Abstract
We synthesized a pentamine (3-3-3-3) and two hexamine (3-3-3-3-3 and 3-4-3-4-3) analogues of the natural polyamine, spermine (3-4-3) and studied their effectiveness in condensing pGL3 plasmid DNA, using light scattering and atomic force microscopic (AFM) techniques. The midpoint concentration of the polyamines on pGL3 condensation (EC50) was 11.3, 10.6, 1.5, 0.49 and 0.52 µM, respectively, for 3-4-3, norspermine (3-3-3), 3-3-3-3, 3-3-3-3-3 and 3-4-3-4-3 in 10 mM Na cacodylate buffer. Dynamic laser light scattering study showed a decrease in hydrodynamic radii of plasmid DNA particles as the number of positive charges on the polyamines increased. AFM data showed the presence of toroids with outer diameter of 117–191 nm for different polyamines, and a mean height of 2.61 ± 0.77 nm. AFM results also revealed the presence of intermediate structures, including those showing circumferential winding of DNA to toroids. The dependence of the EC50 on Na+ concentration suggests different modes of binding of spermine and its higher valent analogues with DNA. Our results show a 20-fold increase in the efficacy of hexamines for DNA condensation compared to spermine, and provide new insights into the mechanism(s) of DNA nanoparticle formation. These studies might help to develop novel nonviral gene delivery vehicles.
INTRODUCTION
The compaction of DNA to nanoparticles is a biologically important phenomenon involved in the packaging of DNA in virus heads and the cell nucleus, as well as in the development of nonviral gene delivery vehicles (1–5). A large number of cationic molecules, including cationic lipids, polyaminolipids, dendrimers and polyethylenimine, are currently under development as nonviral gene delivery vehicles (1,6–10). The first step in the mechanism of action of these molecules is the condensation of DNA to nanoparticles, which can be transported through the cell membrane by endocytosis and/or by other mechanisms that are not well defined at present (1,8,10–13).
Detailed studies of DNA condensation were first conducted with the natural polyamines, spermidine and spermine using electron microscopy (EM) and light scattering techniques (14–18). Polyamines are ubiquitous cellular components and are believed to play important roles in cellular functions, including the packaging of DNA in virus heads (19,20). Polyamine–DNA interactions have been modeled on the basis of electrostatic interactions between the positively charged amino and imino groups of polyamines and the negatively charged phosphate groups of DNA (21). Liquid crystalline textures, including fingerprint textures of the cholesteric phase and columnar hexagonal phase, have been identified in DNA–polyamine complexes formed at high (>1 mg/ml) concentrations of DNA (2,22–24). At very low (∼1 µg/ml) DNA concentrations, nanometric DNA particles are formed in the presence of spermidine, and these particles appear as toroids, spheroids and rods when visualized by EM (14–17,25). Dynamic light scattering (DLS) studies showed a hydrodynamic radius of 40–60 nm for these particles in solution (1,16,18,26). EM and atomic force microscopy (AFM) revealed the presence of intermediates, including flower-like and spaghetti-like structures during the formation of toroids and spheroids as end products of DNA condensation (1,27–31). A recent investigation using freeze-fracture EM showed a hexagonal packaging arrangement of DNA in toroids (32). However, this study was done with Co(NH3)63+ as the condensing agent. Although investigations with Co(NH3)63+ have provided valuable information on the physical chemical aspects of DNA condensation (1,18,32), cationic molecules used in developing gene delivery vehicles are organic cations, such as polyamine-derivatized lipids, polyethylenimine, and dendrimers (1). In addition, most of the reported EM and AFM studies were performed on dried DNA samples that could introduce artifacts on drying as well as by uranyl acetate staining of DNA for EM.
Investigations into the ionic and structural effects of oligoamines (polyamine analogues with a higher positive charge than that of spermine) on the condensation of DNA are lacking at present. In addition, long chain polyamines, such as polyethylenimine are being studied for their ability to promote DNA condensation and gene delivery, although oligoamines of intermediate charge density have received little attention (1). Such molecules might find application in gene delivery vehicles, including the transport of therapeutic oligonucleotides in cancer cells (33). In this study, we synthesized a pentamine and two hexamine analogues of spermine and examined their efficacy in provoking the condensation of a plasmid DNA, pGL-3, using light scattering and AFM techniques. Our results show that the higher valent polyamine analogues are more efficient than spermine in the induction and stabilization of DNA nanoparticles. AFM studies demonstrate the presence of toroidal condensates in the presence of polyamine analogues.
MATERIALS AND METHODS
Plasmid DNA and polyamines
PGL-3 luciferase control vector (5256 bp), which contains the SV40 promoter and enhancer sequences, was purchased from Promega (Madison, WI). The integrity of the plasmid DNA was checked with 1% agarose gel electrophoresis followed by ethidium bromide staining. The results indicated that the plasmid was >90% supercoiled. The plasmid DNA was dissolved in 10 mM Na cacodylate buffer (pH 7.2) containing 0.5 mM EDTA (Na cacodylate buffer), dialyzed from the same buffer, and stored at 4°C. The concentration of the plasmid DNA was determined by measuring the absorbance at 260 nm. Molar DNA phosphate concentration was calculated using a molar extinction coefficient of 6900 M–1 cm–1. Buffer solutions were filtered through 0.22 µm Millipore filters.
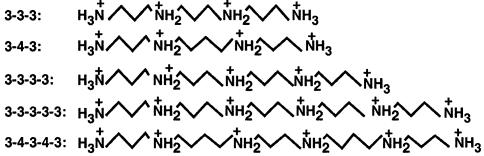
Spermidine.3HCl and spermine.4HCl were purchased from Sigma Chemical Co. (St Louis, MO). Polyamine analogues, 1,11-diamino-4,8-diazaundecane (3-3-3), 1,15-diamino-4,8,12- triazapentadecane (3-3-3-3), 1,19-diamino-4,8,12,16-tetraazanonadecane (3-3-3-3-3), and 1,21-diamino-4,9,13,18-tetraazaheneicosane (3-4-3-4-3) were synthesized by us, as described previously (33). Elemental analysis, NMR, HPLC and mass spectrometry confirmed the chemical structure and purity of polyamine analogues. The protonated chemical structures of the polyamines used in this study are shown in Figure 1. Stock solutions of these polyamine analogues were prepared in double distilled water and appropriate dilutions made in 10 mM Na cacodylate buffer, pH 7.2.
Figure 1.
Chemical structures of polyamines used in this study. The protonated structures are shown here because these molecules are positively charged under the ionic and pH conditions used in this study. 3-4-3 is spermine and 3-3-3 is norspermine.
Total intensity light scattering
DNA condensation experiments were performed in 10 mM Na cacodylate buffer, as described previously (26). The monovalent salt concentrations of the buffer were adjusted using 1 M NaCl stock solutions. Polyamine stock (1–10 mM) solutions were added to DNA solution (1.5 µM) to obtain the desired polyamine concentration in a total volume of 2 ml. The samples were mixed with a vortex mixer for several seconds, and kept undisturbed for a period of 1 h at 22°C to attain equilibrium. The solution was then centrifuged at 500 g for 30 min at 22°C to avoid variations due to aggregates or dust particles. The centrifugation step had no effect on the DNA concentration, as confirmed by measuring the absorbance of DNA at 260 nm before and after centrifugation. Light scattering experiments were performed using a Fluoromax-2 spectrofluorometer. Light from a 150 W Xenon lamp was filtered through double monochromators. The excitation and emission monochromators were set to the same wavelength of 305 nm with a 5 nm bandpass, the integration time set to 5 s, and the scattered light collected at a 90° angle with respect to the incident beam.
Dynamic laser light scattering
We used DynaPro dynamic laser light scattering equipment (Protein Solutions, Lakewood, NJ) with a temperature controlled micro sampler for determining the diffusion coefficients of DNA nanoparticles formed in the presence of different polyamines. DNA solutions were prepared, mixed with polyamine solutions, and 50 µl of the mixture was transferred to the standard quartz cuvette, and scattered light measured at a 90° angle to the incident beam. For monodisperse particles much smaller than the incident beam, the autocorrelation function is given by the equation:
g(1)(τ) = exp[–Dq2(τ)]
where g(1)(τ) is the autocorrelation function, (τ) is the decay time and q is the scattering vector [= (4π n/λ0) sinθ/2], which is a function of solvent refractive index n, the wavelength of the incident beam λ0, the scattering angle θ, and the diffusion coefficient D. The hydrodynamic radius (Rh) is calculated from the diffusion coefficient using the Stokes-Einstein equation:
Rh = kT/6πηD
where k is the Boltzmann constant and T is the absolute temperature in Kelvin. Variability of the data was controlled by programing the software to reject runs with standard deviations higher than 5%.
Atomic force microscopy
The AFM images were acquired on a Nanoscope III microscope (Digital Instruments, Santa Barbara, CA). Plasmid DNA was incubated with the appropriate concentrations of polyamine analogues in 10 mM Na cacodylate buffer for 1 h and deposited onto a freshly cleaved mica surface. The AFM head, equipped with a fluid cell, was placed on top of the scanner (J-scanner, Digital Instruments) and more samples added through the fluid cell. Imaging was done in tapping mode in liquid. Cantilevers (NP-S, Digital Instruments) with a nominal spring constant of 0.32 N/m were used at an oscillation frequency of ∼9 kHz.
Image analysis was performed using Nanoscope software after removing the background slope by flattening images. Height and outer diameter of the toroids were measured using the Nanoscope software. Data are given as mean ± standard error of the mean (S.E.M.).
RESULTS
Static light scattering
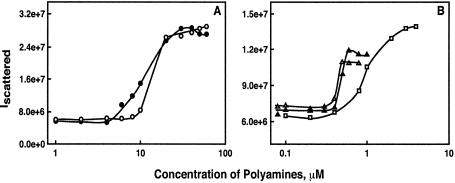
We first determined the efficacy of different polyamines to condense DNA by total intensity light scattering using a Fluoromax-2 spectrofluorometer. Figure 2 shows typical plots of the scattered light intensity of the DNA solution in the presence of different concentrations of polyamines. The scattered light intensity was low in the absence of polyamines; however, a marked increase in the intensity of the scattered light occurred at a critical concentration of each polyamine, due to the formation of condensed DNA particles (26). The increase in the scattered light intensity was concentration-dependent up to a certain concentration of the polyamine, and then leveled off at higher concentrations. Similar scattering profiles were obtained when samples were analyzed after 1–2 h of equilibration. The efficacy of different polyamines on DNA condensation was quantified by determining the EC50 value, the concentration of a polyamine at the midpoint of DNA condensation. In the presence of 10 mM Na+ concentration, the EC50 values were 11.3 ± 0.3, 10.6 ± 1.3, 1.5 ± 0.27, 0.49 ± 0.05 and 0.52 ± 0.1, respectively, for 3-4-3, 3-3-3, 3-3-3-3, 3-3-3-3-3 and 3-4-3-4-3 (Table 1). This result indicates that the pentamine and hexamine analogues are more efficacious than spermine in condensing the plasmid pGL3. However, there was no significant difference in the EC50 values (Table 1) between the two tetramines (3-4-3 and 3-3-3) or between the hexamines (3-3-3-3-3 and 3-4-3-4-3) used in our study, indicating that chemical structural differences of isovalent polyamines may have no significant effect on EC50 values when the charge separation differed by one methylene group.
Figure 2.
Typical plots of the relative intensity of scattered light at 90° plotted against the concentrations of spermine and its analogues. The pGL-3 luciferase plasmid DNA solution had a concentration of 1.5 µM DNA phosphate, dissolved in 10 mM Na cacodylate buffer, pH 7.2. The symbols are as follows: (A) 3-3-3, filled circle; 3-4-3, open circle; (B) 3-3-3-3, open square; 3-3-3-3-3, open triangle; and 3-4-3-4-3, filled triangle.
Table 1. The relative efficacy of spermine and its analogues to condense plasmid DNA.
| Polyamine | Polyamine concentration at midpoint of plasmid DNA condensation (EC50, µM)a |
|---|---|
| 3-4-3 (Spermine) | 11.3 ± 0.3 |
| 3-3-3 (Norspermine) | 10.6 ± 1.3 |
| 3-3-3-3 | 1.5 ± 0.27 |
| 3-3-3-3-3 | 0.49 ± 0.05 |
| 3-4-3-4-3 | 0.52 ± 0.1 |
aAll measurements were made in 10 mM Na cacodylate buffer. EC50 values were determined by plotting the total intensity of scattered light against polyamine concentration (Fig. 1). Data given are mean ± S.E.M. of 3–5 separate measurements.
Hydrodynamic radii of DNA condensates
We next determined the diffusion coefficient of DNA condensates with dynamic laser light scattering equipment and calculated the hydrodynamic radius using the Stokes-Einstein equation. The results are summarized in Table 2. Dynamic light scattering measurements indicated the presence of compact particles with hydrodynamic radii in the range of 50–95 nm. The diffusion coefficient of these particles varied from 2.4 to 4.5 × 10–8 cm2/s. There was a 2-fold decrease in the hydrodynamic radius of the particles as the condensing agent was changed from tetramine (3-4-3) to hexamine (3-4-3-4-3). The nanometric particles produced in the presence of 3-4-3-4-3 were smaller than those produced with 3-3-3-3-3 (67 ± 6.4 versus 51 ± 1.6 nm).
Table 2. Hydrodynamic parameters of DNA nanoparticles formed in the presence of different polyamines.
| Polyamine | Polyamine concentration used for nanoparticle formation (µM) | Diffusion coefficient (cm2/s) | Hydrodynamic radius (nm)a |
|---|---|---|---|
| 3-4-3 (Spermine) | 25 | 2.4 × 10–8 | 94.6 ± 1.6 |
| 3-3-3 | 25 | 2.5 × 10–8 | 91.8 ± 5.3 |
| 3-3-3-3 | 5 | 2.8 × 10–8 | 80 ± 10.4 |
| 3-3-3-3-3 | 2 | 3.4 × 10–8 | 67 ± 6.4 |
| 3-4-3-4-3 | 2 | 4.5 × 10–8 | 51 ± 1.6 |
aAll measurements were made in 10 mM Na cacodylate buffer with the indicated concentrations of polyamines. Data given are mean ± S.E.M. of 3–5 separate measurements.
We also tested the effects of increasing DNA concentration on the hydrodynamic radii of the condensed particles. A 4-fold increase (from 1.5 to 6 µM) in DNA concentration resulted in condensed particles with hydrodynamic radii of 75–105 nm, when spermine was used as the condensing agent. Inter molecular association becomes dominant at higher DNA concentrations, yielding larger condensates. Hence, the majority of our experiments were performed at a low DNA concentration (1.5 µM), to minimize the formation of intermolecular aggregates. Under these conditions, there were no significant differences in the diffusion coefficients of the condensates when measurements were done within 1–2 h of mixing, indicating that the system is equilibrated within this time period.
Structural morphology of DNA condensates by AFM
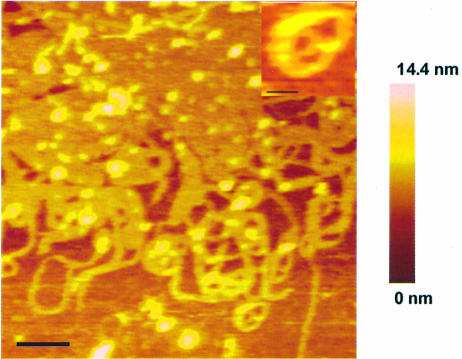
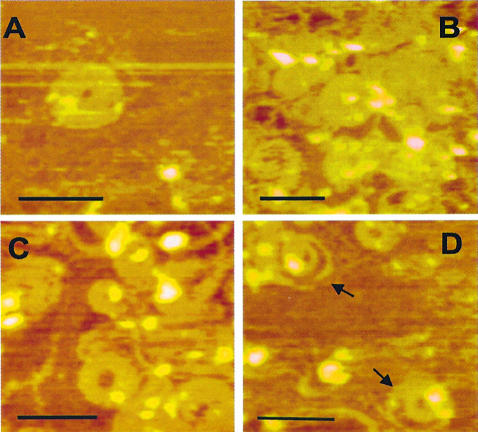
In order to determine the morphology of the DNA condensates formed in the presence of polyamines, we used an atomic force microscope with tapping mode in solution. Figure 3 shows an image of the plasmid DNA condensates formed with 2 µM of 3-3-3-3-3. AFM revealed the presence of individual toroids, circular DNA and a network of toroids. Figure 3 also shows the presence of linear strands and loop-like structures, which are intermediates in the condensation of the DNA. The inset in Figure 3 shows an individual toroid staying on its side, indicating that surface attachment is not a requirement for toroid formation. Similar structures were formed with other polyamines used in this study. Representative toroids, intermediates and toroidal aggregates formed in the presence of spermine (panel A), 3-3-3-3 (panel B) and 3-4-3-4-3 (panel C) are presented in Figure 4. Panel D provides evidence for toroid formation by spooling in the case of 3-4-3-4-3 (marked by arrows).
Figure 3.
Atomic force microscopy images of pGL3 plasmid DNA complexed with 2 µM 3-3-3-3-3. pGL3 plasmid DNA solution (1.5 µM) was incubated with the polyamine solution for 1 h before making the measurements. The imaging was done in tapping mode and in solution. Scale bar is 200 nm. The color bar represents the height of the condensed DNA. The inset shows a single toroid on its side. Scale bar for the inset is 45 nm.
Figure 4.
Scanning force microscopy images showing the toroid structures of pGL3 plasmid DNA formed by incubation with 25 µM spermine (A), 5 µM 3-3-3-3 (B), 2 µM 3-4-3-4-3 (C), and the partly formed toroids, observed in the presence of 2 µM 3-4-3-4-3 (D). Scale bar is 200 nm.
The dimensions of well-developed toroids were measured using the Nanoscope software. The outer diameter varied from 117 to 191 nm (Table 3). There was a decrease in outer diameter of the toroids as the size and charge of the polyamine increased. This result suggests that altering the nature of the polyamine used in condensation can control the size of the DNA nanoparticle. Multimolecular toroids with much larger structures were also found along with the individual toroids (data not shown).
Table 3. AFM measurement of the outer diameter and height of toroids.
| Polyamine | Outer diameter (nm) | Mean height (nm) |
|---|---|---|
| 3-4-3 (Spermine) | 191 ± 12a | 2.61 ± 0.77b |
| 3-3-3-3 | 168 ± 5.4 | |
| 3-3-3-3-3 | 117 ± 8.8 | |
| 3-4-3-4-3 | 118 ± 10.8 |
aMean ± S.E.M. of 5–7 toroids measured in each case.
bThe toroid height given here is the average value for 28 toroids, formed in the presence of polyamines shown in column 1 of this table.
The average height of all toroids, including those formed with spermine to hexamines, was 2.61 ± 0.77 nm (28 toroids were measured). Individual heights varied from 1.5 to 4.2 nm. It is important to note here that the heights of these toroids are significantly lower than that expected from a theoretical calculation, based on the 2 nm theoretical diameter of DNA. This is a limitation of AFM measurements in tapping mode [(34), please see Discussion].
Effect of salt concentration
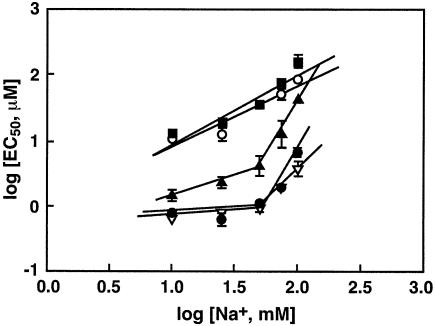
Previous studies by others (16) and us (26) have shown that the EC50 values of tri- and tetravalent polyamines increased with the concentration of monovalent ions, such as Na+, in the medium. We therefore determined the salt dependence of the EC50 using the following concentrations of Na+ in the buffer: 10, 25, 50, 75 and 100 mM. Plots of scattered light intensity versus polyamine concentrations were generated for each polyamine at each of the NaCl concentrations to determine the EC50 values. We then plotted the log[EC50] values against log[Na+] (Fig. 5). Within experimental error, these plots were linear over the entire range of Na+ concentration in the case of spermine and 3-3-3, with slopes of 0.89 and 1.02, respectively. However, two linear regions were observed for the pentamine and hexamines, the slope of the plot at 10–50 mM Na+ concentration was 0.52 for 3-3-3-3 and ∼0.23 for the hexamines. At higher Na+ concentrations, the slope values were 2, 2.5 and 3.2 for 3-3-3-3-3, 3-4-3-4-3 and 3-3-3-3, respectively (Table 4). These slope values, dlog[EC50]/dlog[Na+] are a quantitative measure of the concentration dependence between multivalent and monovalent cations in condensing the plasmid DNA (16,26). In addition, these values provide a measure of the binding affinity of the polyamines with DNA: a lower value of dlog[EC50]/dlog[Na+] indicates a higher binding affinity, according to the equations of the counterion condensation theory [(26), a theoretical description is given in this reference].
Figure 5.
Effect of Na+ in the buffer on the midpoint concentration of polyamines (EC50) to induce DNA condensation. The EC50 values were determined at Na+ concentrations of 10, 25, 50, 75 and 100 mM, as described in Figure 1. The plots of log[Na+] against log[EC50] are in accordance with the equations of the counterion condensation theory (16). A theoretical description of the implications of the slopes of these plots to determine the binding affinity of the cations to DNA is given in a previous publication from this laboratory (26). The symbols represent EC50 values for 3-3-3 (filled square), 3-4-3 (open circle), 3-3-3-3 (filled triangle), 3-3-3-3-3 (inverted open triangle), and 3-4-3-4-3 (filled circle). Error bars indicate standard deviation from three separate experiments. Absence of an error bar indicates that the standard deviation was within the size of the symbol.
Table 4. Dependence of EC50 of polyamines on monovalent ion concentration.
| Polyamine | d[log(EC50)]/d[log(Na+)]a | ||
|---|---|---|---|
| 10–100 mM Na+ | 10–50 mM Na+ | 50–100 Na+ | |
| 3-4-3 (Spermine) | 0.89 | –b | –b |
| 3-3-3 (Norspermine) | 1.02 | –b | –b |
| 3-3-3-3 | – | 0.52 | 3.2 |
| 3-3-3-3-3 | – | 0.24 | 2.03 |
| 3-4-3-4-3 | – | 0.22 | 2.5 |
ad[log(EC50)]/d[log(Na+)] values were determined from the slopes of the lines shown in Figure 5. In the case of spermine and norspermine, a straight line could be constructed over the entire range of Na+ concentrations used. However, in the case of higher polyamine analogues, two straight lines were drawn (Fig. 5), and two slopes calculated.
bNo separate slopes were determined, as a straight line was obtained over the entire range of Na+ concentrations.
DISCUSSION
Results presented in this report demonstrate formation of nanoparticles from a plasmid DNA in the presence of spermine and three higher valent spermine analogues. Previous studies using light scattering, electron microscopic, calorimetric and AFM techniques demonstrated the condensation of closed and linear DNAs in the presence of spermidine and spermine (18,24–28,35–39). Electron microscopic studies have shown the presence of toroids, spheroids and rod-like structures of DNA in the presence of these naturally occurring polyamines. In comparison to distinct toroidal structures of DNA provoked by spermidine and cobalt hexamine [Co(NH3)63+], aggregated structures were formed with spermine (35,40–44), and hence investigations with this tetramine remain limited. In the present study, we examined DNA condensation by novel polyamine analogues by laser light scattering and AFM techniques. Our results show that hexamines are about 20-fold more effective than spermine in compacting the plasmid DNA. Data obtained from dynamic laser light scattering and AFM studies are in excellent agreement.
As can be seen from Table 1, the higher valent polyamines, 3-3-3-3, 3-3-3-3-3 and 3-4-3-4-3 are very efficient in compacting plasmid DNA to nanoparticles. The charge of the condensing agent has a profound influence on the size of the particles formed. The hexamines provoked the formation of more toroidal structures than that by the pentamine, which in turn was better than spermine. The size of the particles decreased with an increase in the charge of the condensing agent. The hexamine analogues condensed DNA to an average hydrodynamic diameter of 118 nm, compared to that of 191 nm for spermine-condensed DNA. In general, the hydrodynamic diameter determined by dynamic laser light scattering and the outer diameter of the toroids measured by the Nanoscope software of the AFM are comparable (Tables 2 and 3), and the data show a consistent decrease in size of the particles as the charge of the polyamine molecule increased.
Our results using AFM are comparable to those reported by other investigators using this method, although different DNA samples and condensing agents were used. For example, Golan et al. (29) measured the dimensions of toroids prepared from pCMV luciferase DNA and poly-L-lysine (PLL) or PLL-asialoorosomucoid (AsOR) conjugate, and reported the height and outer diameter of the toroids as 3.7 ± 0.1 and 142 ± 3.1 nm, respectively. The height of toroids measured in our study is 2.61 ± 0.77 nm. This value is averaged from measurements for all toroids, provoked by spermine to hexamines. The outer diameter of the toroids measured by us varied from 118 to 191 nm, depending on the charge of the polyamine used in our study. Lin et al. (24) found an average outer diameter of 120 ± 15 nm for toroidal structures formed from λ-DNA (48 kb) and spermidine. Golan et al. (29) also found several rods under the conditions of their experiment; the height and widths of these rods were 3.9 ± 0.2 and 50.7 ± 1.7 nm, respectively. These authors proposed an arrangement of DNA in parallel rows in toroids, a mechanism consistent with a folding model for DNA condensation. They also proposed that the folding of rods into toroids could have formed some of the toroids. Such a mechanism has also been proposed by Dunlap et al. (31), who studied the formation of toroids from plasmid DNAs in the presence of didodecylamidoglycylspermine (DOGS) in ethanol or polyethylenimine. The binding affinity and cross-linking tendency of the condensing agent might affect the flexibility of DNA strands and packing density of toroids. Other factors, such as local concentration of condensing agent, temperature, pH and the presence and concentration of monovalent ions may also play important roles for the development of toroids.
It is interesting to note here that the height of toroids measured in this study and that reported by other investigators is lower than that expected on the assumption that toroid formation does not require a surface to initiate the process. For a 6832 bp plasmid DNA, Golan et al. (29) found a toroid height of 3.7 ± 0.1 nm, compared to a height of 0.3 ± 0.1 nm for the uncondensed molecule. Lin et al. (24) found discontinuity in the height distribution of toroids formed from λ-DNA, with heights of 11 ± 4, 20 ± 5, 30 ± 6 and 40 ± 3 nm, measured in multiple Gaussian simulations at the four highest points of the toroids. In a recent study, Sitko et al. (45) reported the width of dense bundles of poly(dG-dC).poly(dG-dC) (∼800 bp) as 22 ± 6 nm, with maximum heights of ∼4–8 nm. They also estimated ∼6–8 parallel molecules of the polynucleotide at the thinner regions of the bundle and 15–20 molecules at the thicker regions of the bundle. These height values are much lower than that expected from condensates composed of several DNA molecules. In fact, the AFM measured height values of single DNA molecules are almost always lower than the theoretical DNA diameter of 2 nm, and the problem associated with this discrepancy has been the topic of a recent publication (34). From a detailed study using non-contact tapping mode in air and jumping mode in aqueous solution, Moreno-Herrero et al. (34) concluded that a major factor contributing to the low height values of DNA is the presence of a salt layer around DNA on the mica surface. In addition, the elastic deformation induced by tip-sample interaction and the compression of the molecule caused by the attractive forces between DNA and the substrate contribute to the reduction in measured height of DNA. The toroid height (2.61 ± 0.77) measured in our study is lower than the theoretical value because of the above mentioned limitations in using AFM for height measurements of biological macromolecules.
Polyamine–DNA interaction has been previously modeled on the basis of the counterion condensation theory developed by Manning (46) and Record et al. (47). This theory predicts a direct relationship between DNA charge neutralization and the concentrations of the monovalent and multivalent ions (16,26,48,49). Assuming a fixed fraction of charge on the DNA is neutralized, Manning’s theory predicted a power law relationship between the concentration of free polyamines in solution and monovalent ion concentration, independent of the nature of the DNA. The salt dependence of the EC50 values of spermine and norspermine (3-3-3) followed a straight line relationship, within experimental error, when log[EC50] was plotted against log[Na+], with slope values of ∼1 (Table 4) over the range of Na+ concentrations studied. In contrast, two linear regions were observed for the pentamine and hexamines. At ionic strengths between 50 and 100 mM Na+, the experimental relationship between polyamine concentration and Na+ concentration obeys the power law with a slope value of 2.5–3.2, which falls between the predicted range of 1 and 6 for the hexamines. But at lower Na+ concentrations, it deviates from the power law. At low salt concentrations, the bound fraction of polyamines is comparable or even larger than free polyamines in solution. Since the relationship holds true only for the concentration of free polyamines in solution, the plots deviate from Manning’s simplified one variable treatment. The deviation was more prominent for the higher valent polyamine analogues than that for the lower valent cations, like spermidine and spermine, probably due to the higher binding affinity of the hexamine and pentamine analogues to DNA.
Several models of polyamine binding to DNA were based on the direct electrostatic interactions between the positive charges of polyamine and the negative charges of DNA (50,51). The binding may depend on the distance between the positive charges on the polyamine relative to the distance between the negative charges of the DNA phosphates. Our previous study (26) on DNA compaction by a series of tetraamine homologues of spermine, differing in the number of methylene groups between the secondary amino groups, demonstrated the structural effects of polyamines on the size of the condensates. Studies on the effect of spermidine on DNA compaction by Baase et al. (52) indicated that cross-linking by polyamine is required for DNA compaction. Polyamine homologues exhibited significant differences in their ability to aggregate and condense genomic DNA and oligonucleotides (53). X-Ray diffraction studies of DNA condensates indicated that the Bragg spacing and interhelical spacing depend on the nature of the counterion (54). The Bragg spacing was in accordance with the model in which the amine groups make contact with the phosphate groups of different helices with fully extended aliphatic chains. Suwalsky et al. (55) proposed a cross-linked model, which assumed that the trimethylene spacing of 0.49 nm is suitable to interact with the adjacent phosphate groups while the tetramethylene spacing of 0.614 nm is suitable to bridge between different duplex strands. Molecular mechanics calculations (56) further predicted folding of DNA strands over the polyamines, while a recent molecular dynamics simulation study predicted differences in the nature of interaction between natural and synthetic polyamines with DNA (57). AFM of DNA complexes with dendronized polymers indicate that DNA wraps around dendronized polymers and the calculated pitch decreased with increase in the linear charge density of the polymer (58). Increasing the number of methylene groups of the polyamine may also enable them to wrap around the DNA molecule instead of being localized in the DNA major groove.
Among the polyamine analogues with the same charges, smaller ions produce more tightly packed condensates (26). Decrease in the size of the condensates with an increase in the charge of the condensing agent can be due to a decrease in the interhelical spacing. Since toroidal volume is proportional to molecularity, a decrease in the size of the condensates can also be attributed to a decrease in the molecularity of the condensates. According to the model proposed by Manning for cation-induced DNA condensation, DNA bends spontaneously at a critical fraction of charge neutralization leading to the formation of toroids (59). Hud et al. (60) proposed a model in which DNA in toroids is organized within a series of equally sized contiguous loops around the toroid axis. Noguchi et al. (61) suggested that formation of toroids is an example of coil–globule transition. Toroid formation is also considered as a self-assembly process in which toroids can be formed by wound DNA or by the bending of rods. Using AFM, Dunlap et al. (31) observed individual DNA strands separated by ∼3.2 nm in the condensed state. Our AFM measurements indicate that polyamine analogues compact DNA into toroidal structures. Non-toroidal forms, including rods and U-shaped structures were also observed along with the toroids. Our results provide evidence for the spooling of DNA to form toroids (Fig. 4D).
In summary, our data demonstrate that higher valent polyamines are capable of provoking DNA compaction into nanometric particles. Among the polyamine analogues, hexamines were more efficient than pentamines, which in turn were more efficient than tetramines in provoking nanoparticle formation, indicating the predominant role played by electrostatic interactions between protonated polyamines and DNA. The charge of the polyamines also influenced the size of the condensates, showing a decrease in the size of the particles as the charge of the polyamine increased. The salt dependence of the midpoint concentration of pentamine and hexamines suggests that the overall charge neutralization of DNA decreases with an increase in monovalent ion concentration in solution. Investigations into the formation and stability of toroids under different ionic conditions are important to optimize conditions under which nonviral gene therapy vehicles can be effective.
Acknowledgments
ACKNOWLEDGEMENTS
T.A. expresses his deep sense of gratitude to Drs Tom Jovin and Vinod Subramaniam for their help and support for conducting the atomic force microscopic experiments during his tenure as an Alexander von Humboldt Fellow at the Max Plank Institute for Biophysical Chemistry. This work was supported by Public Health Service grants CA80163, CA73058 and CA42439 from the National Cancer Institute (to T.J.T. and T.T.), Susan G. Komen Breast Cancer Research Foundation Award (to T.T.), Alexander Von Humboldt Fellowship (to T.A.), and a Grant-in-Aid for Scientific Research from the Ministry of Education, Science and Culture, Japan (to A.S.).
REFERENCES
- 1.Vijayanathan V., Thomas,T. and Thomas,T.J. (2002) DNA nanoparticles and development of DNA delivery vehicles for gene therapy. Biochemistry, 41, 14085–14094. [DOI] [PubMed] [Google Scholar]
- 2.Saminathan M., Thomas,T., Shirahata,A., Pillai,C.K.S. and Thomas,T.J. (2002) Polyamine structural effects on the induction and stabilization of liquid crystalline DNA: potential applications to DNA packaging, gene therapy and polyamine therapeutics. Nucleic Acids Res., 30, 3722–3731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bloomfield V.A. (1997) DNA condensation by multivalent cations. Biopolymers, 44, 269–282. [DOI] [PubMed] [Google Scholar]
- 4.Ibarra B., Caston,J.R., Llorca,O., Valle,M., Valpuesta,J.M. and Carrascosa,J.L. (2000) Topology of the components of the DNA packaging machinery in the phage φ29 prohead. J. Mol. Biol., 298, 807–815. [DOI] [PubMed] [Google Scholar]
- 5.Conwell C.C., Vilfan,I.D. and Hud,N.V. (2003) Controlling the size of nanoscale toroidal DNA condensates with static curvature and ionic strength. Proc. Natl Acad. Sci. USA, 100, 9296–9301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuo P.Y. and Saltzman,W.M. (1996) Novel systems for controlled delivery of macromolecules. Crit. Rev. Eukaryotic Gene Expr., 6, 59–73. [DOI] [PubMed] [Google Scholar]
- 7.Luo D. and Saltzman,W.M. (2000) Synthetic DNA delivery systems. Nat. Biotechnol., 18, 33–37. [DOI] [PubMed] [Google Scholar]
- 8.Zuber G., Dauty,E., Nothisen,M., Belguise,P. and Behr,J.P. (2001) Towards synthetic viruses. Adv. Drug Deliv. Rev., 52, 245–253. [DOI] [PubMed] [Google Scholar]
- 9.Schaffer D.V. and Lauffenburger,D.A. (2000) Targeted synthetic gene delivery vectors. Curr. Opin. Mol. Ther., 2, 155–161. [PubMed] [Google Scholar]
- 10.Blessing T., Remy,J.S. and Behr,J.P. (1998) Monomolecular collapse of plasmid DNA into stable virus-like particles. Proc. Natl Acad. Sci. USA, 95, 1427–1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hansma H.G., Golan,R., Hsieh,W., Lollo,C.P., Mullen-Ley,P. and Kwoh,D. (1998) DNA condensation for gene therapy as monitored by atomic force microscopy. Nucleic Acids Res., 26, 2481–2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu G., Molas,M., Grossmann,G.A., Pasumarthy,M., Perales,J.C., Cooper,M.J. and Hanson,R.W. (2001) Biological properties of poly-L-lysine-DNA complexes generated by cooperative binding of the polycation. J. Biol. Chem., 276, 34379–34387. [DOI] [PubMed] [Google Scholar]
- 13.Montigny W.J., Houchens,C.R., Illenye,S., Gilbert,J., Coonrod,E., Chang,Y.C. and Heintz,N.H. (2001) Condensation by DNA looping facilitates transfer of large DNA molecules into mammalian cells. Nucleic Acids Res., 29, 1982–1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gosule L.C. and Schellman,J.A. (1976) Compact form of DNA induced by spermidine. Nature, 259, 333–335. [DOI] [PubMed] [Google Scholar]
- 15.Eickbush T.H. and Moudrianakis,E.N. (1978) The compaction of DNA helices into either continuous supercoils or folded-fiber rods and toroids. Cell, 13, 295–306. [DOI] [PubMed] [Google Scholar]
- 16.Wilson R.W. and Bloomfield,V.A. (1979) Counterion-induced condensation of deoxyribonucleic acid. A light-scattering study. Biochemistry, 18, 2192–2196. [DOI] [PubMed] [Google Scholar]
- 17.Marx K.A. and Reynolds,T.C. (1982) Spermidine-condensed φX174 DNA cleavage by micrococcal nuclease: torus cleavage model and evidence for unidirectional circumferential DNA wrapping. Proc. Natl Acad. Sci. USA, 79, 6484–6488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thomas T.J. and Bloomfield,V.A. (1983) Collapse of DNA caused by trivalent cations: pH and ionic specificity effects. Biopolymers, 22, 1097–1106. [DOI] [PubMed] [Google Scholar]
- 19.Thomas T. and Thomas,T.J. (2001) Polyamines in cell growth and cell death: molecular mechanisms and therapeutic applications. Cell. Mol. Life Sci., 58, 244–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bode V.C. and Harrison,D.P. (1973) Distinct effects of diamines, polyamines, and magnesium ions on the stability of λ phage heads. Biochemistry, 12, 3193–3196. [DOI] [PubMed] [Google Scholar]
- 21.Bloomfield V.A. (1991) Condensation of DNA by multivalent cations: considerations on mechanism. Biopolymers, 31, 1471–1481. [DOI] [PubMed] [Google Scholar]
- 22.Radler J.O., Koltover,I., Salditt,T. and Safinya,C.R. (1997) Structure of DNA-cationic liposome complexes: DNA intercalation in multilamellar membranes in distinct interhelical packing regimes. Science, 275, 810–814. [DOI] [PubMed] [Google Scholar]
- 23.Livolant F. and Leforestier,A. (1996) Condensed phases of DNA: Structures and phase transitions. Prog. Polym. Sci., 21, 1115–1164. [Google Scholar]
- 24.Lin Z., Wang,C., Feng,X., Liu,M., Li,J. and Bai,C. (1998) The observation of the local ordering characteristics of spermidine-condensed DNA: atomic force microscopy and polarizing microscopy studies. Nucleic Acids Res., 26, 3228–3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arscott P.G., Li,A.Z. and Bloomfield,V.A. (1990) Condensation of DNA by trivalent cations. 1. Effects of DNA length and topology on the size and shape of condensed particles. Biopolymers, 30, 619–630. [DOI] [PubMed] [Google Scholar]
- 26.Vijayanathan V., Thomas,T., Shirahata,A. and Thomas,T.J. (2001) DNA condensation by polyamines: a laser light scattering study of structural effects. Biochemistry, 40, 13644–13651. [DOI] [PubMed] [Google Scholar]
- 27.Liu D., Wang,C., Li,J., Lin,Z., Tan,Z. and Bai,C. (2000) Atomic force microscopy analysis of intermediates in cobalt hexammine-induced DNA condensation. J. Biomol. Struct. Dyn., 18, 1–9. [DOI] [PubMed] [Google Scholar]
- 28.Fang Y. and Hoh,J.H. (1998) Early intermediates in spermidine induced condensation on the surface of mica. J. Am. Chem. Soc., 120, 8903–8909. [Google Scholar]
- 29.Golan R., Pietrasanta,L.I., Hsieh,W. and Hansma,H.G. (1999) DNA toroids: stages in condensation. Biochemistry, 38, 14069–14076. [DOI] [PubMed] [Google Scholar]
- 30.Böttcher C., Endisch,C., Fuhrhop,J.H., Catterall,C. and Eaton,M. (1998) High yield preparation of oligomeric C-type DNA toroids and their characterization by cryoelectron microscopy. J. Am. Chem. Soc., 120, 12–17. [Google Scholar]
- 31.Dunlap D.D., Maggi,A., Soria,M.R. and Monaco,L. (1997) Nanoscopic structure of DNA condensed for gene delivery. Nucleic Acids Res., 25, 3095–3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hud N.V. and Downing,K.H. (2001) Cryoelectron microscopy of λ phage DNA condensates in vitreous ice: the fine structure of DNA toroids. Proc. Natl Acad. Sci. USA, 98, 14925–14930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thomas R.M., Thomas,T., Wada,M., Sigal,L.H., Shirahata,A. and Thomas,T.J. (1999) Facilitation of the cellular uptake of a triplex-forming oligonucleotide by novel polyamine analogues: structure–activity relationships. Biochemistry, 38, 13328–13337. [DOI] [PubMed] [Google Scholar]
- 34.Moreno-Herrero F., Colchero,J. and Baro,A.M. (2003) DNA height in scanning force microscopy. Ultramicroscopy, 96, 167–174. [DOI] [PubMed] [Google Scholar]
- 35.Allison S.A., Herr,J.C. and Schurr,J.M. (1981) Structure of viral φ29 DNA condensed by simple triamines: a light-scattering and electron-microscopy study. Biopolymers, 20, 469–488. [DOI] [PubMed] [Google Scholar]
- 36.Widom J. and Baldwin,R.L. (1983) Inhibition of cation-induced DNA condensation by intercalating dyes. Biopolymers, 22, 1621–1632. [DOI] [PubMed] [Google Scholar]
- 37.Matulis D., Rouzina,I. and Bloomfield,V.A. (2000) Isothermal titration calorimetry and electrostatic mechanism. J. Mol. Biol., 296, 1053–1063. [DOI] [PubMed] [Google Scholar]
- 38.Hansma H.G. (2001) Surface biology of DNA by atomic force microscopy. Annu. Rev. Phys. Chem., 52, 71–92. [DOI] [PubMed] [Google Scholar]
- 39.Fang Y. and Hoh,J.H. (1998) Surface-directed DNA condensation in the absence of soluble multivalent cations. Nucleic Acids Res., 26, 588–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Musso M., Thomas,T., Shirahata,A., Sigal,L.H., Van Dyke,M.W. and Thomas,T.J. (1997) Effects of chain length modification and bis(ethyl) substitution of spermine analogues on purine-purine-pyrimidine triplex DNA stabilization, aggregation, and conformational transitions. Biochemistry, 36, 1441–1449. [DOI] [PubMed] [Google Scholar]
- 41.Newton G.L., Aguilera,J.A., Ward,J.F. and Fahey,R.C. (1996) Polyamine-induced compaction and aggregation of DNA—a major factor in radioprotection of chromatin under physiological conditions. Radiat. Res., 145, 776–780. [PubMed] [Google Scholar]
- 42.Pelta J., Livolant,F. and Sikorav,J.L. (1996) DNA aggregation induced by polyamines and cobalthexamine. J. Biol. Chem., 271, 5656–5662. [DOI] [PubMed] [Google Scholar]
- 43.Osland A. and Kleppe,K. (1977) Polyamine induced aggregation of DNA. Nucleic Acids Res., 4, 685–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Raspaud E., Chaperon,I., Leforestier,A. and Livolant,F. (1999) Spermine-induced aggregation of DNA, nucleosome, and chromatin. Biophys. J., 77, 1547–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sitko J.C., Mateescu,E.M. and Hansma,H.G. (2003) Sequence-dependent DNA condensation and the electrostatic zipper. Biophys. J., 84, 419–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Manning G.S. (1978) The molecular theory of polyelectrolyte solutions with applications to the electrostatic properties of polynucleotides. Q. Rev. Biophys., 11, 179–246. [DOI] [PubMed] [Google Scholar]
- 47.Record M.T. Jr, Anderson,C.F. and Lohman,T.M. (1978) Thermodynamic analysis of ion effects on the binding and conformational equilibria of proteins and nucleic acids: the roles of ion association or release, screening, and ion effects on water activity. Q. Rev. Biophys., 11, 103–178. [DOI] [PubMed] [Google Scholar]
- 48.Li A.Z., Huang,H., Re,X., Qi,L.J. and Marx,K.A. (1998) A gel electrophoresis study of the competitive effects of monovalent counterion on the extent of divalent counterions binding to DNA. Biophys. J., 74, 964–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Subirana J.A. and Vives,J.L. (1981) The precipitation of DNA by spermine. Biopolymers, 20, 2281–2283. [Google Scholar]
- 50.Deng H., Bloomfield,V.A., Benevides,J.M. and Thomas,G.J.,Jr (2000) Structural basis of polyamine-DNA recognition: spermidine and spermine interactions with genomic B-DNAs of different GC content probed by Raman spectroscopy. Nucleic Acids Res., 28, 3379–3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Feuerstein B.G., Williams,L.D., Basu,H.S. and Marton,L.J. (1991) Implications and concepts of polyamine-nucleic acid interactions. J. Cell. Biochem., 46, 37–47. [DOI] [PubMed] [Google Scholar]
- 52.Baase W.A., Staskus,P.W. and Allison,S.A. (1984) Precollapse of T7 DNA by spermidine at low ionic strength: a linear dichroism and intrinsic viscosity study. Biopolymers, 23, 2835–2851. [DOI] [PubMed] [Google Scholar]
- 53.Saminathan M., Antony,T., Shirahata,A., Sigal,L.H., Thomas,T. and Thomas,T.J. (1999) Ionic and structural specificity effects of natural and synthetic polyamines on the aggregation and resolubilization of single-, double-, and triple-stranded DNA. Biochemistry, 38, 3821–3830. [DOI] [PubMed] [Google Scholar]
- 54.Schellman J.A. and Parthasarathy,N. (1984) X-ray diffraction studies on cation-collapsed DNA. J. Mol. Biol., 175, 313–329. [DOI] [PubMed] [Google Scholar]
- 55.Suwalsky M., Traub,W., Shmueli,U., Subirana,J.A. (1969) An X-ray study of the interaction of DNA with spermine. J. Mol. Biol., 42, 363–373. [DOI] [PubMed] [Google Scholar]
- 56.Feuerstein B.G., Pattabiraman,N. and Marton,L.J. (1986) Spermine-DNA interactions: a theoretical study. Proc. Natl Acad. Sci. USA, 83, 5948–5952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Korolev N., Lyubartsev,A.P., Laaksonen,A. and Nordenskiold, L. (2003) A molecular dynamics simulation study of oriented DNA with polyamine and sodium counterions: diffusion and averaged binding of water and cations. Nucleic Acids Res., 31, 5971–5981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gössl I., Shu,L., Schluter,A.D. and Rabe,J.P. (2002) Molecular structure of single DNA complexes with positively charged dendronized polymers. J. Am. Chem. Soc., 124, 6860–6865. [DOI] [PubMed] [Google Scholar]
- 59.Manning G.S. (1980) Thermodynamic stability theory for DNA doughnut shapes induced by charge neutralization. Biopolymers, 19, 37–59. [DOI] [PubMed] [Google Scholar]
- 60.Hud N.V., Downing,K.H. and Balhorn,R. (1995) A constant radius of curvature model for the organization of DNA in toroidal condensates. Proc. Natl Acad. Sci. USA, 92, 3581–3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Noguchi H., Saito,S., Kidoaki,S. and Yoshikawa,K. (1996) Self-organized nanostructures constructed with a single polymer chain. Chem. Phys. Lett., 261, 527–533. [Google Scholar]