M., Blunt,T., Priestley,A., Demengeot,J., Mizuta,R., Lehmann,A.R., Alt,F.W., Jackson,S.P. and Jeggo,P.A. (1994) Ku 80: product of the XRCC5 gene and its role in DNA repair and V(D)J recombination. Science, 265, 1442–1445. [DOI] [PubMed] [Google Scholar]
- 5.Blier P.R., Griffith,A.J., Craft,J. and Hardin. J.A. (1993) Binding of Ku protein to DNA. J. Biol. Chem., 268, 7594–7601. [PubMed] [Google Scholar]
- 6.Tuteja R. and Tuteja,N. (2000) Ku autoantigen: a multifunctional DNA-binding protein. Crit. Rev. Biochem. Mol. Biol., 35, 1–33. [DOI] [PubMed] [Google Scholar]
- 7.Dynan W.S. and Yoo,S. (1998) Interaction of Ku protein and DNA-dependent protein kinase catalytic subunit with nucleic acids. Nucleic Acids Res., 26, 1551–1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tuteja N., Tuteja,R., Ochem,A., Taneja,P., Huang,N.W., Simoncsits,A., Susic,S., Rahman,K., Marusic,L., Chen,J. et al. (1994) Human DNA helicase II: a novel DNA unwinding enzyme identified as the Ku autoantigen. EMBO J., 13, 4991–5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith G.C.M. and Jackson,S.P. (1999) The DNA-dependent protein kinase. Genes Dev., 13, 916–934. [DOI] [PubMed] [Google Scholar]
- 10.Zhang S. and Grosse,F. (1991) Purification and characterization of two DNA helicases from calf thymus. J. Biol. Chem., 266, 20483–20490. [PubMed] [Google Scholar]
- 11.Lee C.-G. and Hurwitz,J. (1992). A new RNA helicase isolated from HeLa cells that catalytically translocates in the 3′ to 5′ direction. J. Biol. Chem., 267, 4398–4407. [PubMed] [Google Scholar]
- 12.Nakajima T., Uchida,C., Anderson,S.F., Lee,C.G., Hurwitz,J., Parvin,J.D. and Montminy,M. (1997) RNA helicase A mediates association of CBP with RNA polymerase II. Cell, 90, 1107–1112. [DOI] [PubMed] [Google Scholar]
- 13.Tang H., Gaietta,G.M., Fischer,W.H., Ellisman,M.H. and Wong-Staal,F. (1997) A cellular cofactor for the constitutive transport element of type D retrovirus. Science, 276, 1412–1415. [DOI] [PubMed] [Google Scholar]
- 14.Zhang S., Buder,K., Burkhardt,C., Schlott,B., Görlach,M. and Grosse,F. (2002) Nuclear DNA helicase II/RNA helicase A binds to filamentous actin. J. Biol. Chem., 277, 843–853. [DOI] [PubMed] [Google Scholar]
- 15.Zhang S., Herrmann,C. and Grosse,F. (1999) Nucleolar localization of murine nuclear DNA helicase II (RNA helicase A). J. Cell Sci., 112, 2693–2703. [DOI] [PubMed] [Google Scholar]
- 16.Lee C.-G. and Hurwitz,J. (1993) Human RNA helicase A is homologous to the maleless protein of Drosophila. J. Biol. Chem., 268, 16822–16830. [PubMed] [Google Scholar]
- 17.Kuroda M.I., Kernan,M.J., Kreber,R., Ganetzky,B. and Baker,B.S. (1991) The maleless protein associates with the X chromosome to regulate dosage compensation in Drosophila. Cell, 66, 935–947. [DOI] [PubMed] [Google Scholar]
- 18.Yoo S. and Dynan,W.S. (1998) Characterization of the RNA binding properties of Ku protein. Biochemistry, 37, 1336–1343. [DOI] [PubMed] [Google Scholar]
- 19.Kaczmarski W. and Khan,S.A. (1993) Lupus autoantigen Ku protein binds HIV-1 TAR RNA in vivo. Biochem. Biophys. Res. Commun., 196, 935–942. [DOI] [PubMed] [Google Scholar]
- 20.Reeves W.H. (1985) Use of monoclonal antibodies for the characterization of novel DNA-binding proteins recognised by human autoimmune sera. J. Exp. Med., 161, 18–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang S., Maacke,H. and Grosse,F. (1995) Molecular cloning of the gene encoding nuclear DNA helicase II. J. Biol. Chem., 270, 16422–16427. [DOI] [PubMed] [Google Scholar]
- 22.Pinol-Roma S., Choi,Y.D., Matunis,M.J. and Dreyfuss,G. (1988) Immunopurification of heterogeneous nuclear ribonucleoprotein particles reveals an assortment of RNA-binding proteins. Genes Dev., 2, 215–227. [DOI] [PubMed] [Google Scholar]
- 23.Zhang S. and Grosse,F. (1997) Domain structure of human nuclear DNA helicase II (RNA helicase A). J. Biol. Chem., 272, 11487–11494. [DOI] [PubMed] [Google Scholar]
- 24.Zhang S. and Grosse,F. (1994) Nuclear DNA helicase II unwinds both DNA and RNA. Biochemistry, 33, 3906–3912. [DOI] [PubMed] [Google Scholar]
- 25.Cary R.B., Peterson,S.R., Wang,J., Bear,D.G., Bradbury,E.M. and Chen,D.J. (1997) DNA looping by Ku and the DNA-dependent protein kinase. Proc. Natl Acad. Sci. USA, 94, 4267–4272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Suwa A., Hirakata,M., Takeda,Y., Jesch,S.A., Mimori,T. and Hardin,J.A. (1994) DNA-dependent protein kinase (Ku protein-p350 complex) assembles on double-stranded DNA. Proc. Natl Acad. Sci. USA, 91, 6904–6908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maldonado E., Shiekhattar,R., Sheldon,M., Cho,H., Drapkin,R., Rickert,P., Lees,E., Anderson,C.W., Linn,S. and Reinberg,D. (1996) A human RNA polymerase II complex associated with SRB and DNA-repair proteins. Nature, 381, 86–89. [DOI] [PubMed] [Google Scholar]
- 28.Mo X. and Dynan,W.S. (2002) Subnuclear localization of Ku protein: functional association with RNA polymerase II elongation sites. Mol. Cell. Biol., 22, 8088–8099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bertinato J., Tomlinson,J.J., Schild-Poulter,C. and Hache,R.J.G. (2003) Evidence implicating Ku antigen as a structural factor in RNA polymerase II-mediated transcription. Gene, 302, 53–64. [DOI] [PubMed] [Google Scholar]
- 30.Huang J., Nueda,A., Yoo,S. and Dynan,W.S. (1997) Heat shock transcription factor 1 binds selectively in vivo to Ku protein and the catalytic subunit of the DNA-dependent protein kinase. J. Biol. Chem., 272, 26009–26016. [DOI] [PubMed] [Google Scholar]
- 31.Barlev N.A., Poltoratsky,V., Owen-Hughes,T., Ying,C., Liu,L., Workman,J.L. and Berger,S.L. (1998) Repression of GCN5 histone acetyltransferase activity via bromodomain-mediated binding and phosphorylation by the Ku-DNA-dependent protein kinase complex. Mol. Cell. Biol., 18, 1349–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Willis D.M., Loewy,A.P., Charlton-Kachigian,N., Shao,J.-S., Ornitz,D.M. and Towler,D.A. (2002) Regulation of osteocalcin gene expression by a novel Ku antigen transcription factor complex. J. Biol. Chem., 277, 37280–37291. [DOI] [PubMed] [Google Scholar]
- 33.Aguilera A. (2001) The connection between transcription and genomic instability. EMBO J., 21, 195–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Labhart P. (1995) DNA-dependent protein kinase specifically represses promoter-directed transcription initiation by RNA polymerase I. Proc. Natl Acad. Sci. USA, 92, 2934–2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kuhn A., Gottlieb,T.M., Jackson,S.P. and Grummt,I. (1995) DNA-dependent protein kinase: a potent inhibitor of transcription by RNA polymerase I. Genes Dev., 9, 193–203. [DOI] [PubMed] [Google Scholar]
- 36.Woodard R.L. Anderson,M.G. and Dynan,W.S. (1999) Nuclear extracts lacking DNA-dependent protein kinase are deficient in multiple round transcription. J. Biol. Chem., 274, 478–485. [DOI] [PubMed] [Google Scholar]
- 37.Torrance H., Giffin,W., Rodda,D.J., Pope,L. and Hache,R.J.G. (1998) Sequence specific binding of Ku autoantigen to single-stranded DNA. J. Biol. Chem., 273, 20810–20819. [DOI] [PubMed] [Google Scholar]
- 38.Takeda Y., Caudell,P., Grady,G., Wang,G., Suwa,A., Sharp,G.C., Dynan,W.S. and Hardin,J.A. (1999) Human RNA helicase A is a lupus autoantigen that is cleaved during apoptosis. J. Immunol., 163, 6269–6274. [PubMed] [Google Scholar]
- 39.Dreyfuss G., Matunis,M.J., Pinol-Roma,S. and Burd,C.G. (1993) hnRNP proteins and the biogenesis of mRNA. Annu. Rev. Biochem., 62, 289–321. [DOI] [PubMed] [Google Scholar]
- 40.Pinol-Roma S. and Dreyfuss,G. (1992) Shuttling of pre-mRNA binding proteins between nucleus and cytoplasm. Nature, 355, 730–732. [DOI] [PubMed] [Google Scholar]
- 41.Matunis M.J., Xing,J. and Dreyfuss,G. (1994) The hnRNP F protein: unique primary structure, nucleic acid-binding properties and subcellular localization. Nucleic Acids Res., 22, 1059–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Krecic A.M. and Swanson,M.S. (1999) hnRNP complexes: composition, structure and function. Curr. Opin. Cell. Biol., 11, 363–371. [DOI] [PubMed] [Google Scholar]
- 43.Idriss H., Kumar,A., Casas-Finet,J.R., Guo,H., Damuni,Z. and Wilson,S.H. (1994) Regulation of in vivo nucleic acid strand annealing activity of heterogeneous nuclear ribonucleoprotein A1 by reversible phosphorylation. Biochemistry, 33, 11382–11390. [DOI] [PubMed] [Google Scholar]
- 44.Hamilton B.J., Burns,C.M., Nichols,R.C. and Rigby,W.F.C. (1997) Modulation of AUUUA response element binding by heterogeneous nuclear ribonucleoprotein A1 in human T lymphocytes. J. Biol. Chem., 272, 28732–28741. [DOI] [PubMed] [Google Scholar]
- 45.Stone J.R., Maki,J.L. and Collins,T. (2003) Basal and hydrogen peroxide stimulated sites of phosphorylation in heterogeneous nuclear ribonucleoprotein C1/C2. Biochemistry, 42, 1301–1308. [DOI] [PubMed] [Google Scholar]
- 46.Fung P.A., Labrecque,R. and Pederson,T. (1997) RNA-dependent phosphorylation of a nuclear RNA binding protein. Proc. Natl Acad. Sci. USA, 94, 1064–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]