Abstract
The transcription factors STAT3 and STAT5 play important roles in the regulation of mammary gland function during pregnancy, lactation, and involution. Given that STAT3 and STAT5 regulate genes involved in proliferation and survival, it is not surprising that inappropriate activation of STAT3 and STAT5 occurs commonly in breast cancer. Although these proteins are structurally similar, they have divergent and opposing effects on gene expression and cellular phenotype. Notably, when STAT5 and STAT3 are activated simultaneously, STAT5 has a dominant effect, and leads to decreased proliferation and increased sensitivity to cell death. Similarly, in breast cancer, activation of both STAT5 and STAT3 is associated with longer patient survival than activation of STAT3 alone. Pharmacological inhibitors of STAT3 and STAT5 are being developed for cancer therapy, though understanding the activation state and functional interaction of STAT3 and STAT5 in a patient's tumor may be critical for the optimal use of this strategy.
Keywords: Signal transduction, transcription factors, gene expression, oncology
1.1 STATs in cancer
Cancer occurs due to the subversion of the normal processes that provide cues for cells to grow, survive, or differentiate. No matter the primary molecular defect, one of the downstream effects is aberrant activation of transcription factors, leading to the changes in gene expression that underlie the malignant phenotype. One family of transcription factors that is often activated in cancer is the Signal Transducer and Activator of Transcription (STAT) family, comprised of seven related proteins. STATs are latent transcription factors that mainly reside in the cytoplasm in an inactive state. STATs become activated by phosphorylation on a critical tyrosine residue. Many upstream kinases can activate STATs, though the most common is the Jak family of kinases, which associate with various cell surface receptors. Additional receptor and non-receptor tyrosine kinases also have the ability to phosphorylate STATs. Tyrosine phosphorylation allows for STATs to form active dimers [1], which can be homodimers or heterodimers with another STAT family member [2] This dimer conformation triggers translocation to the nucleus where the STAT dimer binds to the minor groove of the DNA at cognate binding sites. STATs then recruit additional cofactors to modulate expression of key target genes involved in growth, survival, and differentiation. While most STAT target genes identified to date display increased expression with STAT binding, it is becoming increasingly clear that STATs not only activate the expression of genes, but also can repress gene expression [3,4]. Given the critical nature of the genes they control, the regulation of STAT function is normally tightly controlled. In non-cancerous cells, STAT activation is rapid and transient, with the STAT becoming inactivated largely by tyrosine phosphatases [5] . In addition, upstream kinases are often inhibited by negative regulators such as SOCS proteins, which themselves are STAT target genes [6]. In cancer, by contrast, STATs can become activated constitutively due to aberrant activation of an upstream kinase and/or loss of a negative regulator.
1.2 STAT3 in breast cancer
In the mammary gland, STAT3 poses an interesting paradox. While STAT3 promotes involution of the mammary gland after cessation of lactation [7-9], STAT3 activation in cancer is oncogenic [10], and STAT3 is constitutively activated in about 70% of breast tumors [11]. STAT3 activation is found in all classes of breast cancer, but is most often associated with triple negative breast tumors, which lack expression of the estrogen receptor (ER) or progesterone receptor (PR), and do not display amplification of Her2 [12,13]. In a panel of fourteen breast cancer cell lines, constitutively active STAT3 was only found in the triple-negative cells; importantly, it was activated in five of the six triple negative breast lines tested [12]. In normal mammary cells, STAT3 is activated by leukemia inhibitor factor (LIF) to promote involution [14,15]; however, in breast cancer, STAT3 is most often activated by interleukin-6 (IL-6) [16,17]. In fact, many breast cancer cell lines produce IL-6, which activates STAT3 in an autocrine fashion by signaling through the IL-6 receptor and Jak kinases [12,17]. The continued activation of STAT3 target genes can then lead to malignant cellular behavior. Specifically, STAT3 can upregulate genes promoting protection from apoptosis (such as bcl-xl and mcl1), proliferation (cyclin d1), pluripotency (bcl6 and klf4), invasion (mmp1), and angiogenesis (vegf) [11,18]. IL-6 and STAT3 have also been shown to promote invasion and epithelial-to-mesenchymal transition in part by upregulating the expression of the EMT inducer Twist [19-21]. In addition to upregulating genes promoting cancer phenotypes, STAT3 has additional non-transcriptional functions. For example, STAT3 can interact with microtubules, and may alter cellular shape and motility [22,23]. Moreover, in addition to the activating tyrosine phosphorylation, STAT3 can be phosphorylated on a serine residue near the carboxy-terminus [24]. STAT3 is serine phosphorylated in about 60% of breast tumors and is associated with estrogen receptor negative tumors .[25] Recent evidence suggests that serine phosphorylated STAT3 can be found in the mitochondria and can promote the survival of breast cancer cells solely through its effects on mitochondrial function [26]. Taken together, it is clear that constitutively activated STAT3 can promote the pathogenesis of aggressive breast tumors. Since STAT3 can be inhibited in normal cells with relatively few consequences [27], targeting STAT3 may be an important therapeutic approach in breast cancer.
1.3 STAT5 in breast cancer
The related STAT family member STAT5 is in fact encoded by two closely related genes, STAT5a and STAT5b. The majority of the functions of STAT5a and STAT5b overlap, and thus they are often grouped together as STAT5 [3,28,29]. STAT5 promotes both survival and terminal differentiation of the mammary gland [30-33]. Prolactin is the cytokine which is the main physiological trigger for activating STAT5 in the mammary gland, and this occurs late in pregnancy and during lactation [34]. STAT5 upregulates pro-survival genes, such as bcl-xl, and also genes encoding proteins found in milk such as, beta-casein and whey acidic protein [32,35-37]. Reflecting the fact that STATs can inhibit transcription as well as activate it, STAT5 also represses genes such as BCL6 that prevent terminal breast differentiation [4,13,38]. STAT5 has been found to be constitutively activated in breast tumors, and this occurs more frequently in hormone responsive tumors [13,39]. Higher circulating levels of prolactin are associated with cancer risk in ER/PR positive tumors [40]. Additionally, mice overexpressing a constitutively activated form of STAT5 develop mammary tumors [41]. However, STAT5 activation in breast cancer patients was shown to be a favorable marker for lymph node-negative breast cancer [42], In addition, prolactin and STAT5 have been shown to prevent invasion [43,44]. Activating the prolactin receptor in mesenchymal-like breast cancer cells reduced invasion and the expression of mesenchymal markers [43], suggesting that prolactin and STAT5 can be protective against invasion. Therefore, like STAT3, STAT5 can be activated in breast cancer, though it is often associated with a different tumor subtype than STAT3.
1.4 Co-activation of STAT5 and STAT3 in breast cancer
Despite their structural similarity, STAT5 and STAT3 have opposing functions in normal mammary development. In mice that express constitutively activated STAT5 in the mammary gland, involution is delayed and STAT3 activation does not occur [41]. Furthermore, LIF-mediated STAT3 activation induces apoptosis of mammary epithelial cells; however, STAT5 activation prevents this process [45]. This demonstrates that STAT5 and STAT3, though highly related, have both distinct and opposing functions in normal mammary development. Therefore, it is not surprising that STAT5 and STAT3 also have distinct and opposing functions in breast cancer.
In examining the roles of STAT3 and STAT5 in primary breast cancer, the first interesting observation reflects the disparity in frequency with which these transcription factors are found to be activated. Using immunohistochemistry to the tyrosine phosphorylated form of each protein, it was found that only 7% of breast tumors have activation of STAT5 alone, while 40% of breast tumors have activation of STAT3 alone. Notably, approximately 30% of breast tumors have activation of both STAT5 and STAT3 [13]. The low frequency of tumors displaying only activated STAT5 may reflect the fact that in addition to promoting survival, target genes of STAT5 also drive differentiation. Thus, while activation of STAT5 can contribute to malignancy, it may require the activation of other cooperating pathways, including STAT3. The low frequency of finding STAT5 activated on its own in primary breast cancers also parallels the infrequency with which breast cancer cell lines are found to have activated STAT5 [12].
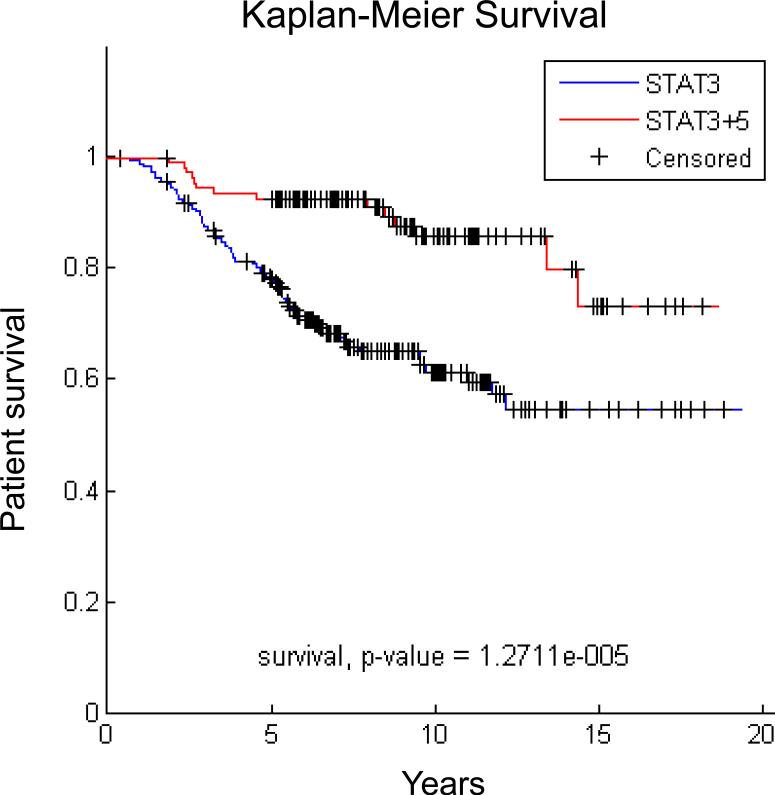
The small number of tumors displaying only STAT5 activation limits the ability to analyze the clinical characteristics of these tumors. However, it is possible to compare the characteristics of tumors with activation of STAT3 alone versus those displaying activation of both STAT3 and STAT5. Tumors with both STATs activated are more likely to be ER+ and less likely to be triple negative than tumors with only activated STAT3. In addition, the cancers with activation of STAT5 and STAT3 are more likely to be more highly differentiated grade I tumors, whereas tumors with activation of STAT3 alone are more likely to be less differentiated grade III. This suggests that tumors with activation of both STATs have better prognostic features than tumors with activation of STAT3 alone. Analyzing the genes that are differentially expressed in tumors with activation of STAT3 alone versus activation of both STATs identified specific gene signatures for these two groups. Utilizing these gene signatures, we examined survival in a cohort [46] of 295 patients with primary breast cancer. We found that patients whose tumors displayed the signature for activation of both STATs had better survival than patients whose tumors displayed the signature of only STAT3 activation (Figure 1). These findings suggest that STAT5 activation can modulate the biology of breast cancers that contain activated STAT3.
Figure 1.
Breast cancers with activation of STAT5 and STAT3 are associated with better survival than tumors with activation of STAT3 alone. Gene expression data for 295 breast cancer patients [46] were classified according to the STAT3+STAT5 vs. STAT3 gene signatures [13] using unsupervised hierarchical clustering [70].
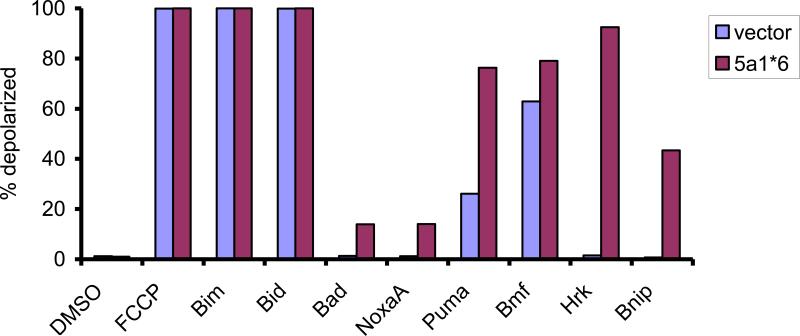
While patients whose tumors display activation of both STATs have a better prognosis than patients whose tumors have activation of STAT3 alone, this difference could reflect differences in the cell of origin or breast tumor subtype. To address this question in a model system, isogenic cell lines were generated with constitutive activation of STAT3 alone or activation of both STATs [13]. The triple negative breast cancer cell line MDA-MB-468, which contains constitutively active STAT3, was stably transfected with a constitutively active form of STAT5 (STAT5a1*6) or an empty vector. These paired cell lines then had activation of STAT3 only, or activation of both STATs. Cells that had activation of both STATs grew more slowly than the cells containing only STAT3 activation. In addition, cells with activation of both STATs were more sensitive to chemotherapeutic agents targeting microtubules, such as vinorelbine and paclitaxel, which are commonly used to treat breast cancer patients. These findings suggested that the intrinsic tendency of cells to undergo chemotherapy-induced apoptosis was enhanced when STAT5 was activated in addition to STAT3. To test this hypothesis, we measured the apoptotic priming of these cells using the technique of BH3 profiling, [47,48]. In this cell-based assay, the ability of mitochondria to become depolarized in the presence of pro-apoptotic peptides is measured. We found that the cells that contained activation of both STAT5 and STAT3 were more primed for cell death than cells that only contained activation of STAT3 (Figure 2). Specifically, increased mitochondrial permeability was induced in the cells with activation of both STAT5 and STAT3 to peptides that inactivate pro-survival proteins such as Mcl-1 (NoxaA), Bcl-xL (Hrk), and others. These data demonstrate that STAT5 opposes the protumorigenic functions of STAT3, reducing the growth and survival of these cells and increasing their drug sensitivity. These differences in cellular phenotype may underlie the survival differences seen in patients whose tumors have activation of STAT3 and STAT5 compared to STAT3 alone.
Figure 2.
Breast cancer cells with activation of both STAT5 and STAT3 are more primed for death than breast cancer cells with activation of STAT3 alone. MDA-MB-468 cells, which have constitutive STAT3 activation, were stably transfected with a vector expressing constitutively activated STAT5 (5a1*6) or an empty vector, and their apoptotic potential was analyzed by BH3 profiling. The indicated peptides were added to mitochondria from the two cell lines and the depolarization of the mitochondrial membrane was measured. FCCP served as a positive control.
Since STAT5 and STAT3 are highly related transcription factors that can bind to and regulate many of the same target genes, the question arises as to how does STAT5 oppose STAT3 in breast cancer? One mechanism by which STAT5 opposes STAT3 function is by directly opposing the regulation of target genes [13]. For example, STAT3 upregulates the transcriptional repressor BCL6, while STAT5 represses BCL6 expression. When both STATs are activated, STAT5 is dominant over STAT3 and represses BCL6 expression. Another mechanism by which STAT5 has this effect on STAT3 may be through upregulation of negative regulators of STATs, particularly SOCS family members [6,49]. Although SOCS proteins are generally target genes of STATs, some family members, including CIS, are particularly responsive to STAT5 [50]. Thus, it is not surprising that constitutively active STAT5 results in upregulation of CIS and SOCS3 in the isogenic MDA-MB-468 cells [13]. There is a reduction in the tyrosine phosphorylation of STAT3 in these cells with activated STAT5, suggesting that this upregulation of SOCS family members may modulate STAT3 phosphorylation. Reducing STAT3 activation would also have important effects on the growth and survival of breast cancer cells. Finally, STAT5 and STAT3 also regulate a distinct set of genes that may promote opposing functions in breast cancer [45]. Therefore, in the case of breast cancer, STAT5 and STAT3 clearly are not redundant transcription factors, but promote distinct and opposing functions.
2.1 Targeting STATs for the treatment of breast cancer
2.1.1 Inhibiting STAT3
STATs are attractive targets for cancer therapy since they are activated in many different cancers and are downstream of various different oncogenic tyrosine kinases. Importantly, with STAT3 being activated in 70% of breast tumors, particularly in the less treatable triple-negative tumors, inhibiting STAT3 might be a potentially important new form of breast cancer therapy. To this end, there have been a number of strategies employed to target STATs, in particular STAT3.
2.1.2 Jak inhibitors
One of the key mechanisms by which STAT3 becomes constitutively activated in triple negative breast cancer is through autocrine production of cytokines, particularly IL-6. While this suggests a number of therapeutic strategies, including blocking antibodies to IL-6 or the IL-6 receptor, there has been great interest in using small molecule inhibitors of the Jak family kinases that mediate STAT3 tyrosine phosphorylation triggered by IL-6. Activating mutations have been found in Jak2 in myeloproliferative disorders, and inhibitors of this kinase were developed and introduced into clinical practice for these diseases [51-53]. Although the Jaks are not known to be mutated in breast cancer, they play an integral role in this oncogenic pathway [12,54,55]. A variety of molecules targeting Jak2 have been shown to be effective at killing cancer cell lines containing activated STAT3 [56,57]. Recently, the Jak2 inhibitor NVP-BSK805 has shown efficacy against the growth of triple negative breast tumor xenografts containing activation of STAT3 [12]. This suggests that targeting the upstream Jak kinases can be effective in targeting breast cancers with STAT3 activation.
2.1.3 STAT3 dimerization inhibitors
Another method to inhibit STAT3 is to target the activating dimerization. In a STAT3 dimer, the phosphorylated tyrosine 705 residue of one monomer binds in a reciprocal fashion to the SH2 domain of its partner. In addition, the SH2 domain of STATs is needed for recruitment to activated, tyrosine phosphorylated, receptor-kinase complexes. Thus, targeting the SH2 domain is an appealing strategy. Over the past few years, a number of dimerization inhibitors have been developed, many of which have been identified using docking modeling and virtual screening. These molecules such as STA-21 and CJ-1383 inhibit the SH2 domain, preventing both STAT3 phosphorylation and dimerization [58] [59]. Importantly, these inhibitors induce apoptosis of breast cancer cell lines with activated STAT3 suggesting that targeting STAT3 dimerization may be a worthwhile therapeutic strategy.
2.1.4 Screening strategies to identify STAT3 inhibitors
Given that a key function of STAT3 is inducing gene transcription, another method to identify STAT3 inhibitors is to develop a cell-based screening system whose read out is STAT3-dependent expression of a reporter gene, such as luciferase. Such an assay system, with an appropriate counter-screen, enables rapid identification of cell-permeable compounds that can inhibit STAT3-dependent transcriptional function without affecting other related transcription factors [60]. This strategy can be applied to a wide range of structurally diverse chemical libraries, including libraries of compounds that are either known to be safe in humans or even approved for use in humans for other indications. This has the potential to greatly accelerate the initiation of proof-of-concept clinical trials in patients with cancer. From such a transcription based screen, three drugs were identified to be potent and specific STAT3 inhibitors: nifuroxazide, pimozide, and pyrimethamine [60-62].
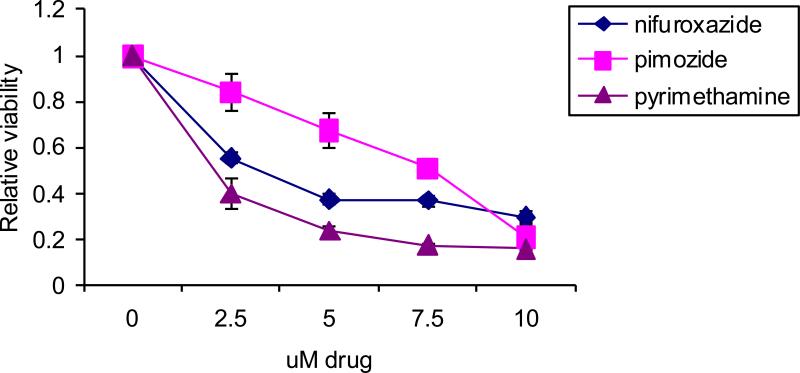
Nifuroxazide is not currently approved for use in the United States, but is used elsewhere as an anti-diarrheal agent. Nifuroxazide decreases STAT3 tyrosine phosphorylation and inhibits Jak2 and Tyk2 kinases [60]. Nifuroxazide can kill cancer cells with activated STAT3, though it has little effect on cells lacking STAT3 activation including non-transformed cells. The second drug to emerge from this screen was pyrimethamine, an anti-microbial with a good safety profile that is FDA-approved for the treatment of toxoplasmosis and malaria. The mechanism of action of pyrimethamine-mediated inhibition of STAT3 is currently unknown. It can decrease STAT3-dependent gene expression independent of effects on STAT3 phosphorylation, though with prolonged high doses pyrimethamine can inhibit STAT3 tyrosine phosphorylation [61,63]. Pyrimethamine is currently in a clinical trial for chronic lymphocytic leukemia (CLL) at the Dana- Farber Cancer Institute (ClinicalTrials.gov Identifier: NCT01066663). Finally, pimozide inhibits STAT3 function by decreasing its activating tyrosine phosphorylation [61]. In contrast to nifuroxazide, pimozide does not directly inhibit upstream kinases [62,64,65]. Rather, it seems that pimozide may be affecting negative regulators of STATs to modulate phosphorylation. Analysis of the effects of pimozide and pyrimethamine on the viability of a large panel of cell lines suggested that these agents might have therapeutic benefit in breast cancer cells [61]. Therefore, we analyzed the effects of these three agents in a triple negative breast cancer cell line, MDA-MB-468, characterized by constitutive STAT3 activation (Figure 3). All three agents led to a dose dependent decrease in viability of this cell line. In particular, the concentrations or pyrimethamine needed for this effect can readily be achieved in patients, suggesting that this transcription-based screening approach can identify drugs that may be suitable for clinical testing.
Figure 3.
Breast cancer cells with activated STAT3 are sensitive to STAT3 inhibitors. MDA-MB-468 cells were treated for 48h with the indicated drugs and cell viability was analyzed by ATP dependent luminescence.
Interestingly, some chemotherapy agents that are commonly used for the treatment of breast cancer may act, at least in part, through inhibition of STAT3. As noted, STAT3 can associate with the cytoskeleton in cells, which raised the hypothesis that agents targeting cytoskeletal components might interfere with STAT3 function. In fact, the microtubule targeting agents paclitaxel and vinorelbine both inhibit STAT3 tyrosine phosphorylation in triple negative breast cancer cell lines [22]. Paclitaxel can decrease the association between STAT3 and microtubules, and also appears to induce a negative regulator of STAT3 tyrosine phosphorylation. Interestingly, some breast cancer cell lines are relatively resistant to this STAT3-inhibitory effect of paclitaxel, although sensitivity can be restored by treating the cells with an inhibitor of DNA methylation [66]. This suggests that negative regulators of STAT3, which can be induced by microtubule-targeted agents, may be silenced by methylation as a mechanism of resistance. Finally, the combination of microtubule targeting agents with drugs that inhibit STAT3 by other mechanisms shows increased efficacy in killing STAT3-dependent tumor cells [66], suggesting this may be a useful combinational strategy in breast cancers with activated STAT3.
2.1.5 STAT5 inhibitors
Given that elevated prolactin levels and STAT5 activation is associated with breast cancer, targeting prolactin and the prolactin receptor has been of great interest, and strategies employing both small molecules and antibodies are being pursued [67,68]. Prolactin binding to its receptor triggers STAT5 phosphorylation through activation of Jak family kinases [69]. As noted, STAT3 activation in many breast cancers (particularly triple negative breast cancer) occurs through an IL-6-mediated autocrine loop, which also is dependent on Jaks. Thus, kinase inhibitors targeting Jaks may have the dual benefit of blocking both of these pathways. Finally, many negative regulators of STAT signaling, such as SOCS proteins, can decrease signaling to both STAT3 and STAT5. Therefore, strategies that work through negative regulators may decrease the effects of both prolactin and IL-6. In this context, the STAT3 inhibitor pimozide, which appears to work through negative regulators, also inhibits STAT5 [62]. Therefore, this drug, or similar agents, might be particularly useful in breast cancer cells that contain activation of both STAT5 and STAT3.
2.1.6 STAT5 inhibitors in the context of STAT3 activation
Evidence from both patients and animal models suggests that STAT5 activation may be associated with breast cancer pathogenesis [39,41]. However, approximately one-third of primary breast cancers display activation of both STAT5 and STAT3, and STAT5 activation in this context appears to contribute to less aggressive tumors [13]. This raises potential concerns about the use of STAT5 inhibitors in patients whose tumors have activation of both pathways. It is conceivable that targeting STAT5 in breast tumors with activation of both STATs may in fact make the tumor more aggressive and less responsive to chemotherapy. In cell culture systems, activated STAT5 reduces the growth of tumor cells with activated STAT3, primes these cells for apoptosis, and makes them more responsive to at least some cytotoxic agents. Therefore, targeting STAT5 alone could remove the suppressive effect that STAT5 has on STAT3. This does not suggest that STAT5 inhibitors would not be useful. However, if they were to be used, knowing the activation status of both STAT3 and STAT5 would be important. Given this interplay between STAT3 and STAT5, dual inhibitors of STAT5 and STAT3 might be particularly beneficial.
3.1 Conclusion
Although STAT5 and STAT3 are structurally similar, they play distinct roles in mammary development and in breast cancer, and thus they are clearly not redundant transcription factors. In fact, STAT5 opposes many of the functions of STAT3 on both a molecular and biological level, and may reduce the aggressiveness of tumors. Targeting STATs is an attractive rational molecular strategy for breast cancer therapy, though understanding the functional relationships between STAT3 and STAT5 will be critical to optimizing this approach.
Transcription factors STAT3 and STAT5 regulate genes critical to cellular function
STAT3 and STAT5 have divergent and opposing functions in mammary development
Both STAT3 and STAT5 can be activated in breast cancers
Co-activation of STAT5 decreases aggressiveness of tumor cells with activated STAT3
Inhibitors of STAT3 and STAT5 may play an important role in breast cancer therapy
Acknowledgements
We would like to thank Anthony Letai and Jeremy S. Ryan for help with performing BH3 profiling. This work was supported by grants from the National Cancer Institute (R01-CA160979), Susan G. Komen for the Cure, the Brent Leahey Fund, Friends of the Dana-Farber Cancer Institute, and a BCRF-AACR grant for Translational Breast Cancer Research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kretzschmar AK, Dinger MC, Henze C, Brocke-Heidrich K, Horn F. Analysis of Stat3 (signal transducer and activator of transcription 3) dimerization by fluorescence resonance energy transfer in living cells. Biochem J. 2004;377:289–97. doi: 10.1042/BJ20030708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cella N, Groner B, Hynes NE. Characterization of Stat5a and Stat5b homodimers and heterodimers and their association with the glucocortiocoid receptor in mammary cells. Mol Cell Biol. 1998;18:1783–92. doi: 10.1128/mcb.18.4.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nelson EA, Walker SR, Alvarez JV, Frank DA. Isolation of unique STAT5 targets by chromatin immunoprecipitation-based gene identification. J Biol Chem. 2004;279:54724–30. doi: 10.1074/jbc.M408464200. [DOI] [PubMed] [Google Scholar]
- 4.Walker SR, Nelson EA, Frank DA. STAT5 represses BCL6 expression by binding to a regulatory region frequently mutated in lymphomas. Oncogene. 2007;26:224–33. doi: 10.1038/sj.onc.1209775. [DOI] [PubMed] [Google Scholar]
- 5.Gu F, Dube N, Kim JW, Cheng A, Ibarra-Sanchez Mde J, Tremblay ML, Boisclair YR. Protein tyrosine phosphatase 1B attenuates growth hormone-mediated JAK2-STAT signaling. Mol Cell Biol. 2003;23:3753–62. doi: 10.1128/MCB.23.11.3753-3762.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sutherland KD, Lindeman GJ, Visvader JE. Knocking off SOCS genes in the mammary gland. Cell Cycle. 2007;6:799–803. doi: 10.4161/cc.6.7.4037. [DOI] [PubMed] [Google Scholar]
- 7.Chapman RS, Lourenco PC, Tonner E, Flint DJ, Selbert S, Takeda K, Akira S, Clarke AR, Watson CJ. Suppression of epithelial apoptosis and delayed mammary gland involution in mice with a conditional knockout of Stat3. Genes Dev. 1999;13:2604–16. doi: 10.1101/gad.13.19.2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Groner B, Hennighausen L. Linear and cooperative signaling: roles for Stat proteins in the regulation of cell survival and apoptosis in the mammary epithelium. Breast Cancer Res. 2000;2:149–53. doi: 10.1186/bcr47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Humphreys RC, Bierie B, Zhao L, Raz R, Levy D, Hennighausen L. Deletion of Stat3 blocks mammary gland involution and extends functional competence of the secretory epithelium in the absence of lactogenic stimuli. Endocrinology. 2002;143:3641–50. doi: 10.1210/en.2002-220224. [DOI] [PubMed] [Google Scholar]
- 10.Burke WM, Jin X, Lin HJ, Huang M, Liu R, Reynolds RK, Lin J. Inhibition of constitutively active Stat3 suppresses growth of human ovarian and breast cancer cells. Oncogene. 2001;20:7925–34. doi: 10.1038/sj.onc.1204990. [DOI] [PubMed] [Google Scholar]
- 11.Alvarez JV, Febbo PG, Ramaswamy S, Loda M, Richardson A, Frank DA. Identification of a genetic signature of activated signal transducer and activator of transcription 3 in human tumors. Cancer Res. 2005;65:5054–62. doi: 10.1158/0008-5472.CAN-04-4281. [DOI] [PubMed] [Google Scholar]
- 12.Marotta LL, Almendro V, Marusyk A, Shipitsin M, Schemme J, Walker SR, Bloushtain-Qimron N, Kim JJ, Choudhury SA, Maruyama R, Wu Z, Gonen M, Mulvey LA, Bessarabova MO, Huh SJ, Silver SJ, Kim SY, Park SY, Lee HE, Anderson KS, Richardson AL, Nikolskaya T, Nikolsky Y, Liu XS, Root DE, Hahn WC, Frank DA, Polyak K. The JAK2/STAT3 signaling pathway is required for growth of CD44+CD24-stem cell-like breast cancer cells in human tumors. J Clin Invest. 2011 doi: 10.1172/JCI44745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Walker SR, Nelson EA, Zou L, Chaudhury M, Signoretti S, Richardson A, Frank DA. Reciprocal effects of STAT5 and STAT3 in breast cancer. Mol Cancer Res. 2009;7:966–76. doi: 10.1158/1541-7786.MCR-08-0238. [DOI] [PubMed] [Google Scholar]
- 14.Kritikou EA, Sharkey A, Abell K, Came PJ, Anderson E, Clarkson RW, Watson CJ. A dual, non-redundant, role for LIF as a regulator of development and STAT3-mediated cell death in mammary gland. Development. 2003;130:3459–68. doi: 10.1242/dev.00578. [DOI] [PubMed] [Google Scholar]
- 15.Schere-Levy C, Buggiano V, Quaglino A, Gattelli A, Cirio MC, Piazzon I, Vanzulli S, Kordon EC. Leukemia inhibitory factor induces apoptosis of the mammary epithelial cells and participates in mouse mammary gland involution. Exp Cell Res. 2003;282:35–47. doi: 10.1006/excr.2002.5666. [DOI] [PubMed] [Google Scholar]
- 16.Berishaj M, Gao SP, Ahmed S, Leslie K, Al-Ahmadie H, Gerald WL, Bornmann W, Bromberg JF. Stat3 is tyrosine-phosphorylated through the interleukin-6/glycoprotein 130/Janus kinase pathway in breast cancer. Breast Cancer Res. 2007;9:R32. doi: 10.1186/bcr1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lieblein JC, Ball S, Hutzen B, Sasser AK, Lin HJ, Huang TH, Hall BM, Lin J. STAT3 can be activated through paracrine signaling in breast epithelial cells. BMC Cancer. 2008;8:302. doi: 10.1186/1471-2407-8-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hsieh FC, Cheng G, Lin J. Evaluation of potential Stat3-regulated genes in human breast cancer. Biochem Biophys Res Commun. 2005;335:292–9. doi: 10.1016/j.bbrc.2005.07.075. [DOI] [PubMed] [Google Scholar]
- 19.Sullivan NJ, Sasser AK, Axel AE, Vesuna F, Raman V, Ramirez N, Oberyszyn TM, Hall BM. Interleukin-6 induces an epithelial-mesenchymal transition phenotype in human breast cancer cells. Oncogene. 2009;28:2940–7. doi: 10.1038/onc.2009.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lo HW, Hsu SC, Xia W, Cao X, Shih JY, Wei Y, Abbruzzese JL, Hortobagyi GN, Hung MC. Epidermal growth factor receptor cooperates with signal transducer and activator of transcription 3 to induce epithelial-mesenchymal transition in cancer cells via up-regulation of TWIST gene expression. Cancer Res. 2007;67:9066–76. doi: 10.1158/0008-5472.CAN-07-0575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yadav A, Kumar B, Datta J, Teknos TN, Kumar P. IL-6 promotes head and neck tumor metastasis by inducing epithelial-mesenchymal transition via the JAK-STAT3-SNAIL signaling pathway. Mol Cancer Res. 2011;9:1658–67. doi: 10.1158/1541-7786.MCR-11-0271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Walker SR, Chaudhury M, Nelson EA, Frank DA. Microtubule-targeted chemotherapeutic agents inhibit signal transducer and activator of transcription 3 (STAT3) signaling. Mol Pharmacol. 2010;78:903–8. doi: 10.1124/mol.110.066316. [DOI] [PubMed] [Google Scholar]
- 23.Ng DC, Lin BH, Lim CP, Huang G, Zhang T, Poli V, Cao X. Stat3 regulates microtubules by antagonizing the depolymerization activity of stathmin. J Cell Biol. 2006;172:245–57. doi: 10.1083/jcb.200503021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang X, Blenis J, Li HC, Schindler C, Chen-Kiang S. Requirement of serine phosphorylation for formation of STAT-promoter complexes. Science. 1995;267:1990–4. doi: 10.1126/science.7701321. [DOI] [PubMed] [Google Scholar]
- 25.Yeh YT, Ou-Yang F, Chen IF, Yang SF, Wang YY, Chuang HY, Su JH, Hou MF, Yuan SS. STAT3 ser727 phosphorylation and its association with negative estrogen receptor status in breast infiltrating ductal carcinoma. Int J Cancer. 2006;118:2943–7. doi: 10.1002/ijc.21771. [DOI] [PubMed] [Google Scholar]
- 26.Gough DJ, Corlett A, Schlessinger K, Wegrzyn J, Larner AC, Levy DE. Mitochondrial STAT3 supports Ras-dependent oncogenic transformation. Science. 2009;324:1713–6. doi: 10.1126/science.1171721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Holland SM, DeLeo FR, Elloumi HZ, Hsu AP, Uzel G, Brodsky N, Freeman AF, Demidowich A, Davis J, Turner ML, Anderson VL, Darnell DN, Welch PA, Kuhns DB, Frucht DM, Malech HL, Gallin JI, Kobayashi SD, Whitney AR, Voyich JM, Musser JM, Woellner C, Schaffer AA, Puck JM, Grimbacher B. STAT3 mutations in the hyper-IgE syndrome. N Engl J Med. 2007;357:1608–19. doi: 10.1056/NEJMoa073687. [DOI] [PubMed] [Google Scholar]
- 28.Teglund S, McKay C, Schuetz E, van Deursen JM, Stravopodis D, Wand D, Brown M, Bodner S, Grosveld G, Ihle JN. Stat5a and Stat5b proteins have essential and nonessential, or redundant, roles in cytokine responses. Cell. 1998;93:841–850. doi: 10.1016/s0092-8674(00)81444-0. [DOI] [PubMed] [Google Scholar]
- 29.Grimley PM, Dong F, Rui H. Stat5a and Stat5b: fraternal twins of signal transduction and transcriptional activation. Cytokine Growth Factor Rev. 1999;10:131–57. doi: 10.1016/s1359-6101(99)00011-8. [DOI] [PubMed] [Google Scholar]
- 30.Liu X, Robinson GW, Hennighausen L. Activation of Stat5a and Stat5b by tyrosine phosphorylation is tightly linked to mammary gland differentiation. Mol Endocrinol. 1996;10:1496–506. doi: 10.1210/mend.10.12.8961260. [DOI] [PubMed] [Google Scholar]
- 31.Liu X, Robinson GW, Wagner K-U, Garrett L, Wynshaw-Boris A, Hennighausen L. Stat5a is mandatory for adult mammary gland development and lactogenesis. Genes Dev. 1997;11:179–186. doi: 10.1101/gad.11.2.179. [DOI] [PubMed] [Google Scholar]
- 32.Cui Y, Riedlinger G, Miyoshi K, Tang W, Li C, Deng CX, Robinson GW, Hennighausen L. Inactivation of Stat5 in mouse mammary epithelium during pregnancy reveals distinct functions in cell proliferation, survival, and differentiation. Mol Cell Biol. 2004;24:8037–47. doi: 10.1128/MCB.24.18.8037-8047.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miyoshi K, Shillingford JM, Smith GH, Grimm SL, Wagner KU, Oka T, Rosen JM, Robinson GW, Hennighausen L. Signal transducer and activator of transcription (Stat) 5 controls the proliferation and differentiation of mammary alveolar epithelium. J Cell Biol. 2001;155:531–42. doi: 10.1083/jcb.200107065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wakao H, Gouilleux F, Groner B. Mammary gland factor (MGF) is a novel member of the cytokine regulated transcription factor gene family and confers the prolactin response. EMBO J. 1994;13:2182–2191. doi: 10.1002/j.1460-2075.1994.tb06495.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tourkine N, Schindler C, Larose M, Houdebine LM. Activation of STAT factors by prolactin, interferon-gamma, growth hormones, and a tyrosine phosphatase inhibitor in rabbit primary mammary epithelial cells. J Biol Chem. 1995;270:20952–61. doi: 10.1074/jbc.270.36.20952. [DOI] [PubMed] [Google Scholar]
- 36.Jolivet G, L'Hotte C, Pierre S, Tourkine N, Houdebine LM. A MGF/STAT5 binding site is necessary in the distal enhancer for high prolactin induction of transfected rabbit alpha s1-casein-CAT gene transcription. FEBS Lett. 1996;389:257–62. doi: 10.1016/0014-5793(96)00598-4. [DOI] [PubMed] [Google Scholar]
- 37.Chughtai N, Schimchowitsch S, Lebrun JJ, Ali S. Prolactin induces SHP-2 association with Stat5, nuclear translocation, and binding to the beta-casein gene promoter in mammary cells. J Biol Chem. 2002;277:31107–14. doi: 10.1074/jbc.M200156200. [DOI] [PubMed] [Google Scholar]
- 38.Logarajah S, Hunter P, Kraman M, Steele D, Lakhani S, Bobrow L, Venkitaraman A, Wagner S. BCL-6 is expressed in breast cancer and prevents mammary epithelial differentiaiton. Oncogene. 2003;22:5572–8. doi: 10.1038/sj.onc.1206689. [DOI] [PubMed] [Google Scholar]
- 39.Cotarla I, Ren S, Zhang Y, Gehan E, Singh B, Furth PA. Stat5a is tryosine phosphorylated and nuclear localized in a high proportion of human breast cancers. Int. J. Cancer. 2004;108:665–671. doi: 10.1002/ijc.11619. [DOI] [PubMed] [Google Scholar]
- 40.Tworoger SS, Sluss P, Hankinson SE. Association between plasma prolactin concentrations and risk of breast cancer among predominately premenopausal women. Cancer Res. 2006;66:2476–82. doi: 10.1158/0008-5472.CAN-05-3369. [DOI] [PubMed] [Google Scholar]
- 41.Iavnilovitch E, Groner B, Barash I. Overexpression and forced activation of Stat5 in mammary gland of transgenic mice promotes cellular proliferation, enhances differentiation, and delays postlactational apoptosis. Mol. Cancer Res. 2002;1:32–47. [PubMed] [Google Scholar]
- 42.Nevalainen MT, Xie J, Torhorst J, Bubendorf L, Haas P, Kononen J, Sauter G, Rui H. Signal transducer and activator of transcription-5 activation and breast cancer prognosis. J. Clin. Oncol. 2004;22:2053–60. doi: 10.1200/JCO.2004.11.046. [DOI] [PubMed] [Google Scholar]
- 43.Nouhi Z, Chughtai N, Hartley S, Cocolakis E, Lebrun JJ, Ali S. Defining the role of prolactin as an invasion suppressor hormone in breast cancer cells. Cancer Res. 2006;66:1824–32. doi: 10.1158/0008-5472.CAN-05-2292. [DOI] [PubMed] [Google Scholar]
- 44.Sultan AS, Xie J, LeBaron MJ, Ealley EL, Nevalainen MT, Rui H. Stat5 promotes homotypic adhesion and inhibits invasive characteristics of human breast cancer cells. Oncogene. 2005;24:746–60. doi: 10.1038/sj.onc.1208203. [DOI] [PubMed] [Google Scholar]
- 45.Clarkson RW, Boland MP, Kritikou EA, Lee JM, Freeman TC, Tiffen PG, Watson CJ. The genes induced by signal transducer and activators of transcription (STAT)3 and STAT5 in mammary epithelial cells define the roles of these STATs in mammary development. Mol Endocrinol. 2006;20:675–85. doi: 10.1210/me.2005-0392. [DOI] [PubMed] [Google Scholar]
- 46.van de Vijver MJ, He YD, van't Veer LJ, Dai H, Hart AA, Voskuil DW, Schreiber GJ, Peterse JL, Roberts C, Marton MJ, Parrish M, Atsma D, Witteveen A, Glas A, Delahaye L, van der Velde T, Bartelink H, Rodenhuis S, Rutgers ET, Friend SH, Bernards R. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 47.Ryan JA, Brunelle JK, Letai A. Heightened mitochondrial priming is the basis for apoptotic hypersensitivity of CD4+ CD8+ thymocytes. Proc Natl Acad Sci U S A. 107:12895–900. doi: 10.1073/pnas.0914878107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ni Chonghaile T, Sarosiek KA, Vo TT, Ryan JA, Tammareddi A, Moore Vdel G, Deng J, Anderson KC, Richardson P, Tai YT, Mitsiades CS, Matulonis UA, Drapkin R, Stone R, Deangelo DJ, McConkey DJ, Sallan SE, Silverman L, Hirsch MS, Carrasco DR, Letai A. Pretreatment mitochondrial priming correlates with clinical response to cytotoxic chemotherapy. Science. 334:1129–33. doi: 10.1126/science.1206727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Barclay JL, Anderson ST, Waters MJ, Curlewis JD. SOCS3 as a tumor suppressor in breast cancer cells, and its regulation by PRL. Int J Cancer. 2009;124:1756–66. doi: 10.1002/ijc.24172. [DOI] [PubMed] [Google Scholar]
- 50.Matsumoto A, Masuhara M, Mitsui K, Yokouchi M, Ohtsubo M, Misawa H, Miyajima A, Yoshimura A. CIS, a cytokine inducible SH2 protein, is a target of the JAK-STAT5 pathway and modulates STAT5 activation. Blood. 1997;89:3148–3154. [PubMed] [Google Scholar]
- 51.Levine RL, Wadleigh M, Cools J, Ebert BL, Wernig G, Huntly BJ, Boggon TJ, Wlodarska I, Clark JJ, Moore S, Adelsperger J, Koo S, Lee JC, Gabriel S, Mercher T, D'Andrea A, Frohling S, Dohner K, Marynen P, Vandenberghe P, Mesa RA, Tefferi A, Griffin JD, Eck MJ, Sellers WR, Meyerson M, Golub TR, Lee SJ, Gilliland DG. Activating mutation in the tyrosine kinase JAK2 in polycythemia vera, essential thrombocythemia, and myeloid metaplasia with myelofibrosis. Cancer Cell. 2005;7:387–97. doi: 10.1016/j.ccr.2005.03.023. [DOI] [PubMed] [Google Scholar]
- 52.Levine RL, Gilliland DG. JAK-2 mutations and their relevance to myeloproliferative disease. Curr Opin Hematol. 2007;14:43–7. doi: 10.1097/00062752-200701000-00009. [DOI] [PubMed] [Google Scholar]
- 53.Wernig G, Kharas MG, Okabe R, Moore SA, Leeman DS, Cullen DE, Gozo M, McDowell EP, Levine RL, Doukas J, Mak CC, Noronha G, Martin M, Ko YD, Lee BH, Soll RM, Tefferi A, Hood JD, Gilliland DG. Efficacy of TG101348, a selective JAK2 inhibitor, in treatment of a murine model of JAK2V617F-induced polycythemia vera. Cancer Cell. 2008;13:311–20. doi: 10.1016/j.ccr.2008.02.009. [DOI] [PubMed] [Google Scholar]
- 54.Caffarel MM, Zaragoza R, Pensa S, Li J, Green AR, Watson CJ. Constitutive activation of JAK2 in mammary epithelium elevates Stat5 signalling, promotes alveologenesis and resistance to cell death, and contributes to tumourigenesis. Cell Death Differ. 2012;19:511–22. doi: 10.1038/cdd.2011.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sakamoto K, Triplett AA, Schuler LA, Wagner KU. Janus kinase 2 is required for the initiation but not maintenance of prolactin-induced mammary cancer. Oncogene. 2010;29:5359–69. doi: 10.1038/onc.2010.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hedvat M, Huszar D, Herrmann A, Gozgit JM, Schroeder A, Sheehy A, Buettner R, Proia D, Kowolik CM, Xin H, Armstrong B, Bebernitz G, Weng S, Wang L, Ye M, McEachern K, Chen H, Morosini D, Bell K, Alimzhanov M, Ioannidis S, McCoon P, Cao ZA, Yu H, Jove R, Zinda M. The JAK2 inhibitor AZD1480 potently blocks Stat3 signaling and oncogenesis in solid tumors. Cancer Cell. 2009;16:487–97. doi: 10.1016/j.ccr.2009.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Blaskovich MA, Sun J, Cantor A, Turkson J, Jove R, Sebti SM. Discovery of JSI-124 (Cucurbitacin I), a selective Janus kinase/Signal transducer and activator of transcription 3 signaling pathway inhibitor with potent antitumor activity against human and murine cancer cells in mice. Cancer Res. 2003;63:1270–1279. [PubMed] [Google Scholar]
- 58.Song H, Wang R, Wang S, Lin J. A low-molecular-weight compound discovered through virtual database screening inhibits Stat3 function in breast cancer cells. Proc Natl Acad Sci U S A. 2005;102:4700–5. doi: 10.1073/pnas.0409894102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen J, Bai L, Bernard D, Nikolovska-Coleska Z, Gomez C, Zhang J, Yi H, Wang S. Structure-Based Design of Conformationally Constrained, Cell-Permeable STAT3 Inhibitors. ACS Med Chem Lett. 2010;1:85–89. doi: 10.1021/ml100010j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nelson EA, Walker SR, Kepich A, Gashin LB, Hideshima T, Ikeda H, Chauhan D, Anderson KC, Frank DA. Nifuroxazide inhibits survival of multiple myeloma cells by directly inhibiting STAT3. Blood. 2008;112:5095–102. doi: 10.1182/blood-2007-12-129718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nelson EA, Sharma SV, Settleman J, Frank DA. A chemical biology approach to developing STAT inhibitors: molecular strategies for accelerating clinical translation. Oncotarget. 2011;2:518–24. doi: 10.18632/oncotarget.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nelson EA, Walker SR, Weisberg E, Bar-Natan M, Barrett R, Gashin LB, Terrell S, Klitgaard JL, Santo L, Addorio MR, Ebert BL, Griffin JD, Frank DA. The STAT5 inhibitor pimozide decreases survival of chronic myelogenous leukemia cells resistant to kinase inhibitors. Blood. 2011;117:3421–9. doi: 10.1182/blood-2009-11-255232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Takakura A, Nelson EA, Haque N, Humphreys BD, Zandi-Nejad K, Frank DA, Zhou J. Pyrimethamine inhibits adult polycystic kidney disease by modulating STAT signaling pathways. Hum Mol Genet. 20:4143–54. doi: 10.1093/hmg/ddr338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bar-Natan M, Nelson EA, Walker SR, Kuang Y, Distel RJ, Frank DA. Dual inhibition of Jak2 and STAT5 enhances killing of myeloproliferative neoplasia cells. Leukemia. 2012;26:1407–10. doi: 10.1038/leu.2011.338. [DOI] [PubMed] [Google Scholar]
- 65.Nelson EA, Walker SR, Xiang M, Weisberg E, Bar-Natan M, Barrett R, Liu S, Kharbanda S, Christie AL, Nicolais M, Griffin JD, Stone RM, Kung AL, Frank DA. The STAT5 Inhibitor Pimozide Displays Efficacy in Models of Acute Myelogenous Leukemia Driven by FLT3 Mutations. Genes Cancer. 2012;3:503–11. doi: 10.1177/1947601912466555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Walker SR, Chaudhury M, Frank DA. STAT3 Inhibition by Microtubule-Targeted Drugs: Dual Molecular Effects of Chemotherapeutic Agents. Mol Cell Pharmacol. 2011;3:13–19. [PMC free article] [PubMed] [Google Scholar]
- 67.Damiano JS, Rendahl KG, Karim C, Embry MG, Ghoddusi M, Holash J, Fanidi A, Abrams TJ, Abraham JA. Neutralization of Prolactin Receptor Function by Monoclonal Antibody LFA102, A Novel Potential Therapeutic for the Treatment of Breast Cancer. [DOI] [PubMed]
- 68.Clevenger CV, Zheng J, Jablonski EM, Galbaugh TL, Fang F. From bench to bedside: future potential for the translation of prolactin inhibitors as breast cancer therapeutics. J Mammary Gland Biol Neoplasia. 2008;13:147–56. doi: 10.1007/s10911-008-9074-8. [DOI] [PubMed] [Google Scholar]
- 69.Canbay E, Norman M, Kilic E, Goffin V, Zachary I. Prolactin stimulates the JAK2 and focal adhesion kinase pathways in human breast carcinoma T47-D cells. Biochem J. 1997;324(Pt 1):231–6. doi: 10.1042/bj3240231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chang HY, Nuyten DS, Sneddon JB, Hastie T, Tibshirani R, Sorlie T, Dai H, He YD, van't Veer LJ, Bartelink H, van de Rijn M, Brown PO, van de Vijver MJ. Robustness, scalability, and integration of a wound-response gene expression signature in predicting breast cancer survival. Proc Natl Acad Sci U S A. 2005;102:3738–43. doi: 10.1073/pnas.0409462102. [DOI] [PMC free article] [PubMed] [Google Scholar]