Abstract
Sequence-specific binding in the minor groove of DNA by small molecules is a growing area of research with possible therapeutic relevance. By selectively binding to DNA sequences required by critical transcription factors, these small molecules could potentially modulate the expression levels of disease-causing genes. Precise targeting of a critical transcription factor of a selected gene requires an understanding of the preferred sequence of the DNA binding compound. As new compounds are being synthesized, there is a need to evaluate their DNA recognition profile. We sought to establish a procedure to determine sequence preference of compounds with previously unknown binding properties. A novel procedure for determining the optimal DNA binding sequence of minor groove binding compounds is described here. The assay also allows for determination of the binding affinity to a particular sequence.
INTRODUCTION
The targeting of disease-causing proteins with small molecule drugs is a proven means of therapeutic intervention. Alternate strategies such as the targeting of specific mRNA, however, have encountered numerous problems (1). One under-explored alternative approach is the displacement of critical transcription factors in the promoters of therapeutic genes by DNA binding small molecules (2,3). This approach requires the development of DNA binding compounds capable of recognizing specific sequences which overlap the binding site of the relevant transcription factor (3–5). Although the compound of interest may indeed bind the targeted site, the danger is that it might bind with higher affinity to sequences not being targeted. This could have deleterious consequences. To ensure the selectivity of small molecule binding to target sites, it is necessary to understand the relative affinity of the intended target versus all other sequences. However, with novel compounds never before characterized, rank ordering of targeted versus other sequences poses a practical challenge. Therefore the approach of sequence-specific targeting by DNA binding compounds requires the establishment of an assay capable of identifying the preferred DNA binding site of newly synthesized compounds. This assay needs to allow for unbiased examination of all sequences, given that each new compound will have undetermined binding properties.
Serious problems exist with the most obvious methodological choices for such an assay. Standard DNase I footprinting suffers from low throughput and an inability to assay numerous potential binding sites simultaneously. Other assays such as compound-mediated displacement of ethidium bromide (6) lack the sensitivity to accurately distinguish DNA binders with low nanomolar affinities due to the micromolar DNA concentrations required by the assay. Neither of these methods allows for unbiased determination of binding preference by high-affinity DNA binding compounds. Methods for the positive selection of the optimal sequences would be a particularly effective way to understand the binding properties of a DNA binder. For example, Restriction Endonuclease Protection Selection and Amplification (REPSA) relies on bound DNA molecules being protected from restriction endonuclease cleavage so that they can subsequently be amplified (7). Incomplete DNA cleavage by the type II restriction enzymes used in this assay, however, leads to a high background of undigested template and a requirement for many rounds of selection. Systematic Evolution of Ligands by EXponential Enrichment (SELEX) is another powerful assay designed to select high-affinity nucleic acid ligands, but requires that the target or ligand be immobilized on a column (8).
We sought to establish an assay that would allow us to query numerous potential sequences simultaneously in an unbiased manner. A selection scheme utilizing uracil DNA glycosylase (UDG) is described here that will do just that. The assay rapidly determines the DNA binding specificity as well as the binding site size of minor groove binding compounds at low DNA and compound concentrations. The robust nature of the assay allows for clean selection with low background, and its PCR-based readout allows for sequence preference determination using compounds with low binding constants. Furthermore, the assay format can be slightly modified so as to determine binding affinity between compound and DNA.
MATERIALS AND METHODS
Sequence of oligonucleotides
UDG selection oligonucleotide. Top strand: TCT TGG CAG CAG GAU UGT CCU UGG ATC CAC TAC GCA GCG CCT CCC TCC ACT NNN NNN NNN AAA AGG GCT GGA GAC GTG GC. Double-stranded selection oligonucleotide was generated using this top strand and the following oligonucleotide primer: GCC ACG TCT CCA GCC CTT UU. This oligonucleotide was also used for PCR amplification of DNA protected from UDG digestion. The other primer used during amplification of protected DNA was TCT TGG CAG CAG GAU UGT CCU U.
UDG affinity oligonucleotide. Top strand: TCT TGG CAG CAG GAU UGT CCU UGG ATC CAC TAC GCA GCG CCT CCC TCC ACT CGC ACC TAA CAA TCC TAC GCG CAC CTA ATA ATC CTA CGC GCA CCT AAC CAT CCT ACG GGG CTG GAG ACG TGG C. Match and mismatch sites are indicated in bold. Double-stranded UDG affinity oligonucleotide was generated using this top strand and the following oligonucleotide primer for the match site (ATTGTTA): GCCA CGT CTC CAG CCC CGT AGG ATG GTT AGG TGC GCG TAG GAT TAT TAG GTG CGC GTA GGA UUG TTA GG. Double-stranded UDG affinity oligonucleotide for the mismatch site (ATTATTA) was generated using the top strand and following oligonucleotide primer: GCCA CGT CTC CAG CCC CGT AGG ATG GTT AGG TGC GCG TAG GAU UAT TAGG. Amplification of UDG affinity oligonucleotide DNA protected from UDG digestion by compound utilized the same primers that were used for amplification of the UDG selection oligonucleotide.
TaqMan oligonucleotide probe. The sequence of the TaqMan oligonucleotide probe was AC TAC GCA GCG CCT CCC TCC ACT.
Primer extension
Primer extension reactions contained 10 mM Tris–HCl pH 8.3, 50 mM KCl, 1.5 mM MgCl2, 0.001% gelatin (PCR buffer), 200 µM each dNTP, 4 µg top strand, 2 µg bottom strand primer and 10 U Jump Start Taq DNA polymerase (Sigma). Primer extension proceeded at 95°C for 10 min, followed by 3 min at 60°C and 15 min at 72°C (100 µl reactions).
UDG selection
Double-stranded selection oligonucleotide (2.6 ng) was incubated with compound in PCR buffer (total volume 25 µl). Three to four different drug concentrations were typically chosen. Binding proceeded at room temperature for 2–4 h. Samples were then transferred to 37°C for 10 min. One unit of UDG was then incubated with the mixture for 1 min before being stopped with an equal volume of 0.2% SDS. Six to seven samples were processed per round of reactions. Digestion products were then diluted 250-fold (2 µl into 498 µl) with 0.2 µg/ml tRNA before proceeding to PCR.
PCR
PCR was in 1× PCR buffer (25 µl reactions) and contained 1.2 µM primers (165 ng each oligonucleotide), 200 nM TaqMan probe, 1 U Taq DNA polymerase, 200 µM dNTP and 2 µl of the diluted UDG digestion mixture. TaqMan PCR proceeded for 27 cycles of 15 s at 95°C, 1 min at 60°C (first cycle denaturation was for 10 min at 95°C). Following PCR, primers were removed using a PCR purification kit (Qiagen) and sequenced using an Applied Biosystems 373A sequencing machine.
DNase I footprinting
DNA oligonucleotides were labeled with [γ-32P]ATP and polynucleotide kinase. Unincorporated nucleotide was removed by Sephadex G25 spin column chromatography according to manufacturer’s specifications (Roche). DNA (2 ng, 50 000 c.p.m.) was incubated with compound in PCR buffer (50 µl reaction) at room temperature for 2 h prior to addition of DNase I (44 U/ml, Roche). Digestion with DNase I proceeded for 1 min. DNase I digestion was stopped by addition of an equal volume of stop buffer (20 mM Tris–HCl pH 7.5, 20 mM EDTA, 5 mM EGTA and 5 mg/ml yeast tRNA). Following extraction with phenol, the mixture was precipitated with 0.1 M NaCl and 2 volumes of ethanol. Pellets were resuspended in 10 µl of sequencing loading buffer. Reaction products were separated on 6–10% sequencing gels. Quantitation was via PhosphorImager analysis.
RESULTS
Design of the selection scheme
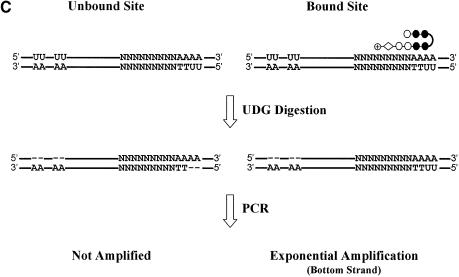
We invented a novel enzyme-based selection scheme using UDG as a reagent to select from a degenerate pool of sequences those that are recognized by a DNA binding compound of interest. UDG interacts with DNA in the minor groove and catalyzes the cleavage of uracil from double-stranded DNA (9). We reasoned that upon binding uracil-tagged DNA, a minor groove binding compound might sterically interfere with this cleavage and protect the DNA from UDG digestion. The protected sequences could then serve as templates in subsequent PCRs and could be identified following amplification and sequence determination. Sequences not bound by the compound are susceptible to UDG digestion and cannot be amplified.
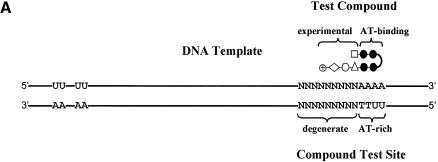
The selection scheme is designed for compounds that contain a well-defined portion known to bind AT-rich DNA (AT-binding cassette) joined to an experimental portion of unknown specificity. The AT-binding cassette guides the compound to a 2–4 bp AT-rich region in the 80 bp oligonucleotide template DNA (Fig. 1A). At least one uracil (U) residue is embedded in this region. A 9 bp degenerate region together with the adjacent AT-rich region represents the test site for the compounds. The size of the degenerate region was limited to 9 bp because compounds binding to sites larger than this would most likely be too large to enter cells and modulate transcription of therapeutic genes. The guided binding of the AT-binding cassette to the AT-rich region of DNA results in positioning the experimental portion of the compound to the degenerate region of the test site. The assay therefore queries the sequence requirement in the degenerate region in order to allow high-affinity binding under stringent conditions such as low compound concentration. The remainder of the oligonucleotide is GC rich in order to restrict binding of the AT-binding cassette to the AT-rich region in the test site. The GC-rich nature of the oligonucleotide also helps prevent ‘sliding’ of the AT-binding cassette upstream or downstream of the AT-rich region, thus registering the compound within the compound test site. If binding of the AT-binding cassette is not restricted exactly to the AT-rich region, but instead slides upstream or downstream, selected nucleotides would appear in multiple positions.
Figure 1.
UDG selection scheme. (A) The DNA template for the UDG selection assay is GC-rich and 80 bp in length. An AT-rich region juxtaposed with a degenerate region constitute the compound test site. Correspondingly, a test compound consists of an AT-binding cassette (black circles) juxtaposed with an experimental portion of unknown specificity (white shapes). Typically, two T residues in the bottom strand of the test site are substituted by U. This serves as a UDG-sensitive probe for compound binding. The duplex oligonucleotide with the sequence drawn here is written as N(9)AAAA·UUTT. Separately, several U residues are embedded in the top strand near the 5′ end of the template to prevent the top strand from being amplified. The upstream U residues are embedded in a GC-rich region which constitutes a poor compound binding site. The drawing is not to scale. (B) The U residues are maintained through successive rounds of selection by including them in the PCR primers. (C) U residues in the template are removed by UDG except those protected by compound binding. Templates with abasic sites, such as the top and bottom unbound strands, are not amplified.
UDG was chosen as the agent to eliminate unbound sequences from the selection assay for several reasons. First of all, the ability to substitute U for T residues in the AT-rich region of the test site conveniently provides a means for test sites to be assayed for protection by a bound compound. Secondly, UDG locates uracil residues by scanning the minor groove of double-stranded DNA, and so should be susceptible to inhibition by minor groove binding compounds. Thirdly, published data support the feasibility of using UDG in a sequence selection assay. Utilizing a U-containing DNA template, UDG was used as the DNA modifying agent in the footprinting of DNA–protein complexes (10).
Compounds that are highly specific will only bind a small percentage of the input degenerate DNA template. Determination of binding preferences may therefore require several rounds of selection at times. This requires that the U residues within the compound-binding site of the template be maintained following PCR amplification. This is accomplished by incorporating them into PCR primer 1 (Fig. 1B). The lack of U residues on the top strand within the test site would potentially allow its PCR amplification independent of compound binding, resulting in a high background. To prevent such amplification, several U residues were incorporated upstream from the test site (near the 5′ end) in a GC-rich region predicted to poorly bind the AT-binding cassette of the compound. An important feature of the selection scheme is that UDG treatment will always destroy the top strand regardless of whether a compound binds at the test site. Therefore, even if the test site is fully protected by high-affinity binding of a compound, UDG digestion will still result in a 50% reduction in the level of PCR product following amplification compared with DNA not treated with UDG. Like the U residues in PCR primer 1, the U residues in the top strand are directly incorporated into PCR primer 2 and are therefore maintained in the template DNA through multiple rounds of selection. Rapid quantitation of PCR results is enabled by utilization of a TaqMan probe corresponding to nucleotides 20–40 of the template DNA.
In summary, the scheme involves the following steps. (i) Binding of compound to the template oligonucleotide which protects the test sites of optimal sequences from UDG. (ii) Treatment with UDG to remove uracil residues from unprotected sequences. (iii) PCR amplification of the protected fragments. Unprotected fragments that contain abasic sites following UDG digestion are not amplified (Fig. 1C). (iv) Multiple rounds of steps (i)–(iii) as needed. (v) Sequence determination of the resulting DNA.
Assay conditions and validation
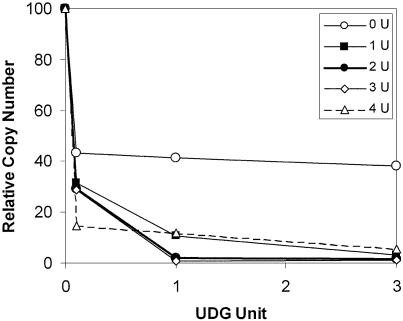
Success of the assay relies on a low background through robust removal of uracil in the unbound template DNA by UDG. This ensures preferential PCR only of those templates that have been protected from UDG by compound binding. First, the optimal number of U residues in the test site of DNA was determined. DNA with one, two, three or four U residues imbedded at the test site were digested with increasing amounts of UDG, and the amount of undigested template was quantitated by TaqMan PCR analysis. Figure 2 shows that incubation of one unit of UDG with a template containing either two or three U residues was optimal since these conditions resulted in the most efficient and complete digestion of template by UDG. Addition of more than one unit of UDG did not lead to increased digestion of template DNA (data not shown). Under these conditions (one unit UDG, two U in test site), UDG digestion increases the number of PCR rounds required for detection of TaqMan product (Ct) by 5–7 cycles. This corresponds to 96.9–99.2% digestion of the starting template by UDG.
Figure 2.
Optimization of UDG digestion. Double-stranded oligonucleotides in which one, two, three or four T residues in a 4 bp AT region were replaced by U residues were made. This AT region was adjacent to the degenerate region. The compound binding sites in these four oligonucleotides were therefore composed of N(9)AAAA·TTTU, N(9)AAAA·TTUU, N(9)AAAA·TUUU and N(9)AAAA·UUUU. Each oligonucleotide was digested with increasing amounts of UDG for 1 min. After the reaction was stopped with SDS, the amount of undigested DNA oligonucleotide amenable to PCR amplification was quantified by TaqMan PCR. Relative copy number indicates the percentage of starting DNA which remains undigested and therefore PCR amplified. The relative copy number of the oligonucleotide with no U residues adjacent to the degenerate region is reduced by 50% because the U residues on the top strand are digested. Therefore the theoretical minimal amount of DNA remaining undigested is 50%.
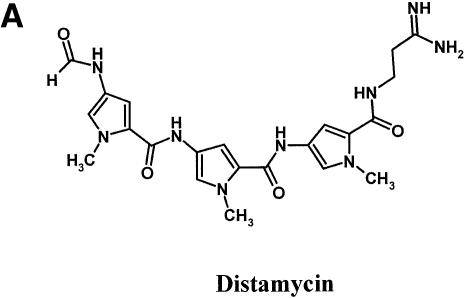
To validate our selection scheme, we first tested distamycin (Fig. 3A), which is a well characterized DNA minor groove binding compound known to bind to 4–5 bp AT-rich regions (11). This compound was taken through a single round of the selection scheme using a template DNA containing a 9 bp degenerate region adjacent to a 2 AU base pair template N(9)AA·UU. The placement of the U residues immediately adjacent to the degenerate region is necessary for a test molecule whose site size is 4–5 bp since the molecule needs to span from the U-containing AT-rich region into the degenerate region in order to affect selection. The template DNA was incubated with compound of increasing concentrations and then digested with UDG. TaqMan PCR amplification of protected templates was followed by oligonucleotide purification and sequencing. Figure 3B shows the sequence of the 9 bp degenerate region of the top strand of the amplified DNA. In the absence of compound, the sequence of the region remains degenerate as expected. As distamycin concentrations increased, a reduction of peaks representing G (black) and C (blue) residues was clearly evident concomitant to the enhancement of A (green) and T (red) peaks. The selection for AT residues was localized to the two to three residues (positions –1, –2 and –3) at the 3′ end of the degenerate region and it was observed with 50 nM distamycin, the lowest concentration tested. Combined with the two U residues in the AT-rich region, the preferred binding site of distamycin is determined to be a 4–5 bp AT-rich sequence. In addition to the site size, the assay also appears to distinguish between variable modes of compound binding to DNA template. Published reports on crystallography and DNase I footprinting studies demonstrated that distamycin binding to DNA varies between compound:template stiochiometry of 1:1 or 2:1 according to compound concentration and the sequence (12). The sequence specificity in binding is largely determined by the width of the minor groove. Sequences with AAAAA have the highest affinity for distamycin because this stretch of DNA has an unusually narrow minor groove. Binding in this case is in the 1:1 mode. The preferred mode of binding at higher distamycin concentrations to sequences with a wider minor groove, such as TATAT, is in the 2:1 mode where the two distamycin molecules are ‘stacked’ (12). The data shown in Figure 3B are consistent with these observations. While a 2–3 bp region of A residues is selected by 150 nM of distamycin, at higher concentrations (450 and 1350 nM), the 2–3 bp region selected is still AT rich but now more mixed in A and T content. The testing of distamycin in the UDG selection scheme produced data on site size, sequence preference and mode of binding exactly as expected from previously reported data using conventional methods.
Figure 3.
Binding site preference for distamycin. (A) Structure of distamycin. (B) Selection oligonucleotide with the sequence of N(9)AA·UU was incubated with 0, 50, 150, 450 or 1350 nM distamycin for 2 h, followed by digestion with UDG and then PCR amplification. PCR product was then purified and sequenced. Positions relative to the AA·UU are indicated at the bottom of each panel by –1 to –9. Black, G; blue, C; green, A; red, T.
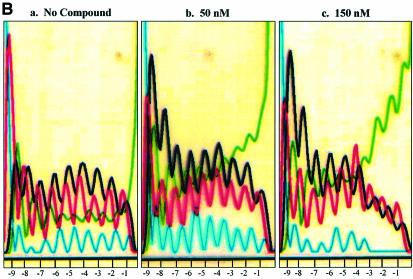
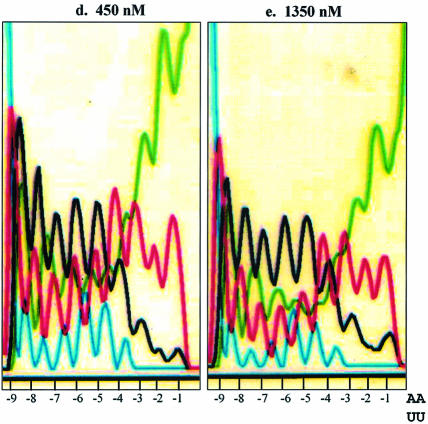
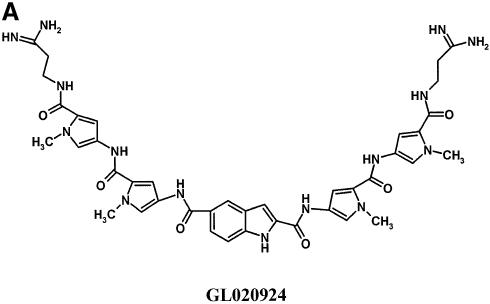
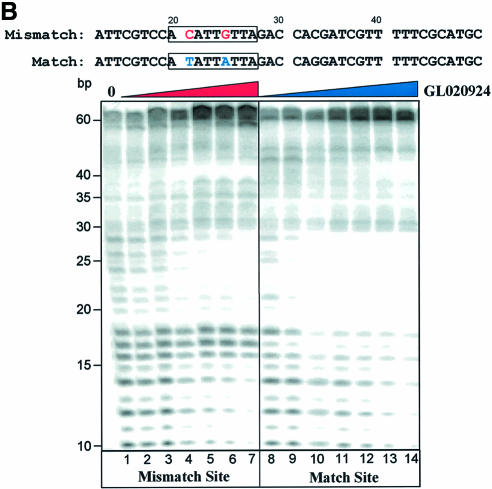
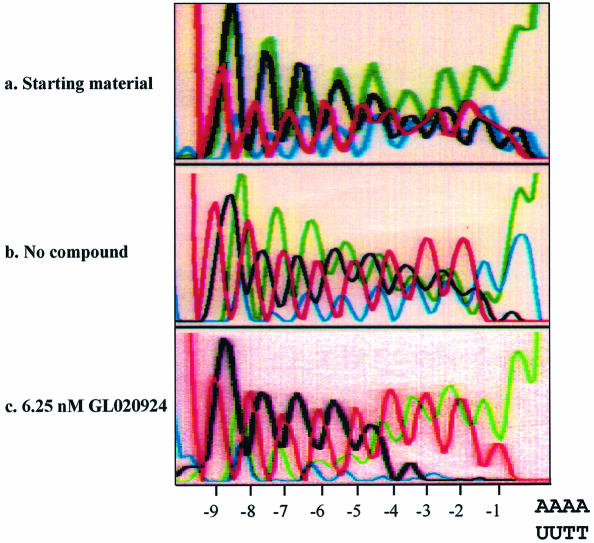
The next compound tested was GL020924 (Fig. 4A), an indole-linked polyamide previously shown using a variety of biophysical methods to bind with strong preference to AT sequences 8 to 10 bp in length (13). To demonstrate AT specificity of GL020924, DNase I footprinting analysis was performed using a template containing a 9 bp AT-rich sequence. Figure 4B confirms that the compound has high affinity towards the AT-only (match) site with an estimated Kd of <50 nM. Disruption of the AT-rich region with G or C residues decreases the affinity to an estimated Kd of ∼300 nM (mismatch site). GL020924 was tested in the UDG selection scheme using the N(9)AAAA·TTUU template DNA. Relative to sequences in the starting oligonucleotide as well as those generated by the no compound control, the compound effectively selected AT sequences at concentrations in the low nanomolar level (Fig. 5). After a single round of selection with 6.25 nM compound, a 4 bp AT stretch was selected. Together with the 4 bp AT-rich region provided by the test site, GL020924 therefore was shown to bind an ∼8 bp AT-rich region by the UDG assay. This result was the one predicted according to previous work (13). We conclude that the UDG selection assay reliably determines both sequence preference and binding site size for DNA binding compounds with AT-binding cassettes.
Figure 4.
Characterization of GL020924 DNA binding by DNase I footprinting. (A) Structure of GL020924. (B) Labeled oligonucleotides, each containing a GL020924 match or mismatch site (boxed area), were incubated with increasing amounts of GL020924 and partially digested with DNase I. Digestion products were separated on a 10% sequencing gel, and results were visualized and quantitated with a PhosphorImager. Compound concentrations were 0, 50, 150, 300, 450, 600 and 750 nM for the mismatch oligonucleotide (lanes 1–7), and 25, 50, 150, 300, 450, 600 and 750 nM for the match oligonucleotide (lanes 8–14).
Figure 5.
Binding site preference for compound GL020924. Selection oligonucleotide with the sequence of N(9)AAAA·TTUU was incubated with either 0 or 6.25 nM GL020924 for 2 h, followed by digestion with UDG and then PCR amplification. PCR product was then purified and sequenced. Starting material is selection oligonucleotide without any treatment. Positions relative to the AAAA·TTUU are indicated at the bottom of each panel by –1 to –9.
GC recognition
The successful therapeutic use of DNA binding compounds to displace critical transcription factors will require the specific recognition of GC as well as AT base pairs. A family of polyamides consisting of pyrrole and imidazole amino acid units is the best characterized group of minor groove binders that can confer sequence-specific recognition properties. For example, with methyl pyrrole units binding either 1:1 or 2:1 (compound:template), the DNA recognition is AT specific (5). When the pyrrole units are replaced with imidazole, hydrogen bonding between the exocyclic amino group of G residues and the N3 position of imidazole allows GC recognition (14). This specificity occurs in the 2:1 stacked binding mode in which imidazole–pyrrole (im–py) pairs recognize GC, py–im pairs recognize CG and py–py pairs are degenerate for AT and TA (14). It has been established that covalent linkage of two stacked polyamides can improve binding. For example, when two polyamides containing three units each are covalently attached using a diaminobutyric acid (DAB) linker, the resulting ‘3 + 3’ compound is improved 2000-fold with respect to affinity and 29-fold with respect to specificity (5). The 2:1 stacked mode of binding is accomplished by binding of the 3 + 3 compound in a hairpin conformation. Additional improvement in specificity and ease of synthesis can be accomplished by adding a β-alanine (β) adjacent to a dimethylaminopropyl (Dp) group to the C-terminus of the polyamide. The DAB linker preferably binds 1 to 2 and the β-Dp tail binds to two AT base pairs (14). Based on these published data, an imidazole-containing 3 + 3 compound was synthesized in order to test the ability of the UDG scheme in selecting GC-containing sequences.
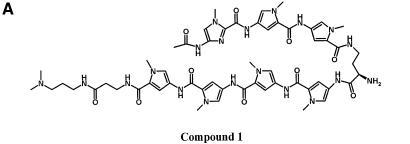
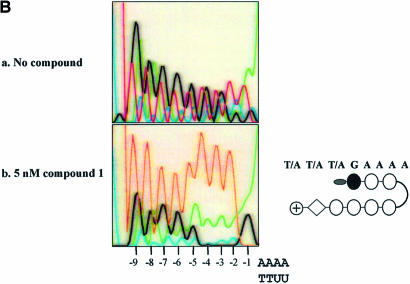
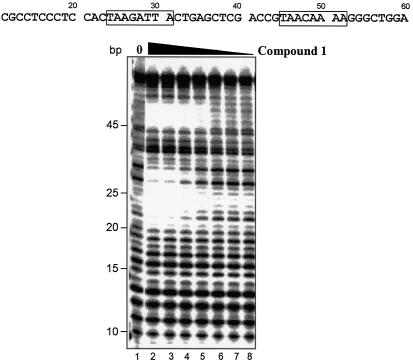
Compound 1 [acetyl-im1-py2-py3-DAB-py4-py5-py6-py7-β-Dp (Fig. 6A)] was used to test the binding preference of a compound with potential specificity for sequences comprised of GC as well as AT base pairs. According to the published reports, the sequence preference of compound 1 is predicted to be WWWGWWWW where W is either A or T. UDG selection assay shows that the im1-py6 pair of compound 1 indeed strongly selects G immediately adjacent to the register-fixing AAAA·UUTT sequences (Fig. 6B). A 2 to 3 AT base pair region is selected immediately 5′ of the G by the β-Dp tail. These observations are consistent with the expected binding and protection of the AAAA·UUTT stretch at the test site by the internal py2-py3-DAB-py4-py5 portion of the polyamide. This sequence preference was obtained by a single round of selection using 5 nM of compound 1. To validate this result, two sequences matching the preference (WWWGWWWW) were incorporated into an oligonucleotide and tested by DNase I footprinting. Compound 1 protected both sequences predicted to be preferred binding sites from DNase I digestion (Fig. 7). The approximate Kd values for these match sites are estimated to be between 50 and 100 nM, and between 25 and 50 nM for the upstream and downstream sites, respectively.
Figure 6.
Binding preference for compound 1. (A) Structure of compound 1. (B) Selection oligonucleotide with the sequence of N(9)AAAA·UUTT was incubated with either 0 or 5 nM compound 1 for 2 h, followed by digestion with UDG and then PCR amplification and sequencing. Positions relative to the AAAA·UUTT are indicated at the bottom of each panel by –1 to –9. A schematic representation of the compound is shown on the right. Individual amino acids of the polyamide are represented by circles. The black circle represents imidazole. White circles represent pyrrole-containing amino acids and the acetyl group at the N-terminus of the polyamide is represented by the gray oval. The DAB linker and β-Dp tail of the polyamide are represented by a curved line and a diamond, respectively. The predicted binding preference of compound 1 is indicated above the schematic representation of the structure.
Figure 7.
Characterization of compound 1 DNA binding by DNase I footprinting. Labeled oligonucleotide containing two compound 1 match sites (boxed areas) was incubated with increasing amounts of compound 1, and was partially digested with DNase I and analyzed as described in the legend to Figure 4B. Compound concentrations were 0, 300, 200, 100, 50, 25, 12.5 and 5 nM (lanes 1–8).
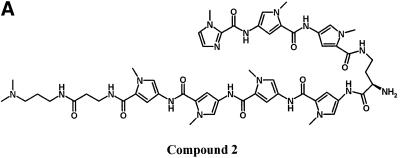
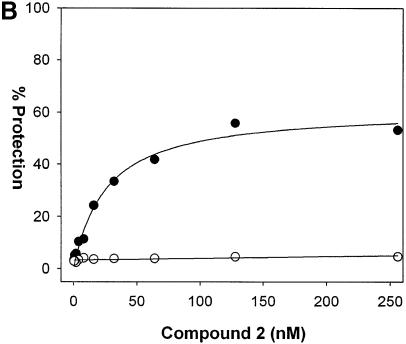
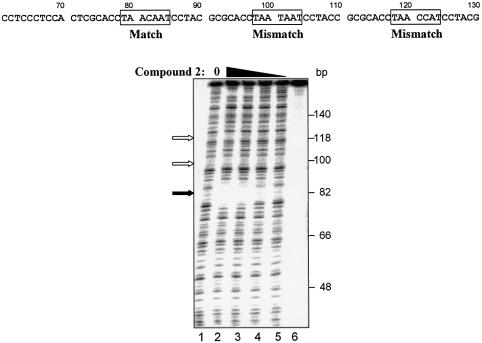
Although the UDG selection scheme determines binding site preference of DNA binders, it is also important to determine the binding affinity for this preferred site. By incorporating one to two U residues in specific regions of the preferred binding site and determining the compound concentration required to protect half of the template from UDG digestion, binding affinity can be quantified. The design of the assay is such that the top strand contains U residues in a poor compound-binding region to ensure digestion of this strand. Since the top strand will always be digested, the dissociation constant is defined as the compound concentration required for 25% protection of starting template. Such an approach was utilized for compound 2 [im1-py2-py3-DAB-py4-py5-py6-β-Dp (Fig. 8A)]. Using a template DNA containing the sequence TAACAAT·AUUGTTA (match site) in place of the degenerate sequence in the test site, compound 2 afforded protection from UDG in a concentration-dependent manner. The apparent Kd, the concentration of compound required for half the maximum level of protection, is 26.0 ± 4.1 nM (Fig. 8B). In parallel, template DNA containing a mismatch sequence TAATAAT·AUUATTA (mismatch site) was not protected. These results were reproducible and agreed with DNase I footprinting experiments (Fig. 9).
Figure 8.
Binding affinity determination of compound 2. (A) Structure of compound 2. (B) A template DNA containing the sequence TAACAAT·AUUGTTA (match site) in place of the degenerate N(9) sequence was incubated with increasing concentrations of compound 2 and then digested with UDG for 1 min. The extent of protection from UDG was determined by TaqMan PCR analysis as described in Materials and Methods. As a negative control, an oligonucleotide containing a mismatch site TAATAAT·AUUATTA was processed in parallel. Closed circles represent data for the match oligonucleotide and open circles represent data for the mismatch oligonucleotide. The Kd of compound 2 for the match site is determined by fitting the data using Sigma Plot to the equation %Protection = (a × [compound])/(b + [compound]) where a is the maximum %Protection and b is Kd.
Figure 9.
Characterization of compound 2 DNA binding by DNase I footprinting. Labeled oligonucleotide containing one match site and two mismatch sites to compound 2 (boxed areas) was incubated with increasing amounts of the compound, and was partially digested with DNase I and analyzed as described in the legend to Figure 4B. Compound concentrations were 270, 90, 30 and 10 nM (lanes 2–5). No compound control is shown in lane 1. No compound, no DNase I control is shown in lane 6. The protected match site is shown by the filled arrow and the positions of the mismatch sites are indicated by the open arrows.
DISCUSSION
The UDG selection scheme determines the preferred binding site of minor groove binding compounds. By incorporating the required U residues into the PCR primers, the assay is designed so that multiple rounds of selection are possible. As the sequence specificity and DNA affinity of designed compounds increases, this will be a necessary requirement. For example, only one in 1024 oligonucleotides will be protected by a compound which binds one specific 5 bp sequence (1 in 45). If the background of undigested U-containing template is 0.8–3.1%, this background will be higher than the signal resulting from specifically bound and protected oligonucleotide. However, with a second round of selection the specificity of such compounds will become evident.
This assay takes advantage of the fact that UDG is a minor groove binding protein recognizing a DNA modification (lack of a 5-methyl group) which is actually in the major groove and should not have a significant effect on minor groove binding compounds. Recognition of dU residues in DNA resulting from either cytosine deamination or misincorporation of dUMP by DNA polymerase (9) is achieved through flipping out the dU residue into the U recognition pocket of UDG. Importantly, uracil residues within the recognition site of DNA ligands do not affect binding unless interaction with the thymine methyl group is required (10).
Optimal results for UDG selection are obtained with 1 min digestion time. Digestion times significantly shorter than this are insufficient for complete UDG digestion, which is required for a low background. Likewise, the fact that the assay is under UDG excess conditions leads to a less robust signal at longer time points. This decrease in signal most likely results from digestion by excess UDG upon compound–DNA dissociation (data not shown).
Final readout of binding preference by sequencing total PCR amplified DNA relies on register fixing by the AT-binding cassette of the compound. Some compounds will potentially bind in either of the two possible orientations and may furthermore be able to ‘slide’ 5′ or 3′. Cloning and sequencing of PCR products may therefore be necessary for a less ambiguous readout of binding preference for some compounds. In order to reduce assumptions regarding binding specificity and orientation, the selection oligonucleotide could be synthesized with one to two central AU base pair(s) flanked by degenerate regions on both sides. When utilized as such, the UDG selection scheme could be used to elucidate the optimal binding site of proteins with minor groove contacts such as TATA binding protein (15). Finally, once having determined DNA ligand binding site preference, the assay can be modified to determine binding affinity. Because the procedure is PCR based, DNA concentrations can be lowered as far as necessary. In studying subnanomolar DNA binding compounds we have routinely dropped DNA concentrations into the low picomolar range (data not shown).
This overall strategy may be applicable towards other glycosylases. Two important factors will determine the usefulness of particular glycosylases in determining the binding preferences for DNA binders of interest. First, the glycosylase should scan the same DNA groove as the ligand of interest. Secondly, the modification recognized by the glycosylase might alter the binding affinity for the compound of interest. The modification should therefore reside in the groove opposite that bound by the compound and glycosylase (see below for exception, however). For example, the modification in 8-oxo G is recognized and removed from DNA by formamidopyrimidine-DNA glycosylase (Fpg) (16) and may be useful in determining the sequence specificity of compound binding the minor groove of GC rich DNA. Whether Fpg interacts with DNA through the minor groove, as would be required in this scenario, is not presently known. Determination of binding preferences of DNA major groove binders would optimally require major groove binding glycosylases which recognize modifications which are either in the minor groove or in the major groove but are known not to effect DNA binding by the factor of interest. No such glycosylases have definitively been identified to date. Despite the fact that UDG recognizes a modification residing in the major groove, it may still be of significant utility in binding site selection assays of major groove binding compounds. This would require steric occlusion between the DNA major groove binder and UDG, despite the fact that UDG is predominantly a minor groove binding protein (17). Such a competitive inhibition occurs in the case of lambda repressor, for example, which utilizes the helix–turn–helix motif to make specific contacts with the major groove and sugar–phosphate backbone (18). When bound to its operator site, lambda repressor efficiently protects U residues within OR1 and OR2 from UDG digestion (18). As described above, one caveat of using the UDG protection assay for DNA major groove binders is that binding interactions requiring the T methyl group will be missed using the U-containing templates. Such an interaction occurs for lambda repressor at the OR3 site, resulting in a more modest protection from UDG (18).
In summary, we have developed a novel assay designed to determine both binding affinity and specificity of DNA minor groove binding compounds. The assay is designed for compounds which have an AT-binding portion in order to guide the compound to an AU-containing region, as well as a second experimental portion with unknown specificity. If desired, the assay can readily be modified so as to reveal the approximate dissociation constants of DNA minor groove binding compounds.
REFERENCES
- 1.Ma D.D., Rede,T., Naqvi,N.A. and Cook,P.D. (2000) Synthetic oligonucleotides as therapeutics: the coming of age. Biotechnol. Annu. Rev., 5, 155–196. [DOI] [PubMed] [Google Scholar]
- 2.Dervan P.B. and Burli,R.W. (1999) Sequence-specific DNA recognition by polyamides. Curr. Opin. Chem. Biol., 3, 688–693. [DOI] [PubMed] [Google Scholar]
- 3.Laurance M.E., Starr,D.B., Michelotti,E.F., Cheung,E., Gonzalez,C., Tam,A.W., Deikman,J., Edwards,C.A. and Bardwell,A.J. (2001) Specific down-regulation of an engineered human cyclin D1 promoter by a novel DNA-binding ligand in intact cells. Nucleic Acids Res., 29, 652–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Khalaf A.I., Pitt,A.R., Scobie,M., Suckling,C.J., Urwin,J., Waigh,R.K., Fishleigh,R.V. and Young,S.C. (2000) Synthesis of novel DNA binding agents: indole containing analogues of bis-netropsin. J. Chem. Res., 58, 264–265. [Google Scholar]
- 5.White S., Baird,E.E. and Dervan,P.B. (1997) On the pairing rules for recognition in the minor groove of DNA by pyrrole-imidazole polyamides. Chem. Biol., 4, 569–578. [DOI] [PubMed] [Google Scholar]
- 6.Boger D.L., Fink,B.E. and Hedrick,M.P. (2000) A solution phase combinatorial approach to the discovery of new, bioactive DNA binding agents and development of a rapid, high throughput screen for determining relative DNA binding affinity or DNA binding sequence selectivity. J. Am. Chem. Soc., 122, 6382–6394. [Google Scholar]
- 7.Hardenbol P., Wang,J.C. and Van Dyke,M.W. (1997) Identification of preferred hTBP DNA binding sites by the combinatorial method REPSA. Nucleic Acids Res., 25, 3339–3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Klug S.J. and Famulok,M. (1994) All you wanted to know about SELEX. Mol. Biol. Rep., 20, 97–107. [DOI] [PubMed] [Google Scholar]
- 9.Lindahl T. and Wood,R.D. (1999) Quality control by DNA repair. Science, 286, 1897–1905. [DOI] [PubMed] [Google Scholar]
- 10.Devchand P.R., McGhee,J.D. and Van de Sande,J.H. (1993) Uracil-DNA glycosylase as a probe for protein–DNA interactions. Nucleic Acids Res., 21, 3437–3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Walker W.L., Kopka,M.L. and Goodsell,D.S. (1997) Progress in the design of DNA sequence-specific lexitropsins. Biopolymers, 44, 323–334. [DOI] [PubMed] [Google Scholar]
- 12.Pelton J.G. and Wemmer,D.E. (1989) Structural characterization of a 2:1 distamycin A.d(CGCAAATTGGC) complex by two-dimensional NMR. Proc. Natl Acad. Sci. USA, 86, 5723–5727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang W., Dai,Y., Schmitz,U. and Bruice,T.W. (2001). A novel dicationic polyamide ligand binds in the DNA minor groove as a dimer. FEBS Lett., 509, 85–89. [DOI] [PubMed] [Google Scholar]
- 14.Geierstanger B.H., Mrksich,M., Dervan,P.B. and Wemmer,D.E. (1994) Design of a GC-specific DNA minor groove-binding peptide. Science, 266, 646–650. [DOI] [PubMed] [Google Scholar]
- 15.Burley S.K. and Roeder,R.G. (1996) Biochemistry and structural biology of transcription factor IID (TFIID). Annu. Rev. Biochem., 65, 769–799. [DOI] [PubMed] [Google Scholar]
- 16.David-Cordonnier M.H., Laval,J. and O′Neill,P. (2000) Clustered DNA damage, influence on damage excision by XRS5 nuclear extracts and Escherichia coli Nth and Fpg proteins. J. Biol. Chem., 275, 11865–11873. [DOI] [PubMed] [Google Scholar]
- 17.Parikh S.S., Mol,C.D., Slupphaug,G., Bharati,S., Krokan,H.E. and Tainer,J.A. (1998) Base excision repair initiation revealed by crystal structures and binding kinetics of human uracil-DNA glycosylase with DNA. EMBO J., 17, 5214–5226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jordan S.R. and Pabo,C.O. (1988) Structure of the lambda complex at 2.5 Å resolution: details of the repressor–operator interactions. Science, 242, 893–899. [DOI] [PubMed] [Google Scholar]