Abstract
CarD, an essential transcription regulator in Mycobacterium tuberculosis, directly interacts with the RNA polymerase (RNAP). We used a combination of in vivo and in vitro approaches to establish that CarD is a global regulator that stimulates the formation of RNAP-holoenzyme open promoter (RPo) complexes. We determined the X-ray crystal structure of Thermus thermophilus CarD, allowing us to generate a structural model of the CarD/RPo complex. On the basis of our structural and functional analyses, we propose that CarD functions by forming protein/protein and protein/DNA interactions that bridge the RNAP to the promoter DNA. CarD appears poised to interact with a DNA structure uniquely presented by the RPo: the splayed minor groove at the double-stranded/single-stranded DNA junction at the upstream edge of the transcription bubble. Thus, CarD uses an unusual mechanism for regulating transcription, sensing the DNA conformation where transcription bubble formation initiates.
Keywords: mycobacteria, ribosomal RNA (rRNA), transcription activator, initiation
At least 30% of the world’s population is infected with latent Mycobacterium tuberculosis, which in some individuals will reactivate and cause an estimated 1.4 million deaths a year [World Health Organization (WHO) Global Tuberculosis Report 2012, www.who.int/tb/publications/global_report/en/index.html]. Significant obstacles in controlling the epidemic result from the chronic nature of M. tuberculosis infection, which necessitates prolonged treatment and generates a large reservoir of latently infected people. This health crisis is exacerbated by the alarming emergence of drug-resistant strains. The inadequacies of present tuberculosis therapies demand the discovery of new agents to treat M. tuberculosis infection, which requires insight into the pathways used by the pathogen to survive in the host.
During earlier investigations aimed at better understanding mycobacterial stress responses, we identified CarD as an essential, highly expressed protein that is also transcriptionally induced by multiple types of stress (1). Transient depletion of CarD revealed that mycobacteria lacking CarD are sensitive to killing by reactive oxygen species, ciprofloxacin, and starvation and are unable to replicate and persist in mice. Despite the importance of CarD in mycobacteria, the function of this protein and its mechanism of action are still unknown.
The N-terminal 64 residues of CarD contain sequence similarity to the RNA polymerase-interacting domain (RID) of transcription-repair coupling factor (TRCF) (2) and, like the TRCF-RID, the CarD-RID makes a direct protein/protein interaction with the RNA polymerase (RNAP) β-subunit on the β1-lobe (1, 3, 4). In other work, Garcia-Moreno et al. have shown that a Myxococcus xanthus CarD homolog, called CdnL, also interacts with this region of the RNAP and is essential for viability (5). Furthermore, depletion of Mycobacterium smegmatis CarD affects the mRNA levels of hundreds of genes, suggesting that CarD is as an essential global regulator of transcription in mycobacteria (1). We recently isolated CarD mutants (CarDR25E and CarDR47E) with weakened interactions with the RNAP β1 domain and showed that this interaction is required for M. tuberculosis viability, stress resistance, rifampicin resistance, and persistent infection in mice (4). Therefore, essential functions of CarD are mediated through its association with the RNAP. The CarD C-terminal 98 residues are also critical for M. tuberculosis viability (1), but the structure and function of this region are unknown. Thus, CarD, which is widely distributed among bacteria (Fig. S1) (1), represents a distinct class of RNAP binding proteins that regulate transcription and essential processes in the bacterium. Here we used a combination of in vitro structural and biochemical approaches, as well as in vivo approaches, to uncover the unique mechanistic basis of CarD function.
Results
CarD Regulates Transcription Initiation.
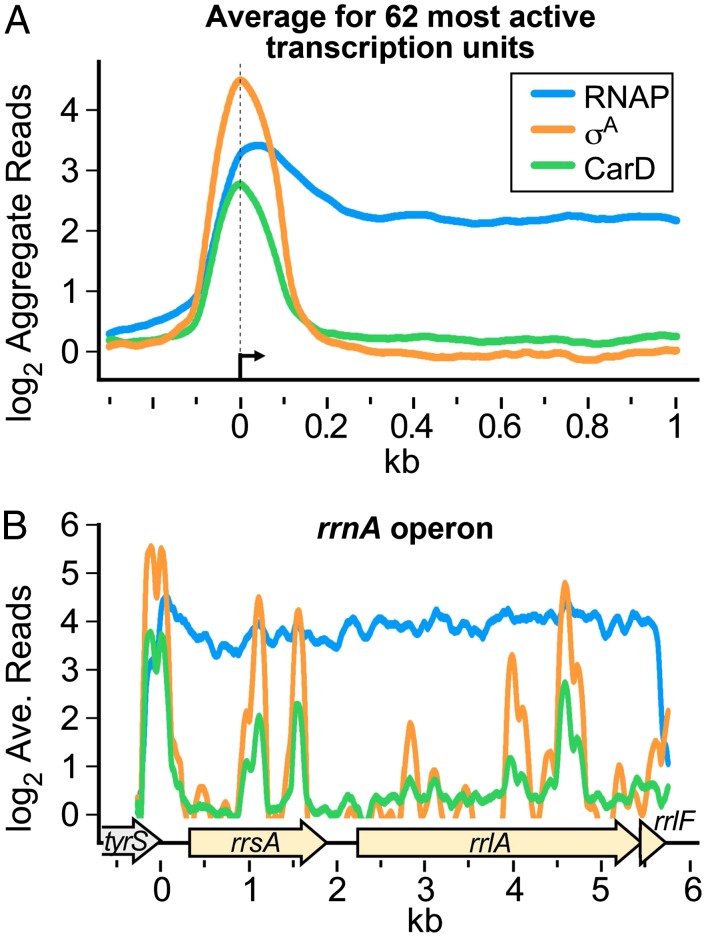
CarD modulates transcription through its direct interaction with RNAP (1, 4). To determine in which stage of the transcription cycle (initiation, elongation, or termination) CarD acts, we used chromatin immunoprecipitation sequencing (ChIP-seq) (6) to survey the distribution of CarD throughout the M. smegmatis chromosome. Specific antibodies targeting core RNAP, σA, or a hemagglutinin (HA) epitope fused to CarD (CarD-HA) were used to coimmunoprecipitate associated DNA that was then sequenced. We found that CarD was never present on the genome in the absence of RNAP. However, whereas RNAP core enzyme was found throughout transcribed regions of the genome, CarD was primarily associated with promoter regions and highly correlated with σA (Fig. 1A and Table S1). For example, at the rrnA operon, CarD and σA colocalized at the promoter region (Fig. 1B). Compilation of the ChIP-seq data and previous microarray expression profiling analyses (1) indicated that CarD was broadly distributed on promoters of most transcription units regardless of whether they were deregulated during CarD depletion. The colocalization of σA and CarD lead us to propose that in vivo, CarD associates with RNAP initiation complexes at most promoters and is therefore a global regulator of transcription initiation.
Fig. 1.
Normalized log2 of ChIP-seq reads from M. smegmatis DNA coimmunoprecipitated with RNAP β, σA, or CarD-HA. (A) Aggregate profiles averaged over 62 highly active transcription units. Protein–DNA complexes containing CarD-HA, RNAP β, and RNAP σ were immunoprecipitated from M. smegmatis lysates. The coprecipitated DNA was sequenced, and the number of sequence reads per base pair was normalized to total reads per sample and expressed as a log2 value. Normalized reads per base pair from DNA precipitated from cells expressing only the HA epitope were used as background and subtracted from the other samples. The 62 transcription units were selected on the basis of high signal and isolation from surrounding transcription units. (B) Profiles for the rrnA operon (complement of M. smegmatis 3819731–3825749 with 3825475 set as 0). ORFs are denoted by arrows underneath. Traces are colored as in A.
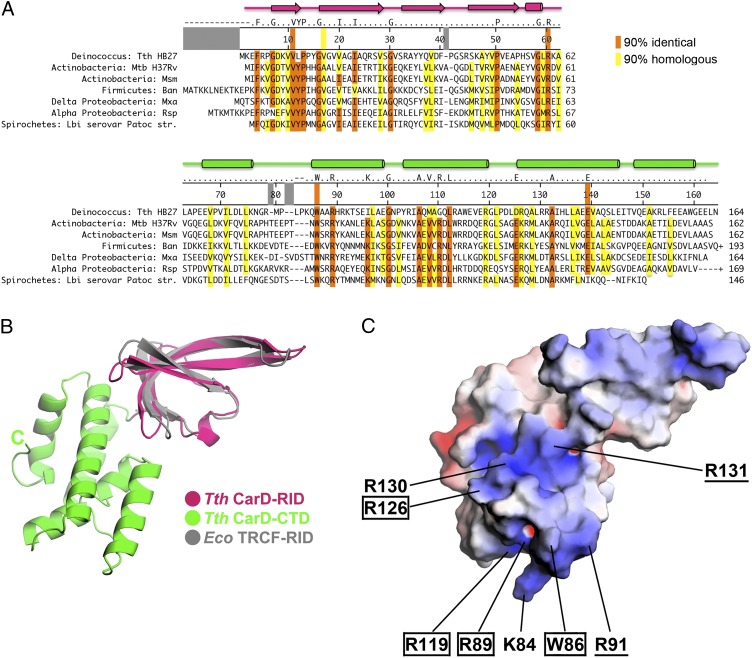
The X-Ray Crystal Structure of CarD Reveals a Surface-Exposed Tryptophan Residue with a Surrounding Basic Patch.
To provide a structural framework to understand the molecular mechanism by which CarD modulates RNAP function, we set out to determine the X-ray crystal structure of CarD. We were unable to obtain suitable crystals of M. tuberculosis CarD, but we determined the structure of CarD from Thermus thermophilus to 2.4 Å resolution (Table S2 and Fig. S2 A and B). The CarD proteins from T. thermophilus and Mycobacterium sp. are highly similar in sequence (44% homologous, 25% identical; Fig. 2A), so we proceeded to characterize CarD from T. thermophilus and M. tuberculosis in parallel. The T. thermophilus CarD structure, which comprises two domains (Fig. 2B and Fig. S2C), confirms the Tudor-like fold of the N-terminal domain (Fig. 2B, T. thermophilus CarD-RID, magenta), in common with the Escherichia coli TRCF-RID (Fig. 2B, gray) (2, 3, 7). The CarD C-terminal domain (CarD-CTD) is a compact, all α-helical fold with no apparent structural similarity to any previously described fold (Fig. 2B, green).
Fig. 2.
Primary and 3D structural features of T. thermophilus CarD. (A) Representative CarD sequences from an alignment of 452 sequences. At least one sequence from each major bacterial group containing CarD is shown (Fig. S1). The numbering along the top of the sequences denotes T. thermophilus CarD. Positions that are identical or homologous in ≥90% of the sequences are shaded orange or yellow, respectively. Groups of residues considered homologous were (DE), (HKR), (AILVM), (NQ), (FWY), (ST), (P), (C), and (G). The orange or yellow stripes that extend up into the numbering bar denote residues discussed in the text. The T. thermophilus CarD secondary structure elements are denoted schematically at the top (β-strands shown as arrows and α-helices as cylinders), with the N-terminal RID colored magenta and the CTD colored green (see B). (B) Crystal structure of T. thermophilus CarD, shown in ribbon format. The N-terminal CarD-RID (magenta) is superimposed with the E. coli TRCF-RID (gray), illustrating the structural conservation. (C) Crystal structure of T. thermophilus CarD (same view as in B), shown as a molecular surface and colored according to the electrostatic surface potential [red, −5 kT; white, neutral; blue, +5 kT; where k is the Boltzmann constant, T is temperature (22)]. Surface-exposed W86 is denoted, along with basic (K or R) residues contributing to the surrounding basic patch. Residues conserved in ≥90% of sequences from the full alignment are boxed; residues conserved between T. thermophilus and Mycobacterium sp. are underlined.
T. thermophilus CarD crystallized with two molecules in the asymmetric unit, so two crystallographically independent structures were refined (Table S2). Despite having unique crystal packing environments, the two molecules are nearly identical in structure (rmsd of 0.965 Å over 158 α-carbon positions), indicating that the relative orientation of the two CarD structural domains (CarD-RID and CarD-CTD, Fig. 2B) is rigidly maintained (Fig. S3A). This is likely due to a small but significant interface between the two domains (∼810 Å2 buried accessible surface area) that includes a network of conserved interactions (Fig. 2A and Fig. S3B). Two conserved charged residues (CarD-RID-R60 and CarD-CTD-E139) form a partially buried, interdomain salt bridge. CarD-RID-R60 is also in position to hydrogen bond with the carbonyl-oxygen of conserved CarD-CTD-G100. Conserved hydrophobic residues V11 and V17 make van der Waals interactions with the alkyl chains of R60 and E139 as well as with the side chain of CarD-CTD residue 142 (Q in T. thermophilus CarD, but conserved as a hydrophobic residue in the larger family of CarD proteins) (Fig. 2A).
A notable feature of the CarD-CTD is a nearly universally conserved tryptophan (T. thermophilus CarD-W86) with a largely solvent exposed side chain (Fig. 2). The tryptophan is located at the end of the CarD-CTD distal to the CarD-RID and is surrounded by a basic electrostatic surface formed from a cluster of basic residues. Within this basic surface patch, R89, R119, and R126 are universally conserved; R91 and R131 are conserved between T. thermophilus and Mycobacterium sp.; whereas K84 and R130 are found in T. thermophilus CarD but not Mycobacterium sp. CarD (Fig. 2 A and C and Fig. S4). Analysis of M. smegmatis and M. tuberculosis CarD homology models revealed that the surface-exposed tryptophan and surrounding basic surface patch are conserved features (Fig. S4).
Structural Modeling Points to a Role for W86 in CarD/Promoter DNA Contacts.
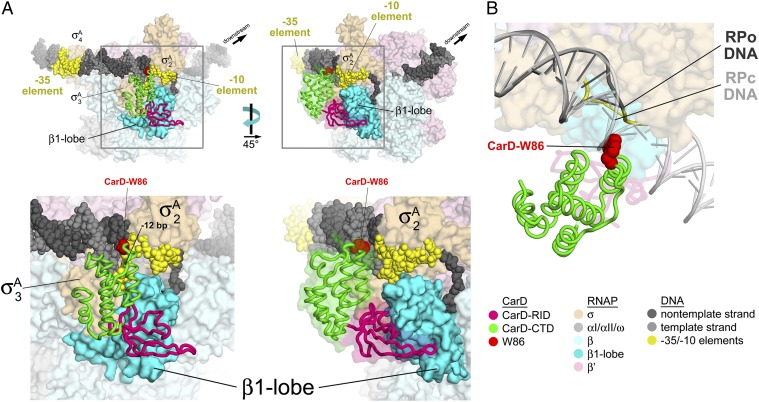
To elucidate the possible roles for W86 and the surrounding basic surface patch (Fig. 2C and Fig. S4) in the direct modulation of transcription initiation by CarD (Fig. 1), we generated a structural model of the complex between CarD and the RNAP open promoter (RPo) complex. The CarD-RID shares sequence and structural homology with the TRCF-RID (Fig. 2B) (3, 7), as well as a common binding mode to the RNAP β1-lobe (4). Therefore, the crystal structure of the Thermus TRCF-RID/RNAP β1-lobe complex [Protein Data Bank (PDB) ID 3MLQ] (3) provided a starting point to generate a model of the CarD/β1-lobe complex by superimposition of the corresponding RID domains (Fig. S5A). The resulting CarD/β1-lobe model was then placed into the context of a model of RPo (8, 9) by superimposition of the corresponding β1-lobe domains (Fig. S5B). Additional reorientation of structural elements was not required as there were no significant steric clashes in the resulting CarD/RPo model (Fig. 3).
Fig. 3.
Structural model of the Thermus CarD/RPo complex. The model was generated as described in Fig. S5. The color coding of the structural elements is denoted in the key at Lower Right. (A) Two views of the Thermus CarD/RPo model are shown. The RNAP holoenzyme (EσA) is shown as a molecular surface. The σA structural domains (σA2, σA3, σA4) (23) are labeled. The DNA is shown as CPK atoms. CarD is shown as an α-carbon worm, except the side chain of W86 is shown as CPK atoms (colored red). (Right) A transparent molecular surface of CarD is also shown (omitted from the Left view for clarity). (Lower) The boxed regions of the overall views (Upper) are magnified. (B) Magnified view showing the close approach of the CarD-CTD (and W86) to the upstream fork junction of the transcription bubble in RPo (dark gray) and compared with a model for the RNAP closed promoter complex (RPc, light gray) (8). The DNAs are shown as phosphate-backbone worms. The RPc DNA sterically clashes with the CarD-CTD around W86.
In the CarD/RPo model, the interaction of the CarD-RID with the RNAP β1-lobe places the CarD-CTD in a position to interact directly with the promoter DNA at the upstream edge of the −10 element on the opposite face of the DNA as σ (Fig. 3). Consistent with a role for the CarD-CTD in DNA interaction, CarD alone, at high concentration, is capable of non-sequence-specific protein/DNA interactions (Fig. S6A), and these interactions are mediated by the CarD-CTD (Fig. S6B). The CarD-CTD/DNA interactions are centered on W86 and the surrounding basic patch, both conserved structural features of CarD (Fig. 2A and Fig. S4). The upstream edge of the transcription bubble at most promoters (and in our model) is at position −11 (with respect to the most common transcription start site) (10), which is strand separated, whereas position −12 is base paired (11). CarD potentially interacts with the DNA from about −11 to −15. The minor groove of the DNA at the upstream edge of the transcription bubble, which faces away from σ, is highly distorted (widened) due to the strand separation beginning at −11 and extending downstream (Fig. 3). As a consequence, the basic surface of the CarD-CTD closely approaches the DNA from the widened minor groove, with CarD-W86 most proximal to the DNA, roughly aligned with the −12 base pair (Fig. 3). This interaction mode appears to be uniquely possible in the widened minor groove at the upstream fork of the transcription bubble; B-form DNA of an RNAP closed promoter complex (RPc) model (8) severely clashes with W86 and other elements of the CarD-CTD (Fig. 3B).
CarD Functions as a Transcription Activator.
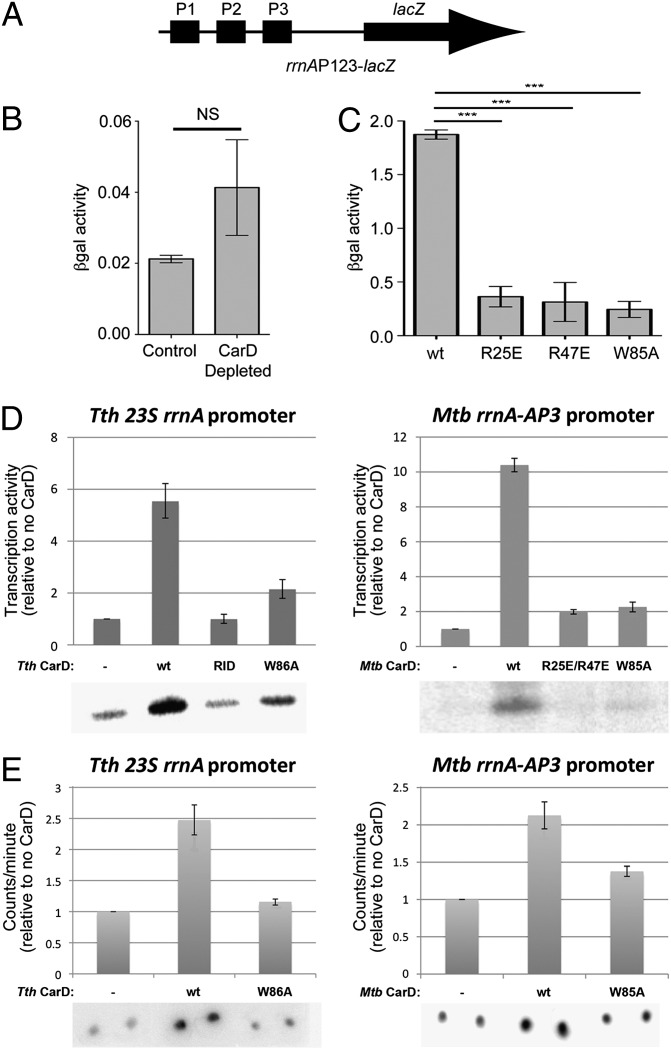
CarD was proposed to repress M. smegmatis rRNA transcription, using a mechanism functionally similar to but structurally distinct from DksA repression of E. coli rRNA transcription, in part because depletion of CarD led to a twofold increase in M. smegmatis 16S rRNA abundance and CarD could complement the elevated rRNA levels of an E. coli dksA mutant (1). To directly test the effect of CarD on rRNA promoter activity in vivo, we measured β-galactosidase (βgal) activity from lacZ fused to the M. smegmatis rrnA control region [comprising promoters rrnA-P1, -P2, and -P3 (rrnAP123)] (Fig. 4A) and transformed into M. smegmatis. Depletion of CarD resulted in increased βgal activity (Fig. 4B), comparable to the increase in 16S rRNA amount observed previously during CarD depletion (1). However, weakening the CarD/RNAP protein/protein interaction (using M. smegmatis strains expressing CarDR25E or CarDR47E mutants) (4) resulted in decreased βgal activity (Fig. 4C). Consistent with the decreased rrnA promoter activity, the 16S rRNA levels in the M. smegmatis-CarDR25E strain were found to be lower than in the presence of CarDwt (Fig. S6C), despite similar levels of CarD protein expression in the strains (Fig. S6D). The findings that weakening the CarD/RNAP protein/protein interaction decreases promoter activity (Fig. 4C) and leads to lower 16S rRNA levels (Fig. S6C) suggest that CarD may activate M. smegmatis rRNA transcription initiation, in contrast to the proposal that CarD acts as a repressor of rRNA transcription. It is possible that depletion of CarD, a global regulator that is essential for M. smegmatis viability (1) and present at most M. smegmatis promoters (Fig. 1), might give rise to pleiotropic effects that indirectly increase M. smegmatis rRNA promoter activity.
Fig. 4.
CarD activates transcription initiation at rRNA promoters. (A) Schematic illustrating the M. smegmatis rrnAP123-lacZ cassette used for B and C. (B and C) Normalized βgal units of activity in M. smegmatis containing rrnAP123-lacZ (A). The mean of four to five replicates is graphed and the error bars represent the SEM. NS, not significant; ***P value ≤ 0.005 in t tests. (B) Depletion of CarD (1) resulted in increased (approximately twofold) βgal activity. (C) Disruption of the CarD/RNAP protein/protein interaction (R25E or R47E substitutions) (4), as well as the W85A substitution, resulted in decreased (more than fourfold in each case) βgal activity. (D) CarD stimulates transcription in vitro (Left, Thermus transcription system; Right, Mycobacterial transcription system). In each panel, transcription activity (determined by phosphorimagery of 32P-labeled transcripts from denaturing polyacrylamide gels such as those shown below), normalized to no CarD (−), is shown as a histogram. “RID” denotes CarD-RID only (T. thermophilus CarD residues 1–64). The error bars represent the SEM of three replicates. (E) CarD stimulates RPo formation, as determined using filter binding (Left, Thermus transcription system; Right, Mycobacterial transcription system). RNAP and 32P-labeled promoter DNA were incubated to form RPo, challenged with a large excess of unlabeled competitor DNA, and then washed through a nitrocellulose filter to remove unbound DNAs. Bound DNA was quantitated by phosphorimagery. In each panel, binding activity, normalized to no CarD (−), is shown as a histogram. The error bars represent the SEM of four replicates.
To better understand the direct effect of CarD on rRNA promoters, we chose to test the effect of CarD on in vitro transcription, using T. thermophilus and mycobacterial transcription systems. In both systems, CarD robustly activated transcription from a corresponding rRNA promoter (Fig. 4D). The activation occurred through stimulation of promoter binding (Fig. 4E) and required the CarD/RNAP interaction as well as the CarD-CTD (Fig. 4D). Thus, in contrast to E. coli DksA, which represses rRNA promoter activity in vitro and in vivo (12), CarD is an activator of rRNA promoters.
CarD Function Depends on the Conserved, Solvent-Exposed Tryptophan.
Due to the prominent position of W86 at the CarD/DNA interface (Fig. 3), we examined the effect of an Ala substitution of this conserved residue (Fig. 2A). M. tuberculosis CarDW85A retained its ability to bind RNAP in vivo (Fig. S6E), but both T. thermophilus CarDW86A and M. tuberculosis CarDW85A were defective in stimulating in vitro transcription (activating transcription only about 2-fold compared with 5- to 10-fold activation for CarDwt; Fig. 4D) and promoter binding (stimulating promoter binding 1.1- to 1.3-fold rather than 2.7- to 3-fold for CarDwt; Fig. 4E). M. smegmatis CarDW85A was also inactive in in vivo assays (Fig. 4C and Fig. S6C).
Discussion
In summary, we have shown that in vitro CarD activates transcription initiation (Fig. 4D) by stimulating the formation of RPo (Fig. 4E), consistent with its in vivo colocalization with the RNAP-σA-holoenzyme (EσA) at promoter regions (Fig. 1). The CarD structure comprises two domains, an N-terminal RID domain that makes a protein/protein interaction with the RNAP β1-lobe in the same manner as the TRCF-RID (3, 4) and a CTD that harbors two conserved structural features, a solvent-exposed tryptophan residue (T. thermophilus CarDW86/M. tuberculosis CarDW85) and a surrounding basic patch (Fig. 2 and Fig. S4). A model of the CarD/RPo complex indicates that the CarD-RID/RNAP protein/protein interaction positions the CarD-CTD to interact with the promoter DNA at the upstream fork of the transcription bubble through the CarD-CTD basic patch, with the conserved tryptophan residue centrally positioned to insert into the minor groove directly over the −12 base pair (Fig. 3). Mutagenesis of T. thermophilus CarDW86/M. tuberculosis CarDW85 confirms the central role of this residue in the CarD functions of stimulating promoter binding and activating transcription (Fig. 4 C–E).
On the basis of these results, we propose that CarD stimulates promoter binding and activates transcription by forming favorable protein/protein (CarD-RID/RNAP β1-lobe) and protein/DNA (CarD-CTD/promoter DNA) interactions that bridge the RNAP to the promoter DNA (Fig. 3). Although CarD localization on the M. smegmatis chromosome is highly correlated with σA (Fig. 1), there are no significant CarD/σA protein/protein interactions predicted by our CarD/RPo model (Fig. 3). With its positioning with respect to the DNA by the CarD-RID/RNAP β1-lobe interaction, the CarD-CTD appears disposed to interact most favorably with the splayed minor groove found at the double-stranded/single-stranded DNA junction at the upstream edge of the transcription bubble formed by EσA (Fig. 3). Thus, CarD appears to interact most favorably with a DNA structure uniquely presented by EσA at promoters (Fig. 3B), as well as at σ-dependent pauses (13), likely explaining the correlated σA/CarD spikes observed within some transcription units (Fig. 1B).
Almost all previously reported prokaryotic activators of EσA (Eσ70 in E. coli) transcription act through a single paradigm whereby the activators function as sequence-specific DNA binding proteins that bind operators just upstream of the core promoter elements and establish favorable protein/protein contacts either with the RNAP α-C-terminal domains or with σA (14–16). An exception is the MerR family of activators that bind a specific operator located in the spacer between the −10 and −35 elements of limited stress response regulons and alter the conformation of the promoter DNA to enable RPo formation by EσA (17). The work presented here establishes CarD as a global regulator that represents an additional exception to the major prokaryotic activation paradigm. The CarD-RID binds to the RNAP β-subunit (like TRCF, a transcription terminator), positioning the CarD-CTD to sense the unique DNA conformation presented at the upstream edge of the transcription bubble in RPo (Fig. 3).
Our findings establish CarD as the founding member of a unique class of regulators that bind RNAP via the RID-domain/β1-lobe interaction and activate transcription initiation. As an essential global regulator in mycobacteria, CarD plays a complex role in vivo that will require further work to clarify. The work presented here delineates the basic molecular mechanism for the direct modulation of RNAP transcription activity by CarD, providing an essential framework for understanding its in vivo role. Our work also provides a structural basis for probing the mechanism more deeply. Of particular interest for future in vitro studies is to determine which step(s) of the complex promoter opening pathway (18) CarD affects and whether CarD activation of transcription initiation shows any dependence on promoter sequence.
Materials and Methods
Full details of the methods used are presented in SI Materials and Methods.
ChIP-Seq Analysis.
ChIP was performed as previously described (1). Coprecipitated DNA was sequenced using a SOLiD sequencer (Life Technologies), which provided sufficient reads for 100-fold coverage of the genome. The normalized, background-corrected log2 reads per base pair were then smoothed over a 20-bp window and σA and CarD peaks were identified as described previously (19, 20). To calculate average ChIP signals for the aggregate profiles (Fig. 1A), we selected a subset of 62 genes meeting the following criteria: (i) ≥300 bp in gene length, (ii) average RNAP log2 ChIP signal ≥1.6/bp, (iii) associated with a σA peak with log2 ChIP signal ≥3/bp, (iv) absence of other σA peaks within 500 bp upstream or 1,000 bp downstream of the associated σA peak, (v) absence of an oppositely oriented gene with an average RNAP log2 ChIP signal ≥1 upstream from the gene (because an oppositely oriented gene could create a divergent promoter region with potential for overlapping σA and CarD ChIP signals), and (vi) absence of an upstream gene with average RNAP log2 ChIP signal >0 within 100 bp upstream from the gene (because such an arrangement would indicate the gene is an internal member of an operon). The RNAP, CarD, and σA signals from the 62 genes were then averaged using the distance from the center of the associated σA peaks to align the genes.
Expression, Purification, and Crystallization of T. thermophilus CarD.
The gene encoding T. thermophilus CarD was amplified from T. thermophilus HB8 genomic DNA, cloned into the overexpression vector pET SUMO (Invitrogen), and transformed into BL21(DE3) (Novagen). Transformed cells were grown to an ODλ600 of ∼0.6 in the presence of 50 µg/mL kanamycin at 37 °C. Expression was then induced with 1 mM isopropyl β-d-thiogalactopyranoside (IPTG) for 4 h at 28 °C. Cells were harvested by centrifugation, resuspended, and then lysed using a continuous-flow French press (Avestin). The overexpressed protein was purified by Ni2+-affinity chromatography; overnight digestion of the SUMO His-tag using His-tagged ULP-1 protease (Invitrogen); removal of the His-tagged Ulp-1, SUMO-His-tag, and uncleaved protein by subtractive Ni2+-affinity chromatography; and size-exclusion chromatography. For overproduction of selenomethionyl-substituted protein, protein was overexpressed and labeled as previously described (21) and then purified using the same protocol as for the wild-type protein.
X-Ray Structure Determination of CarD.
T. thermophilus CarD crystals were grown at 22 °C, using hanging-drop vapor diffusion. The structure was solved by single-wavelength anomalous dispersion with data collected from selenomethionyl-substituted CarD (Table S2).
β-Galactosidase Assays.
To perform βgal assays during CarD depletion, an M. smegmatis strain expressing CarDwt from the attB site (mgm1849) (4) or WT M. smegmatis Mc2155 containing the empty pDB19 vector at the attB site (control strain) was transformed with pTE-2MOX (expresses WT TetR) (1) and the plasmid pCLS15-AP123-lacZ (low-copy KanR episomal plasmid that expresses lacZ from the M. smegmatis rrnA promoters in tandem as present in the genome). CarD was depleted as previously described and depletion was confirmed by Western blot (4). M. smegmatis ΔcarD attB::tetcarDWT, ΔcarD attB::tetcarDR25E, ΔcarD attB::tetcarDR47E, and ΔcarD attB::tetcarDW85A strains (engineered as described) (1) were transformed with pHMG147-AP123-lacZ (HygR episomal plasmid that expresses lacZ from the M. smegmatis rrnA promoters) to perform βgal assays in strains expressing CarD mutants. βgal activity was determined from log-phase cultures, using standard procedures (see SI Materials and Methods for full details).
In Vitro Transcription Assays.
In vitro transcription assays were performed using standard procedures (see SI Materials and Methods for full details).
Promoter Binding Assays.
Reactions were prepared as for the in vitro transcription assays. Briefly, core RNAP (100 nM) was incubated with CarD for 10 min before the addition of σA (500 nM). Labeled linear promoter DNA (25 nM) was added and the reactions were incubated at 65 °C (T. thermophilus) or 37 °C (Mycobacterium bovis) for 10 min to form open complexes. Once open complexes were formed, they were challenged with competitor DNA (double-stranded FullCon promoter DNA fragment, 1 μM) and a 10-μL aliquot was removed and bound to prewashed filters (MF-membrane filters; Millipore) and immediately washed with 4 mL wash buffer (10 mM Tris⋅HCl, pH 8.0, 200 mM NaCl). Radioactive signal, corresponding to labeled promoter DNA fragment bound to RNAP, was quantified by phosphorimagery.
Native Gel Electrophoresis Mobility Shift Assays.
Native gel electrophoresis mobility shift assays were performed using standard procedures (see SI Materials and Methods for full details).
qRT-PCR.
RNA was prepared from 5–10 mL of log-phase M. smegmatis ΔcarD attB::tetcarDwt, ΔcarD attB::tetcarDR25E, ΔcarD attB::tetcarDR47E, and ΔcarD attB::tetcarDW85A cultures and 16S rRNA levels were measured and normalized to sigA transcript levels as previously described (1).
Coimmunoprecipitation.
Cell lysates were prepared from 50 mL of log-phase M. smegmatis ΔcarD attB::tetcarDWT-HA, ΔcarD attB::tetcarDR25E-HA, ΔcarD attB::tetcarDR47E-HA, and ΔcarD attB::tetcarDW85A-HA (engineered as described in ref. 1) cultures. HA-tagged CarD proteins were immunoprecipitated from each lysate as previously described (4). For the Western blot analyses, CarD-HA and RNAP β were detected using mouse monoclonal antibodies specific for CarD (4) and RNAP β (clone 8RB13; Neoclone), respectively, and goat anti-mouse secondary antibodies conjugated to IRDye 800CW (LI-COR). The amount of signal was measured for fluorescent intensity, using the LI-COR Odyssey Scanner.
Supplementary Material
Acknowledgments
We thank R. Gourse, W. Ross, and R. Saecker for helpful discussions and A. Weixlbaumer for help with synchrotron data collection. We thank A. Heroux at the National Synchrotron Light Source (NSLS) beamline X25 for support with synchrotron data collection. We also thank the Genomics Core Laboratory at Memorial Sloan–Kettering Cancer Center (MSKCC) for performing the next-generation sequencing for ChIP-seq experiments and N. Socci and A. Krek at the MSKCC Bioinformatics Core Facility for processing the sequencing data. This work was based, in part, on research conducted at the NSLS, supported by the US Department of Energy, Office of Basic Energy Sciences. M.S.G. is supported by Grant AI64693 from the National Institutes of Health. C.L.S. is supported by a Biomedical Research Grant from the American Lung Association.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors for T. thermophilus CarD have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 4L5G). The ChiP-Seq data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE48164).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1308270110/-/DCSupplemental.
References
- 1.Stallings CL, et al. CarD is an essential regulator of rRNA transcription required for Mycobacterium tuberculosis persistence. Cell. 2009;138(1):146–159. doi: 10.1016/j.cell.2009.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Deaconescu AM, et al. Structural basis for bacterial transcription-coupled DNA repair. Cell. 2006;124(3):507–520. doi: 10.1016/j.cell.2005.11.045. [DOI] [PubMed] [Google Scholar]
- 3.Westblade LF, et al. Structural basis for the bacterial transcription-repair coupling factor/RNA polymerase interaction. Nucleic Acids Res. 2010;38(22):8357–8369. doi: 10.1093/nar/gkq692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weiss LA, et al. Interaction of CarD with RNA polymerase mediates Mycobacterium tuberculosis viability, rifampin resistance, and pathogenesis. J Bacteriol. 2012;194(20):5621–5631. doi: 10.1128/JB.00879-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.García-Moreno D, et al. CdnL, a member of the large CarD-like family of bacterial proteins, is vital for Myxococcus xanthus and differs functionally from the global transcriptional regulator CarD. Nucleic Acids Res. 2010;38(14):4586–4598. doi: 10.1093/nar/gkq214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316(5830):1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- 7.Gallego-García A, Mirassou Y, Elías-Arnanz M, Padmanabhan S, Jiménez MA. NMR structure note: N-terminal domain of Thermus thermophilus CdnL. J Biomol NMR. 2012;53(4):355–363. doi: 10.1007/s10858-012-9648-z. [DOI] [PubMed] [Google Scholar]
- 8.Feklistov A, Darst SA. Structural basis for promoter-10 element recognition by the bacterial RNA polymerase σ subunit. Cell. 2011;147(6):1257–1269. doi: 10.1016/j.cell.2011.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Murakami KS, Masuda S, Campbell EA, Muzzin O, Darst SA. Structural basis of transcription initiation: An RNA polymerase holoenzyme-DNA complex. Science. 2002;296(5571):1285–1290. doi: 10.1126/science.1069595. [DOI] [PubMed] [Google Scholar]
- 10.Shultzaberger RK, Chen Z, Lewis KA, Schneider TD. Anatomy of Escherichia coli sigma70 promoters. Nucleic Acids Res. 2007;35(3):771–788. doi: 10.1093/nar/gkl956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo Y, Gralla JD. Promoter opening via a DNA fork junction binding activity. Proc Natl Acad Sci USA. 1998;95(20):11655–11660. doi: 10.1073/pnas.95.20.11655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haugen SP, Ross W, Gourse RL. Advances in bacterial promoter recognition and its control by factors that do not bind DNA. Nat Rev Microbiol. 2008;6(7):507–519. doi: 10.1038/nrmicro1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perdue SA, Roberts JW. Σ(70)-dependent transcription pausing in Escherichia coli. J Mol Biol. 2011;412(5):782–792. doi: 10.1016/j.jmb.2011.02.011. [DOI] [PubMed] [Google Scholar]
- 14.Benoff B, et al. Structural basis of transcription activation: The CAP-alpha CTD-DNA complex. Science. 2002;297(5586):1562–1566. doi: 10.1126/science.1076376. [DOI] [PubMed] [Google Scholar]
- 15.Browning DF, Busby SJW. The regulation of bacterial transcription initiation. Nat Rev Microbiol. 2004;2(1):57–65. doi: 10.1038/nrmicro787. [DOI] [PubMed] [Google Scholar]
- 16.Jain D, Nickels BE, Sun L, Hochschild A, Darst SA. Structure of a ternary transcription activation complex. Mol Cell. 2004;13(1):45–53. doi: 10.1016/s1097-2765(03)00483-0. [DOI] [PubMed] [Google Scholar]
- 17.Brown NL, Stoyanov JV, Kidd SP, Hobman JL. The MerR family of transcriptional regulators. FEMS Microbiol Rev. 2003;27(2-3):145–163. doi: 10.1016/S0168-6445(03)00051-2. [DOI] [PubMed] [Google Scholar]
- 18.Saecker RM, Record MT, Jr, Dehaseth PL. Mechanism of bacterial transcription initiation: RNA polymerase - promoter binding, isomerization to initiation-competent open complexes, and initiation of RNA synthesis. J Mol Biol. 2011;412(5):754–771. doi: 10.1016/j.jmb.2011.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mooney RA, et al. Regulator trafficking on bacterial transcription units in vivo. Mol Cell. 2009;33(1):97–108. doi: 10.1016/j.molcel.2008.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reppas NB, Wade JT, Church GM, Struhl K. The transition between transcriptional initiation and elongation in E. coli is highly variable and often rate limiting. Mol Cell. 2006;24(5):747–757. doi: 10.1016/j.molcel.2006.10.030. [DOI] [PubMed] [Google Scholar]
- 21.Doublié S. Preparation of selenomethionyl proteins for phase determination. Methods Enzymol. 1997;276:523–530. [PubMed] [Google Scholar]
- 22.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: Application to microtubules and the ribosome. Proc Natl Acad Sci USA. 2001;98(18):10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Campbell EA, et al. Structure of the bacterial RNA polymerase promoter specificity sigma subunit. Mol Cell. 2002;9(3):527–539. doi: 10.1016/s1097-2765(02)00470-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.