Abstract
Telomerase is a ribonucleoprotein reverse transcriptase responsible for the maintenance of one strand of telomere terminal repeats. Telomerase-mediated sequence addition is dictated by a short ‘template’ region of the RNA component. Despite the short template segment, telomerases from many organisms have been shown to mediate the synthesis of long extension products. This synthesis presumably depends on two types of translocation events: simultaneous translocation of the RNA–DNA duplex relative to the active site after each nucleotide incorporation (type I or nucleotide addition processivity), and translocation of the RNA relative to the DNA product after each round of repeat synthesis (type II or repeat addition processivity). In contrast, telomerases from yeasts have been shown to synthesize mostly short products, implying a defect in one or both types of translocation. In this report, we analyzed the processivity of yeast telomerase in vitro, and identified two position-specific elongation barriers within the 5′ region of the RNA template that can account for the synthesis of incomplete first round products. These barriers respond differently to variations in nucleotide concentration, primer sequence and mutations in the catalytic protein subunit, consistent with their having distinct mechanistic bases. In addition, by using optimal primers and high concentrations of dGTP, we were able to detect significant type II translocation by the yeast enzyme. Thus, the difference between the elongation property of yeast and other telomerases appears to be quantitative rather than qualitative. Our results suggest that yeast may be a useful system for investigating the physiologic significance of repeat addition processivity.
INTRODUCTION
Telomerase is a ribonucleoprotein (RNP) that is responsible for maintaining the terminal repeats of telomeres in most organisms (1). It acts as an unusual reverse transcriptase (RT), using a small segment of an integral RNA component as template for the synthesis of the dG-rich strand of telomeres (2).
Telomerase activity has been characterized from a wide range of organisms, and genes encoding both the RNA and protein components of the enzyme complex have been identified (reviewed in 3–7). Telomerase RNAs from ciliated protozoa, in addition to having a short templating segment, share a common secondary structure. Telomerase RNAs from yeast and mammals are considerably larger, and within each group conserved structural elements can be identified based on phylogenetic and mutational analysis (8,9). The catalytic reverse transcriptase protein subunit (TERT), initially purified from Euplotes aediculatus as p123, was subsequently found to be homologous to Est2p, a yeast protein required for telomere maintenance (10–12). Both proteins possess RT-like motifs, alterations in which render telomerase inactive both in vitro and in vivo. Subsequently, homologues of TERT were identified in diverse organisms (13–20). Additional mutational analysis of the RT-motifs in these latter proteins supports a role for TERT in directly mediating catalysis (21,22). Because co-expression of TERT and telomerase RNA in vitro in the rabbit reticulocyte lysate system (RRL) suffices to re-constitute enzyme activity (21,23), these two subunits probably constitute the core of the enzyme complex. Many telomerase-associated polypeptides have been identified using either biochemical or genetic tools. Preliminary studies suggest that these factors may participate in either telomerase assembly or function (24–28).
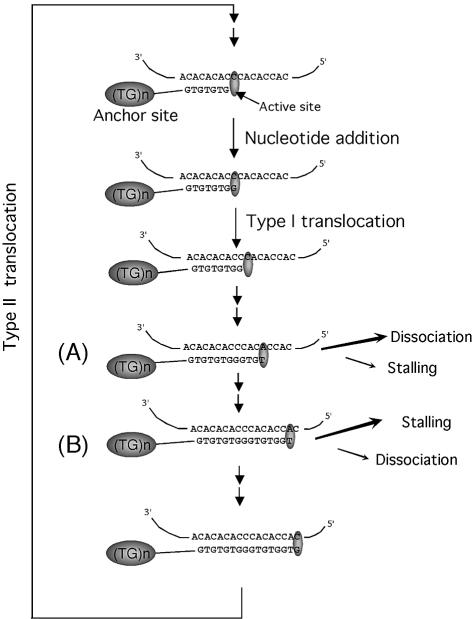
A striking feature of telomerase is its ability to add long stretches of nucleotides to a given primer despite the short length of the template. Telomerase isolated from ciliated protozoa and humans is especially efficient at primer elongation, being capable of adding hundreds of nucleotides (involving multiple rounds of copying the template) to the starting primer without dissociating (29,30). These processive telomerases must be competent at carrying out two types of translocation reaction: simultaneous movement of the RNA–DNA duplex relative to the active site after each nucleotide incorporation (type I or nucleotide addition processivity), and unpairing of the RNA–DNA hybrid followed by re-alignment of the DNA 3′ end to the 3′ region of the RNA template (type II or repeat addition processivity) (Fig. 1). Processive elongation is believed to require interactions between a 5′ region of the DNA primer (away from the RNA–DNA hybrid) and a second site in the TERT subunit. This second site has also been referred to as the ‘product site’ or ‘anchor site’ (31–35). Thus, when DNA primers without such a 5′ region are used in Tetrahymena telomerase assays, predominantly short products are generated (31,32). Anchor site interaction is not absolutely essential for multiple repeat additions; even with very short primers or primers with non-telomeric sequences, a small percentage of the products can become extremely elongated (36,37). Nevertheless, existing data suggest that interaction of telomerase with primer nucleotides immediately upstream of the base-pairing region can substantially enhance the efficiency of type II translocation (37).
Figure 1.
Type I and II translocation by telomerase. The putative reaction steps for the S.cerevisiae telomerase, including alignment, nucleotide addition and type I and II translocations are illustrated (see Introduction for a detailed explanation). In addition, a model for the differential response of the two elongation barriers to primer, nucleotides and TERT mutations is presented (see Discussion for a more detailed explanation). Briefly, at position A, the telomerase/substrate complex is prone to dissociation. Low nucleotide concentrations and the reduced binding caused by the primer grip mutation can therefore impair processive synthesis. In contrast, at position B, the telomerase/substrate complex is stable but prone to stalling. Because translocation is limiting, the addition of more nucleotides would not be expected to improve processivity. In addition, the stability conferred by the long RNA–DNA hybrid at this position may override the effect of primer grip mutations such that no processivity change occurs as a consequence of the mutations.
Structural elements of the telomerase enzyme that control nucleotide and repeat addition processivity have been identified in a variety of systems. For example, several conserved RT motifs in yeast, human and Tetrahymena TERTs have been shown to modulate nucleotide addition processivity (38–41). Alterations in the C-terminal extension of yeast and human TERTs can also have significant impacts on either nucleotide or repeat addition processivity (39,42,43). More over, template mutations in yeast, human and Tetrahymena telomerase RNA, and non-template mutations in Tetrahymena telomerase RNA were demonstrated to have diverse effects on either the efficiency of nucleotide or repeat addition (44–47). Thus, telomerase processivity appears to be governed by a complex set of interactions between components of the enzyme and the primer and nucleotide substrate.
Remarkably, telomerases isolated from fungi appear to be naturally non-processive, generating predominantly short products in vitro (12,48–52). The non-processive telomerases are likely to be less efficient in mediating at least one type of translocation reaction. Furthermore, fungal telomerases have been shown to bind stably to their elongation products (50,53). In such instances, termination of DNA synthesis cannot be attributed to dissociation of the enzyme from the elongated primer. These observations raise the interesting possibility that yeast telomerase may possess fundamentally distinct structural and mechanistic features (e.g. lacking the ability to unpair RNA–DNA hybrids) (50). It should be stressed, however, that the ‘in vitro’ processivity of telomerase may not accurately reflect its ‘in vivo’ processivity; Tetrahymena telomerase, for example, appears to be much less processive in vivo than in vitro as revealed by analysis using mutant RNAs (54).
The budding yeast Saccharomyces cerevisiae (Sc) represents an ideal model system for analyzing telomerase structures and functions because the organism is amenable to both genetic and biochemical studies. In earlier experiments, we identified residues of the yeast TERT subunit (named Est2p) required for optimal nucleotide addition processivity. However, because of the paucity of long extension products, it was difficult to assess the existence and efficiency of type II translocation for either the wild-type or mutant enzymes. In this study, we quantitatively evaluated Sc telomerase elongation properties in the 5′ region of the RNA template. Our analysis uncovered two elongation barriers in this region, which can partly account for the propensity of yeast telomerase to synthesize incomplete first round products. Interestingly, the two elongation barriers are differentially affected by alterations in primer sequence, dGTP concentration and TERT mutations, suggesting that they have distinct mechanistic bases. In addition, the effects of hybrid-disrupting mutations in the DNA primer on enzyme processivity and enzyme–substrate stability imply that yeast telomerase can accumulate long RNA–DNA hybrids. Finally, by using optimal primers and nucleotide combinations, we were able to detect unequivocally and assess the efficiency of type II translocation for the yeast enzyme. Our results suggest that the difference between the elongation property of yeast and other telomerases is quantitative rather than qualitative, and that yeast may be a useful model system for investigating the physiologic significance of repeat addition processivity.
MATERIALS AND METHODS
Yeast strains
The haploid S.cerevisiae strain W303-a (MATa ade2-1 trp1-1 leu2-3,112 his3-11,15 ura3-1 can1-100) was used for the preparation of unmodified telomerase. The construction of the strain with protein A-tagged Est2p has been described (55). The strains bearing the C721A and M723A mutations in TERT have also been described (39).
Primers
All oligodeoxynucleotide primers were purchased from Sigma-Genosis or Qiagen-Operon and purified by denaturing gel electrophoresis prior to use.
Purification and assay of yeast telomerase
Whole-cell extracts, active diethylaminoethyl (DEAE) fractions, and IgG-Sepharose enriched protein A-tagged telomerase were prepared as previously described (48,49,55). A typical reaction using DEAE-enriched telomerase was carried out in 30 µl containing the following: 50 mM Tris–HCl, pH 8.0, 2 mM magnesium acetate, 1 mM spermidine, 1 mM DTT, 5% glycerol (contributed by the protein fraction), variable concentrations of unlabeled dTTP and dGTP, 20 µCi of [α-32P]dGTP (3000 Ci/mmol), 5 µM of primer oligodeoxynucleotides, and 3 µl of column fractions. For IgG-Sepharose bound telomerase, the assay is initiated by the addition of a 15 µl cocktail containing 100 mM Tris–HCl, pH 8.0, 4 mM magnesium chloride, 2 mM DTT, 2 mM spermidine, 100 µM dTTP, 10 µM primer oligodeoxynucleotides, and 20 µCi [α-32P]dGTP (3000 Ci/mmol) to 20 µl of IgG-Sepharose pretreated with 4 mg of protein extract. Primer extension products were processed and analyzed by gel electrophoresis as previously described (49,56).
For determination of processivity, the signal for each band was determined by Imagequant software and normalized to the amount of transcript by dividing against the number of labeled residues. All primers used in this study end with 3 Gs and are expected to align with the RNA template such that their 3′ most residues are positioned opposite the 10th nucleotide of the template, allowing the sequence TGTGGTG to be added. For nucleotides beyond the +7 position, we assume a sequence of TGTG… (calculations were made assuming other compatible sequences, and the conclusions were not altered.)
![]()
where Ti denotes the amount of transcript calculated for the primer + i position and N is the highest number such that a visible signal can be discerned in the PhosphorImager file for the primer + N product.
For measurement of enzyme–DNA stability, telomerase bound to IgG-Sepharose was assayed using the appropriate primers. Upon completion of the reaction, the beads were washed three times with 150 µl each of TMG-10(150), and the supernatants pooled. The supernatants and the beads were then separately processed by RNase and proteinase K digestion, ethanol precipitation and gel electrophoresis.
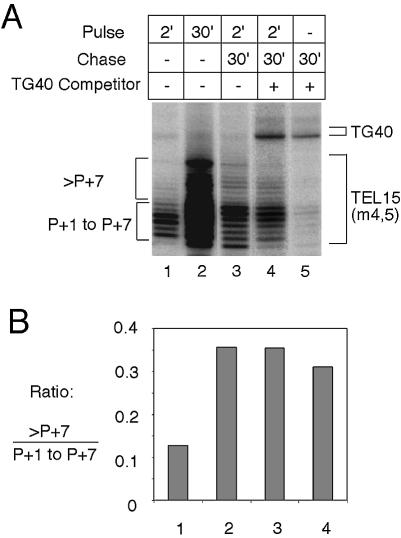
For the pulse–chase experiment, telomerase was incubated with 2 µM of TEL15(m4,5) at 22°C for 5 min and the reaction initiated with 0.4 µM 32P-dTTP (3000 Ci/mmol) and 50 µM dGTP. After 2 min of this pulse reaction, 50 µM unlabeled dTTP was added to start the chase reaction, which lasted for 30 min at 22°C. The chase reaction was performed either in the presence or absence of 10 µM competitor primer [(TGTGTGGG)5, named TG40 in Fig. 8].
Figure 8.
Pulse–chase analysis of yeast telomerase. (A) Yeast telomerase was subjected to pulse–chase analysis using the TEL15(m4,5) primer as described in Materials and Methods. The duration of the pulse and chase reactions and the presence or absence of the TG40 competitor primer are indicated at the top. Products due to TEL15(m4,5) and TG40 are indicated by brackets at the right. First and second round products for TEL15(m4,5) are indicated by brackets at the left (‘P + 1 to P + 7’ and ‘>P + 7’, respectively). (B) The ratios of the second round products (>P + 7) to the first round products (P + 1 to P + 7) were determined for assays 1–4 shown in part (A) and plotted.
RESULTS
Identification of two elongation barriers in the 5′ region of telomerase RNA template
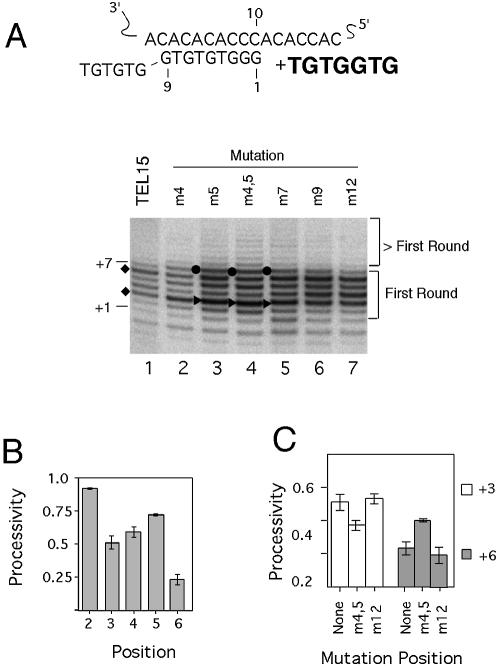
The elongation properties of the Sc telomerase were examined in standard primer extension assays initially using a 15-nt fully telomeric primer with three tandem G residues at its 3′ ends (TEL15). Such a primer can be expected to align precisely with the RNA template such that its 3′ most residue is positioned opposite the 10th nucleotide of the template, allowing quantitative analysis of the reaction products (Fig. 2A, note that both the RNA template and the DNA primer residues are numbered from 3′ to 5′). To obtain a detailed view of telomerase elongation, we determined the processivity of Sc telomerase for the primer at each position along the reverse transcript. For TEL15, there appear to be two elongation barriers, at the primer + 3 position and the primer + 6 position (henceforth abbreviated as P + 3 and P + 6) (Fig. 2A, lane 1, indicated by diamonds; Fig. 2B). The processivities of telomerase at these positions are ∼0.5 and 0.25, respectively.
Figure 2.
Effects of primer mutations on yeast telomerase elongation. (A) An alignment of the template region of yeast telomerase RNA with the 15-nt canonical yeast primer (TEL15) is shown at the top. The template residues and the primer residues are both numbered from 3′ to 5′. The 3′ most residue of the primer should be positioned opposite the 10th nucleotide of the template. The sequence that is expected to be added by telomerase is shown in bold. Results of primer extension assays that were performed using TEL15 and related primers, 0.2 µM dGTP and 50 µM dTTP are shown in the bottom panel. The positions of the mutations (either T to A or G to C) in the DNA primers are indicated at the top of the panel. The locations of the P + 1 and P + 7 products are indicated at the left side of the panel. The diamonds point to the locations of the two elongation barriers for TEL15, at the P + 3 and P + 6 positions. The filled triangles and circles point to changes in the relative intensities of the P + 3 and P + 7 products, respectively. (B) The processivity of telomerase for the TEL15 primer from the P + 2 to P + 6 positions are plotted. (C) The processivity of telomerase for the TEL15, TEL15(m4,5) and TEL15(m12) primers at the P + 3 and P + 6 positions are plotted.
The effect of primer sequence alterations on telomerase processivity at the two elongation barriers
To assess the effect of primer sequence on telomerase processivity, we tested variant primers containing mutations at defined positions. All mutations are either G-to-C or T-to-A transversion mutations and are designated by the mutation position(s) (e.g. in Fig. 2A, m4 designates a T-to-A mutation at primer position 4). Two evident effects of mutations on telomerase-mediated extension can be discerned. First, the overall level of DNA synthesis is increased for most non-canonical primers, possibly due to increased enzyme turnover (Fig. 2A, compare lanes 2–7 with lane 1). This finding is consistent with an earlier study of primer mutations (49). Second, mutations located near the center of the potential RNA–DNA hybrid [in TEL15(m4), TEL15(m5) and TEL15(m4,5)] significantly altered the distribution of the reaction products. In particular, the relative amounts of the P + 3 and P + 7 products were increased (Fig. 2A, compare the bands marked by filled triangles and circles in lanes 2–4 with the others). Quantitative analysis revealed position-specific alterations in processivity for the TEL15(m4), (m5) and m(4,5) primers. Because the processivities are very similar for all three primers, only the values for the TEL15(m4,5) primer are presented in Figure 2C. As shown in the plot, relative to TEL15, telomerase exhibited a 30% decrease in processivity at the P + 3 position for TEL15(m4,5), which can account for the increased intensity of this product. TEL15(m4,5) also supported a 2-fold increase in processivity at the P + 6 position, which can account for the increased intensity of the P + 7 product. These results suggest that mutations in DNA primer that disrupt the RNA–DNA hybrid can cause complex changes in the efficiencies of type I translocations. In contrast, mutations outside the RNA–DNA hybrid [e.g. in TEL15(m12)], though capable of altering the overall level of DNA synthesis, did not result in significant changes in processivity (Fig. 2A and C). The same results were obtained using either unmodified telomerase purified by DEAE chromatography or protein A-tagged telomerase enriched by IgG-Sepharose affinity-adsorption, suggesting that the alterations in banding pattern were due to the intrinsic properties of Sc telomerase (data not shown).
Differential effects of dGTP concentration on telomerase processivity at the two elongation barriers
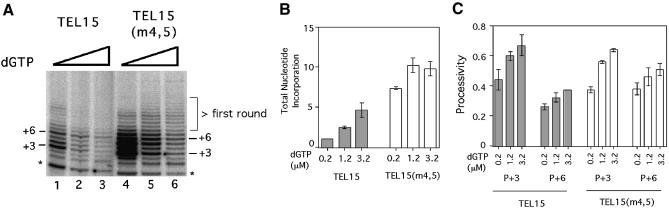
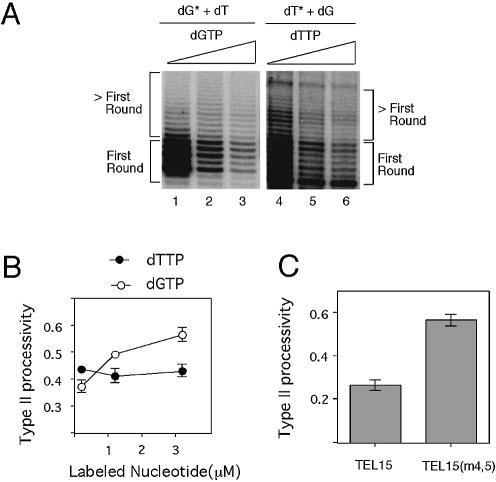
The elongation barriers appear to occur preferentially before the incorporation of labeled dGTP, which is present at 0.2 µM in standard assays. Specifically, extensions beyond P + 3 and P + 6 of TEL15 (and related primers) both require the incorporation of dGTP. To determine how the barriers are affected by nucleotide concentrations, we assayed TEL15 and TEL15(m4,5) in the presence of varying concentrations of dGTP (Fig. 3A). Because the dGTP concentrations were altered by adding unlabeled dGTP to standard reactions, the intensities of the products appeared to diminish with increasing dGTP concentration. However, total DNA synthesis for each primer was in fact increased when the reduced specific activity of the label was taken into account (Fig. 3B).
Figure 3.
Effects of dGTP concentration on telomerase activity and processivity. (A) Primer extension assays were performed using either TEL15 (lanes 1–3) or TEL15(m4,5) (lanes 4–6) and telomerase that was purified by DEAE chromatography. All reactions contained in addition to 0.2 µM of labeled dGTP and 50 µM of unlabeled dTTP, either 0 µM (lanes 1 and 4), 1 µM (lanes 2 and 5), or 3 µM (lanes 3 and 6) of unlabeled dGTP. The positions of the P + 3 and P + 6 products are indicated at both sides of the panel. A low molecular weight RNase-insensitive product (indicated by asterisks) can be observed in this set of assays. (B) The total levels of nucleotide incorporation for the TEL15 and TEL15(m4,5) primer are plotted at different dGTP concentrations. The dGTP concentrations (in µM) are indicated at the bottom. (C) The processivity of telomerase at the P + 3 and P + 6 positions for TEL15 and TEL15(m4,5) is plotted at different dGTP concentrations. The dGTP concentrations (in µM) are indicated at the bottom of the diagram.
Interestingly, the two major barriers respond differently to the change in dGTP concentration. The relative intensity of the P + 3 product for TEL15 diminished progressively when the total dGTP concentration was raised from 0.2 to 1.2 and 3.2 µM, consistent with a relief in elongation block (Fig. 3A, lanes 1–3). In contrast, the relative intensity of the P + 6 product for TEL15 remained strong in comparison with surrounding bands, consistent with a retention of the barrier. Quantitative determination of processivity confirmed the differential effects of nucleotide concentration increase on elongation barriers. Thus, the processivity for TEL15 at the P + 3 position was increased by ∼60% in the presence of high dGTP concentrations, while that at the P + 6 position was only increased by ∼20% (Fig. 3C).
A mutant primer that gave rise to an altered elongation pattern [TEL15(m4,5)] was also tested using increasing concentrations of dGTP, and the results subjected to the same quantitative analysis (Fig. 3A and C). For this primer, the elongation barrier at the P + 3 position was again relieved by increasing the dGTP concentration, such that the processivity increased by ∼70% (Fig. 3C). The processivity for TEL15(m4,5) at the P + 6 position was higher than the TEL15, and was increased by only ∼20% in response to higher dGTP concentrations (Fig. 3C). Together, these results indicate that dGTP is not as limiting for telomerase elongation at the 5′ boundary barrier (P + 6 position) as at the internal barrier (P + 3 position), suggesting that the two barriers may have different mechanistic bases.
Lack of strict correlation between telomerase processivity and enzyme–product complex stability
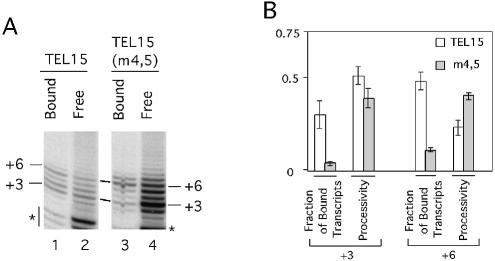
In prior analysis of retroviral RT processivity, mutations that affect processivity have often been found to affect enzyme–substrate stability (57,58). To determine whether alterations in telomerase–primer stability correlate with changes in telomerase processivity, we took advantage of the stable association between protein A-tagged telomerase and IgG-Sepharose. Reactions were performed in triplicate using two 15-nt primers [TEL15 and TEL15(m4,5)] and IgG-bound telomerase. Upon termination of the reactions, the labeled products were separated into soluble and Sepharose-bound fractions before analysis in denaturing gels (Fig. 4A). The fraction of enzyme-bound DNA and enzyme processivity at the two elongation barriers (P + 3 and P + 6) were then quantified and plotted (Fig. 4B).
Figure 4.
Analysis of the stability of telomerase–DNA complexes. (A) Primer extension assays were performed using either TEL15 (lanes 1 and 2) or TEL15(m4,5) (lanes 3 and 4) and telomerase that was bound to IgG-Sepharose. The assays also include 0.2 µM dGTP and 50 µM dTTP. Following the reaction, the IgG-Sepharose was washed three times with 150 µl each of TMG-10(150). Both the Sepharose fraction (Bound; lanes 1 and 3) and the combined washes (Free; lanes 2 and 4) were analyzed for the presence of labeled products. The positions of the P + 3 and P + 6 products are indicated at both sides of the panels. Some low molecular weight RNase-insensitive products (indicated by asterisks) can be observed in this set of assays. (B) The fraction of enzyme-bound product and enzyme processivity at the P + 3 and P + 6 position for TEL15 and TEL15(m4,5) are determined in duplicate assays and the averages and deviations plotted for comparison. Enzyme processivity at the P + 3 and P + 6 position was determined by assuming that the amount of reverse transcript at each position was the sum of the bound and free product, and by using the formula described in Materials and Methods.
Several interesting observations can be made from this comparative analysis. First, a higher percentage of the longer product (P + 6) was found in the bound fraction for each primer than the shorter product (P + 3), suggesting that increasing duplex length can increase the overall stability of the enzyme–DNA complex (Fig. 4B) (53). Second, for the hybrid-disrupting TEL15(m4,5) primer, the stability of the enzyme–DNA complex relative to that for TEL15 was markedly lower at each position, again consistent with the importance of the hybrid. Third, the processivity at the P + 3 position, but not at the P + 6 position, correlated with enzyme–product stability; while the stability at both positions was reduced by the TEL15(m4,5) mutations, only the processivity at P + 3 was reduced by the same mutations. This last observation provides further support for the notion that the two elongation barriers are due to different factors.
Differential effects of TERT mutations on processivity at the two elongation barriers
A number of structural elements in conserved RT motifs have been shown to influence processivity in retroviral enzymes. These elements include the conserved motif 1, 2, C and E. In the crystal structure of HIV-1 RT/substrate complex, motif E (also known as the ‘primer grip’) comprises a beta hairpin that is in close proximity to the 3′ end of the DNA primer. We have identified two residues (Cys721 and Met723) at the tip of this hairpin of yeast TERT that affect the elongation properties of yeast telomerase (39). As shown in Figure 5A, either amino acid residue, when substituted by alanine, caused telomerase to synthesize slightly shorter products on average using the TEL15 primer as a substrate (compare lanes 3–6 with lanes 1 and 2). Both mutations also resulted in marked telomere shortening in vivo, suggesting that the seemingly moderate reduction in elongation ability is physiologically significant (39). To determine quantitatively how the mutations affected elongation at the two major barriers, we compared processivity at the P + 3 and P + 6 positions for the wild-type and mutant enzymes from duplicate assays. As shown in Figure 5B, both mutations significantly reduced processivity at the P + 3 position without greatly affecting processivity at the P + 6 position. This result is again consistent with the notion of distinct underlying mechanisms for the two elongation barriers.
Figure 5.
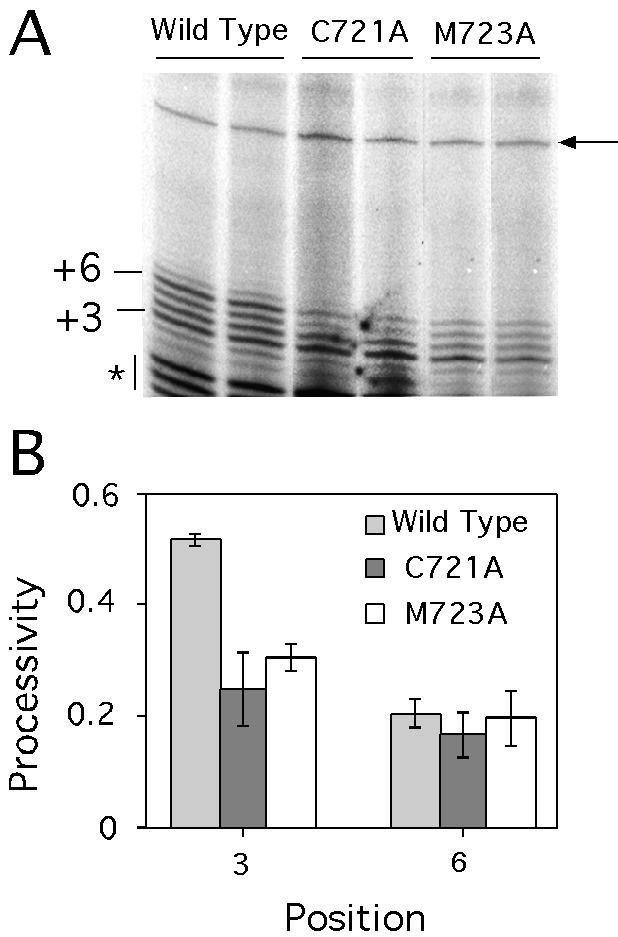
Effects of primer grip mutations on telomerase elongation. (A) Primer extension assays were performed using TEL15 and telomerases that have been purified by IgG-Sepharose adsorption. The assays also include 0.2 µM dGTP and 50 µM dTTP. The positions of the P + 3 and P + 6 products are indicated at the left side. Several low molecular weight RNase-insensitive products (indicated by asterisks) can be observed in this set of assays. A prelabeled 46-nt primer was added before ethanol precipitation as a recovery control (indicated by an arrow at the right). (B) The processivity of telomerase at the P + 3 and P + 6 positions are plotted for both the wild-type and mutant enzymes.
Yeast telomerase is capable of type II translocation
In the course of analyzing type I translocation by yeast telomerase, we noticed that a significant amount of >P + 7 products can be detected in assays utilizing TEL15(m4,5) (Figs 2A and 3A). The synthesis of these products was abolished by pre-treatment of the extracts with RNase A, consistent with their being due to telomerase (data not shown). Based on the putative alignment position of this primer, the >P + 7 products can arise only if yeast telomerase is capable of re-positioning the 3′ end of the DNA relative to the RNA template following one complete round of template copying (type II translocation). A direct measure of type II translocation efficiency is the processivity at the P + 7 position. To determine whether this processivity is modulated by nucleotide concentrations, we performed assays using different combinations of labeled and unlabeled nucleotides (Fig. 6A). The labeled nucleotide was included at 0.2, 1.2 and 3.2 µM, while the unlabeled nucleotide at 50 µM. Quantitative analysis indicates that the processivity at the P + 7 position was increased from 0.37 to 0.56 by increasing the dGTP concentration from 0.2 to 3.2 µM (while keeping dTTP constant at 50 µM) (Fig. 6A and B). Similar titration of labeled dTTP (while keeping dGTP constant at 50 µM) had little effect on type II translocation efficiency (Fig. 6A and B). In contrast to TEL15(m4,5), the canonical yeast primer TEL15 supported the synthesis of very low levels of >P + 7 products (Figs 2A and 3A). Quantitative analysis confirmed that type II translocation for TEL15 is ∼50% that of TEL15(m4,5) (Fig. 6C).
Figure 6.
Analysis of type II translocation efficiency. (A) Primer extension assays were performed using TEL15(m4,5) and different combinations of labeled nucleotide (indicated by an asterisk) and unlabeled nucleotide. The positions of the first round and higher order products are indicated by brackets. The total concentration of the labeled nucleotide was 0.2 µM (lanes 1 and 4), 1.2 µM (lanes 2 and 5) and 3.2 µM (lanes 3 and 6). (B) The processivity of telomerase at the P + 7 position as a function of dGTP and dTTP concentration is plotted. (C) The processivity of telomerase at the P + 7 position for the TEL15 and TEL15(m4,5) was determined in assays containing 3.2 µM dGTP and 50 µM dTTP and plotted.
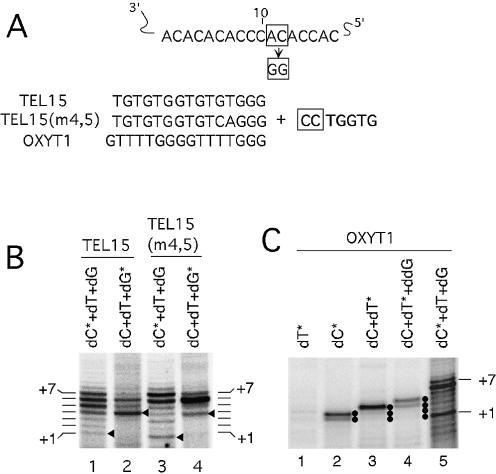
One alternative interpretation of the >P + 7 products is that these products are due to misalignment of the primer to a more 3′ region of the TLC1 RNA template and still no more than one round of polymerization. However, detectable signals were obtained for products up to the P + 19 position, arguing that at least one type II translocation must have occurred. Furthermore, no misalignment of the primer can be detected when assays were performed using a telomerase bearing two point mutations at position 11 and 12 along the RNA template (Fig. 7A). With this particular mutant RNA, telomerase is expected to add the sequence ‘CCTGGTG’ in the first round of polymerization for both the TEL15 and TEL15(m4,5) primer. Consistent with this expectation, dGTP incorporation was not detectable until the P + 4 position (Fig. 7B, lanes 2 and 4), whereas dCTP incorporation was detected at the P + 1 position (Fig. 7B, lanes 1 and 3). The use of another primer with the same 3′ end sequence (OXYT1) also gave expected results (Fig. 7C).
Figure 7.
The primers align to the RNA template in the predicted manner. (A) The predicted alignment of the template region of the mutated yeast telomerase RNA with the 15-nt yeast primers [TEL15 and TEL15(m4,5)] and a heterologous primer that also ends in 3Gs (OXYT1) is shown. The template residues are numbered from 3′ to 5′. The 3′ most residue of the primers are predicted to be positioned opposite the 10th nucleotide of the template. The sequence that is expected to be added by the mutant telomerase is also shown with the first two dC residues boxed. (B) Primer extension assays were performed using either TEL15 (lanes 1 and 2) or TEL15(m4,5) (lanes 3 and 4) and telomerase that was purified by DEAE chromatography. All reactions contain the three nucleotides that are necessary for copying the entire mutant template (dCTP, dGTP and dTTP). The labeled nucleotides (indicated by asterisks) and the unlabeled nucleotides are present at 0.2 and 50 µM, respectively. The positions of the P + 1 to P + 7 products are indicated at both sides of the panel. (C) Primer extension assays were performed using OXYT1 as primer, different combinations of labeled nucleotides (specified by asterisks) and unlabeled nucleotides as indicated at the top, and telomerase that was purified by DEAE chromatography. The positions of the P + 1 and P + 7 products are indicated on the right side.
To rule out further the possibility that the second round of synthesis was due to dissociation and rebinding of the enzyme to completed first round product, we performed a pulse–chase experiment based on previously described procedures for the Tetrahymena and human telomerase (30,45). Yeast telomerase was allowed to bind and extend TEL15(m4,5) in the presence of labeled dTTP. After a 2-min pulse, a large excess of unlabeled dTTP was added to chase the reaction. The chase reaction was performed either in the absence or presence of a competitor oligonucleotide (TG40). As shown in Figure 8, only a small fraction of the product became longer than P + 7 at the end of the 2-min pulse (Fig. 8A, lane 1; Fig. 8B). However, a higher percentage of the product became longer than P + 7 upon completion of the chase, regardless of the presence or absence of the TG40 competitor (Fig. 8A, lanes 3 and 4; Fig. 8B). When TG40 was added at the start of the reaction, it was capable of suppressing almost all of the TEL15(m4,5) extension products (Fig. 8A, lane 5). Thus, the first round extension products must be processively extended to become longer than P + 7. We conclude that yeast telomerase must be capable of carrying out at least one type II translocation reaction.
DISCUSSION
Distinct mechanisms of elongation block for Sc telomerase
Our studies indicate that under the standard reaction conditions, there are two apparent elongation barriers along the Sc telomerase RNA template, specifically at the 13th and the 16th nucleotide positions (Position A and B in Fig. 1). These two elongation barriers appear to have different mechanistic basis, as implied by the differential effects of primer–template mismatches, nucleotide concentrations and catalytic subunit mutations. Notably, the elongation block at the 13th nucleotide is markedly relieved by increasing the dGTP concentration, but exacerbated by primer mutations that disrupt the RNA–DNA duplex and by mutations in the ‘primer grip’ of TERT. In contrast, the block at the 16th nucleotide is only slightly altered by increasing the dGTP concentration and TERT mutations, but is suppressed by duplex-disrupting primer mutations.
What are the differences between the structures of the enzyme–substrate complex at these two elongation barriers that are responsible for their differential response to primer, nucleotides and TERT mutations? One plausible candidate is the length of RNA–DNA hybrid. We imagine that at the 13th template position, the shorter RNA–DNA hybrid might result in a less-stable complex that is prone to dissociation (position A in Fig. 1). Thus, the rate of dissociation might compete with the rate of translocation and nucleotide addition. Increasing dGTP concentration may then favor processive elongation instead of dissociation. The effect of TERT mutation at this position can be rationalized if the Cys721 and Met723 residues mediate tighter binding of the enzyme to the primer, as has been demonstrated for the primer grip of HIV-1 RT (59–61). In contrast, at the 16th template position, the longer RNA–DNA hybrid may result in a fairly stable complex that is prone to stalling (position B in Fig. 1). If translocation is limiting, then adding more nucleotides would not be expected to improve processivity. In addition, the stability conferred by the long RNA–DNA hybrid at this position may override the effect of primer grip mutations such that no processivity change occurs as a consequence of the mutations.
The block at the 16th nucleotide of the Sc telomerase RNA template exhibits many features reported for a K.lactis telomerase elongation block (50). Both are located near the 5′ template boundary, are resistant to increased nucleotide concentration, and are suppressed by reducing the length of the putative RNA–DNA duplex. Thus, a hybrid length-dependent elongation barrier near the 5′ RNA template boundary may be a general property of yeast telomerase, regardless of the overall template length (30 nt for K.lactis and 17 nt for S.cerevisiae telomerase). Conversely, our findings also suggest that yeast telomerase does not maintain a constant length of RNA–DNA hybrid, but rather extends the hybrid as elongation progresses.
The physiologic function for the 5′-boundary proximal elongation block is unclear. A reasonable hypothesis, previously proposed for K.lactis telomerase, is that inefficient elongation may serve to limit the degree of telomere extension, thereby maintaining telomere length homeostasis (50). We have recently shown that a Sc telomerase with increased processivity can cause an increase in the equilibrium lengths of telomeres, indicating that telomerase processivity is indeed a determinant of telomere length (39). Another potential role of this elongation block is to minimize the ability of telomerase to copy beyond the template boundary. An RNA stem located immediately adjacent to the 5′ template boundary of K.lactis telomerase has been shown to be crucial to telomerase template definition; in the absence of such a structure, the TERT protein is capable intrinsically of copying beyond the 5′ template boundary (8). Conceivably, by reducing the probability of the catalytic center to even reach the 5′ boundary, the elongation blocks can provide an alternative mechanism for minimizing the enzyme’s copying of non-template residues. The relevance of these proposals for ciliate and mammalian telomerases awaits further investigation.
Type II translocation by yeast telomerase
Despite the accumulation of long hybrid and stalling near the 5′ template boundary, we have shown that Sc telomerase is capable of limited type II translocation and repeat addition processivity in assays containing optimal primers and nucleotides. Notably, the efficiency of type II translocation is enhanced by disrupting the potential RNA–DNA hybrid through DNA mutations, consistent with the idea that unwinding is a critical first step in this reaction.
Interestingly, in two respects, the properties of the native yeast enzyme appear to resemble those of the recombinant Tetrahymena enzyme (consisting only of telomerase RNA and TERT). Both enzymes are relatively non-processive. In addition, type II translocation by both enzymes appears to be stimulated by high concentrations of dGTP (17,62). The latter observation suggests that some mechanistic features of type II translocation may be conserved between yeast and other telomerases. In contrast to the recombinant Tetrahymena telomerase, the native Tetrahymena enzyme is very processive, and this high degree of processivity is observed even with low dGTP concentrations (30,63). This discrepancy in properties of the recombinant and native enzyme has been attributed to the effects of auxiliary factors or conformational differences (17). It is not known if the ‘native’ yeast telomerase isolated from cell extracts is in fact missing important auxiliary factors or exhibits a different conformation in comparison with the ‘in vivo’ complex.
The discovery of repeat addition processivity by yeast telomerase lays the groundwork for investigating the physiologic significance of this unique telomerase property. Earlier mutational analysis of the nucleotide addition processivity of yeast telomerase indicates that this processivity is important for telomere maintenance in vivo (39,44). Identification of yeast mutants with specific alterations in repeat addition processivity should be equally informative.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Dr Liz Blackburn for comments on an early version of the manuscript. This work was supported by an American Cancer Society Research Project Grant and an RO1 from the National Institute of Health. The Department of Microbiology and Immunology gratefully acknowledges the support of the William Randolph Hearst Foundation.
REFERENCES
- 1.Greider C.W. and Blackburn,E.H. (1985) Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell, 43, 405–413. [DOI] [PubMed] [Google Scholar]
- 2.Greider C.W. and Blackburn,E.H. (1989) A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature, 337, 331–337. [DOI] [PubMed] [Google Scholar]
- 3.Nugent C.I. and Lundblad,V. (1998) The telomerase reverse transcriptase: components and regulation. Genes Dev., 12, 1073–1085. [DOI] [PubMed] [Google Scholar]
- 4.Collins K. (1999) Ciliate telomerase biochemistry. Annu. Rev. Biochem., 68, 187–218. [DOI] [PubMed] [Google Scholar]
- 5.Kelleher C., Teixeira,M., Forstemann,K. and Lingner,J. (2002) Telomerase: biochemical considerations for enzyme and substrate. Trends Biochem. Sci., 27, 572–579. [DOI] [PubMed] [Google Scholar]
- 6.Harrington L. (2003) Biochemical aspects of telomerase function. Cancer Lett., 194, 139–154. [DOI] [PubMed] [Google Scholar]
- 7.Blackburn E. (2000) The end of the (DNA) line. Nat. Struct. Biol., 7, 847–850. [DOI] [PubMed] [Google Scholar]
- 8.Tzfati Y., Fulton,T.B., Roy,J. and Blackburn,E.H. (2000) Template boundary in a yeast telomerase specified by RNA structure. Science, 288, 863–867. [DOI] [PubMed] [Google Scholar]
- 9.Chen J.L., Blasco,M.A. and Greider,C.W. (2000) Secondary structure of vertebrate telomerase RNA. Cell, 100, 503–514. [DOI] [PubMed] [Google Scholar]
- 10.Lendvay T.S., Morris,D.K., Sah,J., Balasubramanian,B. and Lundblad,V. (1996) Senescence mutants of Saccharomyces cerevisiae with a defect in telomere replication identify three additional EST genes. Genetics, 144, 1399–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lingner J. and Cech,T.R. (1996) Purification of telomerase from Euplotes aediculatis: requirement of a primer 3′ overhang. Proc. Natl Acad. Sci. USA, 93, 10712–10717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lingner J., Hughes,T.R., Shevchenko,A., Mann,M., Lundblad,V. and Cech,T.R. (1997) Reverse transcriptase motifs in the catalytic subunit of telomerase. Science, 276, 561–567. [DOI] [PubMed] [Google Scholar]
- 13.Nakamura T.M., Morin,G.B., Chapman,K.B., Weinrich,S.L., Andrews,W.H., Lingner,J., Harley,C.B. and Cech,T.R. (1997) Telomerase catalytic subunit homologs from fission yeast and human. Science, 277, 955–959. [DOI] [PubMed] [Google Scholar]
- 14.Meyerson M., Counter,C.M., Eaton,E.N., Ellisen,L.W., Steiner,P., Caddle,S.D., Ziaugra,L., Beijersbergen,R.L., Davidoof,M.J. and Liu,Q. et al. (1997) hEST2, the putative human telomerase catalytic subunit gene, is up-regulated in tumor cells and during immortalization. Cell, 90, 785–795. [DOI] [PubMed] [Google Scholar]
- 15.Kilian A., Bowtell,D.D., Abud,H.E., Hime,G.R., Venter,D.J., Keese,P.K., Duncan,E.R., Reddel,R.R. and Jefferson,R.A. (1997) Isolation of a candidate human telomerase catalytic subunit gene, which reveals complex splicing patterns in different cell types. Hum. Mol. Genet., 6, 2011–2019. [DOI] [PubMed] [Google Scholar]
- 16.Bryan T.M., Sperger,J.M., Chapman,K.B. and Cech,T.R. (1998) Telomerase reverse transcriptase genes identified in Tetrahymena thermophila and Oxytrica trifallax. Proc. Natl Acad. Sci. USA, 95, 8479–8484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Collins K. and Gandhi,L. (1998) The reverse transcriptase component of the Tetrahymena telomerase ribonucleoprotein complex. Proc. Natl Acad. Sci. USA, 95, 8485–8490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Greenberg R.A., Allsopp,R.C., Chin,L., Morin,G.B. and DePinho,R.A. (1998) Expression of mouse telomerase reverse transcriptase during development, differentiation and proliferation. Oncogene, 16, 1723–1730. [DOI] [PubMed] [Google Scholar]
- 19.Oguchi K., Liu,H., Tamura,K. and Takahashi,H. (1999) Molecular cloning and characterization of AtTERT, a telomerase reverse transcriptase homolog in Arabidopsis thaliana. FEBS Lett., 457, 465–469. [DOI] [PubMed] [Google Scholar]
- 20.Fitzgerald M.S., Riha,K., Gao,F., Ren,S., McKnight,T.D. and Shippen,D.E. (1999) Disruption of the catalytic telomerase subunit gene from Arabidopsis inactivates telomerase and leads to a slow loss of telomeric DNA. Proc. Natl Acad. Sci. USA, 96, 14813–14818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weinrich S.L., Pruzan,R., Ma,L., Ouellette,M., Tesmer,V.M., Holt,S.E., Bodnar,A.G., Lichtsteiner,S., Kim,N.W., Trager,J.B., Taylor,R.D., Carlos,R., Andrews,W.H., Wright,W.E., Shay,J.W., Harley,C.B. and Morin,G.B. (1997) Reconstitution of human telomerase with the template RNA component hTR and the catalytic protein subunit hTRT. Nature Genet., 17, 498–502. [DOI] [PubMed] [Google Scholar]
- 22.Harrington L., Zhou,W., McPhail,T., Oulton,R., Yeung,D.S.K., Mar,V., Bass,M.B. and Robinson,M.O. (1997) Human telomerase contains evolutionarily conserved catalytic and structural subunits. Genes Dev., 11, 3109–3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beattie T.L., Zhou,W., Robinson,M.O. and Harrington,L. (1998) Reconstitution of human telomerase activity in vitro. Curr. Biol., 8, 177–180. [DOI] [PubMed] [Google Scholar]
- 24.Collins K., Kobayashi,R. and Greider,C.W. (1995) Purification of Tetrahymena telomerase and cloning of genes encoding the two protein components of the enzyme. Cell, 81, 677–686. [DOI] [PubMed] [Google Scholar]
- 25.Gandhi L. and Collins,K. (1998) Interaction of recombinant Tetrahymena telomerase proteins p80 and p95 with telomerase RNA and telomeric DNA substrates. Genes Dev., 12, 721–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harrington L., McPhail,T., Mar,V., Zhou,W., Oulton,R., Program,A.E., Bass,M.B., Arruda,I. and Robinson,M.O. (1997) A mammalian telomerase-associated protein. Science, 275, 973–977. [DOI] [PubMed] [Google Scholar]
- 27.Nakayama J.-I., Saito,M., Nakamura,H., Matsuura,A. and Ishikawa,F. (1997) TLP1: a gene encoding a protein component of mammalian telomerase is a novel member of WD repeats family. Cell, 88, 875–884. [DOI] [PubMed] [Google Scholar]
- 28.Holt S.E., Aisner,D.L., Baur,J., Tesmer,V.M., Dy,M., Ouellette,M., Trager,J.B., Morin,G.B., Toft,D.O., Shay,J.W., Wright,W.E. and White,M.A. (1999) Functional requirement of p23 and Hsp90 in telomerase complexes. Genes Dev., 13, 817–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morin G. (1989) The human telomere terminal transferase enzyme is a ribonucleoprotein that synthesizes TTAGGG repeats. Cell, 59, 521–529. [DOI] [PubMed] [Google Scholar]
- 30.Greider C. (1991) Telomerase is processive. Mol. Cell. Biol., 11, 4572–4580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee M. and Blackburn,E.H. (1993) Sequence-specific DNA primer effects on telomerase polymerization activity. Mol. Cell. Biol., 13, 6586–6599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Collins K. and Greider,C.W. (1993) Tetrahymena telomerase catalyzes nucleolytic cleavage and nonprocessive elongation. Genes Dev., 7, 1364–1376. [DOI] [PubMed] [Google Scholar]
- 33.Hammond P.W., Lively,T.N. and Cech,T.R. (1997) The anchor site of telomerase from Euplotes aediculatus revealed by photo-cross-linking to single- and double-stranded DNA primers. Mol. Cell. Biol., 17, 296–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Melek M., Davis,B.T. and Shippen,D.E. (1994) Oligonucleotides complementary to the Oxytrica nova telomerase RNA delineate the template domain and uncover a novel mode of primer utilization. Mol. Cell. Biol., 14, 7827–7838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harrington L.A. and Greider,C.W. (1991) Telomerase primer specificity and chromosome healing. Nature, 353, 451–454. [DOI] [PubMed] [Google Scholar]
- 36.Wang H. and Blackburn,E.H. (1997) De novo telomere addition by Tetrahymena telomerase in vitro. EMBO J., 16, 866–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baran N., Haviv,Y., Paul,B. and Manor,H. (2002) Studies on the minimal lengths required for DNA primers to be extended by the Tetrahymena telomerase: implications for primer positioning by the enzyme. Nucleic Acids Res., 30, 5570–5578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miller M.C., Liu,J.K. and Collins,K. (2000) Template definition by Tetrahymena telomerase reverse transcriptase. EMBO J., 19, 4412–4422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peng Y., Mian,I.S. and Lue,N.F. (2001) Analysis of telomerase processivity: mechanistic similarity to HIV-1 reverse transcriptase and role in telomere maintenance. Mol. Cell, 7, 1201–1211. [DOI] [PubMed] [Google Scholar]
- 40.Bosoy D. and Lue,N. (2001) Functional analysis of conserved residues in the putative ‘finger’ domain of telomerase reverse transcriptase. J. Biol. Chem., 276, 46305–46312. [DOI] [PubMed] [Google Scholar]
- 41.Bryan T.M., Goodrich,K.J. and Cech,T.R. (2000) A mutant of Tetrahymena telomerase reverse transcriptase with increased processivity. J. Biol. Chem., 275, 24199–24207. [DOI] [PubMed] [Google Scholar]
- 42.Hossain S., Singh,S. and Lue,N. (2002) Functional analysis of the C-terminal extension of telomerase reverse transcriptase. A putative ‘thumb’ domain. J. Biol. Chem., 277, 36174–36180. [DOI] [PubMed] [Google Scholar]
- 43.Huard S., Moriarty,T. and Autexier,C. (2003) The C terminus of the human telomerase reverse transcriptase is a determinant of enzyme processivity. Nucleic Acids Res., 31, 4059–4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Forstemann K., Zaug,A., Cech,T. and Lingner,J. (2003) Yeast telomerase is specialized for C/A-rich RNA templates. Nucleic Acids Res., 31, 1646–1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen J. and Greider,C. (2003) Determinants in mammalian telomerase RNA that mediate enzyme processivity and cross-species incompatibility. EMBO J., 22, 304–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ware T.L., Wang,H. and Blackburn,E.H. (2000) Three telomerases with completely non-telomeric template replacements are catalytically active. EMBO J., 19, 3119–3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lai C., Miller,M. and Collins,K. (2003) Roles for RNA in telomerase nucleotide and repeat addition processivity. Mol. Cell, 11, 1673–1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cohn M. and Blackburn,E.H. (1995) Telomerase in yeast. Science, 269, 396–400. [DOI] [PubMed] [Google Scholar]
- 49.Lue N.F. and Peng,Y. (1998) Negative regulation of yeast telomerase activity through an interaction with an upstream region of the DNA primer. Nucleic Acids Res., 26, 1487–1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fulton T.B. and Blackburn,E.H. (1998) Identification of Kluyveromyces lactis telomerase: discontinuous synthesis along the 30-nucleotide-long templating domain. Mol. Cell. Biol., 18, 4961–4970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lue N.F. and Peng,Y. (1997) Identification and characterization of a telomerase activity from Schizosaccharomyces pombe. Nucleic Acids Res., 25, 4331–4337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Prowse K.R., Avilion,A.A. and Greider,C.W. (1993) Identification of a nonprocessive telomerase activity from mouse cells. Proc. Natl Acad. Sci. USA, 90, 1493–1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Prescott J. and Blackburn,E.H. (1997) Functionally interacting telomerase RNAs in the yeast telomerase complex. Genes Dev., 11, 2790–2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yu G.-L., Bradley,J.D., Attardi,L.D. and Blackburn,E.H. (1990) In vivo alteration of telomere sequences and senescence caused by mutated Tetrahymena telomerase RNAs. Nature, 344, 126–132. [DOI] [PubMed] [Google Scholar]
- 55.Xia J., Peng,Y., Mian,I.S. and Lue,N.F. (2000) Identification of functionally important domains in the N-terminal region of telomerase reverse transcriptase. Mol. Cell. Biol., 20, 5196–5207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lue N.F. and Xia,J. (1998) Species-specific and sequence-specific recognition of the dG-rich strand of telomeres by yeast telomerase. Nucleic Acids Res., 26, 1495–1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Beard W.A., Stahl,S.J., Kim,H.R., Bebenek,K., Kumar,A., Strub,M.P., Becerra,S.P., Kunkel,T.A. and Wilson,S.H. (1994) Structure/function studies of human immunodeficiency virus type 1 reverse transcriptase. Alanine scanning mutagenesis of an alpha-helix in the thumb subdomain. J. Biol. Chem., 269, 28091–28097. [PubMed] [Google Scholar]
- 58.Bebenek K., Beard,W.A., Casas-Finet,J.R., Kim,H.R., Darden,T.A., Wilson,S.H. and Kunkel,T.A. (1995) Reduced frameshift fidelity and processivity of HIV-1 reverse transcriptase mutants containing alanine substitutions in helix H of the thumb subdomain. J. Biol. Chem., 270, 19516–19523. [DOI] [PubMed] [Google Scholar]
- 59.Wohrl B.M., Krebs,R., Thrall,S.H., Le Grice,S.F.J., Scheidig,A.J. and Goody,R.S. (1997) Kinetic analysis of four HIV-1 reverse transcriptase enzymes mutated in the primer grip region of p66. Implications for DNA synthesis and dimerization. J. Biol. Chem., 272, 17581–17587. [DOI] [PubMed] [Google Scholar]
- 60.Ghosh M., Williams,J., Powell,M.D., Levin,J.G. and Le Grice,S.F. (1997) Mutating a conserved motif of the HIV-1 reverse transcriptase palm subdomain alters primer utilization. Biochemistry, 36, 5758–5768. [DOI] [PubMed] [Google Scholar]
- 61.Jacques P.S., Wohrl,B.M., Ottmann,M., Darlix,J.L. and Le Grice,S.F. (1994) Mutating the ‘primer grip’ of p66 HIV-1 reverse transcriptase implicates tryptophan-229 in template-primer utilization. J. Biol. Chem., 269, 26472–26478. [PubMed] [Google Scholar]
- 62.Hardy C.D., Schultz,C.S. and Collins,K. (2000) Requirements for the dGTP-dependent repeat addition processivity of recombinant Tetrahymena telomerase. J. Biol. Chem., 28, 4863–4871. [DOI] [PubMed] [Google Scholar]
- 63.Collins K. and Greider,C.W. (1995) Utilization of ribonucleotides and RNA primers by Tetrahymena telomerase. EMBO J., 14, 5422–5432. [DOI] [PMC free article] [PubMed] [Google Scholar]