Abstract
The DNA methylation status of the protozoan parasite Entamoeba histolytica was heretofore unknown. In the present study, we developed a new technique, based on the affinity of methylated DNA to 5-methylcytosine antibodies, to identify methylated DNA in this parasite. Ribosomal DNA and ribosomal DNA circles were isolated by this method and we confirmed the validity of our approach by sodium bisulfite sequencing. We also report the identification and the characterization of a gene, Ehmeth, encoding a DNA methyltransferase strongly homologous to the human DNA methyltransferase 2 (Dnmt2). Immunofluorescence microscopy using an antibody raised against a recombinant Ehmeth showed that Ehmeth is concentrated in the nuclei of trophozoites. The recombinant Ehmeth has a weak but significant methyltransferase activity when E.histolytica genomic DNA is used as substrate. 5-Azacytidine (5-AzaC), an inhibitor of DNA methyltransferase, was used to study in vivo the role of DNA methylation in E.histolytica. Genomic DNA of trophozoites grown with 5-AzaC (23 µM) was undermethylated and the ability of 5-AzaC-treated trophozoites to kill mammalian cells or to cause liver abscess in hamsters was strongly impaired.
INTRODUCTION
Entamoeba histolytica is a gastrointestinal protozoan parasite that poses a serious health problem, with 50 million annual infections throughout the world (1). The amoebic trophozoites normally reside in the human large bowel and occasionally invade the intestinal mucosa, disseminating to other organs, mainly the liver (2,3).
DNA methylation is an epigenetic modification that occurs in a wide range of eukaryotic and prokaryotic organisms (4). DNA methylation occurs at the C-5 or N-4 positions of cytosine and at the N-6 position of adenine and is catalyzed by enzymes known as DNA methyltransferases (5). All DNA methyltransferases use S-adenosyl methionine as a methyl donor. DNA methylation was first associated with the DNA restriction–modification system, thought to be important in protecting bacteria from foreign DNAs such as transposons and viral DNAs (reviewed in 6,7). Dam methylation at adenine residues in GATC sequences controls DNA replication, repair of replication errors, and pathogenicity of a number of bacteria (8). Dam-deficient or Dam-overproducing Salmonella strains are dysregulated for the expression of virulence factors like the cytotoxin SpvB (8,9).
In higher eukaryotes, DNA methylation regulates a number of important biological functions including chromatin structure (10), silencing of gene expression (11), parental imprinting and chromosome X inactivation in females (12), and development and protection from selfish genetic elements (13). Methylation occurs in cytosine C5 at the CG sequences and ∼60–90% of CG sequences are methylated. Methylation of CG sites in the promoter regions of genes usually leads to a reduction of gene expression. Repression of gene expression occurs at three levels of control: (i) several transcription factors are not able to bind to methylated target sites; (ii) DNA methylation recruits 5-methylcytosine (m5C) binding proteins that act as repressors of gene transcription; and (iii) DNA methylation triggers histone deacetylation and thereby induces chromatin condensation, which leads to a strong and stable repression of gene expression (14–17).
The methylation status of the DNA from E.histolytica was heretofore unknown. In the present study, we provide evidence of m5C in E.histolytica ribosomal DNA (rDNA) and of an active DNA methyltransferase (Ehmeth) that could be involved in the regulation of virulence.
MATERIALS AND METHODS
Parasite and cell culture conditions
Trophozoites of the E.histolytica strain HM1:IMSS were grown under axenic conditions in Diamond’s TYI-S-33 medium (18) at 37°C. Trophozoites in the log phase of growth were used in all experiments.
Escherichia coli strains
The phenotypes of the E.coli strains used in this study are as following: XL1-Blue (Stratagene): recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac[F′proAB lacIqZΔM15 Tn10 (tetr)]; GM2163 (New England Biolabs): F– ara-14 leuB6 fhuA31 lacY1tsx78 glnV44 galk2 gal t22 mcrA dcm-6 hisG4 rfbD1 rpsL136 dam13::Tn9 xylA5 mtl-1 thi-1 mcrB1; M15 (Qiagen): Nals Strs rifs, lac– ara– gal– mtl– F– recA+ uvr+; BL-21 (DE3) (Stratagene): E.coli B F– dcm ompT hsdS (r–B m–B) gal λ (DE3).
Preparation of E.histolytica genomic DNA
Genomic DNA free of RNA contamination was prepared with the DNAeasy Tissue Kit (Qiagen) according to the manufacturer’s instructions. The RNase A treatment is crucial to ensure that the methyl groups recognized by the antibody to m5C described below are not from residual RNA.
Genomic/antibody blot analysis
DNA (0.5 µg) was denatured by boiling the sample for 5 min followed by a quick cooling on ice. The DNA was spotted on Protran BA85 nitrocellulose paper (Schleicher and Schuell) pre-soaked in 10× SSC, baked for 2 h at 80°C and processed as followed. The blot was blocked with 5% naïve rabbit serum and incubated overnight with sheep polyclonal antibody to m5C (1/10 000) (MBS), washed in phosphate-buffered saline (PBS) and subjected to interaction with an HRP-conjugated goat anti-rabbit antibody (1/5000) (Jackson), and then developed by enhanced chemiluminescence. At the concentration of m5C antibody used, we observed a linear relationship between the amounts of E.histolytica DNA spotted to the membrane and the emission of light read with an ImageMaster™ VDS-CL apparatus (Amersham Biosciences) (data not shown).
Affinity chromatography using m5C antibodies as ligand
Sheep polyclonal antibody to m5C (260 µg; MBS) was cross-linked to a 0.2 ml column of immobilized protein A (Seize-X Protein A Immunoprecipitation Kit; Pierce Biotechnology) in accordance with the manufacturer’s instructions. Entamoeba histolytica genomic DNA (2 µg) was cleaved with DpnII, and ligated overnight with the adaptors R-Bgl-24 oligo and R-Bgl-12 oligo (Table 1). The 12mer adaptor was melted away by heating the reaction for 3 min at 72°C and the ends were filled in with Taq DNA polymerase (5u; Promega) for 5 min at 72°C. The ligation was diluted by adding 300 µl of binding/wash buffer (0.14 M NaCl, 0.008 M Na2PO4, 0.002 M potassium phosphate and 0.01 M KCl, pH 7.4). The DNA was denatured by heat and incubated overnight at room temperature with the affinity column prepared above. The column was washed extensively with the binding/wash buffer, resuspended in 50 µl of the same buffer, and 5 µl of the suspension was directly used for the PCR. DNA bound to the column was amplified with the R-Bgl-24 primer (Table 1). A program of 1 min at 95°C and 3 min at 72°C for a total of 25 cycles was used. The PCR product was then cloned in the pGEM-T Easy System (Promega) and sequenced at the DNA Facility (Faculty of Medicine, Technion, Haifa, Israel).
Table 1. Primers used in this study.
| Primer | Locationa | Primer sequence | Direction | Underlined restriction site |
|---|---|---|---|---|
| Ehmethstart | 1682 | 5′-atgcaacagaaacaagtaaatgttat-3′ | Sense | |
| EhmethBglII | 2820 | 5′-tatagatctttattcttttaagtcatc-3′ | Antisense | BglII |
| EhmethBamHI | 1686 | 5′-tatggatccaacagaaacaagtaaatg-3′ | Sense | BamHI |
| GSTEhmethBamHI | 1682 | 5′-ggcggatccatgcaacagaaacaagta-3′ | Sense | BamHI |
| R-Bgl-12 | (adaptor) | 5′-gatctgcggtga-3′ | ||
| R-Bgl-24 | (adaptor) | 5′-agcactctccagcctctcaccgca-3′ | ||
| Seneb5′ | 5868 | 5′-tatttgattaaataattgaataa-3′ | Sense | |
| Seneb3′ | 6197 | 5′-atttaaaacatattaaatttatc-3′ | Antisense | |
| Sene5′ | 5868 | 5′-tatttgatcaaataattgaataa-3′ | Sense | |
| Sene3′ | 6197 | 5′-atttaaaacatattagatttatc-3′ | Antisense | |
| T7 | pBKCMV | 5′-taatacgactcactataggg-3′ | Antisense |
aThe position of the primers used to amplify Ehmeth is defined according to their location in the contig TIGR 316636 read in anti-parallel. The position of primers Seneb and Sene is defined according to their location in GenBank (accession no. X65163).
Sodium bisulfite reaction and strand-specific PCR
Sodium bisulfite treatment of E.histolytica genomic DNA was performed according to the method described by Warnecke et al. (19). The set of primers used to amplify rDNA that encodes SRP before (Sene5′ and Sene3′) and after (Seneb5′ and Seneb3′) treatment with sodium bisulfite are described in Table 1.
Amplification and cloning of Ehmeth
General molecular biology techniques were used according to Sambrook et al. (20). Ehmeth was amplified from an excised trophozoite cDNA library cloned in the pBK-CMV phagemid (a gift from Dr Tomo Nozaki, National Institute of Infectious Diseases, Tokyo, Japan) with the primers Ehmethstart and T7 (Table 1) and cloned in the pGEM-T vector (Promega) to give the pGEM-meth vector.
Northern blot and dot blot hybridization
For northern blot hybridization, total RNA was prepared using a TRI-reagent solution (Sigma). Total RNA (10 µg) was size fractionated on 4% polyacrylamide denaturing gel containing 8 M urea under denaturing conditions and subsequently blotted electrophoretically onto a nylon membrane. Hybridization was carried out with DNA probes randomly labeled using the Random Primer DNA Labeling Mix Kit (Beit Haemeck). Membranes were washed after overnight hybridization using non-stringent (0.1% SDS, 2× SSC) followed by stringent conditions (0.1% SDS, 0.1× SSC). Detection was done by autoradiography.
Construction of Ehmeth expression vector
A recombinant Ehmeth was prepared from the prokaryotic expression vector system pQE32 (Qiagen). The pQE32 plasmid is characterized by a 6× His affinity tag coding sequence that is located at the N-terminal end of a multiple cloning site. Ehmeth was amplified by PCR from the plasmid pGEM-meth with the primers EhmethBamHI and EhmethBglII (Table 1). The PCR product was digested with BamHI and BglII and cloned into the pQE32 plasmid that was previously linearized with BamHI to give the vector pQE32-Ehmeth.
A recombinant Ehmeth was also prepared from the prokaryotic expression vector system PGEX-4T-1 (Pharmacia Biotech). This vector allows the expression of a protein fused to a glutathione-S-transferase (GST) tag. Ehmeth was amplified by PCR from the plasmid pGEM-Ehmeth with the primers GSTEhmethBamHI and EhmethBglII (Table 1). The PCR product was digested with BamHI and BglII and cloned into the PGEX-4T-1 plasmid previously linearized with BamHI. Verification of the proper ligations and orientations in the resulting hybrid plasmids was performed by digestion with restriction endonucleases. One of the vectors that carry Ehmeth fused to the GST tag in the correct orientation (PGEX-4T-1-Ehmeth) was sequenced to confirm that no mutations have been introduced into Ehmeth during the construction.
Expression and purification of recombinant Ehmeth
For expression of His-Ehmeth, E.coli M15 transfected with the pQE32-Ehmeth vector was grown overnight in Luria broth (LB) containing kanamycin (25 µg/ml) and ampicillin (100 µg/ml). The pre-culture was used to inoculate (1:10) Terrific broth medium (450 ml) supplemented with kanamycin (25 µg/ml) and ampicillin (100 µg/ml) and grown for ∼40 min at 37°C until OD600 reached 0.4. Synthesis of the fusion protein was initiated by adding isopropyl-beta-d-thiogalactopyranoside (IPTG) at a final concentration of 2 mM to the growing culture. After overnight incubation in the presence of IPTG at room temperature, the bacteria were harvested and lysed in BugBuster protein extraction reagent (Novagen) supplemented with 10 mM imidazole and 0.15 M NaCl. The fusion protein containing 6× His tag residues at its N-terminal end was purified under native conditions on Ni-NTA resin (Qiagen). The protein was then eluted with elution buffer (50 mM NaH2PO4, pH 8.0, 300 mM NaCl and 250 mM imidazole). His-Ehmeth protein was quantitated by Bradford’s method (21).
For expression of GST-Ehmeth, E.coli BL-21 (DE3) transfected with the PGEX-4T-1-Ehmeth vector were grown overnight in LB medium containing ampicillin (100 µg/ml). The pre-culture was used to inoculate (1:100) 2× YT medium supplemented with ampicillin (100 µg/ml) and grown for ∼3 h at 30°C until OD600 reached 0.8. Synthesis of the fusion protein was initiated by adding IPTG at a final concentration of 0.5 mM to the growing culture. After 16 h of incubation (overnight) in the presence of IPTG at room temperature, the bacteria were harvested and lysed in BugBuster protein extraction reagent (Novagen). The recombinant GST-Ehmeth protein was purified under native conditions on gluthatione–agarose resin (Sigma). The protein was then eluted with glutathione elution buffer [50 mM Tris–HCl pH 8.0, 10 mM glutathione (Sigma)]. The recombinant GST-Ehmeth protein was quantitated by Bradford’s method (21).
The cleavage with thrombin (Cleancleave Kit; Sigma) of the GST tag in the recombinant Ehmeth led systematically to the precipitation of Ehmeth.
Preparation of a polyclonal anti-Ehmeth antibody
GST-Ehmeth (0.5 mg) was emulsified in 1 ml of complete Freund’s adjuvant (Sigma) completed to 1.5 ml with PBS. The mixture was injected subcutaneously into a 2 kg rabbit. Injection of GST-Ehmeth (0.2 mg) emulsified with 1 ml of incomplete Freund’s adjuvant and completed with PBS to 1.5 ml was repeated three more times at 4 week intervals. Two days prior to each injection, a blood sample was taken from the rabbit in order to check the level of antibodies produced against the GST-Ehmeth (anti-Ehmeth antibodies). The serum used for the different experiments was obtained from the third bleeding of the rabbit. Ehmeth antibody was purified by affinity chromatography on a column with immobilized protein A. The concentration of Ehmeth antibody after purification was 3.7 µg/µl.
SDS–PAGE and western immunoblotting
The purified recombinant Ehmeth were resolved under reducing conditions (22) on the 10% SDS–polyacrylamide gel developed with the GelCode Blue staining reagent (Pierce). Entamoeba histolytica trophozoites (106) were solubilized with 1 ml of 1% Nonidet P-40 (Sigma) in PBS in the presence of 0.2 mM cysteine proteinases inhibitor N-(trans-epoxysucciniyl)-l-leucine 4-guanidobutylamide (E-64) (Sigma) and 0.5 mM serine proteinases inhibitor 4-(2-aminoethyl)benzenesulfonyl fluoride (AEBSF) (Sigma). Proteins from whole trophozoites (25 µg per lane) were resolved under reducing conditions (22) on 10% SDS–polyacrylamide gel and transferred electrophoretically to nitrocellulose membranes. The blots were reacted with a rabbit polyclonal anti-Ehmeth (1/500). After incubation with the first antibody, the blots were subjected to interaction with an HRP-conjugated goat anti-rabbit antibody (1/5000) (Jackson) and developed by enhanced chemiluminescence.
Microscopic localization of Ehmeth in trophozoites
To record the distribution of Ehmeth in trophozoites, trophozoites (106 cells/ml) were washed in PBS buffer, resuspended in cold acetone for 10 s, and washed twice with PBS buffer. Fixed trophozoites were incubated for 1 h in 1.5% normal goat serum in PBS, reacted with anti-Ehmeth antibody (1:400) for 1 h, washed with PBS and reacted for 1 h with anti rabbit/goat Ig fluorescein 5(6)-isothiocyanate (FITC) labeled antibody (Sigma) diluted 1:100. Trophozoites were then washed with PBS and, to observe the nuclei, they were reacted with 4′,6-diamino-2-phenylindole dihydrochloride (DAPI; Sigma). A stock solution of DAPI (1 mg/ml in ethanol) was prepared at a concentration of 5 µg/ml in 50:50 EtOH/0.1 M HCl. DAPI staining was performed by pipetting 5 µl into each sample (200 µl), which was then incubated for 5 min at room temperature. Phase contrast and fluorescent images were taken using an Axioscop2 (Zeiss) epifluorescence microscope with a 63×/1.25 Plan Neofluar oil immersion objective and a differential interference contrast filter. Images were captured using a CCD camera and computed with ImagePro@Plus software (Media Cyberneticx, USA).
DNA methylation activity
DNA methyltransferase activity was measured by the incorporation of tritium from AdoMet in a reaction buffer [50 mM NaCl, 10 mM Tris–HCl, 10 mM MgCl2, 1 mM dithiothreitol (pH 7.9)] containing purified GST-Ehmeth (10 µg), E.histolytica genomic DNA (5 µg) and 3 µl of [3H]AdoMet (15 Ci/mmol; Amersham). After incubation at 37°C for 2 and 16 h, the reaction volume was diluted with water to a final volume of 100 µl, an equivalent volume of phenol was then added and, following mixing and centrifugation, unreacted AdoMet was removed from the aqueous phase by gel filtration on a Bio-Rad A-05 M resin. Incorporation of tritium was estimated by scintillation counting.
Cytopathic activity
The destruction rate of cultured CHO cell monolayers by trophozoites grown with or without 5-azacytidine (5-AzaC) (23 µM) was performed as described previously (23,24).
Adhesion assay to CHO cells
The adhesion assay to CHO monolayers was performed as described previously (24).
Hemolytic activity
The hemolytic activity was performed as described previously (25).
Induction of amoebic liver abscesses
Syrian golden hamsters (6-week-old) were inoculated intra-hepatically with: (i) 5 × 105 trophozoites; (ii) 5 × 105 trophozoites grown in the presence of 10 µM 5-AzaC; and (iii) 5 × 105 trophozoites grown in the presence of 23 µM 5-AzaC. Before inoculation, 5-AzaC was removed from the culture by washing the trophozoites twice with TYI-S-33 medium. Hamsters (six for each group) were killed 1 week after intra-hepatic inoculation, and the formation of lesions was evaluated.
RESULTS
Entamoeba histolytica genomic DNA contains m5C
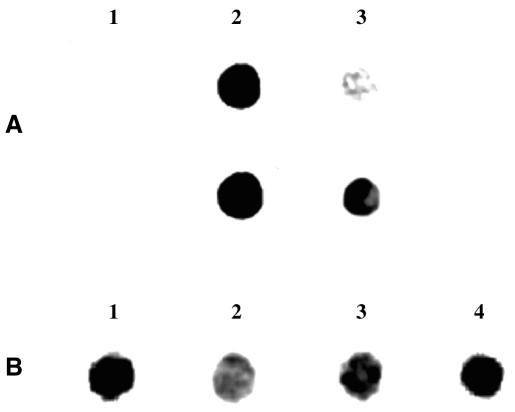
Methylation restriction sensitive assays, including McrBC analysis (26,27), failed to demonstrate the presence of m5C in E.histolytica DNA (data not shown). The presence of m5C in E.histolytica genomic DNA was demonstrated using an immunological method. This very sensitive technique detected the presence of m5C in the genomic DNA of Drosophila melanogaster where m5C is approximately 50 times lower than in mammals (28,29). In the DNA spot assay (Fig. 1A), PCR-synthesized DNA and PCR-synthesized DNA treated in vitro with M.SssI methyltransferase were used, respectively, as negative and positive control. The commercial antibody against m5C reacts specifically with synthetic DNA treated in vitro with M.SssI methyltransferase, but does not react with the untreated PCR product. We then looked for a possible reaction with E.histolytica genomic DNA. Entamoeba histolytica showed a clearly positive reaction with the m5C antibody, which indicates that this modified base is present in the parasite (Fig. 1A and B). The specificity of the reaction between m5C antibody and the genomic DNA of E.histolytica was demonstrated by competition experiments involving m5C (10–5 M) (Fig. 1B, lane 2).
Figure 1.
Immunological detection of m5C in E.histolytica genomic DNA. (A) PCR-synthesized DNA (lane 1), PCR-synthesized DNA treated in vitro with M.SssI DNA methyltransferase (lane 2) and E.histolytica genomic DNA (lane 3) were spotted on nitrocellulose paper and incubated with antibodies to m5C. Ehmeth DNA was used as template for the production of PCR-synthesized DNA. The time of exposure for the upper line was 2 ms and for the lower line 12 ms. (B) The specificity of the reaction between m5C antibody and the genomic DNA of E.histolytica (lane 1) was demonstrated by competition experiments involving m5C (10–5 M) (lane 2). 5-AzaC is an inhibitor of 5-cytosine DNA methyltransferase currently used as a potent demethylating agent. Genomic DNA of trophozoites grown with 5-AzaC (23 µM) for 48 h (lane 3) and then cultivated for 2 weeks without 5-AzaC (lane 4) were also spotted on nitrocellulose paper and incubated with antibodies to m5C. The time of exposure was 12 ms. For both experiments (A and B), 0.5 µg of DNA was systematically spotted on the nitrocellulose paper.
Identification of methylated regions in E.histolytica genome by affinity chromatography
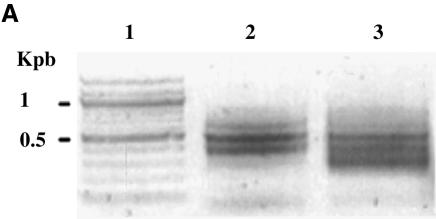
Purification of CpG islands using the methyl binding domain of MeCP2 was previously described (30). The absence of knowledge about the methylation pattern in E.histolytica encouraged us to develop our own method to identify the methylated region in this parasite. Based on the previous results, we constructed a methylated DNA binding column that contains m5C antibodies as ligand. Genomic DNA was digested with DpnII, bound to adaptors and loaded to the affinity chromatography column. DNA purified by the column was amplified using the adaptors as primers (Table 1 and Fig. 2A), cloned in pGEM-T Easy vector and sequenced. Among the 25 clones analyzed, 20 clones contained rDNA (accession no. X65163, from nucleotide 5872 to 6092 and from nucleotide 10 to 178) and five clones contained rDNA circle (accession no. X75436, from nucleotide 1808 to 1935). One of the rDNA sequence includes an open-reading frame (ORF) identified as a protein of 65 amino acids homologous (73% identities) with a senescence-associated protein (SRP) expressed by Pisum sativum (accession no. BAB33421) (Fig. 2B).
Figure 2.
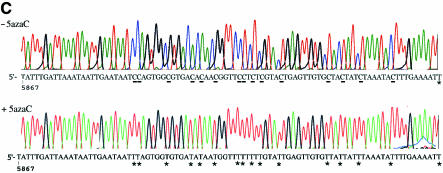
Purification and analysis of E.histolytica methylated DNA regions. (A) E.histolytica genomic DNA was restricted with DpnII, ligated to R-Bgl-24 adaptors and amplified with primers R-Bgl-24 before (lane 2) and after (lane 3) m5C antibody affinity chromatography. Lane 1, 100 bp ladder (New England Biolabs). Note the difference of pattern before and after affinity chromatography. (B) Sequence analysis of rDNA region (accession no. X65163). The DpnII fragment isolated by m5C antibody affinity chromatography is underlined. The ORF for the senescence-related protein is represented in bold, and its putative amino acid sequence represented below. (C) Genomic sodium bisulfite analysis sequencing of the rDNA region that includes SRP (accession no. X65163) in trophozoites grown without 5-AzaC (upper chromatogram) or with 5-AzaC for 1 week (lower chromatogram). The methylated C residues are resistant to the bisulfite treatment (underlined). The unmethylated C residues are converted by the bisulfite treatment (designated by a star). The same distribution of methylated cytosine was found following the sequencing of four clones coming from two separate experiments with sodium bisulfite.
To confirm the specificity of the m5C antibodies affinity chromatography, we focused our analysis on the rDNA region that encodes SRP. We searched for methylated cytosine by sodium bisulfite reaction and strand-specific PCR. This procedure converts all cytosine residues to uracil, giving rise to thymine after amplification by PCR. Only methylated cytosines are refractory to the deamination. The rDNA region that encodes SRP was amplified from genomic DNA and from sodium bisulfite-treated genomic DNA, cloned in pGEM-T Easy vector and sequenced. The distribution of cytosines found at the place of cytosine versus cytosine found at the place of thymine indicates that methylated cytosines are clustered upstream to the putative start codon of SRP (Fig. 2C). Interestingly, methylation occurred not only to the CG sites but also to all the cytosine present in this cluster.
Isolation and characterization of a cDNA for Ehmeth, a DNA methyltransferase homologous to human Dnmt2
Based on the previous experiment that showed the presence of m5C in the genomic DNA of E.histolytica, we hypothesized that a DNA 5-cytosine methyltransferase, is also present in the parasite. We searched the E.histolytica genome database (www.tigrblast.tigr.org) for the presence of genes homologous to the human DNA methyltransferase 1 (accession no. P26358) using the BLAST algorithm (31). We identified a gene, Ehmeth, which encodes a protein with strong homologies to Dnmt2 (accession no. AAC39764), a human DNA methyltransferase of unknown function (28% identities and 47% positives). Like Dnmt2, Ehmeth is devoid of the regulatory regions present at the N-terminal part of human Dnmt1 and Dnmt3 (32) (data not shown). No gene encoding a DNA methyltransferase other than Ehmeth was detected in the E.histolytica genome database.
The ORF of Ehmeth is included in the contig (316 636 reads in anti-parallel) from nucleotide 1682 to nucleotide 2820 (Supplementary Material Fig. 1). This sequence was used as a template for the synthesis of Ehmethstart, a primer that includes the putative start codon of Ehmeth (Table 1). Ehmeth was amplified from an E.histolytica cDNA library cloned in the PBK-CMV plasmid (a gift from Dr Tomo Nogazaki), with primers Ehmethstart and T7 (Table 1). The sequence comparison of Ehmeth from the genome database and from the amplified cDNA library shows the presence of three introns localized, respectively, between nucleotides 1796 and 1852, 1906 and 1965, and 2451 and 2505 (Supplementary Material Fig. 1). This observation is interesting in and of itself, since only a small number of genes show the presence of introns in this parasite (33).
The computer analysis was performed using the ExPASy proteomics tools (www.expasy.ch/tools). Ehmeth encodes a 322 amino acid protein devoid of putative signal peptide. The theoretical molecular weight of Ehmeth is 37.4 kDa, and its isoelectric point is 7.96. PSORTII analysis based on Reinhardt’s method for cytoplasmic/nuclear discrimination (34) predicted a nuclear localization for Ehmeth.
ClustalW alignment (Supplementary Material Fig. 2) (35) of Ehmeth and human Dnmt2 shows the presence of conserved motifs that correspond, respectively, to the S-adenosyl methionine binding domain (from amino acid 5 to 22), the catalytic site (from amino acid 72 to 92), and the PROSITE signature (http://www.expasy.org/prosite) to cytosine-specific DNA methyltransferases C-terminal (PS00095) (from amino acid 298 to 316).
Transcription of Ehmeth
In order to establish the transcription of Ehmeth in trophozoites, a series of northern hybridization experiments were carried out (Fig. 3). Northern hybridization experiments detected the presence of an mRNA species of ∼1 kb with a probe specific to Ehmeth. This band was observed after 120 h of exposure. In contrast, <5 h of exposure was needed to detect the 0.38 kb band of the rRNA small subunit. This result suggests that the amount of Ehmeth mRNA in trophozoites is very low. Similarly, Dnmt2 mRNA is hardly detected in human tissue (36).
Figure 3.
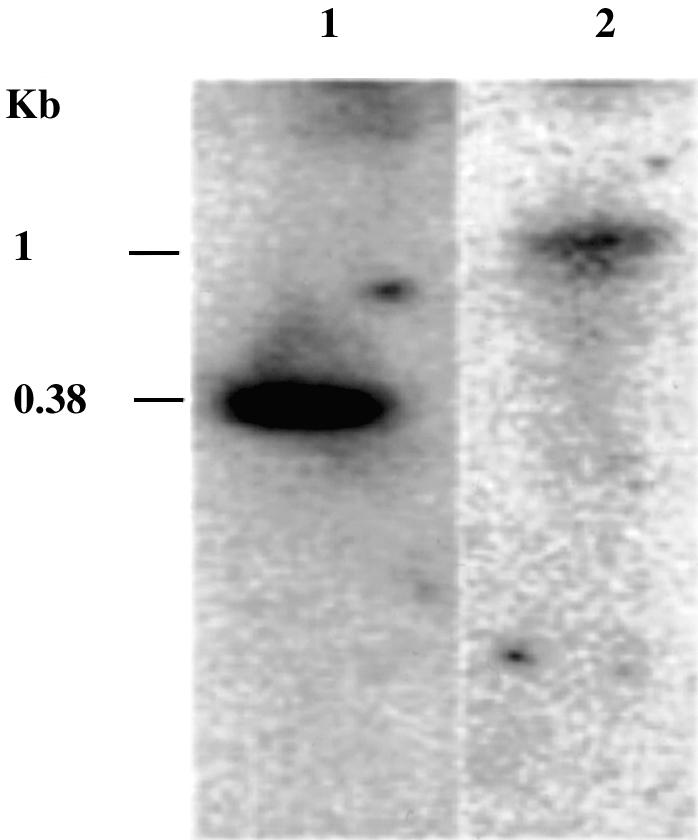
Northern blot analysis of Ehmeth. Total RNA (10 µg) was size fractionated on 4% polyacrylamide denaturing gel containing 8 M urea under denaturing conditions and subsequently blotted electrophoretically onto a nylon membrane. Hybridization was carried out by a probe for the small subunit of E.histolytica ribosomal RNA (ssrRNA) (lane 1) or for the Ehmeth ORF (lane 2). The exposure time of X-ray film was 120 h for Ehmeth and 5 h for ssrRNA.
Expression of Ehmeth in E.coli and purification of the recombinant protein
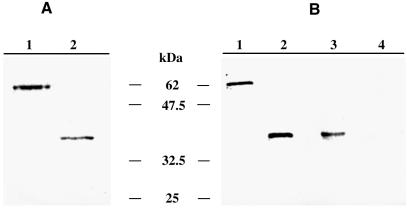
Ehmeth was first expressed as a fusion protein containing an N-terminal hexahistidine tag (His-Ehmeth). His-Ehmeth has an apparent molecular mass of 37 kDa which is in good agreement with the theoretical molecular weight of Ehmeth (37.4 kDa) (Fig. 4A). Because over-expression of His-Ehmeth in bacteria resulted in very low expression of the recombinant protein in soluble material (<0.1 µg/µl) and in the formation of inactive inclusion bodies (data not shown), we used another expression system.
Figure 4.
SDS–PAGE and western blot analysis of Ehmeth. (A) GelCode Blue staining of GST-Ehmeth (1 µg; lane 1) and His-Ehmeth (1 µg; lane 2) resolved under reducing conditions on a 10% SDS–polyacrylamide gel. (B) Western blot analysis under reducing conditions of trophozoites total lysate probed with the anti-Ehmeth antibody (1/500). Lane 1, GST-Ehmeth (10 ng); lane 2, His-Ehmeth (10 ng); lane 3, total lysate of trophozoites (25 µg); lane 4, trophozoites total lysate (25 µg) probed with rabbit pre-immune serum (1/500).
The Ehmeth cDNA ORF was subcloned into the PGEX-4T-1 expression vector, and recombinant Ehmeth was expressed as a fusion protein containing an N-terminal glutathione-S-transferase (GST) tag (GST-Ehmeth). The expression of recombinant fusion protein was expressed at high levels on induction with IPTG, and soluble material was purified by affinity purification over glutathione–Sepharose. GST-Ehmeth (0.7 µg/µl) had an apparent molecular mass of 64 kDa on SDS–PAGE (Fig. 4A), which is in good agreement with the expected size for a fusion between GST (28 kDa) and the theoretical molecular weight of Ehmeth (37.4 kDa). Cleavage of the GST tag with thrombin leads systematically to the precipitation of Ehmeth (data not shown).
Western blot analysis
The presence of Ehmeth in total lysate of trophozoites was determined by Western immunoblotting under reducing conditions using a specific antibody obtained from rabbit immunized with GST-Ehmeth. Immunoblot analysis shows that this antibody reacts specifically with GST-Ehmeth (10 ng) and His-Ehmeth (10 ng) and it recognizes an expected band of 37 kDa in the total lysate of trophozoites (Fig. 4B). In addition, a band of 25 kDa is sometimes detected in the total lysate of trophozoites but this band is not reproducible and could be the result of degradation (data not shown). No bands were observed when rabbit pre-immune serum was used as the first antibody (Fig. 4B).
Cellular localization of Ehmeth
Ehmeth was detected by immunofluorescence microscopy using antibodies raised against GST-Ehmeth (Fig. 5). Computer-assisted image overlay analysis of the signal given by Ehmeth antibody and by DAPI, a specific nuclear stain, shows that Ehmeth is concentrated in the nuclei of trophozoites. This result confirms the bioinformatics prediction about the nucleus localization of Ehmeth, and supports a role for Ehmeth in the methylation of E.histolytica rDNA. Remarkably, the spatial distribution of Ehmeth is similar to the distribution of the rDNA observed by Willhoeft and Tannich (37). No signal was observed when rabbit pre-immune serum was used as first antibody (data not shown).
Figure 5.
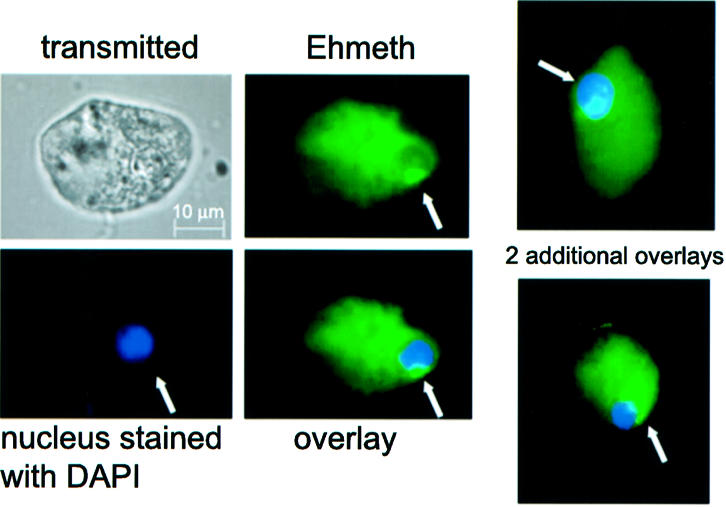
Cellular localization of Ehmeth in E.histolytica trophozoites. Ehmeth was detected by immunofluorescence microscopy using anti-Ehmeth antibody. Ehmeth distribution is shown in green using a primary anti-Ehmeth antibody and a secondary antibody conjugated with FITC. Nuclei (blue) were stained by DAPI. Computer-assisted image overlay analysis of the signal given by Ehmeth antibody and by DAPI, shows that Ehmeth is concentrated in the nuclei of trophozoites. Two additional examples showing a nuclear localization of Ehmeth are presented on the right.
Methyltransferase activity of GST-Ehmeth
DNA methyltransferase activity of GST-Ehmeth is followed by incorporation into E.histolytica genomic DNA (5 µg), of a methyl group provided by [3H]S-adenosyl-l-methionine. After 2 h of incubation at 37°C, no significant DNA methyltransferase activity was detected with GST-Ehmeth (10 µg) compared with the background level measured with GST alone (10 µg) (data not shown). In contrast, M.SssI methyltransferase (5 U; New England Biolabs) efficiently catalysed the transfer of methyl residues to E.histolytica genomic DNA (159 000 ± 2000 c.p.m. in 2 h). However, following 16 h of incubation at 37°C, a low but significant activity was observed with GST-Ehmeth (2450 ± 45 c.p.m.) compared with the background level measured with GST alone (450 ± 10 c.p.m.). No significant activity was measured in the same experimental condition with GST-Ehmeth incubated without genomic DNA (410 ± 30 c.p.m.).
5-AzaC induces DNA demethylation and inhibits virulence of E.histolytica
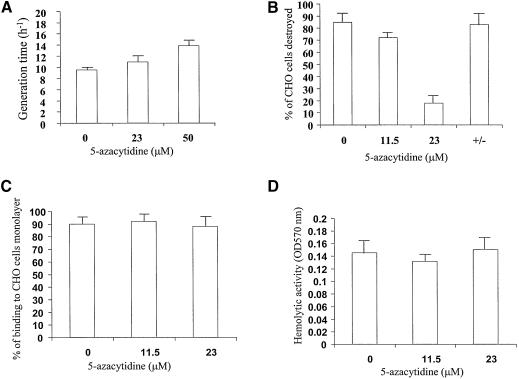
5-AzaC is an inhibitor of 5-cytosine DNA methyltransferase (38) currently used as a potent demethylating agent. Once incorporated in the DNA of an organism, this drug forms covalent complexes between the DNA and DNA methyltransferases that require recombination for repair. We first determined the concentration of 5-AzaC which was appropriated for the growth of E.histolytica. 5-AzaC (100 µM) totally inhibits the growth of E.histolytica, which reinforces our demonstration of an active DNA (m5C) methyltransferase in the parasite. 5-AzaC (23 µM) does not interfere significantly with the growth rate (Fig. 6A) of the parasite; therefore, we chose 5-AzaC (23 µM) as our working concentration.
Figure 6.
Growth, virulence and adherence assays in trophozoites grown with 5-AzaC. (A) Dose–response analysis of 5-AzaC concentration on the growth of E.histolytica. Data represent the mean and standard deviation of three independent experiments. (B) Cytopathic activity measuring the in vitro ability of amoeba grown in the presence of 5-AzaC (23 µM) to destroy a monolayer of CHO cells. The column +/– shows the cytopathic activity of trophozoites grown for 2 days with 5-AzaC (23 µM) and then cultivated without 5-AzaC for 2 weeks. Data represent the mean and standard deviation of three independent experiments done in duplicate. (C) Adherence ability of trophozoites grown in the presence of 5-AzaC (23 µM) to a formaldehyde-fixed monolayer of CHO cells. Data represent the mean and standard deviation of three independent experiments done in duplicate. (D) Hemolytic assay measuring the ability of trophozoites grown in the presence of 5-AzaC (23 µM) to lyse human red blood cells. Data represent the mean and standard deviation of three independent experiments done in duplicate.
5-AzaC induces a significant demethylation in the genomic DNA of trophozoites (Fig. 1B, lane 3). This demethylation was confirmed in the rDNA region that encodes SRP by bisulfite sequencing analysis (Fig. 2C). DNA demethylation induced by 5-AzaC is correlated with an inhibition of cytopathic activity (Fig. 6B). Interestingly, binding activity (Fig. 6C) and hemolytic activities (Fig. 6D) are not affected by 5-AzaC.
Trophozoites that were grown for 2 days with 5-AzaC (23 µM) and then cultivated without 5-AzaC for 2 weeks, recovered a methylation pattern (Fig. 1B, lane 4) and cytopathic activity (Fig. 6B) similar to the untreated trophozoites. This result suggests that the effect of 5-AzaC on E.histolytica virulence is reversible.
The effect of 5-AzaC on the ability of trophozoites to induce the formation of liver abscesses in hamster was also determined (Table 2). Hamsters (6/6) injected with E.histolytica trophozoites presented extensive necrotic lesions (>2 cm). In contrast, hamsters (4/6) injected with trophozoites grown in the presence of 5-AzaC (12 µM) developed smaller lesions (<1 cm), whereas only one among six hamsters injected with trophozoites grown in the presence of 5-AzaC (23 µM) developed a very small abscess (<0.3 cm). These results taken together suggest that DNA methylation is involved in the control of E.histolytica virulence.
Table 2. Hamster liver abscess formation by transfected trophozoites grown with various concentration of 5-AzaC.
| 5-AzaC (µM) | |||
|---|---|---|---|
| 0 | 11.5 | 23 | |
| Number of animals with abscess | 6/6 | 4/6 | 1/6 |
| Mean size of abscess (cm) | >2 | 1–2 | <0.3 |
DISCUSSION
DNA methylation is an epigenetic modification that affects gene transcription and chromatin formation. This epigenetic modification is widely distributed, from prokaryotic organisms to higher eukaryotes. In this work, we have demonstrated the presence of m5C in a number of regions in the E.histolytica genome using affinity chromatography with m5C antibody as ligand and sodium bisulfite reaction. Remarkably, only rDNA-related sequences were isolated by our technique. Although we cannot rule out the presence of other methylated regions in the parasite, rDNA could constitute the main target for methylation. Alternatively, the rDNA which is on an episome with a copy number of ∼200 per genome (39), could constitute the most abundant sequence targeted for methylation.
The pattern of methylation upstream to SRP is unconventional compared with the pattern of methylation in other eukaryotics where CG dinucleotide is the primary target of methylation (17). Such a non-conventional pattern of methylation was recently observed in the genome of Drosophila expressing Dnmt2 (40). Methylation of rDNA is related to aging in higher eukaryotes (41–44). In the mouse, ∼40% of rDNA repeats are methylated, probably to protect the genes against unwanted homologous recombination events (45). The biological significance of rDNA methylation in E.histolytica remains to be elucidated. In addition, the technique we have developed to identify methylated DNA regions can be applied to other organisms like Drosophila, where the presence of m5C is rare and where CG dinucleotide is not the primary target of methylation.
The DNA methyltransferase present in E.histolytica belongs to a very intriguing family. So far, three active DNA methyltransferases (Dnmt1, Dnmt3a, Dnmt3b) and one candidate protein (Dnmt2) have been identified in mammals (5,46). These enzymes share a C-terminal component that forms the catalytic domain and contains all the amino acid sequence motifs characteristic for prokaryotic DNA cytosine-C5 methyltransferases (47). Dnmt1 shows a significant preference for hemimethylated DNA (48–50) and Dnmt3 methyltransferases are able to methylate both hemimethylated DNA and non-methylated DNA (51). Both families have an N-terminal component that determine the specific functions played by these methyltransferases in the cells. Dnmt2 does not have such an N-terminal domain (32).
The function of Dnmt2 in the cell is unknown and only recently has its weak, non-conventional DNA methyltransferase activity been established (40,52). DNA methyltransferase activity of Ehmeth shares a number of characteristics with Dnmt2. Ehmeth does not contain in its N-terminal domains some regulatory elements which are present in Dnmt1 and Dnmt3 (53,54); the amount of mRNA in the cell is low, and GST-Ehmeth has a weak DNA methyltransferase activity. A number of hypotheses could explain this low activity in vitro. One of them is the presence of the GST tail that could hamper the activity of Ehmeth. Although we were not able to rule out this hypothesis because the cleavage of the GST tail caused the precipitation of Ehmeth, a number of DNA methyltransferases have yet to be expressed as a GST-fusion protein without loss of their enzymatic activity (54). Another hypothesis is that, since our DNA substrate came from wild-type E.histolytica, many of the methyltransferase substrate sites were presumably already methylated. Some proteins that interact with DNA methyltransferases, like histone deacetylase or the small ubiquitin-like modifier 1 (55–58), have been proposed as plausible mechanisms that regulate DNA methyltransferases function. Our failure to efficiently express Ehmeth as His-tagged recombinant protein in E.coli, and the loss of solubility when the GST tag is cleaved, suggest that additional proteins or cofactors may be required for the stabilization and in vivo activity of Ehmeth.
Our knowledge of the role of DNA methylation in unicellular eukaryotes is scanty. Kinetoplastid flagellates such as the parasite Trypanosoma brucei contain a β-d-glucosyl(hydroxymethyl)uracil (called J), a modified base that replaces a fraction of thymine in their DNA. J is involved in the transcriptional repression of silent variant surface glycoprotein gene expression sites (59). This example directly links the involvement of an epigenetic modification to virulence in a protozoan parasite. A similar link was previously demonstrated in prokaryote cells, where Dam methylation regulates the virulence of Salmonella strains (9). We searched for such a correlation between DNA methylation and virulence in E.histolytica by using 5-AzaC. This inhibitor of DNA methyltransferase which is currently used as a DNA demethylating agent, is used at a non-toxic concentration to induce, for example, the expression of tumor suppressor genes that have been silenced by DNA methylation in cancer cells (60).
5-AzaC in E.histolytica caused a decrease in m5C present in the cell, and consequently a shift from a virulent to a non-virulent phenotype. Interestingly, the effect of 5-AzaC is reversible, as a recovery of the virulent phenotype measured in vitro is observed once 5-AzaC is removed for 2 weeks from the growth media. This observation is in agreement with the definition of ‘epigenetic’ which allows a modification of the genome to be reversible.
In conclusion, this work is the first report of the existence of a modified base, m5C, in specific regions of the E.histolytica genome. This work also demonstrates the presence of an active DNA methyltransferase in the parasite. Our results, based on 5-AzaC inhibition of DNA methylation, suggest that Ehmeth could regulate the virulence of the parasite. Work is in progress to more specifically inhibit this enzyme by antisense and RNAi techniques and to identify genes that are regulated by Ehmeth. Several other roles of cytosine methylation have been suggested in mammals, such as epigenetic control of gene expression during development and protection against the selfish element (46). In the future, it will also be interesting to determine the significance of epigenetics for other aspects of the biology of this parasite, such as differentiation to cysts and inactivation of selfish elements like retrotransposons that have recently been identified recently in the genome of E.histolytica (61–63).
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENT
This research was supported by Grant 370/00-1 from the Israel Science Foundation.
REFERENCES
- 1.World Health Organization (1997) Amoebiasis. WHO Weekly Epidemiologic Record, 72, 97–100. [Google Scholar]
- 2.Espinosa-Cantellano M. and Martinez-Palomo,A. (2000) Pathogenesis of intestinal amebiasis: from molecules to disease. Clin. Microbiol. Rev., 13, 318–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stanley S.L. (2001) Pathophysiology of amoebiasis. Trends Parasitol., 17, 280–285. [DOI] [PubMed] [Google Scholar]
- 4.Lewin B., Stanley,S.L.,Jr and Reed,S.L. (1998) The mystique of epigenetics. Cell, 93, 301–303. [DOI] [PubMed] [Google Scholar]
- 5.Bestor T.H., Verdine,G.L., Santi,D.V., Norment,A., Garrett,C.E., Gilchrist,C.A., Petri,W.A., Singh,V.K., Moskovitz,J., Wilkinson,B.J. et al. (1994) DNA methyltransferases. Curr. Opin. Cell Biol., 6, 380–389. [DOI] [PubMed] [Google Scholar]
- 6.Pingoud A. and Jeltsch,A. (2001) Structure and function of type II restriction endonucleases. Nucleic Acids Res., 29, 3705–3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wilson G.G. and Murray,N.E. (1991) Restriction and modification systems. Annu. Rev. Genet., 25, 585–627. [DOI] [PubMed] [Google Scholar]
- 8.Low D.A., Weyand,N.J., Mahan,M.J., Goto,T., Monk,M., Gilchrist,C.A., Petri,W.A., Singh,V.K., Moskovitz,J., Wilkinson,B.J. et al. (2001) Roles of DNA adenine methylation in regulating bacterial gene expression and virulence. Infect. Immun., 69, 7197–7204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garcia-Del Portillo F., Pucciarelli,M.G. and Casadesus,J. (1999) DNA adenine methylase mutants of Salmonella typhimurium show defects in protein secretion, cell invasion and M cell cytotoxicity. Proc. Natl Acad. Sci. USA, 96, 11578–11583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Robertson K.D., Gilchrist,C.A., Petri,W.A., Singh,V.K., Moskovitz,J., Wilkinson,B.J. and Jayaswal,R.K. (2002) DNA methylation and chromatin—unraveling the tangled web. Oncogene, 21, 5361–5379. [DOI] [PubMed] [Google Scholar]
- 11.Attwood J.T., Yung,R.L., Richardson,B.C., Gilchrist,C.A., Petri,W.A., Singh,V.K., Moskovitz,J., Wilkinson,B.J. and Jayaswal,R.K. (2002) DNA methylation and the regulation of gene transcription. Cell. Mol. Life Sci., 59, 241–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goto T., Monk,M., Gilchrist,C.A., Petri,W.A., Singh,V.K., Moskovitz,J., Wilkinson,B.J. and Jayaswal,R.K. (1998) Regulation of X-chromosome inactivation in development in mice and humans. Microbiol. Mol. Biol. Rev., 62, 362–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yoder J.A., Walsh,C.P. and Bestor,T.H. (1997) Cytosine methylation and the ecology of intragenomic parasites. Trends Genet., 13, 335–340. [DOI] [PubMed] [Google Scholar]
- 14.Dobosy J.R. and Selker,E.U. (2001) Emerging connections between DNA methylation and histone acetylation. Cell. Mol. Life Sci., 58, 721–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nan X., Cross,S. and Bird,A. (1998) Gene silencing by methyl-CpG-binding proteins. Novartis Found. Symp., 214, 6–16; discussion 16–21, 46–50. Review. [DOI] [PubMed] [Google Scholar]
- 16.Tate P.H. and Bird,A.P. (1993) Effects of DNA methylation on DNA-binding proteins and gene expression. Curr. Opin. Genet. Dev., 3, 226–231. [DOI] [PubMed] [Google Scholar]
- 17.Meehan R., Lewis,J., Cross,S., Nan,X., Jeppesen,P. and Bird,A. (1992) Transcriptional repression by methylation of CpG. J. Cell Sci. Suppl., 16, 9–14. [DOI] [PubMed] [Google Scholar]
- 18.Diamond L.S., Harlow,D.R. and Cunnick,C.C. (1978) A new medium for the axenic cultivation of Entamoeba histolytica and other Entamoeba. Trans. R. Soc. Trop. Med. Hyg., 72, 431–432. [DOI] [PubMed] [Google Scholar]
- 19.Warnecke P.M., Stirzaker,C., Song,J., Grunau,C., Melki,J.R. and Clark,S.J. (2002) Identification and resolution of artifacts in bisulfite sequencing. Methods, 27, 101–107. [DOI] [PubMed] [Google Scholar]
- 20.Sambrook J., Fritsch,E.F.M. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Edn. Cold Spring Harbor Laboratory Press, Plainview, NY. [Google Scholar]
- 21.Bradford M.M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem., 72, 248–254. [DOI] [PubMed] [Google Scholar]
- 22.Laemmli U.K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature, 227, 680–685. [DOI] [PubMed] [Google Scholar]
- 23.Bracha R. and Mirelman,D. (1984) Virulence of Entamoeba histolytica trophozoites. Effects of bacteria, microaerobic conditions and metronidazole. J. Exp. Med., 160, 353–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ankri S., Padilla-Vaca,F., Stolarsky,T., Koole,L., Katz,U. and Mirelman,D. (1999) Antisense inhibition of expression of the light subunit (35 kDa) of the Gal/GalNac lectin complex inhibits Entamoeba histolytica virulence. Mol. Microbiol., 33, 327–337. [DOI] [PubMed] [Google Scholar]
- 25.Ankri S., Stolarsky,T. and Mirelman,D. (1998) Antisense inhibition of expression of cysteine proteinases does not affect Entamoeba histolytica cytopathic or haemolytic activity but inhibits phagocytosis. Mol. Microbiol., 28, 777–785. [DOI] [PubMed] [Google Scholar]
- 26.Oakeley E.J. (1999) DNA methylation analysis: a review of current methodologies. Pharmacol. Ther., 84, 389–400. [DOI] [PubMed] [Google Scholar]
- 27.Gast F.U., Brinkmann,T., Pieper,U., Kruger,T., Noyer-Weidner,M. and Pingoud,A. (1997) The recognition of methylated DNA by the GTP-dependent restriction endonuclease McrBC resides in the N-terminal domain of McrB. Biol. Chem., 378, 975–982. [DOI] [PubMed] [Google Scholar]
- 28.Achwal C.W., Iyer,C.A. and Chandra,H.S. (1983) Immunochemical evidence for the presence of 5mC, 6mA and 7mG in human, Drosophila and mealybug DNA. FEBS Lett., 158, 353–358. [DOI] [PubMed] [Google Scholar]
- 29.Gowher H., Leismann,O. and Jeltsch,A. (2000) DNA of Drosophila melanogaster contains 5-methylcytosine. EMBO J., 19, 6918–6923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cross S.H., Charlton,J.A., Nan,X. and Bird,A.P. (1994) Purification of CpG islands using a methylated DNA binding column. Nature Genet., 6, 236–244. [DOI] [PubMed] [Google Scholar]
- 31.Altschul S.F., Gish,W., Miller,W., Myers,E.W. and Lipman,D.J. (1990) Basic local alignment search tool. J. Mol. Biol., 215, 403–410. [DOI] [PubMed] [Google Scholar]
- 32.Okano M., Xie,S. and Li,E. (1998) Dnmt2 is not required for de novo and maintenance methylation of viral DNA in embryonic stem cells. Nucleic Acids Res., 26, 2536–2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wilihoeft U., Campos-Gongora,E., Touzni,S., Bruchhaus,I., Tannich,E., Gilchrist,C.A., Petri,W.A., Singh,V.K., Moskovitz,J., Wilkinson,B.J. et al. (2001) Introns of Entamoeba histolytica and Entamoeba dispar. Protist, 152, 149–156. [DOI] [PubMed] [Google Scholar]
- 34.Reinhardt A. and Hubbard,T. (1998) Using neural networks for prediction of the subcellular location of proteins. Nucleic Acids Res., 26, 2230–2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thompson J.D., Higgins,D.G. and Gibson,T.J. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res., 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Van den Wyngaert I., Sprengel,J., Kass,S.U. and Luyten,W.H. (1998) Cloning and analysis of a novel human putative DNA methyltransferase. FEBS Lett., 426, 283–289. [DOI] [PubMed] [Google Scholar]
- 37.Willhoeft U. and Tannich,E. (2000) Fluorescence microscopy and fluorescence in situ hybridization of Entamoeba histolytica nuclei to analyse mitosis and the localization of repetitive DNA. Mol. Biochem. Parasitol., 105, 291–296. [DOI] [PubMed] [Google Scholar]
- 38.Santi D.V., Norment,A., Garrett,C.E., Gilchrist,C.A., Petri,W.A., Singh,V.K., Moskovitz,J., Wilkinson,B.J. and Jayaswal,R.K. (1984) Covalent bond formation between a DNA-cytosine methyltransferase and DNA containing 5-azacytosine. Proc. Natl Acad. Sci. USA, 81, 6993–6997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bagchi A., Bhattacharya,A. and Bhattacharya,S. (1999) Lack of a chromosomal copy of the circular rDNA plasmid of Entamoeba histolytica. Int. J. Parasitol., 29, 1775–1783. [DOI] [PubMed] [Google Scholar]
- 40.Tang L.Y., Reddy,M.N., Rasheva,V., Lee,T.L., Lin,M.J., Hung,M.S. and Shen,C.K. (2003) The eukaryotic DNMT2 genes encode a new class of cytosine-5 DNA methyltransferases. J. Biol. Chem., 19, 19. [DOI] [PubMed] [Google Scholar]
- 41.Oakes C.C., Smiraglia,D.J., Plass,C., Trasler,J.M. and Robaire,B. (2003) Aging results in hypermethylation of ribosomal DNA in sperm and liver of male rats. Proc. Natl Acad. Sci. USA, 100, 1775–1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.de Carvalho C.V., Payao,S.L. and Smith,M.A. (2000) DNA methylation, ageing and ribosomal genes activity. Biogerontology, 1, 357–361. [DOI] [PubMed] [Google Scholar]
- 43.Machwe A., Orren,D.K. and Bohr,V.A. (2000) Accelerated methylation of ribosomal RNA genes during the cellular senescence of Werner syndrome fibroblasts. FASEB J., 14, 1715–1724. [DOI] [PubMed] [Google Scholar]
- 44.Swisshelm K., Disteche,C.M., Thorvaldsen,J., Nelson,A. and Salk,D. (1990) Age-related increase in methylation of ribosomal genes and inactivation of chromosome-specific rRNA gene clusters in mouse. Mutat. Res., 237, 131–146. [DOI] [PubMed] [Google Scholar]
- 45.Santoro R. and Grummt,I. (2001) Molecular mechanisms mediating methylation-dependent silencing of ribosomal gene transcription. Mol. Cell, 8, 719–725. [DOI] [PubMed] [Google Scholar]
- 46.Jeltsch A. (2002) Beyond Watson and Crick: DNA methylation and molecular enzymology of DNA methyltransferases. Chembiochem, 3, 274–293. [DOI] [PubMed] [Google Scholar]
- 47.Bestor T., Laudano,A., Mattaliano,R. and Ingram,V. (1988) Cloning and sequencing of a cDNA encoding DNA methyltransferase of mouse cells. The carboxyl-terminal domain of the mammalian enzymes is related to bacterial restriction methyltransferases. J. Mol. Biol., 203, 971–983. [DOI] [PubMed] [Google Scholar]
- 48.Tollefsbol T.O. and Hutchison,C.A.,III (1998) Analysis in Escherichia coli of the effects of in vivo CpG methylation catalyzed by the cloned murine maintenance methyltransferase. Biochem. Biophys. Res. Commun., 245, 670–678. [DOI] [PubMed] [Google Scholar]
- 49.Tollefsbol T.O. and Hutchison,C.A.,III. (1997) Control of methylation spreading in synthetic DNA sequences by the murine DNA methyltransferase. J. Mol. Biol., 269, 494–504. [DOI] [PubMed] [Google Scholar]
- 50.Tollefsbol T.O. and Hutchison,C.A.,III (1995) Mammalian DNA (cytosine-5-)-methyltransferase expressed in Escherichia coli, purified and characterized. J. Biol. Chem., 270, 18543–18550. [DOI] [PubMed] [Google Scholar]
- 51.Okano M., Xie,S. and Li,E. (1998) Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nature Genet., 19, 219–220. [DOI] [PubMed] [Google Scholar]
- 52.Hermann A., Schmitt,S. and Jeltsch,A. (2003) The human Dnmt2 has residual DNA-(cytosine-C5)-methyltransferase activity. J. Biol. Chem., 10, 10. [DOI] [PubMed] [Google Scholar]
- 53.Bacolla A., Pradhan,S., Larson,J.E., Roberts,R.J. and Wells,R.D. (2001) Recombinant human DNA (cytosine-5) methyltransferase. III. Allosteric control, reaction order and influence of plasmid topology and triplet repeat length on methylation of the fragile X CGG.CCG sequence. J. Biol. Chem., 276, 18605–18613. [DOI] [PubMed] [Google Scholar]
- 54.Aoki A., Suetake,I., Miyagawa,J., Fujio,T., Chijiwa,T., Sasaki,H. and Tajima,S. (2001) Enzymatic properties of de novo-type mouse DNA (cytosine-5) methyltransferases. Nucleic Acids Res., 29, 3506–3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fuks F., Burgers,W.A., Godin,N., Kasai,M. and Kouzarides,T. (2001) Dnmt3a binds deacetylases and is recruited by a sequence-specific repressor to silence transcription. EMBO J., 20, 2536–2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kang E.S., Park,C.W. and Chung,J.H. (2001) Dnmt3b, de novo DNA methyltransferase, interacts with SUMO-1 and Ubc9 through its N-terminal region and is subject to modification by SUMO-1. Biochem. Biophys. Res. Commun., 289, 862–868. [DOI] [PubMed] [Google Scholar]
- 57.Burgers W.A., Fuks,F. and Kouzarides,T. (2002) DNA methyltransferases get connected to chromatin. Trends Genet., 18, 275–277. [DOI] [PubMed] [Google Scholar]
- 58.Deplus R., Brenner,C., Burgers,W.A., Putmans,P., Kouzarides,T., de Launoit,Y. and Fuks,F. (2002) Dnmt3L is a transcriptional repressor that recruits histone deacetylase. Nucleic Acids Res., 30, 3831–3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cross M., Kieft,R., Sabatini,R., Wilm,M., de Kort,M., van der Marel,G.A., van Boom,J.H., van Leeuwen,F. and Borst,P. (1999) The modified base J is the target for a novel DNA-binding protein in kinetoplastid protozoans. EMBO J., 18, 6573–6581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Christman J.K. (2002) 5-Azacytidine and 5-aza-2′-deoxycytidine as inhibitors of DNA methylation: mechanistic studies and their implications for cancer therapy. Oncogene, 21, 5483–5495. [DOI] [PubMed] [Google Scholar]
- 61.Van Dellen K., Field,J., Wang,Z., Loftus,B. and Samuelson,J. (2002) LINEs and SINE-like elements of the protist Entamoeba histolytica. Gene, 297, 229–239. [DOI] [PubMed] [Google Scholar]
- 62.Bhattacharya S., Bakre,A. and Bhattacharya,A. (2002) Mobile genetic elements in protozoan parasites. J. Genet., 81, 73–86. [DOI] [PubMed] [Google Scholar]
- 63.Willhoeft U., Buss,H. and Tannich,E. (2002) The abundant polyadenylated transcript 2 DNA sequence of the pathogenic protozoan parasite Entamoeba histolytica represents a nonautonomous non-long-terminal-repeat retrotransposon-like element which is absent in the closely related nonpathogenic species Entamoeba dispar. Infect. Immun., 70, 6798–6804. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.