Abstract
In neurons, soluble N-ethylmaleimide–sensitive factor attachment receptor (SNARE) proteins drive the fusion of synaptic vesicles to the plasma membrane through the formation of a four-helix SNARE complex. Members of the Sec1/Munc18 protein family regulate membrane fusion through interactions with the syntaxin family of SNARE proteins. The neuronal protein Munc18a interacts with a closed conformation of the SNARE protein syntaxin1a (Syx1a) and with an assembled SNARE complex containing Syx1a in an open conformation. The N-peptide of Syx1a (amino acids 1–24) has been implicated in the transition of Munc18a-bound Syx1a to Munc18a-bound SNARE complex, but the underlying mechanism is not understood. Here we report the X-ray crystal structures of Munc18a bound to Syx1a with and without its native N-peptide (Syx1aΔN), along with small-angle X-ray scattering (SAXS) data for Munc18a bound to Syx1a, Syx1aΔN, and Syx1a L165A/E166A (LE), a mutation thought to render Syx1a in a constitutively open conformation. We show that all three complexes adopt the same global structure, in which Munc18a binds a closed conformation of Syx1a. We also identify a possible structural connection between the Syx1a N-peptide and SNARE domain that might be important for the transition of closed-to-open Syx1a in SNARE complex assembly. Although the role of the N-peptide in Munc18a-mediated SNARE complex assembly remains unclear, our results demonstrate that the N-peptide and LE mutation have no effect on the global conformation of the Munc18a–Syx1a complex.
Keywords: membrane trafficking, SM proteins, protein crystallography
Neurons communicate across specialized intercellular junctions called synapses. Arrival of an action potential in a presynaptic neuron triggers a calcium (Ca2+) influx, which leads to the exocytosis of presynaptic vesicles and release of neurotransmitters for uptake via receptors in the postsynaptic neuron. Soluble N-ethylmaleimide–sensitive factor attachment receptor (SNARE) proteins drive the fusion of neurotransmitter-containing vesicles to the presynaptic plasma membrane, a critical step in Ca2+-triggered exocytosis. Three neuronal SNARE proteins—syntaxin 1a (Syx1a) and synaptosomal-associated protein of 25 kDa (SNAP25), located on the synaptic membrane, and vesicle-associated membrane protein 2 (VAMP2, also called synaptobrevin) combine to form a four-helix bundle, a process that drives membrane fusion. Syx1a contains an N-terminal regulatory region in addition to its C-terminal SNARE domain and adopts two conformations: a closed conformation, in which a three-helix bundle (designated Habc) in the N-terminal region binds intramolecularly to the SNARE domain, and an open conformation in which the SNARE domain binds SNAP25 and VAMP2 to form the ternary SNARE complex.
The neuronal Sec1/Munc18a (SM)-like protein Munc18a is essential for synaptic vesicle fusion and associates with the neuronal SNARE proteins in at least two distinct modes. Munc18a interacts tightly with the closed conformation of Syx1a, inhibiting SNARE complex assembly, as well as with the assembled SNARE complex containing Syx1a in the open conformation. Kinetic data suggest that Munc18a remains bound to Syx1a during the closed-to-open transition (1). The crystal structure of the Munc18a–Syx1a complex has been solved (2), but structural details of the Munc18a–SNARE complex binding mode remain unknown. Munc18a domain 3a, which is adjacent to the Syx1a SNARE domain in the Munc18a–Syx1a complex, may be involved in formation of the ternary SNARE complex (3), but the mechanism by which the binary Munc18a–Syx1a complex assembles into the SNARE complex remains unclear.
The highly conserved Syx1a N-peptide (amino acids 1–24) is required for the interaction of Munc18a with the SNARE complex and for stimulation of SNARE-mediated liposome mixing by Munc18a (4–7). Intriguingly, removal of the Syx1a N-peptide (Syx1aΔN) facilitates SNARE complex formation in the presence of Munc18a (1). Understanding the molecular basis of the effects of the N-peptide has been confounded in part by the use of N-terminal affinity tags, the presence of which reduces binding at the Munc18a–Syx1a N-peptide interface (1–3).
The Syx1a mutant L165A/E166A (LE) also promotes SNARE assembly in the presence of Munc18a (1, 8). The LE mutation is located in the hinge region of Syx1a that connects the N-terminal three-helix bundle (Habc) and C-terminal SNARE domain. Affinity pull-down and NMR experiments indicated that the LE mutation abrogates binding of Munc18a and disrupts the closed conformation of Syx1a (9). However, more recent isothermal titration calorimetry (ITC) data revealed strong Munc18a–Syx1a LE binding (1).
To investigate the closed-to-open transition of Munc18a-bound Syx1a, we used X-ray crystallography and small-angle X-ray scattering (SAXS) to study complexes of Munc18a bound to the soluble cytoplasmic portion of Syx1a with its native N terminus (Syx1a WT), Syx1a lacking its N-peptide (Syx1aΔN), and Syx1a LE. We found that Munc18a binds the closed conformation of Syx1a in all three complexes. Even though the structures are very similar overall, subtle differences suggest a connection between the N-peptide and SNARE-binding sites that may be important in regulating SNARE complex formation from Munc18a-bound Syx1a.
Results
Syx1a N-Peptide Does Not Affect the Global Structure of Munc18a–Syx1a.
Based on the observation that Munc18a bound to Syx1a lacking its N-terminal peptide fails to inhibit SNARE complex assembly in solution (1), we hypothesized that the loss of Syx1a N-peptide binding induces a conformational change in the Munc18a–Syx1a complex. We solved the crystal structure of Munc18a bound to Syx1a lacking the N-peptide (amino acids 25–266; ΔN). Moreover, because the N-terminal His6 affinity-tagged Syx1a used in the original crystal structure of the complex (amino acids 1–267; designated TAG herein) binds more weakly to Munc18a compared with Syx1a with a native N terminus (1), we determined the structure of a Syx1a–Munc18a complex containing an untagged Syx1a (designated Syx1a WT) (Fig. 1 and Table 1). In this crystal structure, a truncated syntaxin (amino acids 1–243) was used to optimize crystallization, because residues 244–266 were disordered in the original Munc18a–Syx1a structure. We found the same overall structure in all three Munc18a–Syx1a complexes, in which Munc18a clasps the closed conformation of Syx1a (Fig. 1B). ITC data showed similar binding affinities in Munc18a–Syx1a TAG and Munc18a–Syx1aΔN (Kd = 10.0 ± 0.5 nM and 8.1 ± 1.0 nM, respectively), slightly weaker than the binding affinity of Munc18a–Syx1a WT (amino acids 1–262; Kd = 1.4 ± 0.3 nM) (1). The small contribution of the N-peptide to the free energy of this interaction is consistent with the structural similarities of these complexes.
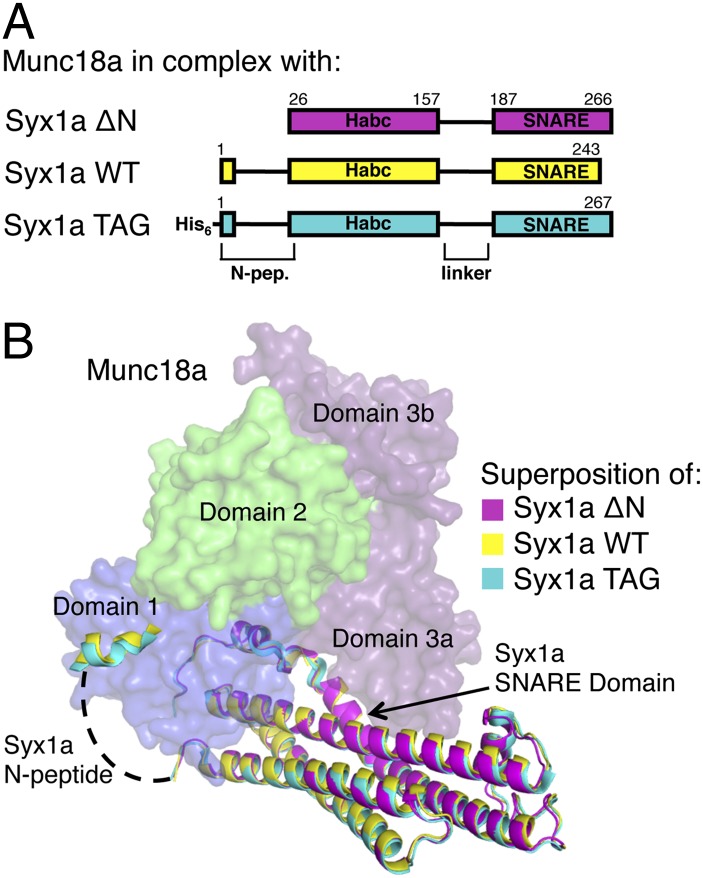
Fig. 1.
Crystal structures of Munc18a bound to Syx1a with and without its native N terminus. (A) Different Syx1a constructs present in three separate Munc18a–Syx1a crystal structures. (B) Superposition of all three Munc18a–Syx1a structures. Munc18a [colored surface representation of domains 1 (blue), 2 (green), and 3a and 3b (purple)] from one structure only is shown for clarity. Reported in this work are structures of Munc18a bound to Syx1a in the presence (WT, yellow) and absence (ΔN, violet) of its native N terminus; the re-refined structure (3C98) of Munc18a bound to Syx1a bearing an N-terminal His6-tag (TAG, cyan) was published previously (1). Syx1a N-peptide residues 10–26 were disordered (dashed line). Structural alignments yielded small rmsd values: WT and TAG (0.61 Å), WT and ΔN (0.61 Å), and TAG and ΔN (0.60 Å).
Table 1.
X-ray crystallography data collection and refinement statistics
| Munc18a–Syx1a (ΔN) | Munc18a–Syx1a (WT) | |
| Data collection | ||
| Space group | I422 | P4212 |
| Cell dimensions | ||
| a, b, c, Å | 154.9, 154.9, 150.3 | 156.8, 156.8, 78.7 |
| α, β, γ, ° | 90, 90, 90 | 90, 90, 90 |
| Resolution, Å | 32.64–2.50 (2.58–2.50)* | 110.9–3.20 (3.42–3.20) |
| Rmerge | 0.100 (0.646) | 0.109 (0.677) |
| CC1/2 | 0.996 (0.812) | 0.998 (0.823) |
| I/σI | 14.4 (1.9) | 14.8 (2.8) |
| Completeness, % | 99.8 (100) | 100 (100) |
| Multiplicity | 4.5 (4.6) | 7.1 (7.3) |
| Refinement | ||
| Resolution, Å | 32.64–2.50 | 110.9–3.20 |
| No. reflections work/test | 29,754/1996 | 15,399/1342 |
| Rwork/Rfree | 0.184/0.223 | 0.199/0.263 |
| Number of atoms | ||
| Protein | 6,248 | 6,303 |
| Solvent | 69 | – |
| Average B-factors | ||
| Wilson | 59.1 | 95.8 |
| Model atoms | 59.5 | 91.9 |
| rmsd | ||
| Bond lengths, Å | 0.010 | 0.010 |
| Bond angles, ° | 1.10 | 1.17 |
Values in parentheses are data for the highest-resolution shell. Munc18a–Syx1a (TAG) crystals belong to space group P4212, with unit cell dimensions a = b = 157.5 Å and c = 80.5 Å (2). Rmerge = Σh Σi|Ii(h) - <I(h)>|/Σh Σi I(h), where Ii(h) is the ith measurement of reflection h, and <I(h)> is the weighted mean of all measurements of h. <I/σ(I)> = mean intensity of reflections divided by standard deviation CC1/2 as defined in ref. 32. R and Rfree = Σh|Fobs(h) – Fcalc(h)|/Σh Fobs(h) for the working and test reflection sets, respectively.
Differences in Syx1a WT and Syx1aΔN Complexes with Munc18a.
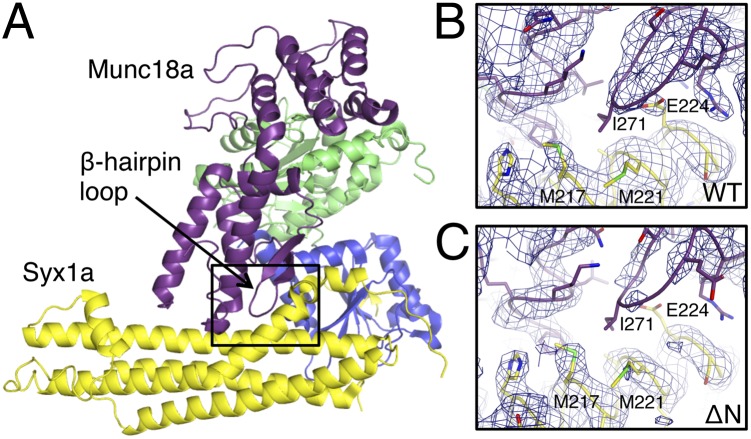
Comparison of the Munc18a–Syx1a structures with and without the Syx1a N-peptide revealed subtle differences in Munc18a. In the presence of the Syx1a N-peptide, an ordered β-hairpin loop (amino acids 269–274) in Munc18a domain 3a interacted with the Syx1a SNARE domain (Fig. 2 A and B). In contrast, in the Munc18a–Syx1aΔN complex, the electron density was very weak around the β-hairpin loop when compared at the same resolution and contour level (Fig. 2C), indicating that the Munc18a domain 3a loop is disordered in the absence of Syx1a N-peptide binding to Munc18a. The absence of electron density was confirmed by comparing real space correlation coefficients (RSCCs), which measure the similarities in electron density maps generated using experimental data or data calculated from the atomic model, of the hairpin loop residues in the Munc18a–Syx1aΔN complex with those of equivalent residues in the other Munc18a–Syx1a complexes. For the hairpin residues (amino acids 269–274), the average RSCC was 0.78 in the absence of Syx1a N-peptide and 0.92 in the presence of Syx1a N-peptide (Fig. S1). These observations suggest coupling between a loss of the N-peptide and disordering of the domain 3a hairpin resulting in exposure of the Syx1a SNARE domain.
Fig. 2.
Munc18a domain 3a β-hairpin loop. (A) A complex of Munc18a (blue, green, purple) and Syx1a (yellow) oriented to show the Munc18a β-hairpin loop and Syx1a SNARE domain. (B and C) Enlargements of the boxed regions showing the electron density (2Fo-Fc, contoured at 1 σ) for the complex of Munc18a and tag-free Syx1a (B) and Syx1aΔN (C). All electron density maps were generated with a 3.2-Å resolution cutoff.
Another difference between the complexes was seen in the interface between Munc18a domains 1 and 2. In the presence of the Syx1a N-peptide, a network of polar interactions that includes D108 and R171 connected the Syx1a N-peptide and SNARE domain binding sites (Fig. S2). In the absence of the N-peptide, D108 and R171 formed a salt bridge that severed this linked network (Fig. S2). We mutated Munc18a R171 as well as R39, which lies at the end of the network and binds to the Syx1a SNARE domain (Fig. S3). Munc18a R39C bound full-length Syx1a with a sixfold weaker affinity, consistent with previous work (10), and partially removed the block to SNARE assembly (Fig. S4 and Table S1). In contrast, Munc18a R171A did not affect Syx1a-binding energetics (Fig. S4 and Table S1). Overall, the data indicate that removal of direct contacts between Munc18a and Syx1a can override the block provided by the N-peptide, but the molecular nature of the coupling between the N-peptide and the SNARE binding site remains unclear.
Native Syx1a and Syx4 N-Peptide Interactions with Munc18a Are Similar.
The homologous and highly conserved Syx1a and Syx4 N-peptides bind Munc18a indiscriminately (3). However, comparison of the crystal structures of Munc18a bound to the N-terminally His6-tagged Syx1a or Syx4 N-peptide indicated that Syx4 N-peptide forms twice as many hydrogen bonds and Van der Waals interactions with Munc18a compared with Syx1a N-peptide bearing the His6 affinity tag (3). The crystal structure of Munc18a bound to Syx1a with a native N terminus (reported here) allows for a direct comparison to evaluate the effect of the Syx1a N-terminal affinity tag on Munc18a binding.
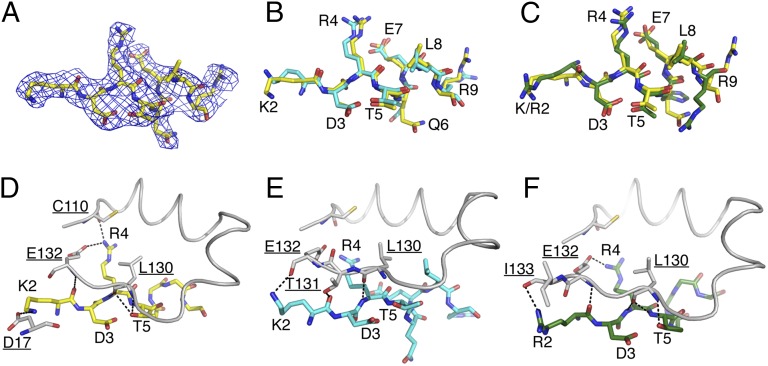
The structural differences observed between the native Syx1a N-peptide and either tagged Syx1a or Syx4 N-peptides bound to Munc18a were subtle (Fig. 3 B and C). Superposition of the N-peptides yielded rmsd values of 0.87 Å for Syx1a WT vs. Syx1a TAG and 1.2 Å for Syx1a WT vs. Syx4. The hydrogen bonding observed in the interaction of the Syx1a N-peptide with Munc18a was closer to that of the Syx4 peptide bound to Munc18a than to that of the N-terminally tagged Syx1a bound to Munc18a. Syx1a with a native N terminus and Syx4 each formed seven potential intermolecular hydrogen bonds with Munc18a, whereas the tagged Syx1a N-peptide formed only three hydrogen bonds with Munc18a (Fig. 3 D–F). These observations support the conclusion that the affinity tag partially disrupts the interaction between the Syx1a N-peptide and Munc18a, and provide a structural basis for the similar binding affinities of Munc18a with Syx1a and Syx4 N-peptides (1, 3).
Fig. 3.
Comparison of Syx1a N-peptides. (A) Electron density (2Fo-Fc, contoured at 1 σ) of native Syx1a N-peptide (amino acids 2–9). (B) Overlay of native Syx1a N-peptide (yellow) with tagged Syx1a N-peptide [cyan, from 3C98 (1)]; rmsd = 0.9 Å. (C) Overlay of native Syx1a N-peptide (yellow) with Syx4 [green, from 3PUJ (3)]; rmsd = 1.2 Å. (D–F) Hydrogen bonds (dashed lines) formed between residues of Munc18a (white) and native Syx1a N-peptide (yellow; D), tagged Syx1a N-peptide (cyan; E), or Syx4 N-peptide (green; F). Side chains for Syx residues 6–9 were removed for clarity; Munc18a residues are underlined.
Syx1a, Syx1aΔN, and Syx1a LE Bind Munc18a via the Same Mode.
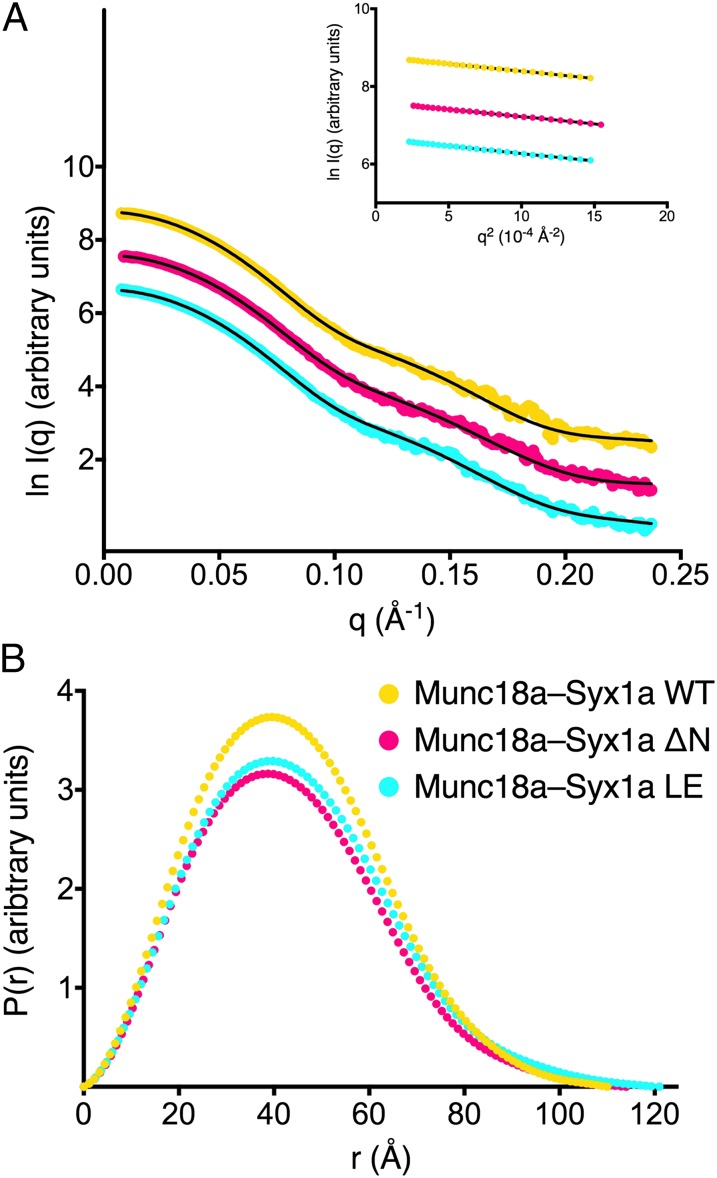
Binding of closed Syx1a to Munc18a, with or without the N-peptide, is the favored conformation observed in crystal structures. However, this does not preclude the possibility that the Syx1a N-peptide influences the Munc18a–Syx1a conformational equilibrium in solution, free of crystal lattice constraints. Thus, we measured SAXS data from Munc18a–Syx1a complexes in the presence and absence of Syx1a N-peptide (amino acids 1–267 or 25–267) to determine whether removal of the Syx1a N-peptide changes the Munc18a–Syx1a conformation in solution. Model-independent parameters, including the radius of gyration, maximum interatomic distance, and the pair-distance distribution function, demonstrated that the complexes were very similar in solution (Fig. 4 and Table 2).
Fig. 4.
Scattering curves and SAXS-derived parameters for Munc18a–Syx1a complexes. (A) SAXS data for Munc18a bound to Syx1a in the presence (WT, yellow) and absence (ΔN, violet) of its native N terminus, and for Munc18a bound to Syx1a L165A/E166A (LE, cyan), offset on the y- axis for clarity. Solid lines represent the scattering curves calculated from the Munc18a–Syx1a structure [fit to the experimental curves for WT (χ2 = 1.6) and LE (χ2 = 1.6)] and from the Munc18a–Syx1aΔN structure [fit to the ΔN experimental curve (χ2 = 1.5)]. Experimental curves were merged from scattering curves at two concentrations, 1 mg/mL and 4 mg/mL. (Inset) Guinier regions for each of the Munc18a–Syx1a complexes. (B) The pair distance distribution function, P(r), calculated using the indirect Fourier transform method in GNOM (31) from the scattering curves in A for each of the Munc18a–Syx1a complexes. P(r) plots are color-coded as in A.
Table 2.
Parameters for Munc18a–Syx1a solution structures
| Munc18a–Syx1a (ΔN) | Munc18a–Syx1a (WT) | Munc18a–Syx1a (LE) | |
| Dmax, Å | 114 | 110 | 121 |
| Rg, Å | |||
| Guinier | 33.3 | 33.6 | 34.4 |
| GNOM | 33.3 | 33.5 | 34.2 |
| Theoretical* | 32.8 | 33.3 | 33.3† |
Derived from CRYSOL (30).
Coordinates for the Munc18a–Syx1a WT structure were used to fit the Munc18a–Syx1a LE experimental data.
Scattering profiles calculated from the Munc18a–Syx1a crystal structures reported here agree well with the solution scattering data for Munc18a–Syx1a (χ2 = 1.6) and Munc18a–Syx1aΔN (χ2 = 1.5) (Fig. 4A and Table 3). The crystal structure data were measured from crystals grown at low pH (5.2–6.0), which were subsequently adapted to pH 7.5 in the case of the TAG crystals (2), whereas the samples used for SAXS were at pH 8.0, ruling out any effect of pH on conformation. We conclude that both Munc18a–Syx1a complexes adopt the same conformation.
Table 3.
Scattering curve fits for Munc18a–Syx1a complexes
| Solution complex | Crystal complex | χ2 |
| Munc18a–Syx1a (ΔN) | Munc18a–Syx1a (ΔN) | 1.5 |
| Munc18a–Syx1a (WT) | Munc18a–Syx1a (WT) | 1.6 |
| Munc18a–Syx1a (LE) | Munc18a–Syx1a (WT) | 1.6 |
Derived from CRYSOL (30).
The finding that Syx1a N-peptide binding has no effect on the global conformation of the Munc18a–Syx1a complex contradicts recently published solution scattering data showing that Syx1a with an intact N terminus forms a complex with Munc18a that differs considerably from the crystal structure (11). In both studies, the scattering experiments were performed using similar buffer conditions and protein concentrations. The most notable differences are the use and placement of protein-affinity tags. The previous study used C-terminally His6-tagged Syx1a and tag-free Munc18a, whereas we used N-terminally His6-tagged Munc18a and tag-free Syx1a in our SAXS experiments. To test whether the tag on Munc18a affected the results, we repeated the SAXS experiment with tag-free Munc18a–Syx1a, and observed no significant change in the structural parameters (Table S2).
Given that Munc18a–Syx1aΔN facilitates SNARE complex assembly (1), we also used SAXS to investigate the structural parameters of Syx1a (amino acids 1–267) L165A/E166A bound to Munc18a (Munc18a–Syx1a LE), a complex that is also compatible with SNARE assembly (1). The Syx1a LE mutant was initially characterized by its inability as a GST fusion protein to capture Munc18a from whole brain lysates (9). We repeated the GST-Syx1a pull-down experiment with recombinant purified Munc18a and observed comparable binding of Munc18a by GST-Syx1a WT and GST-Syx1a LE (Fig. S5). Published ITC data indicate a tight interaction between Munc18a and Syx1a LE (Kd = 7.7 ± 0.6 nM) (1), which we have reproduced here (Fig. S6 and Table S3).
The results of several previous studies support the idea that the Syx1a LE mutant favors an open conformation. NMR data indicate that the LE mutation disrupts intramolecular interactions between the Habc and SNARE domains of closed Syx1a (9), and another study showed that Syx1a LE is more susceptible to proteolysis (12). Direct structural data for the Munc18a–Syx1a LE complex have been lacking, however, and our data show that the LE mutant can adopt a closed conformation that tightly binds Munc18a. Indeed, the scattering profile calculated from the Munc18a–Syx1a WT crystal structure closely agreed with the Munc18a–Syx1a LE solution scattering data (χ2 = 1.6) (Fig. 4A and Table 3). Moreover, the largest interatomic distance Dmax and radius of gyration (Rg) values of all three Munc18a–Syx1a complexes were similar (Fig. 4A and Table 2). The slightly larger Rg and Dmax values for Munc18a–Syx1a LE (Table 2 and Fig. 4 A and B) might be related to a heterogeneous sample in which a small fraction was open Syx1a LE, or to a homogeneous sample with a distinct mode of binding in which the complex was slightly larger than the Munc18a–Syx1a WT complex. The similar thermodynamic properties for the interaction of Munc18a with WT Syx1a (Kd = 1.4 ± 0.3 nM, ΔH = –34.6 ± 0.2 kcal/mol) and Syx1a LE (Kd = 7.7 ± 0.6 nM, ΔH = –34.8 ± 0.2 kcal/mol) (1) favor the first possibility, although it is possible that the LE mutation had a subtle structural effect on the Munc18a–Syx1a complex. Note that although Rg values increased slightly as a function of sample concentration, suggesting a small degree of aggregation, this occurred in all three Syx1a variants (Table S4), ruling out aggregation as a source of the difference between LE and the other complexes. Taken together, the SAXS data indicate that the presumed “constitutively open” Syx1a LE mutation has little to no effect on the Munc18a–Syx1a binding mode.
Discussion
The role of the Syx1a N-peptide in Munc18a-mediated SNARE complex assembly remains unclear. The N-peptide is required for Munc18a stimulation of SNARE-mediated liposome mixing and for the interaction of Munc18a with the assembled SNARE complex (47). Syx1a knockdown experiments in cultured neurons have shown that the N-peptide is essential for synaptic vesicle fusion (13), although another study found that Munc18a bearing mutations at the Syx1a N-peptide binding interface, which impair SNARE complex binding, had no effect on synaptic transmission (14). Moreover, ITC data indicate that the Syx1a N-peptide contributes only minimally to the binding energetics of the Munc18a–Syx1a complex, and that the interactions of cognate and noncognate Munc18 and Syx N-peptide pairs are nonspecific (1, 3). Here we show that the N-peptide has no effect on the Munc18a–closed Syx1a binding mode. We also confirm previous observations that a Syx1a N-terminal affinity tag compromises Munc18a–Syx1a N-peptide binding (1, 3), and that the native Syx1a and Syx4 N-peptides bind Munc18a in a similar manner.
We hypothesized that disruption of Syx1a N-peptide binding shifts the Munc18a–Syx1a conformational equilibrium to favor a conformation that allows binding to other SNARE proteins. Given that the crystal structures of Munc18a–Syx1a in the presence and absence of the N-peptide adopt the same global structure, we considered the possibility that crystallization favors a single conformation. We examined the structural parameters of Munc18a–Syx1a in solution by SAXS, thereby eliminating possible constraints imposed by packing in a crystal lattice, and found that Munc18a was associated with a closed conformation of Syx1a, Syx1aΔN, and Syx1a LE. The structural data are consistent with previously reported ITC data, which show only small differences in the thermodynamics for the interactions of Munc18a with Syx1a, Syx1aΔN, and Syx1a LE (1). These data indicate that removing the Syx1a N-peptide or introducing the LE mutation has no effect on the global conformation of the Munc18a–Syx1a complex.
It is difficult to reconcile the structural data reported in this work with the finding that Munc18a bound to Syx1aΔN or Syx1aLE does not inhibit SNARE complex assembly. However, we did observe that the β-hairpin loop in Munc18a domain 3a became disordered in the absence of the Syx1a N-peptide, possibly exposing residues on the adjacent Syx1a SNARE domain. Domain 3a is disordered in the crystal structure of the unbound Munc18a homolog in squid, indicating that it is a conformationally dynamic region (15). These observations support the hypothesis that displacement of the Syx1a N-peptide leads to a conformational change in Munc18a domain 3a that promotes SNARE complex assembly. Also of interest is our finding that the unbound squid Munc18a showed a slight displacement of domain 1 with respect to domain 2. An electrostatic network that includes residues R171 and R39 runs through the domain 1–domain 2 interface and connects the N-peptide–binding and SNARE-binding sites of Munc18a (Fig. S3). Munc18a R39 directly contacts Syx1a, and the R39C mutation increases evoked transmission in neurons (16) and reduces the binding affinity of Munc18a for Syx1a (10).
We confirmed weaker binding and found reduced inhibition of SNARE assembly by Munc18a R39C (Fig. S4). On the other hand, we found only minimal effects of the R171A mutation (Table S1), suggesting that this residue might not contribute significantly to the energy of the interface, or that multiple pathways couple N-peptide binding to changes in the Syx1a-binding interface.
Other findings suggest allosteric coupling between the Munc18a N-peptide binding site and domain 3a in the Syx1a closed-to-open transition. Syx4 N-peptide binding induces an extended helix in domain 3a of both Munc18a and Munc18c structures that may facilitate SNARE complex formation (3), because the extended helix is incompatible with binding to the closed conformation of Syx1a. However, the Munc18a–Syx1a WT crystal structure is inconsistent with a recent model suggesting that a fully engaged (i.e., tag-free) Syx1a N-peptide leads to an extended helix in Munc18a domain 3a (amino acids 326–359) (3), given the lack of conformational difference in this region in the presence or absence of Syx1a N-peptide. Nonetheless, the subtle differences seen in our crystal structures are consistent with the possibility that Munc18a domain 3a plays a role in the Syx1a closed-to-open transition. Structural data for Munc18a bound to an assembled SNARE complex will be essential to our understanding of the role of the Munc18a–Syx1a complex in SNARE-mediated membrane fusion.
The structure of Syx1a WT bound to Munc18a indicates that the LE mutation disrupts several interactions between the two proteins, consistent with the notion that this mutant favors an open conformation (2, 9). However, SAXS data for the Munc18a–Syx1a LE complex show that Syx1a LE can adopt a closed conformation, which is consistent with the modest fivefold lower affinity of Munc18a for Syx1a LE relative to Syx1a WT (1). Although the LE mutation does not have a strong effect on the Munc18a–Syx1a binding mode, it may affect Munc18a–SNARE complex binding. In Caenorhabditis elegans, the Syx homolog Unc-64 bearing the LE mutation rescues neurotransmitter release in the absence of Unc-13 (Munc13 homolog) (8). Munc13 is essential for synaptic vesicle priming, and its MUN domain accelerates SNARE complex assembly in the presence of Munc18a (17). In Syx1a knockout mice, the isoform Syx1b LE shows reduced localization to the plasma membrane and increased synaptic vesicle fusion (18). It is possible that the LE mutation stabilizes the Munc18a–SNARE complex interaction in vivo, thus mimicking a likely key role of Munc13 (17). Another possibility is that the LE mutation alters the relative proportions of Syx1a associated with its SNARE partners and with Munc18a in the cell. Additional studies are needed to tease apart the binding and conformational dynamics of Syx1a in vivo.
Our structural data contradict recently published SAXS and small-angle neutron scattering data showing differences in the structural dimensions of Munc18a–Syx1a with or without the Syx1a N-peptide (11). It is possible the Syx1a C-terminal tag influences the Munc18a–Syx1a conformation in an N-peptide–dependent manner, but the mechanism by which this occurs is not obvious. NMR data indicate that Syx1a samples a closed conformation that is stabilized by Munc18a (19). In agreement with the NMR results, our X-ray crystal structures and SAXS data conclusively show that Munc18a binds a closed conformation of Syx1a in solution in both the presence and absence of the Syx1a N-peptide.
It is possible that the interaction of Munc18a with the closed conformation of Syx1a is the most energetically favorable binding mode in the absence of the Syx1a transmembrane domain and a membrane environment. Whereas Munc18a inhibits complex formation of soluble SNARE proteins (1), Munc18a facilitates SNARE assembly with Syx1a immobilized through its C terminus to affinity resin (3) and stimulates lipid mixing of liposomes containing full-length SNARE proteins (4–7, 20). Recent work has demonstrated that Munc18a also reduces lipid mixing of SNARE-embedded liposomes, in line with its inhibitory effect on SNARE assembly in solution (7). The limited degrees of freedom and topological orientation afforded by membrane anchors likely play important roles in Munc18a-mediated SNARE assembly; thus, anchoring Syx1a and/or the presence of a membrane could influence the Munc18a–Syx1a conformational equilibrium, which may favor a more SNARE-accessible conformation. Additional reconstitution and in vivo experiments are needed to elucidate the interplay of synaptic proteins in the vesicle fusion cycle.
Methods
Protein Expression and Purification.
All Munc18a–Syx1a complexes, with the exception of Munc18a–Syx1a (amino acids 25–266; ΔN) used for crystallization, were coexpressed from the pET-Duet vector (Novagen) as N- or C-terminally His6-tagged Munc18a and tag-less Syx1a. Syx1aΔN was cloned into the pTWIN1 vector (New England Biolabs) with an intein-mediated self-cleaving chitin-binding domain (CBD) C-terminal affinity tag, and Munc18a was cloned into a pQE9 (Qiagen) vector with an N-terminal His6 tag.
All recombinant proteins were expressed in Escherichia coli BL21 (DE3) RIL+ grown in LB medium at 37 °C to an A600 of 0.6–0.8, induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG), and then grown for another 4 h at 21–25 °C. Pellets were resuspended in lysis buffer [20 mM Na2HPO4/NaH2PO4 (pH 8.0), 300 mM NaCl, 5 mM imidazole) with protease inhibitor mixture set V (Calbiochem) and DNase I (Sigma-Aldrich). Cells were lysed using an EmulsiFlex homogenizer (Avestin) at a maximum pressure of 15,000 psi, after which lysates were centrifuged at 39,000 × g for 30 min at 4 °C to remove insoluble material. Clarified lysates were incubated with TALON metal affinity resin (Clontech), Ni2+-NTA agarose, or chitin-agarose beads and then washed with lysis buffer. Proteins were eluted with 200 mM imidazole. The Syx1aΔN CBD affinity tag was cleaved with 40 mM DTT at room temperature overnight, denatured in 8 M urea, and refolded into 20 mM Tris (pH 8.5), 500 mM NaCl, and 1 mM EDTA.
To generate a tag-free complex, Munc18a–Syx1a was incubated overnight at 4 °C with tobacco etch virus protease in lysis buffer with 10% glycerol to cleave the N-terminal His6 tag from Munc18a. All proteins were dialyzed against 20 mM Tris (pH 8.0), 75 mM NaCl, 1 mM EDTA, and 1 mM DTT for 2 h at 4 °C; loaded on HiTrapQ or MonoQ columns (GE Healthcare); and eluted with a linear NaCl gradient. For crystallization, Syx1aΔN was added at a slight molar excess to Munc18a, and the complex was purified on a preparative S200 gel filtration column (GE Healthcare) in 20 mM Tris (pH 8.0), 150 mM NaCl, 1 mM EDTA, and 1 mM DTT.
Crystallization, Data Collection, and Processing.
Crystals resembling thick plates were obtained with the hanging drop vapor diffusion method for Munc18a–Syx1aΔN and the sitting drop vapor diffusion method for Munc18a–Syx1a WT at 25 °C, using Munc18a–Syx1a concentrations of 145 μM and 116 μM, respectively. Well solutions contained 27% PEG 400, 10 mM EDTA, 10 mM DTT, 224 mM ammonium acetate, and 100 mM sodium acetate, at pH 6.0 for Munc18a–Syx1aΔN and pH 5.2 for Munc18a–Syx1a WT.
Diffraction data for each complex (Table 1) were measured from a single crystal at the Lawrence Berkeley National Laboratory Advanced Light Source beamline 8.2.1 for Munc18a–Syx1aΔN and at the Stanford Synchrotron Radiation Lightsource beamline 11-1 for Munc18a–Syx1a WT. For the Munc18a–Syx1aΔN complex, a total of 60° of data were collected in four segments, using 17.5- to 20-s exposure times and 0.7–1.0° rotations per frame, with a 300-mm crystal-to-detector distance on an ADSC Q315 detector. For the Munc18a–Syx1a WT complex, a total of 89° of data were collected in three segments, using a 15-s exposure time and 1.2° oscillations per frame, with a 450-mm crystal-to-detector distance on a Rayonix 325 detector. Diffraction data were processed and scaled using Mosflm and Scala (21, 22). There was one Munc18a–Syx1aΔN or Munc18a–Syx1a WT complex per asymmetric unit, with a solvent content of 47% or 51%, respectively.
Phasing, Model Building, and Refinement.
Phases for the Munc18a–Syx1aΔN structure were obtained by molecular replacement with Phaser (23), using the rerefined Munc18a–Syx1a structure [Protein Data Bank (PDB) ID code 3C98]. The z-scores for the rotation and translation functions were 27.4 and 73.5, respectively. Rigid-body refinement in Phenix (24) using data to 3.2 Å gave R and Rfree values of 36.8% and 36.5%, respectively. For the Munc18a–Syx1a WT structure, rigid-body refinement in Phenix using data from 3.2 Å yielded R and Rfree values of 42.8% and 42.5%, respectively. The models for each complex were built in Coot (25), and refinement was performed using Phenix (24) and BUSTER (26). For test sets, 6% of the reflections for the Munc18a–Syx1aΔN complex and 8% of the reflections for the Munc18a–Syx1a WT complex were removed before refinement. Bulk solvent and anisotropic temperature factor corrections were applied throughout the refinement. Final refinement statistics are presented in Table 1. Figures were generated using PyMOL version 1.3r1 (Schrodinger).
Structure Calculations (Hydrogen Bonds, rmsd and RSCC Values).
Because the Munc18a–Syx1a WT and TAG structures were solved at different resolutions (3.2 Å and 2.6 Å, respectively), the TAG structure was re-refined with a resolution cutoff of 3.2 Å, and the Munc18a–Syx1a N-peptide interface was compared. At a 3.2-Å cutoff for assigning hydrogen bonds, two of the original three intermolecular hydrogen bonds between tagged Syx1a and Munc18a were observed, confirming reduced hydrogen bonding in the presence of the affinity tag. H-bond geometry was confirmed using the DIMPLOT program in LIGPLOT (27). Syx1a and Syx4 N-peptides (amino acids 2–9) were aligned in PyMOL to calculate rmsd values for all matched atoms. Given the limited reliability of temperature factors refined at this resolution, RSCC values calculated in Phenix (28) (model vs. data tool with high resolution; 3.2 Å) were used to quantify differences in electron density.
SAXS.
SAXS data were measured at the Stanford Synchrotron Radiation Lightsource beamline 4-2 in the range of 0.00680 Å-1 ≤ q ≤ 0.534 Å-1, where q = 4πsin(θ)/λ, from solutions of Munc18a bound to Syx1a with (amino acids 1–267) and without (amino acids 25–267) its N-peptide, or to Syx1a LE (amino acids 1–267), at concentrations of 0.5–6 mg/mL in 20 mM Tris (pH 8.0), 150 mM NaCl, 1 mM EDTA, 1 mM DTT, and 2% glycerol. Monodispersity of the protein samples was confirmed by dynamic light scattering using a DynaPro molecular sizing instrument and Dynamics V6 software (Protein Solutions), and concentrations were determined by A280 measurements using a NanoDrop spectrophotometer (Thermo Scientific) and molar extinction coefficients calculated in ProtParam (29). All samples and buffers were passed through 0.2-μm spin filters (Corning) before being loaded into a 1.5-mm quartz capillary flow cell maintained at 20 °C, and 10 × 1 s exposures were measured for each concentration. Raw scattering data were normalized to the incident beam intensity and corrected for buffer scattering. Scattering curves corresponding to sample concentrations of 1 mg/mL and 4 mg/mL were scaled and merged. X-ray scattering profiles from the crystal structures reported herein were calculated and fit to solution scattering curves using CRYSOL (30) (Table 2). The pair distance distribution function, P(r), was calculated from the scattering curves using the indirect Fourier transform method in GNOM (31) (Table 3).
Supplementary Material
Acknowledgments
We thank Niket Shah for a critical reading of the manuscript. K.N.C. was supported by a fellowship from the National Science Foundation and a Biophysics Training Grant from the National Institutes of Health. This work was supported by National Institutes of Health Grant R01 MH58570. Portions of this research were carried out at the Stanford Synchrotron Radiation Lightsource, a directorate of the SLAC National Accelerator Laboratory and an Office of Science User Facility operated for the US Department of Energy Office of Science by Stanford University. The Stanford Synchrotron Radiation Lightsource Structural Molecular Biology Program is supported by the Department of Energy’s Office of Biological and Environmental Research and by the National Institutes of Health’s National Institute of General Medical Sciences (Grant P41GM103393) and National Center for Research Resources (Grant P41RR001209).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. J.R.-R. is a guest editor invited by the Editorial Board.
Data deposition: Atomic coordinates and structure factors for the Munc18a–Syx1a WT and Munc18a–Syx1aΔN complexes have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 4JEU and 4JEH, respectively).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1303753110/-/DCSupplemental.
References
- 1.Burkhardt P, Hattendorf DA, Weis WI, Fasshauer D. Munc18a controls SNARE assembly through its interaction with the syntaxin N-peptide. EMBO J. 2008;27(7):923–933. doi: 10.1038/emboj.2008.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Misura KM, Scheller RH, Weis WI. Three-dimensional structure of the neuronal-Sec1-syntaxin 1a complex. Nature. 2000;404(6776):355–362. doi: 10.1038/35006120. [DOI] [PubMed] [Google Scholar]
- 3.Hu S-H, et al. Possible roles for Munc18-1 domain 3a and syntaxin1 N-peptide and C-terminal anchor in SNARE complex formation. Proc Natl Acad Sci USA. 2011;108(3):1040–1045. doi: 10.1073/pnas.0914906108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shen J, Tareste DC, Paumet F, Rothman JE, Melia TJ. Selective activation of cognate SNAREpins by Sec1/Munc18 proteins. Cell. 2007;128(1):183–195. doi: 10.1016/j.cell.2006.12.016. [DOI] [PubMed] [Google Scholar]
- 5.Shen J, Rathore SS, Khandan L, Rothman JE. SNARE bundle and syntaxin N-peptide constitute a minimal complement for Munc18-1 activation of membrane fusion. J Cell Biol. 2010;190(1):55–63. doi: 10.1083/jcb.201003148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rathore SS, et al. Syntaxin N-terminal peptide motif is an initiation factor for the assembly of the SNARE-Sec1/Munc18 membrane fusion complex. Proc Natl Acad Sci USA. 2010;107(52):22399–22406. doi: 10.1073/pnas.1012997108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schollmeier Y, Krause JM, Kreye S, Malsam J, Söllner TH. Resolving the function of distinct Munc18-1/SNARE protein interaction modes in a reconstituted membrane fusion assay. J Biol Chem. 2011;286(35):30582–30590. doi: 10.1074/jbc.M111.269886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Richmond JE, Weimer RM, Jorgensen EM. An open form of syntaxin bypasses the requirement for UNC-13 in vesicle priming. Nature. 2001;412(6844):338–341. doi: 10.1038/35085583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dulubova I, et al. A conformational switch in syntaxin during exocytosis: Role of munc18. EMBO J. 1999;18(16):4372–4382. doi: 10.1093/emboj/18.16.4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fisher RJ, Pevsner J, Burgoyne RD. Control of fusion pore dynamics during exocytosis by Munc18. Science. 2001;291(5505):875–878. doi: 10.1126/science.291.5505.875. [DOI] [PubMed] [Google Scholar]
- 11.Christie MP, et al. Low-resolution solution structures of Munc18:Syntaxin protein complexes indicate an open binding mode driven by the syntaxin N-peptide. Proc Natl Acad Sci USA. 2012;109(25):9816–9821. doi: 10.1073/pnas.1116975109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Graham ME, Barclay JW, Burgoyne RD. Syntaxin/Munc18 interactions in the late events during vesicle fusion and release in exocytosis. J Biol Chem. 2004;279(31):32751–32760. doi: 10.1074/jbc.M400827200. [DOI] [PubMed] [Google Scholar]
- 13.Zhou P, et al. Syntaxin-1 N-peptide and Habc-domain perform distinct essential functions in synaptic vesicle fusion. EMBO J. 2013;32(1):159–171. doi: 10.1038/emboj.2012.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meijer M, et al. Munc18-1 mutations that strongly impair SNARE-complex binding support normal synaptic transmission. EMBO J. 2012;31(9):2156–2168. doi: 10.1038/emboj.2012.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bracher A, Perrakis A, Dresbach T, Betz H, Weissenhorn W. The X-ray crystal structure of neuronal Sec1 from squid sheds new light on the role of this protein in exocytosis. Structure. 2000;8(7):685–694. doi: 10.1016/s0969-2126(00)00156-8. [DOI] [PubMed] [Google Scholar]
- 16.Wu MN, Littleton JT, Bhat MA, Prokop A, Bellen HJ. ROP, the Drosophila Sec1 homolog, interacts with syntaxin and regulates neurotransmitter release in a dosage-dependent manner. EMBO J. 1998;17(1):127–139. doi: 10.1093/emboj/17.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ma C, Li W, Xu Y, Rizo J. Munc13 mediates the transition from the closed syntaxin-Munc18 complex to the SNARE complex. Nat Struct Mol Biol. 2011;18(5):542–549. doi: 10.1038/nsmb.2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gerber SH, et al. Conformational switch of syntaxin-1 controls synaptic vesicle fusion. Science. 2008;321(5895):1507–1510. doi: 10.1126/science.1163174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen X, Lu J, Dulubova I, Rizo J. NMR analysis of the closed conformation of syntaxin-1. J Biomol NMR. 2008;41(1):43–54. doi: 10.1007/s10858-008-9239-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tareste DC, Shen J, Melia TJ, Rothman JE. SNAREpin/Munc18 promotes adhesion and fusion of large vesicles to giant membranes. Proc Natl Acad Sci USA. 2008;105(7):2380–2385. doi: 10.1073/pnas.0712125105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Collaborative Computational Project, Number 4 The CCP4 suite: Programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50(Pt 5):760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 22. Leslie AGW, Powell HR (2007) Processing diffraction data with MOSFLM. Evolving Methods for Macromolecular Crystallography: The Structural Path to the Understanding of the Mechanism of Action of CBRN Agents, NATO Science Series, eds Reed RJ, Sussman JL (Springer, New York), Vol 245, pp 41–51.
- 23.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Adams PD, et al. PHENIX: Building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58(Pt 11):1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 25.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 26.Blanc E, et al. Refinement of severely incomplete structures with maximum likelihood in BUSTER-TNT. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2210–2221. doi: 10.1107/S0907444904016427. [DOI] [PubMed] [Google Scholar]
- 27.Wallace AC, Laskowski RA, Thornton JM. LIGPLOT: A program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 1995;8(2):127–134. doi: 10.1093/protein/8.2.127. [DOI] [PubMed] [Google Scholar]
- 28.Afonine PV, et al. phenix.model_vs_data: A high-level tool for the calculation of crystallographic model and data statistics. J Appl Cryst. 2010;43(Pt 4):669–676. doi: 10.1107/S0021889810015608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gasteiger E, et al. Protein identification and analysis tools on the ExPASy server. In: Walker JM, editor. The Proteomics Handbook. Clifton, NJ: Humana Press; 2005. pp. 571–607. [DOI] [PubMed] [Google Scholar]
- 30.Svergun D, Barberato C, Koch MHJ. CRYSOL: A program to evaluate X-ray solution scattering of biological macromolecules from atomic coordinates. J Appl Cryst. 1995;28:768–773. [Google Scholar]
- 31.Semenyuk AV, Svergun DI. GNOM: A program package for small-angle scattering data processing. J Appl Cryst. 1991;24:537–540. [Google Scholar]
- 32.Evans P. Scaling and assessment of data quality. Acta Crystallogr D Biol Crystallogr. 2006;62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.