Abstract
The ability of viruses to control and/or evade the host antiviral response is critical to the establishment of a productive infection. One of the strategies utilized by West Nile virus (WNV) to circumvent the host response is to evade detection by the pathogen recognition receptor RIG-I early in infection. To begin elucidating the mechanisms by which WNV eludes detection, we undertook a systematic analysis of the WNV genome and antigenome to identify RIG-I-specific pathogen associated molecular patterns (PAMPs). Multiple segments of the WNV genome and anitigenome induced a RIG-I-specific antiviral response. However, incorporation of the stimulatory regions into larger RNAs substantially reduced their capacity to activate RIG-I. These results suggested that WNV evades the host response by sequestering RIG-I-specific PAMPs within the complete genome and antigenome at early times post-infection. Furthermore, activation of the RIG-I pathway may require the liberation of PAMPs by the cell’s normal RNA processing pathways.
Keywords: West Nile virus, RIG-I, Pathogen associated molecular patterns, Pathogen recognition receptor
Introduction
West Nile virus (WNV) is a positive-sense, single-stranded RNA virus from the family Flaviviridae. The genome is ~ 11 kb and consists of an open reading frame (ORF) flanked by 5′ and 3′ untranslated regions (UTRs). Within the UTRs, conserved sequences and predicted secondary structures encode the signals for negative strand synthesis, genome amplification, translation, and packaging. The incoming viral genomic RNA functions as a template for both translation, which produces a single polyprotein, and replication. The viral polyprotein is co- and post-translationally cleaved by host and viral proteases to generate ten individual proteins. The structural proteins, core (C), membrane (prM/M), and envelope (E), are involved in viral assembly, host cell binding, and entry. The seven nonstructural (NS) proteins (NS1, NS2A/B, NS3, NS4A/B, and NS5) support viral replication and evasion of the host antiviral response. Following polyprotein synthesis, genomic RNA is transcribed by the viral polymerase, NS5, to generate the complementary minus-strand antigenome, which serves as a template for synthesis of additional genomic RNA. Newly synthesized genomic RNA is either translated by the host cell or packaged into virus particles (Brinton, 2001, 2002; Chambers et al., 1990; Lindenbach et al., 2007).
In areas where WNV is endemic, such as the Middle East, Africa, and Asia, infection is typically asymptomatic or associated with a mild febrile illness known as West Nile fever (Petersen and Roehrig, 2001). In contrast, recent outbreaks in the Western hemisphere have been marked by an increase in disease severity, including meningitis, encephalitis, and acute flaccid paralysis (Hayes and Gubler, 2006; Nash et al., 2001; Klee et al., 2004). Since its introduction into the United States in 1999, WNV has spread to every state within the continental United States, as well as parts of Canada, Mexico and the Caribbean (information found on the CDC website www.cdc.gov/ncidod/dvbid/westnile/index.htm). As of January 2012, yearly outbreaks of WNV have resulted in 13,229 reported cases with neurological complications and 1,263 deaths, making WNV the leading cause of mosquito-borne neuroinvasive disease in the United States.
The ability to rapidly sense an invading pathogen and respond appropriately is a critical factor influencing the outcome of infection. The intracellular pathogen recognition receptor (PRR) RIG-I plays a critical role in sensing a wide variety of viruses, including positive strand viruses such as WNV, Japanese encephalitis virus (JEV), hepatitis C virus (HCV), dengue virus and poliovirus, and negative strand RNA viruses such as paramyxoviruses, influenza, and vesicular stomatitis virus (VSV) (Fredericksen et al., 2008; Loo et al., 2008; Saito et al., 2008; Kato et al., 2006; Rehwinkel et al., 2010). The fact that RIG-I detects a wide variety of viruses spanning multiple families suggests that it is able to interact with multiple substrates. Recent evidence indicates that double stranded structures within viral RNAs function as primary activators of RIG-I during infection (Baum et al., 2010; Berg et al., 2012; Kato et al., 2008; Malathi et al., 2010), though 5′ triphosphate (5′ppp) moieties enhance detection of short dsRNAs (Hornung et al., 2006; Pichlmair et al., 2006; Schlee et al., 2009; Schmidt et al., 2009). Additionally, poly-U/UC motifs in the genomes of HCV, measles, rabies and Ebola viruses are important for activating RIG-I (Saito et al., 2008). Once activated by a viral PAMP, RIG-I initiates a signaling cascade that results in the activation of latent transcription factors such as interferon regulatory factor 3 (IRF-3) (Hiscott, 2007; Stetson and Medzhitov, 2006; Takahasi et al., 2008). Activation of IRF-3 leads to the induction of type-I interferons (IFN) as well as a subset of antiviral effector proteins such as IFN-stimulated gene 56 (ISG56), (Hiscott, 2007; Kawai et al., 2005; Stetson and Medzhitov, 2006; Takahasi et al., 2008). The induction of these IRF-3 target genes results in the establishment of an antiviral state within in the cell, which blocks viral replication.
As eukaryotes evolved strategies to combat invading pathogens, viruses have co-evolved processes to escape them. While many viruses actively impede the RIG-I signaling pathway (Pachler and Vlasak, 2011; Gack et al., 2009; Masatani et al., 2010; Ling et al., 2009; Papon et al., 2009; Barral et al., 2009; Bowie and Unterholzner, 2008), the pathogenic strain of WNV, WNV New York (WNV-NY), eludes detection at early times post-infection (Fredericksen and Gale, 2006). However, the mechanism(s) by which WNV evades detection early during infection is currently unclear. We have previously demonstrated that cells treated with UV-inactivated WNV fail to induce an antiviral response, suggesting that RIG-I senses a product of viral replication (Fredericksen and Gale, 2006). Furthermore, the WNV-NY genome lacks a poly-U/UC region, suggesting that RIG-I senses either dsRNA structures within the WNV genome or anti-genome or an as of yet unidentified stimulatory motif. To distinguish between these possibilities, we undertook a systematic analysis of the WNV-NY genome and antigenome to define RIG-I-specific PAMPs. Multiple RIG-I stimulatory regions were identified throughout the WNV genome and antigenome. However, incorporation of these regions into larger RNAs abolished their stimulatory potential, suggesting that the WNV PAMPs are masked in the context of the full genome and antigenome. This masking of the PAMPs likely accounts for WNV’s ability to evade the innate immune response early during infection.
Results
The 5′ and 3′ UTRs of WNV induce an antiviral response
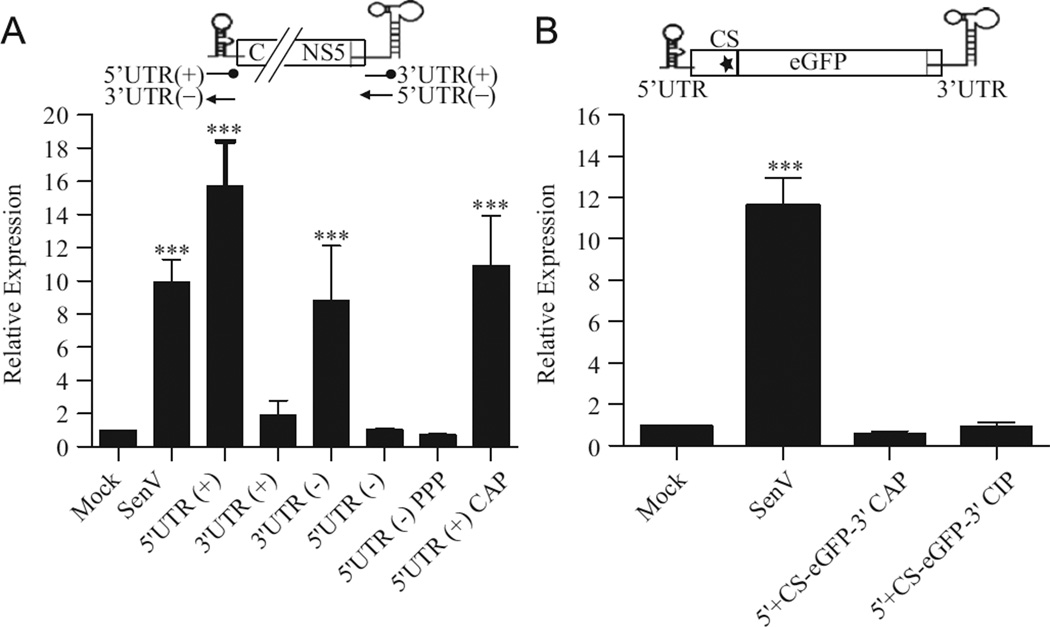
Because secondary structures within RNA are essential for recognition by and activation of RIG-I (Baum et al., 2010; Berg et al., 2012; Kato et al., 2008; Lu et al., 2010; Malathi et al., 2010; Myong et al., 2009; Rehwinkel et al., 2010; Schlee et al., 2009; Schmidt et al., 2009), we hypothesized that the highly structured 5′ and 3′ UTRs of WNV stimulate RIG-I. Therefore, we assessed the capacity of these regions to stimulate a RIG-I-dependent antiviral response using a luciferase reporter assay. Huh7 cells were transfected with a RIG-I-dependent luciferase reporter, ISG56-luc (Grandvaux et al., 2002), and subsequently either infected with Sendai Virus (SenV), a potent RIG-I-specific activator of the innate immune response, or transfected with RNAs corresponding to the genomic (+) or antigenomic (−) 5′ and 3′ UTRs. Cell lysates were recovered and analyzed for luciferase expression 8 h after RNA transfection to ensure that the induction of luciferase expression was due to the primary activation of the RIG-I signal transduction pathway and not subsequent feedback amplification loops. Significant induction of luciferase expression was detected in SenV-infected cells as well as cultures transfected with the WNV 5′ (+) and 3′ (−) UTRs (Fig. 1A). In contrast, neither the WNV 3′ UTR (+) nor WNV 5′ UTR (−) fragments induced luciferase expression. In order to more accurately mimic the RNAs present during a native infection, the stimulatory capacity of the 5′ UTR (+) containing a cap structure and the 5′ UTR (−) containing a 5′ PPP were also examined (Fig. 1A). Neither the presence of a capped structure on the 5′ UTR ( + ) nor a 5′ PPP on the 5′ UTR (−) altered the stimulatory capacity of these fragments.
Fig. 1.
The 5′ UTR( + ) and 3′ UTR( −) region of the WNV-NY genome induce an antiviral response. Huh7 monolayers were transfected with pISG56-luc and pCMV-Renilla 16 h prior to infection with SenV or transfection with 500 ng of the indicated RNAs in triplicate. Cell lysates were prepared 8 h post-transfection and assayed for luciferase activity. Values represent the average luciferase expression compared to mock ( ± standard error) from a minimum of two independent experiments. Statistical analysis was performed using Dunnett’s multiple comparison analysis, *p <0.05, **p <0.01 ***p < 0.001. (A) Cells were transfected with RNA fragments corresponding to the genomic or antigenomic 5′ and 3′ UTRs. (B) Cells were transfected with 5′ UTR+CS-eGFP-3′UTR RNAs.
The robust induction of the antiviral response by the capped 5′ UTR (+) fragment suggested that the incoming viral genome is capable of functioning as a PAMP for RIG-I detection. However, the failure of WNV to initiate a rapid antiviral response indicates that this region is not accessible to RIG-I early in infection (Fredericksen and Gale, 2006). Interactions between the 5′ and 3′ ends of the WNV genome have been shown to be essential for genome replication, suggesting that the 5′ UTR does not function as an independent structural element during infection (Brinton and Dispoto, 1988; Hahn et al., 1987; Khromykh et al., 2001; Zhang et al., 2008). Therefore, we generated a construct consisting of the 5′ UTR to the conserved sequences (CS) element, located in the N-terminal coding region of the C gene, linked to the 3′ UTR by the egfp reporter gene (5’ UTR+CS-eGFP-3′UTR). Neither CIP-treated nor capped 5’UTR+CS-eGFP-3′UTR RNAs stimulated luciferase expression (Fig. 1B), suggesting that the 5′ UTR is not a functional PAMP when presented to the cell in this context.
Multiple regions of the WNV genome and antigenome induce the host antiviral response
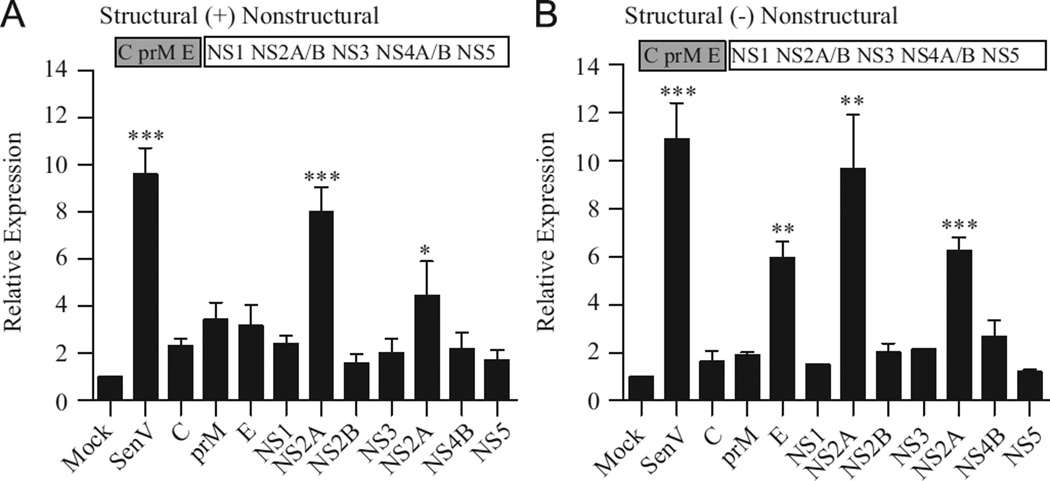
To determine whether other regions of the genome also harbor RIG-I PAMPs, the stimulatory capacity of RNAs corresponding to the individual WNV genes was assessed. While several fragments induced low to moderate levels of luciferase expression, only RNAs corresponding to NS2a (+), E (−), NS2a (−) and NS4a (−) functioned as potent stimulators of the antiviral response (Fig. 2A and B). Thus, multiple regions within the WNV genome and antigenome are capable of stimulating an antiviral response.
Fig. 2.
Multiple regions of the WNV genome and antigenome induce an antiviral response. Cells were transfected with ISG56-luc and pCMV-Renilla as described in Fig. 1. Cultures were subsequently transfected with 500 ng of RNA fragments corresponding to genomic (A) or antigenomic (B) orientation of WNV genes. Luciferase expression was assessed as described in Fig. 1.
The antiviral response to WNV PAMPs is RIG-I-dependent
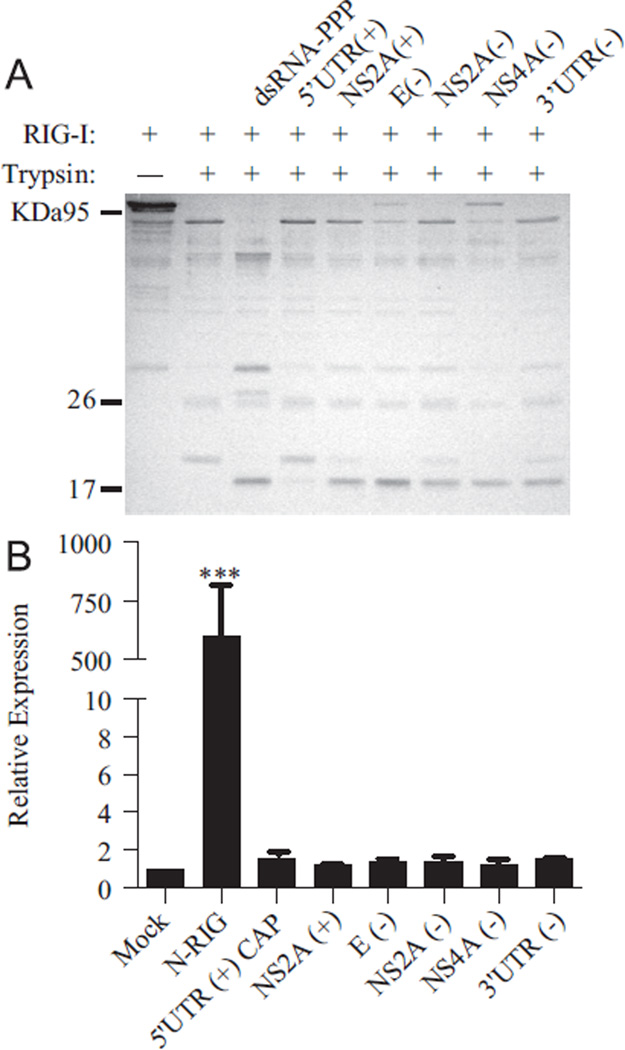
The rapid and robust induction of luciferase expression by the 5′ UTR (+), 3′ UTR (−), NS2a (+), E (−), NS2a (−) and NS4a (−) RNAs was indicative of a RIG-I-dependent response. To confirm the specificity of this response, the ability of the identified PAMPs to form a stable complex with RIG-I was examined using limited trypsin digestion. This assay assesses the ability of a PAMP to induce conformational changes in RIG-I associated with the generation of the signaling-active form (Saito et al., 2007, 2008; Takahasi et al., 2008). Incubation of Escherichia coli-purified RIG-I with each of the WNV PAMPs identified above resulted in the accumulation of a 17-kDa trypsin resistant band indicative of the signaling-active from of RIG-I (Fig. 3A). However, only a weak band was detected in the presence of the 5′ UTR (+) fragment, suggesting that, unlike the other PAMPS, this fragment does not bind tightly to RIG-I. We further verified the specificity of the identified PAMPs using the RIG-I-deficient Huh7.5 cell line (Fig. 3B) (Blight et al., 2002; Sumpter et al., 2005). High levels of luciferase expression were detected in control cells transfected with a constitutively active form of RIG-I, N-RIG. In contrast, WNV RNA fragments did not induce an antiviral response in the absence of functional RIG-I. Together, these findings indicated that the WNV PAMPs identified above induce a RIG-I-dependent antiviral response.
Fig. 3.
WNV PAMPs induce a RIG-I-specific antiviral response. (A) Purified RIG-I was incubated with control dsRNA or the indicated WNV RNA fragments prior to the addition of trypsin. Digestion products were separated on a 12.5% SDS polyacrylamide gel and visualized by Coomassie stain. (B) Huh7.5 monolayers were transfected with luciferase reporter constructs and 500 ng of the indicated RNAs or pEF-flagN-RIG, as described in Fig. 1. Luciferase expression was assessed as described in Fig. 1.
Incorporation of WNV-NY PAMPs into larger RNAs masks their stimulatory capacity
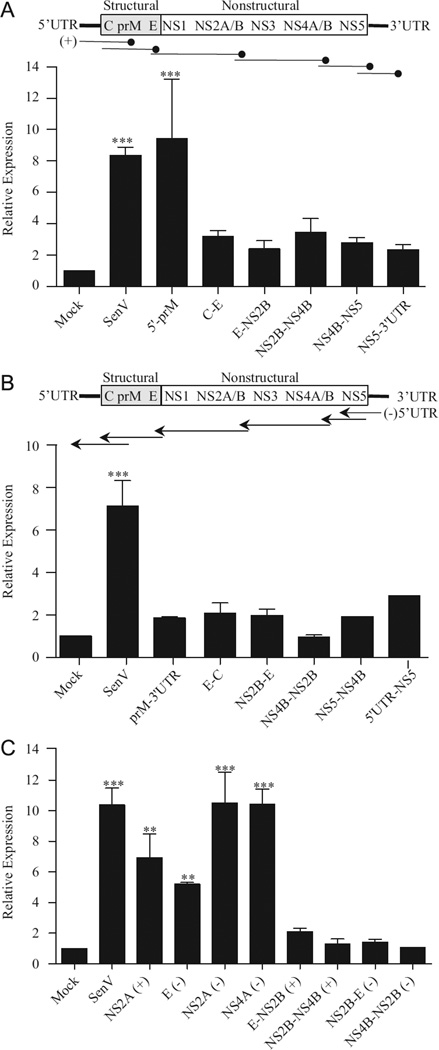
To determine whether WNV RNA sequences or structures spanning the individual genes are capable of stimulating RIG-I, we also tested the stimulatory capacity of overlapping segments of the viral genome and antigenome (Fig. 4A and B). With the exception of the 50′ UTR-prM ( + ) fragment, the overlapping segments failed to induce high levels of luciferase expression, despite the fact that several segments contained the highly stimulatory NS2a ( + ), E (−), NS2a (−) or NS4a (−) regions (Fig. 4A and B). In this experiment, we transfected cells with equivalent amounts of each of the RNAs based on mass. To ensure that the lack of stimulation by the larger fragments was not due to differences in the RNA copy number introduced into the cell, we also assessed the stimulatory capacity of these regions by transfecting equivalent moles of the various fragments. While similar levels of induction were detected for the NS2a (+), E (−), NS2a (−) and NS4a (−) regions under these conditions, equal molar amounts of the larger RNA fragments did not induce luciferase expression (Fig. 4C). This suggests that RIG-I is unable to efficiently detect WNV PAMPs in the context of a larger RNA even when equivalent RNA copy numbers are introduced into the cell.
Fig. 4.
Larger RNAs containing WNV PAMPs fail to activate an antiviral response. Huh7 monolayers were transfected with ISG56-luc and pCMV-Renilla as described in Fig. 1 and subsequently transfected with 500 ng of RNA fragments corresponding to the indicated sections of the WNV genome (A) or antigenome (B) or with equal molar amounts (2 pmol) of the indicated RNAs (C). Luciferase expression was assessed as described in Fig. 1.
WNV genomic and subgenomic RNAs do not induce RIG-I activation
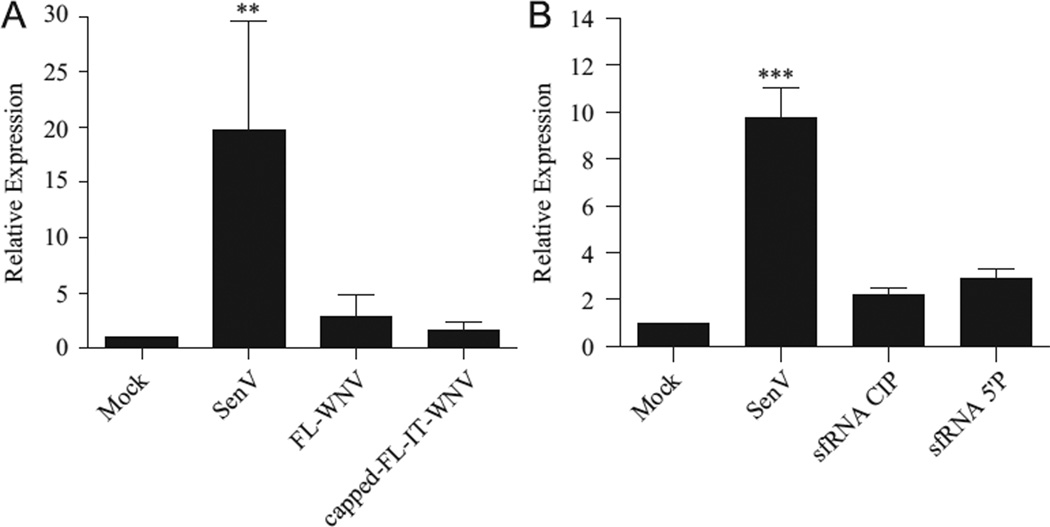
The lack of RIG-I activation by the 50′ UTR+CS-eGFP-30UTR RNAs and the larger overlapping segments of the viral genome suggested that the stimulatory capacity of these PAMPs is masked in the context of full-length viral RNAs. Therefore, we assessed the stimulatory capacity of full-length genomic RNA isolated from culture supernatants (FL-WNV) or produced through in vitro transcription and capping (capped-FL-IT-WNV) (Fig. 5A). Both forms of genomic RNA failed to induce luciferase expression, further supporting the hypothesis that WNV PAMPs are masked in the context of the full genome.
Fig. 5.
WNV genomic and subgenomic RNAs do not induce an antiviral response. Cells were transfected with ISG56-luc and pCMV-Renilla as described in Fig. 1. Cultures were subsequently transfected with 500 ng of: (A) genomic RNA isolated from culture supernatants (FL-WNV) or full length in vitro transcribed and capped WNV genomic RNA (capped-FL-IT-WNV) or (B) the indicated sfRNA constructs. Luciferase expression was assessed as described in Fig. 1 the indicated sfRNA constructs. Luciferase expression was assessed as described in Fig. 1.
Together our findings suggested that processing of the viral genome and antigenome is necessary to expose the identified RIG-I PAMPs. During infection, incomplete digestion of genomic viral RNA by the 50′-30′ exoribonuclease XRN1 results in the accumulation of a highly structured 30′ UTR-derived monophosphorylated subgenomic RNA (sfRNA) (Brinton et al., 1986; Funk et al., 2011; Pijlman et al., 2008). To determine if sfRNA is a PAMP, we tested the stimulatory capacity of both a CIP-treated and a monophosphorylated sfRNA construct. Although luciferase expression in cells transfected with either CIP-treated or monophosphorylated sfRNA was consistently higher than background levels, the increase in luciferase expression did not reach statistical significance for either RNA (Fig. 5B). This finding suggests that sfRNA does not function as a major PAMP during WNV infection and is consistent with previous reports that the sfRNA promotes WNV pathogenesis (Pijlman et al., 2008; Schuessler et al., 2012).
Discussion
In this study we utilized a systematic approach to identify RIG-I-dependent PAMPs present in the WNV genome and antigenome. Our findings demonstrated that WNV, in contrast to many other viruses (Kato et al., 2006; Plumet et al., 2007; Saito et al., 2008; Wang et al., 2008), encodes multiple segments located throughout the genome and antigenome that are capable of stimulating RIG-I. The lack of sequence similarity between the stimulatory regions suggests that RIG-I detects dsRNA structures within these regions rather than a specific sequence motif. Secondary structural predictions indicate that the identified stimulatory regions within the WNV genome are highly structured (Supplemental Figs. S1–5). While these structural predictions need to be confirmed experimentally, they provide a conceptual scaffold to begin defining the structural elements involved in RIG-I detection. Our findings also suggest that the intact WNV genome and antigenome are poor activators of RIG-I. Larger RNA fragments as well as the full-length genomic RNA failed to induce the rapid activation of the antiviral response. These findings are consistent with a recent report demonstrating that RIG-I does not bind to the full-length genome of SenV (Baum et al., 2010).
Multiple factors may play a role in masking WNV PAMPs within full-length viral RNAs. It is possible that the secondary structures detected by RIG-I are altered or abolished in the context of the full genome. Alternatively, tertiary structures within the viral RNA may sterically hinder RIG-I’s ability to bind the PAMPs. Comparison of the secondary structure predictions for the 5′UTR alone and the 5′UTR in the context of the 5′UTR-prM fragment suggests that the structure of this region is not substantially altered when incorporated into larger RNAs (Supplemental Fig. S6). However, in the case of the incoming genome, the stimulatory capacity of the capped 5′ UTR (+) may be obstructed by several additional factors. Upon entering cells, the viral genome undergoes cap-dependent translation to generate the viral polyprotein. Thus, host translation machinery may block RIG-I’s access to this region. The viral genomic RNA also serves as a template for synthesis of the negative sense anti-genome, which requires interactions between the conserved sequence (CS) elements located within the 5′ end of core and the 3′ UTR as well as base pairing between a 5′ UAR (upstream initiation AUG region element) and a 3′ UAR (Brinton and Dispoto, 1988; Zhang et al., 2008; Hahn et al., 1987; Khromykh et al., 2001). Therefore, long distance interactions between the 5′ and 3′ UTRs may sequester the stimulatory signals within the 5′ UTR early in infection. The observation that the 5′UTR-prM fragment stimulated RIG-I, while the 5′UTR+CS-eGFP-3′UTR fragment did not, is consistent with this hypothesis.
Our findings also suggested that WNV evades detection at early times post-infection by sequestering potential PAMPS within the viral genome and antigenome. The mechanism(s) by which WNV PAMPs eventually become accessible to RIG-I during the course of infection are currently under investigation. As previously demonstrated for SenV and influenza virus (Baum et al., 2010), subgenomic defective interfering (DI) particles may play a role in stimulating RIG-I activation. The deletion of large segments from the viral genome may result in the exposure of WNV PAMPs that were previously buried within the full-length viral RNAs, thus providing the proper context for RIG-I recognition. In addition, the normal cellular processes of RNA degradation may be involved in the exposing of WNV PAMPs within the genome during the course of infection. This hypothesis is supported by the observation that large fragments of the WNV genome, which fail to induce the rapid activation of RIG-I, induce an antiviral response approximately 30 h post-transfection (Fredericksen, unpublished results). The fact that the subgenomic sfRNA of WNV failed to substantially activate RIG-I suggested that the exonuclease XRN1 is not involved in liberating RIG-I PAMPs. However, other cellular pathways, such as the “no-go” RNA degradation pathway, which is triggered by stalled ribosomal movement, may be involved in the processing of WNV viral RNAs (Doma and Parker, 2006; Tomecki and Dziembowski, 2010). Furthermore, as has been demonstrated for HCV, activation of the OAS/RNAseL pathway later in infection may liberate additional PAMPs that help sustain and/or amplify the antiviral response (Malathi et al., 2010; Scherbik et al., 2006). In the case of the WNV antigenome, alternative RNA processing pathways may be required for the release of virally encoded PAMPs. The antigenome of WNV exists within the cell in two forms, the double-stranded replicative form (ds-RF) and the replicative intermediate (RI). The ds-RF consists of a nascent antigenome paired with the genome template, while the RI consists of a single copy of the viral antigenome and multiple strands of nascent genomic RNA being synthesized (Gillespie et al., 2010; Westaway et al., 1999). Detection of viral antigenome by the host cell may be limited due to the fact that the RI is sequestrated within membrane invagination. However, processing the ds-RF by either the cell’s RNAi or ADAR/TSN pathway may liberate PAMPs from the WNV antigenome (Scadden, 2005; Umbach and Cullen, 2009). These mechanisms, as well as other as of yet unknown cellular degradation pathways, may contribute, individually or in concert, to the liberation of PAMPs. Further analysis of the WNV RNAs produced both in vitro and in vivo will be necessary to more precisely map the WNV stimulatory regions within the viral genome and antigenome and to elucidate how PAMPs are produced over the course of infection.
Materials and methods
Cells and viruses
Huh7 and Huh7.5 (Apath) cells were propagated in Dulbelco’s modified Eagle’s medium (DMEM) supplemented with 10% FBS, 2 mM L-glutamine, 1 mM sodium pyruvate, 1 mM non-essential amino acids and antibiotic/antimycotic solution (complete DMEM). Sendai virus (SenV), Cantell strain was obtained from Charles River.
Plasmids
pFLWNV (Shi et al., 2002) was used as a template for PCR-amplification of the indicated segments of the WNV-NY genome (Table 1). Primer sequences are available upon request. Amplified segments were cloned into pVL-blunt (a gift from Dr. Vincent Lee), pBluKSM or pWSK29 (a gift from Dr. Sydney Kushner, Wang and Kushner, 1991) and the sequence confirmed. The reporter plasmids pISG56-luc ( a gift from Dr. Ganes Sens, Grandvaux et al., 2002) and pCMV-Renilla (Promega) encode the firefly luciferase gene under transcriptional control of the ISG56 promoter and the Renilla luciferase gene under the constitutively active cytomegalovirus (CMV) early promoter, respectively. pEF-flag-N-RIG encodes the constitutively active N-terminus of RIG-I (Yoneyama et al., 2004).
Table 1.
Position in the WNV-NY genome of RNA used in experiments. The nucleotide positions are based on the sequence from GenBank accession no. AF404756.
| Name | Location in genome |
|---|---|
| 5′ UTR | 1–97 |
| Core (C) | 98–465 |
| Pre-membrane (prM) | 406–966 |
| Envelope (E) | 904–2469 |
| Non-structural 1 (NS1) | 2407–3525 |
| Non-structural 2a (NS2a) | 3526–4218 |
| Non-structural 2b (NS2b) | 4219–4611 |
| Non-structural 3 (NS3) | 4612–6468 |
| Non-structural 4a (NS4a) | 6469–6915 |
| Non-structural 4b (NS4b) | 6916–7680 |
| Non-structural 5 (NS5) | 7681–10395 |
| 3′ UTR | 10396–11029 |
| 5′ UTR to conserved sequence (5′UTR+CS) | 1–196 |
| 5′ UTR-prM | 1–966 |
| C-E | 98–2467 |
| E-NS2b | 979–4102 |
| NS2b–NS4b | 4231–7679 |
| NS4b–NS5 | 6916–10389 |
| NS5-3′UTR | 7681–11029 |
| 5′ UTR+Cs-eGFP-3′UTR | 1–196, 10396–11029 |
RNA fragments
In vitro transcribed RNA fragments were generated according to the manufacturer’s protocol (Ampliscribe T3 and T7 kits; Epicentre). Briefly, a 20 µl reaction mixture containing 1 µg of linearized plasmid encoding the indicated RNAs was incubated at 42 °C for 2 h. DNA template was subsequently removed from the reaction by treating with DNase I. RNA was recovered by phenol/ chloroform extraction followed by ethanol precipitation. Unless otherwise indicated, RNAs were treated with calf alkaline phosphatase (CIP) to remove free 5′ triphosphates (New England Biolabs). The purity of the RNAs was confirmed by polyacrylamide gel electrophoresis. The NS2b fragment was further purified on a 6% polyacrilamide gel containing 8 M urea. The RNA was eluted overnight at 4 °C from the excised gel fragments in buffer containing 200 mM NaCl, 10 mM Tris-HCl pH 7.5, 1 mM EDTA. The eluted RNA was precipitated with ethanol, aliquoted and stored at −80°C. The Amplicap kit (Epicentre) was used to generate the capped 5′ UTR segment and full length genomic RNA. Monophosphorylated sub-genomic WNV RNA (sfRNA) was generated by incubating CIP-treated in vitro transcribed sfRNA with T4 polynucleotide kinase according to the manufacturer’s instructions (NEB). WNV genomic RNA was isolated from culture supernatants recovered from WNV-NY infected cells. Cell debris was removed by low speed centrifugation at 1500 rpm for 5 min and genomic RNA was recovered by Trizol (Invitrogen) extraction.
Luciferase reporter assays
Subconfluent monolayers of Huh7 or Huh7.5 cells in a 48 well plate were transfected with 100 ng of ISG56-luc and 20 ng of pCMV-Renilla using Lipofectamine 2000 transfection reagent (Invitrogen). Where indicated, cells were also transfected with 200 ng of pEF-flagN-RIG. At 16 h post-transfection, cells were mock-transfected, transfected with 500 ng or 2 pmol of the indicated RNAs using TransMessenger transfection reagent (Qiagen), or infected with SenV (100 HA units), in triplicate. Cell lysates were prepared 8 h after RNA transfection or infection with SenV and luciferase levels were detected using a dual luciferase kit according to the manufacturer’s protocol (Promega). Luciferase activity was quantified using a Bert-hold Centro XS3 LB960 luminometer. Normalized luciferase levels were determined by dividing firefly luciferase levels by control Renilla luciferase levels. Values represent the average luciferase expression compared to mock (±standard error) from a minimum of two independent experiments. Statistical analysis was performed using Dunnett’s multiple comparison analysis.
Trypsin digestion
Control dsRNA (Invivogen) or the indicated WNV RNA fragments (30 pmol) were incubated with purified E. coli-produced RIG-I (15 pmol) for 15 min at room temperature. The RNA/RIG-I mixtures were digested with trypsin (0.83 mg) for 15 min at 37 1C. Trypsin was inactivated by the addition of protease inhibitor (Sigma) and the digestion products were separated on a 12.5% SDS polyacrylamide gel. Bands were visualized using Imperial Protein Stain (Thermo scientific).
Supplementary Material
Acknowledgments
This work was supported by NIH Grant AI047990 (RAM) and NIH Grant AI083397 (BLF).
Footnotes
Appendix A. Supporting information
Supplementary data associated with this article can be found in the online version at http://dx.doi.org/10.1016/j.virol.2012.06.009.
References
- Barral PM, Sarkar D, Fisher PB, Racaniello VR. RIG-I is cleaved during picornavirus infection. Virology. 2009;391:171–176. doi: 10.1016/j.virol.2009.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum A, Sachidanandam R, Garcia-Sastre A. Preference of RIG-I for short viral RNA molecules in infected cells revealed by next-generation sequencing. Proc. Natl. Acad. Sci. USA. 2010;107:16303–16308. doi: 10.1073/pnas.1005077107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg RK, Melchjorsen J, Rintahaka J, Diget E, Søby S, Horan KA, Gorelick RJ, Matikainen S, Larsen CS, et al. Genomic HIV RNA induces innate immune responses through RIG-I-dependent sensing of secondary-structured RNA. In: Sommer P, editor. PLoS ONE. Vol. 7. 2012. p. e29291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blight KJ, McKeating JA, Rice CM. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J. Virol. 2002;76:13001–13014. doi: 10.1128/JVI.76.24.13001-13014.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowie AG, Unterholzner L. Viral evasion and subversion of patternrecognition receptor signalling. Nat. Rev. Immunol. 2008;8:911–922. doi: 10.1038/nri2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinton MA. Host factors involved in West Nile virus replication. Ann. NY. Acad. Sci. 2001;951:207–219. doi: 10.1111/j.1749-6632.2001.tb02698.x. [DOI] [PubMed] [Google Scholar]
- Brinton MA. The molecular biology of West Nile Virus: a new invader of the western hemisphere. Annu. Rev. Microbiol. 2002;56:371–402. doi: 10.1146/annurev.micro.56.012302.160654. [DOI] [PubMed] [Google Scholar]
- Brinton MA, Dispoto JH. Sequence and secondary structure analysis of the 5′-terminal region of flavivirus genome RNA. Virology. 1988;162:290–299. doi: 10.1016/0042-6822(88)90468-0. [DOI] [PubMed] [Google Scholar]
- Brinton MA, Fernandez AV, Dispoto JH. The 3′-nucleotides of flavivirus genomic RNA form a conserved secondary structure. Virology. 1986;153:113–121. doi: 10.1016/0042-6822(86)90012-7. [DOI] [PubMed] [Google Scholar]
- Chambers TJ, Hahn CS, Galler R, Rice CM. Flavivirus genome organization, expression, and replication. Ann. Rev. Microbiol. 1990;44:649–688. doi: 10.1146/annurev.mi.44.100190.003245. [DOI] [PubMed] [Google Scholar]
- Doma MK, Parker R. Endonucleolytic cleavage of eukaryotic mRNAs with stalls in translation elongation. Nature. 2006;440:561–564. doi: 10.1038/nature04530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredericksen BL, Gale M. West Nile virus evades activation of interferon regulatory factor 3 through RIG-I-dependent and -independent pathways without antagonizing host defense signaling. J. Virol. 2006;80:2913–2923. doi: 10.1128/JVI.80.6.2913-2923.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredericksen BL, Keller BC, Fornek J, Katze MG, Gale M., Jr Establishment and maintenance of the innate antiviral response to West Nile virus involves both RIG-I and MDA5 signaling through IPS-1. J. Virol. 2008;82:609–616. doi: 10.1128/JVI.01305-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funk A, Truong K, Nagasaki T, Torres S, Floden N, Balmori Melian E, Edmonds J, Dong H, Shi PY, Khromykh AA. RNA structures required for production of subgenomic flavivirus RNA. J. Virol. 2011;84:11407–11417. doi: 10.1128/JVI.01159-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gack MU, Albrecht RA, Urano T, Inn K, Huang I, Carnero E, Farzan M, Inoue S, Jung J, García-Sastre A. Influenza A virus NS1 targets the ubiquitin ligase TRIM25 to evade recognition by the host viral RNA sensor RIG-I. Cell Host and Microbe. 2009;5:439–449. doi: 10.1016/j.chom.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie LK, Hoenen A, Morgan G, Mackenzie JM. The endoplasmic reticulum provides the membrane platform for biogenesis of the flavivirus replication complex. J. Virol. 2010;84:10438–10447. doi: 10.1128/JVI.00986-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandvaux N, Servant MJ, tenOever B, Sen GC, Balachandran S, Barber GN, Lin R, Hiscott J. Transcriptional profiling of interferon regulatory factor 3 target genes: direct involvement in the regulation of interferon-stimulated genes. J. Virol. 2002;76:5532–5539. doi: 10.1128/JVI.76.11.5532-5539.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn CS, Hahn YS, Rice CM, Lee E, Dalgarno L, Strauss EG, Strauss JH. Conserved elements in the 3′ untranslated region of flavivirus RNAs and potential cyclization sequences. J. Mol. Biol. 1987;198:33–41. doi: 10.1016/0022-2836(87)90455-4. [DOI] [PubMed] [Google Scholar]
- Hayes EB, Gubler DJ. West Nile virus: epidemiology and clinical features of an emerging epidemic in the United States. Annu. Rev. Med. 2006;57:181–194. doi: 10.1146/annurev.med.57.121304.131418. [DOI] [PubMed] [Google Scholar]
- Hiscott J. Triggering the innate antiviral response through IRF-3 activation. J. Biol. Chem. 2007;282:15325–15329. doi: 10.1074/jbc.R700002200. [DOI] [PubMed] [Google Scholar]
- Hornung V, Ellegast J, Kim S, Brzozka K, Jung A, Kato H, Poeck H, Akira S, Conzelmann KK, et al. 5′-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314:994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, Uematsu S, Jung A, Kawai T, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- Kato H, Takeuchi O, Mikamo-Satoh E, Hirai R, Kawai T, Matsushita K, Hiiragi A, Dermody TS, Fujita T, Akira S. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J. Exp. Med. 2008;205:1601–1610. doi: 10.1084/jem.20080091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai T, Takahashi K, Sato S, Coban C, Kumar H, Kato H, Ishii KJ, Takeuchi O, Akira S. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat. Immunol. 2005;6:981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- Khromykh AA, Meka H, Guyatt KJ, Westaway EG. Essential role of cyclization sequences in flavivirus RNA replication. J. Virol. 2001;75:6719–6728. doi: 10.1128/JVI.75.14.6719-6728.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klee AL, Maidin B, Edwin B, Poshni I, Mostashari F, Fine A, Layton M, Nash D. Long-term prognosis for clinical West Nile virus infection. Emerg. Infect. Dis. 2004;10:1405–1411. doi: 10.3201/eid1008.030879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindenbach BD, Thiel H, Rice CM. Flaviviridae: The viruses and their replication. In: Knipe DM, Howley PM, editors. Fields Virology. 5th edn. Philadelphia: Lippincott-Raven Publishers; 2007. [Google Scholar]
- Ling Z, Tran KC, Teng MN. Human respiratory syncytial virus nonstructural protein NS2 antagonizes the activation of beta interferon transcription by interacting with RIG-I. J. Virol. 2009;83:3734–3742. doi: 10.1128/JVI.02434-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo YM, Fornek J, Crochet N, Bajwa G, Perwitasari O, Martinez-Sobrido L, Akira S, Gill MA, Garcia-Sastre A, et al. Distinct RIG-I and MDA5 signaling by RNA viruses in innate immunity. J. Virol. 2008;82:335–345. doi: 10.1128/JVI.01080-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Xu H, Ranjith-Kumar CT, Brooks MT, Hou TY, Hu F, Herr AB, Strong RK, Kao CC, Li P. The structural basis of 5′-triphosphate doublestranded RNA recognition by RIG-I C-terminal domain. Structure. 2010;18:1032–1043. doi: 10.1016/j.str.2010.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malathi K, Saito T, Crochet N, Barton DJ, Gale M, Jr, Silverman RH. RNase L releases a small RNA from HCV RNA that refolds into a potent PAMP. RNA. 2010;16:2108–2119. doi: 10.1261/rna.2244210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masatani T, Ito N, Shimizu K, Ito Y, Nakagawa K, Sawaki Y, Koyama H, Sugiyama M. Rabies virus nucleoprotein functions to evade activation of the RIG-I-mediated antiviral response. J. Virol. 2010;84:4002–4012. doi: 10.1128/JVI.02220-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myong S, Cui S, Cornish PV, Kirchhofer A, Gack MU, Jung JU, Hopfner KP, Ha T. Cytosolic viral sensor RIG-I is a 5′-triphosphate-dependent translocase on double-stranded RNA. Science. 2009;323:1070–1074. doi: 10.1126/science.1168352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nash D, et al. The outbreak of West Nile Virus infection in the New York city area in 1999. N. Engl. J Med. 2001;344:1807–1814. doi: 10.1056/NEJM200106143442401. [DOI] [PubMed] [Google Scholar]
- Pachler K, Vlasak R. Influenza C virus NS1 protein counteracts RIG-I-mediated IFN signalling. Virology journal. 2011;8:48. doi: 10.1186/1743-422X-8-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papon L, Oteiza A, Imaizumi T, Kato H, Brocchi E, Lawson TG, Akira S, Mechti N. The viral RNA recognition sensor RIG-I is degraded during encephalomyocarditis virus (EMCV) infection. Virology. 2009;393:311–318. doi: 10.1016/j.virol.2009.08.009. [DOI] [PubMed] [Google Scholar]
- Petersen LR, Roehrig JT. West Nile virus: a reemerging global pathogen. Emerg. Infect. Dis. 2001;7:611–614. doi: 10.3201/eid0704.010401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichlmair A, Schulz O, Tan CP, Näslund TI, Liljeström P, Weber F, Reis e, Sousa C. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science. 2006;314:997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- Pijlman GP, Funk A, Kondratieva N, Leung J, Torres S, van der Aa L, Liu WJ, Palmenberg AC, Shi PY, et al. A highly structured, nuclease-resistant, noncoding RNA produced by flaviviruses is required for pathogenicity. Cell Host Microbe. 2008;4:579–591. doi: 10.1016/j.chom.2008.10.007. [DOI] [PubMed] [Google Scholar]
- Plumet S, Herschke F, Bourhis JM, Valentin H, Longhi S, Gerlier D. Cytosolic 5′-triphosphate ended viral leader transcript of measles virus as activator of the RIG I-mediated interferon response. PLoS One. 2007;2:e279. doi: 10.1371/journal.pone.0000279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehwinkel J, Tan CP, Goubau D, Schulz O, Pichlmair A, Bier K, Robb N, Vreede F, Barclay W, et al. RIG-I detects viral genomic RNA during negative-strand RNA virus infection. Cell. 2010;140:397–408. doi: 10.1016/j.cell.2010.01.020. [DOI] [PubMed] [Google Scholar]
- Saito T, Hirai R, Loo YM, Owen D, Johnson CL, Sinha SC, Akira S, Fujita T, Gale M. Regulation of innate antiviral defenses through a shared repressor domain in RIG-I and LGP2. Proc. Natl. Acad. Sci. USA. 2007;104:582–587. doi: 10.1073/pnas.0606699104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito T, Owen DM, Jiang F, Marcotrigiano J, Gale M., Jr Innate immunity induced by composition-dependent RIG-I recognition of hepatitis C virus RNA. Nature. 2008;454:523–527. doi: 10.1038/nature07106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scadden ADJ. The RISC subunit Tudor-SN binds to hyper-edited double-stranded RNA and promotes its cleavage. Nat. Struct. Mol. Biol. 2005;12:489–496. doi: 10.1038/nsmb936. [DOI] [PubMed] [Google Scholar]
- Scherbik SV, Paranjape JM, Stockman BM, Silverman RH, Brinton MA. RNase L plays a role in the antiviral response to West Nile virus. J. Virol. 2006;80:2987–2999. doi: 10.1128/JVI.80.6.2987-2999.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlee M, Roth A, Hornung V, Hagmann CA, Wimmenauer V, Barchet W, Coch C, Janke M, Mihailovic A, et al. Recognition of 5′-triphosphate by RIG-I helicase requires short blunt double-stranded RNA as contained in panhandle of negative-strand virus. Immunity. 2009;31:25–34. doi: 10.1016/j.immuni.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt A, Schwerd T, Hamm W, Hellmuth JC, Cui S, Wenzel M, Hoffmann FS, Michallet MC, Besch R, et al. 5′-triphosphate RNA requires basepaired structures to activate antiviral signaling via RIG-I. Proc. Natl. Acad. Sci. USA. 2009;106:12067–12072. doi: 10.1073/pnas.0900971106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuessler A, Funk A, Lazear H, Cooper DA, Torres S, Daffis S, Jha BK, Kumagai Y, Takeuchi O, et al. West Nile virus non-coding subgenomic RNA contributes to viral evasion of type I interferon-mediated antiviral response. J. Virol. 2012;86:5708–5718. doi: 10.1128/JVI.00207-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi PY, Tilgner M, Lo MK, Kent KA, Bernard KA. Infectious cDNA Clone of the Epidemic West Nile Virus from New York City. J. Virol. 2002;76:5847–5856. doi: 10.1128/JVI.76.12.5847-5856.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stetson DB, Medzhitov R. Antiviral defense: interferons and beyond. J. Exp. Med. 2006;203:1837–1841. doi: 10.1084/jem.20061377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumpter R, Loo YM, Foy E, Li K, Yoneyama M, Fujita T, Lemon SM, Gale M. Regulating intracellular antiviral defense and permissiveness to hepatitis C virus RNA replication through a cellular RNA helicase, RIG-I. J. Virol. 2005;79:2689–2699. doi: 10.1128/JVI.79.5.2689-2699.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahasi K, Yoneyama M, Nishihori T, Hirai R, Kumeta H, Narita R, Gale M, Jr, Inagaki F, Fujita T. Nonself RNA-sensing mechanism of RIG-I helicase and activation of antiviral immune responses. Mol Cell. 2008;29:428–440. doi: 10.1016/j.molcel.2007.11.028. [DOI] [PubMed] [Google Scholar]
- Tomecki R, Dziembowski A. Novel endoribonucleases as central players in various pathways of eukaryotic RNA metabolism. RNA. 2010;16:1692–1724. doi: 10.1261/rna.2237610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umbach JL, Cullen BR. The role of RNAi and microRNAs in animal virus replication and antiviral immunity. Genes Dev. 2009;23:1151–1164. doi: 10.1101/gad.1793309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Gao X, Barrett JW, Shao Q, Bartee E, Mohamed MR, Rahman M, Werden S, Irvine T, et al. RIG-I mediates the co-induction of tumor necrosis factor and type I interferon elicited by myxoma virus in primary human macrophages. PLoS Pathog. 2008;4:e1000099. doi: 10.1371/journal.ppat.1000099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Kushner S. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene. 1991;100:195–199. [PubMed] [Google Scholar]
- Westaway EG, Khromykh AA, Mackenzie JM. Nascent flavivirus RNA colocalized in situ with double-stranded RNA in stable replication complexes. Virology. 1999;258:108–117. doi: 10.1006/viro.1999.9683. [DOI] [PubMed] [Google Scholar]
- Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, Taira K, Akira S, Fujita T. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 2004;5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- Zhang B, Dong H, Stein DA, Iversen PL, Shi PY. West Nile virus genome cyclization and RNA replication require two pairs of long-distance RNA interactions. Virology. 2008;373:1–13. doi: 10.1016/j.virol.2008.01.016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.