Abstract
p24 proteins are a family of type I membrane proteins localized to compartments of the early secretory pathway and to coat protein I (COPI)- and COPII-coated vesicles. They can be classified, by sequence homology, into four subfamilies, named p24α, p24β, p24γ, and p24δ. In contrast to animals and fungi, plants contain only members of the p24β and p24δ subfamilies, the latter probably including two different subclasses. It has previously been shown that transiently expressed red fluorescent protein (RFP)–p24δ5 (p24δ1 subclass) localizes to the endoplasmic reticulum (ER) at steady state as a consequence of highly efficient COPI-based recycling from the Golgi apparatus. It is now shown that transiently expressed RFP–p24δ9 (p24δ2 subclass) also localizes to the ER. In contrast, transiently expressed green fluorescent protein (GFP)–p24β3 mainly localizes to the Golgi apparatus (as p24β2) and exits the ER in a COPII-dependent manner. Immunogold electron microscopy in Arabidopsis root tip cells using specific antibodies shows that endogenous p24δ9 localizes mainly to the ER but also partially to the cis-Golgi. In contrast, endogenous p24β3 mainly localizes to the Golgi apparatus. By a combination of experiments using transient expression, knock-out mutants, and co-immunoprecipitation, it is proposed that Arabidopsis p24 proteins form different heteromeric complexes (including members of the β and δ subfamilies) which are important for their stability and their coupled trafficking at the ER–Golgi interface. Evidence is also provided for a role for p24δ5 in retrograde Golgi–ER transport of the KDEL-receptor ERD2.
Key words: Arabidopsis, coat protein I (COPI), coat protein II (COPII), ER–Golgi transport, p24 proteins, secretory pathway.
Introduction
p24 proteins constitute a family of small (20–25kDa) type I membrane proteins which localize to compartments of the early secretory pathway and to coat protein I (COPI)- and COPII-coated vesicles (for reviews, see Strating and Martens, 2009; Dancourt and Barlowe, 2010). All p24 proteins consist of a large luminal portion, which includes the GOLD (GOLgi Dynamics) and coiled-coil domains, a single transmembrane domain, and a short cytoplasmic C-terminus which contains motifs for COPI and COPII binding (Supplementary Fig. S1 available at JXB online). Whereas the transmembrane domain seems to recognize a single sphingolipid species (Contreras et al., 2012), the luminal GOLD domain is predicted to be involved in specific protein–protein interactions and has been postulated to interact with putative cargo proteins (Anantharaman and Aravind, 2002; Carney and Bowen, 2004). The coiled-coil domain of p24 proteins enables intermolecular interactions between copies of the same protein, but also between different p24 proteins. Indeed it has been proposed that oligomerization is required for the proper localization of p24 proteins (Füllerkrug et al., 1999; Gommel et al., 1999; Ciufo and Boyd, 2000; Emery et al., 2000; Jenne et al., 2002; Langhans et al., 2008; Montesinos et al., 2012).
p24 proteins have been proposed to play a role in quality control of protein movement through the secretory pathway (Wen and Greenwald, 1999; Belden and Barlowe, 2001), cargo protein selection and packaging into transport vesicles (Schimmöller et al., 1995; Muniz et al., 2000; Takida et al., 2008; Castillon et al., 2011; Fujita et al., 2011), the formation of COPI vesicles and retrograde Golgi–endoplasmic reticulum (ER) transport (Aguilera-Romero et al., 2008), the formation of ER exit sites (ERES) (Lavoie et al., 1999), and the biogenesis and maintenance of the Golgi apparatus (Mitrovic et al., 2008; Koegler et al., 2010). Therefore, p24 proteins are one of the most interesting groups of proteins involved in regulating the structure and function of the organelles of the secretory pathway. In addition, several publications have proposed a role for p24 proteins in early embryonic development in mice (Denzel et al., 2000; Jerome-Majewska et al., 2010), insulin biosynthesis and subsequent secretion in pancreatic beta cells (Zhang and Volchuk, 2010), or amyloid precursor metabolism and pathogenesis of Alzheimer disease (Chen et al., 2006; Vetrivel et al., 2007; Hasegawa et al., 2010). p24 proteins have also been shown to interact with G protein-coupled receptors (GPCRs), including protease-activated receptors (PAR-1 and PAR-2), nucleotide P2Y receptors, and μ-opioid receptors (Luo et al., 2007, 2011).
Over the years, p24 proteins have been proposed to function as cargo receptors, to concentrate cargo within COPI or COPII vesicles, but the trafficking of a putative cargo mediated by p24 proteins has only recently been demonstrated in mammals and yeast. In mammals, p24 proteins have been shown to form hetero-oligomeric complexes that bind to correctly remodelled glycosylphosphatidylinositol (GPI) anchors to concentrate GPI-anchored proteins (GPI-APs) at ERES for their efficient packaging into COPII vesicles and transport to the Golgi (Fujita et al., 2011). In the Golgi, at a lower pH, p24 complexes dissociate from GPI-APs, which are transported to the cell surface, while p24 proteins are recycled to the ER in COPI vesicles (Fujita et al., 2011). In yeast, sorting of GPI-APs appears to be independent of p24 proteins. Instead, the p24 complex appears to act as an adaptor that facilitates vesicle formation by recruiting COPII components to specific ERES already enriched in GPI-APs (Castillon et al., 2011).
Although there has been no general agreement regarding the nomenclature of p24 proteins, it is now clear that they can be classified into four different subfamilies by sequence homology, named p24α, p24β, p24γ and p24δ (Domiguez et al., 1998; Strating et al., 2009). Whereas animals and fungi have representatives in all four subfamilies, plants have only members of the p24δ (nine in Arabidopsis) and the p24β (two in Arabidopsis) subfamilies (Strating et al., 2009). Following this nomenclature, the Arabidopsis p24 proteins have been named p24δ3 to p24δ11 (since the names p24δ1 and 2 have already been used) (Supplementary Fig. S1 at JXB online) (Montesinos et al., 2012). Chen and Zheng (2012) have proposed that the members of the delta subfamily belong to two different subclasses (which correspond to the two main branches of this subfamily), the δ1 subclass (comprising p24δ1a–d; p24δ3–6 in the present study) and the δ2 subclass (comprising p24δ2a–d; p24δ7–11 in the present study). On the other hand, the two Arabidopsis p24 proteins of the beta subfamily have been named p24β2 and p24β3 (since the name p24β1 has already been used) (Supplementary Fig. S1) (Montesinos et al., 2012).
Interestingly, all Arabidopsis p24 proteins of the delta subfamily contain in their C-terminal tail a dilysine motif in the -3,-4 position, which binds COPI subunits (Contreras et al., 2004a ), and a diaromatic/large hydrophobic motif in the -7,-8 position, which binds COPII subunits but also potentiates COPI binding by the dilysine motif (Contreras et al., 2004b ). As a consequence, it has been proposed that p24δ proteins show higher affinity for COPI than for COPII subunits (Contreras et al., 2004b ). Indeed, it has been shown that transiently expressed p24δ5 localizes mainly to the ER at the steady state as a consequence of highly efficient COPI-based recycling from the Golgi apparatus (Langhans et al., 2008; Montesinos et al., 2012). A similar ER localization has been shown for other members of the p24δ1 subclass (comprising p24δ3–p24δ6) (Chen and Zheng, 2012; Montesinos et al., 2012). In contrast, members of the p24δ2 subclass (comprising p24δ7–p24δ11) have been proposed to localize to both the ER and Golgi (Chen and Zheng, 2012).
Using specific antibodies, endogenous p24δ5 and p24δ4 have been localized to the ER and p24β2 to the Golgi apparatus in Arabidopsis root tip cells by immunogold electron microscopy (Montesinos et al., 2012). It has been shown that whereas the dilysine motif in the cytoplasmic tail determines the location of p24δ5 in the early secretory pathway, the luminal domain may contribute to its distribution downstream of the Golgi apparatus (Montesinos et al., 2012). It has also been shown that p24δ5 and p24β2 interact with each other (via their coiled-coil domains) and exhibit coupled trafficking at the ER–Golgi interface. It has been proposed that p24δ5 and p24β2 may interact with each other at ERES for ER exit and coupled transport to the Golgi apparatus. Once in the Golgi, p24δ5 interacts very efficiently with the COPI machinery for retrograde transport back to the ER (Montesinos et al., 2012).
In this study, the analysis has been extended to a second member of the p24δ subfamily (p24δ9, p24δ2 subclass) and to the second member of the p24β subfamily (p24β3). While transiently expressed p24δ9 localizes to the ER at steady state, p24β3 mainly localizes to the Golgi apparatus and exits the ER in a COPII-dependent manner. Immunogold electron microscopy in Arabidopsis root tip cells using specific antibodies shows that endogenous p24δ9 localizes mainly to the ER but also partially to the cis-Golgi. In contrast, endogenous p24β3 mainly localizes to the Golgi apparatus. By a combination of experiments using transient expression, knock-out mutants, and co-immunoprecipitation, it is proposed that Arabidopsis p24 proteins form different heteromeric complexes for their coupled trafficking at the ER–Golgi interface. Evidence is also provided for a role for p24δ5 in retrograde Golgi–ER transport of the KDEL-receptor ERD2.
Materials and methods
Plant material
Arabidopsis thaliana ecotype Columbia (Col-0) and T-DNA mutant plants were grown in growth chambers as previously described (Ortiz-Masia et al., 2007). For immunogold electron microscopy, seedlings were grown on MS (Murashige and Skoog) medium containing 0.5% agar, and the roots were harvested after 5 d. To obtain a membrane fraction from Arabidopsis roots, seedlings were grown in liquid MS medium for 15 d. Arabidopsis thaliana cell suspension cultures (LT87) (Axelos et al., 1992) were cultivated as described (Ortiz-Zapater et al., 2006). Plants of Nicotiana tabacum cv. Petit Havana were grown from surface-sterilized seeds on MS medium with 2% (w/w) sucrose in a controlled room at 25 °C with cycles of 16h light and 8h darkness. Wild-type Nicotiana benthamiana plants were grown from surface-sterilized seeds on soil in a controlled room at 22 °C with a 16h daylength.
Recombinant plasmid production
The coding sequences of red fluorescent protein (RFP)–p24δ9, cyan fluorescent protein/green fluorescent protein (CFP/GFP)–p24β2, or GFP/yellow fluorescent protein (YFP)–p24β3 were synthesized commercially de novo (Geneart AG), based on the sequences of GFP/CFP/YFP/RFP and that of the Arabidopsis p24 proteins At1g26690 (p24δ9), At3g07680 (p24β2), and At3g22845 (p24β3). All RFP-tagged proteins were tagged with monomeric RFP (mRFP) to prevent oligomerization. Similarly, only mGFP5 was used for GFP-tagged proteins. The sequence of the fluorophore was always located behind the coding sequence of the p24 signal sequence and the 5ʹ extreme end of the mature p24 coding sequence (Supplementary Fig. S1 at JXB online). The coding sequences of RFP–p24δ9 or XFP–p24β2/β3 were cloned into the pBP30 vector (carrying the 35S promoter; Nebenführ et al., 1999) through BglII/NotI.
Transient gene expression
Mesophyll protoplasts from N. tabacum var. SR1 leaf cells were isolated and transfected as previously described (Bubeck et al., 2008). Unless otherwise stated, 1–50 μg of plasmid DNA was transfected and expressed for 20h. Protoplasts from A. thaliana (LT87) cell suspension cultures were isolated as previously described (Axelos et al., 1992). Where indicated, inhibitors (50 μM E-64, 100 μM MG-132) were added to the protoplast medium 30min after electroporation, before the 20h overnight incubation to allow for expression of the different constructs. Transient expression mediated by Agrobacterium tumefaciens was performed in 4- to 6-week-old tobacco plants (wild type, N. benthamiana) as described previously (Lerich et al., 2011).
Plasmids encoding marker proteins were: GFP–p24β2 and RFP–p24δ5 (Langhans et al., 2008; Montesinos et al., 2012), Man1–RFP and Man1–GFP (Nebenführ et al., 1999), GFP–HDEL (Nebenführ et al., 2000), ERD2–CFP/YFP (Brandizzi et al., 2002), 6kDa VP–CFP (Wei and Wang, 2008), Sec12 (Pimpl et al., 2003), ARF1(T31N) (Lee et al., 2003), and ARF1(Q71L) (Pimpl et al., 2003).
Generation of antibodies
Rabbit antibodies were generated by Eurogentec (Belgium, http://www.eurogentec.com) using as antigens peptides corresponding to the N-terminus of p24δ9 (LHFELQSGRT) or p24β3 (LSVTVNDEE).
Confocal microscopy and immunofluorescence labelling
Imaging was performed using a Zeiss Axiovert LSM510 Meta confocal laser scanning microsope (CLSM). At the Metadetector, the main beam splitters (HFT) 458/514 and 488/543 were used. The following fluorophores (excited and emitted by frame switching in the multitracking mode) were used: GFP (488nm/496–518nm), CFP (458nm/464–486nm), YFP (514nm/529–550nm), and RFP (543nm/593–636nm). Post-acquisition image processing was performed using the Zeiss LSM 5 image Browser (4.2.0.121) and CorelDrawX4 (14.0.0.567) or ImageJ (v.1.45m).
Immunogold electron microscopy
Root tips from Arabidopsis were high pressure frozen, freeze substituted, embedded, labelled, and post-stained as previously described (Bubeck et al., 2008). Antibodies were used at the following dilutions: Nt-p24β3 (1:100) and Nt-p24δ9 (1:100). Micrographs were taken with a JEM1400 transmission elctron microscope operating at 80kV using a TVIPS F214 digital camera.
Preparation of membrane extracts, co-immunoprecipitation, pull-down experiments, and western blotting
Membrane fractions were obtained from Arabidopsis cell suspension cultures (LT87), Arabidopsis roots, or tobacco protoplasts as described previously (Montesinos et al., 2012). Protein extracts were used for SDS–PAGE followed by western blot analysis, co-immunoprecipitation, or pull-down experiments. Co-immunoprecipitation experiments from Arabidopsis cultures were performed using magnetic beads (Dynal, Invitrogen), as described previously (Montesinos et al., 2012). Pull-down experiments from tobacco protoplasts expressing RFP-tagged proteins were performed using RFP-Trap magnetic beads (Chromotek), following the recommendations of the manufacturer. For western blot analysis, nitrocellulose membranes were blocked with 5% non-fat dry milk/0.1 % Tween-20, incubated for 1h at room temperature with the primary antibodies, washed, and incubated with peroxidase-labelled sheep anti-rabbit antibodies (GE Healthcare) for 1h at room temperature. After washing, the immune complexes were detected by the SuperSignal West Pico chemiluminiscent Substrate (Pierce, Thermo Scientific). The intensity of the bands obtained after western blot was quantified using the Quantity One software (Bio-Rad Laboratories). Western blot with an antibody against the plasma membrane ATPase (Montesinos et al., 2012) was used as a loading control. Antibodies against RFP and GFP were obtained from Clontech and Life Technologies, respectively.
Mutant characterization
A line (Columbia, background) containing a T-DNA insertion in p24δ10 (SALK_144586C, p24δ10–1) was identified from the SALK T-DNA collection (http://signal.salk.edu/cgi-bin/tdnaexpress). It was characterized by PCR as previously described (Ortiz-Masia et al., 2007). The primers used for this mutant were the following: 5ʹ-CCGGTAACAATTACCATCACG-3ʹ and 5ʹ-ACGAAGTACCC AAGGTTCCAC-3ʹ. The T-DNA left border and Actin7 (ACT7, At5g09810) primers used were described previously (Ortiz-Masia et al., 2007). Reverse transcription–PCR (RT–PCR) analysis of the p24δ10–1 mutant was performed as described (Ortiz-Masia et al., 2007) to show the absence of p24δ10, and the primers used were 5ʹ-CAAAGTGTATCGCCGAAGACATC-3ʹ and 5ʹ-GCATCC CTGCAACTCCTATGCAGA-3ʹ. p24δ 4–1, p24δ 5–1, and p24δ4δ5 mutant lines have been described previously (Montesinos et al., 2012).
Due to the lack of p24β2 and p24β3 knock-out T-DNA insertion mutants in mutant collections, artificial microRNAs (amiRNAs) were used to knock down the expression of the genes. The β2-directed amiRNA construct was designed using a Web-based program (http://wmd2.weigelworld.org) (Schwab et al., 2006; Ossowski et al., 2008). The pRS300 plasmid was used as a template to create the amiRNA (Ossowski et al., 2008). Primer sequences were the following: I, 5ʹ-ATAATCAGTGCA AACGACGCGATCTCTCTTTTGTATTCC-3ʹ; II, 5ʹ-GATCGCG TCGTTTGCACTGATTATCAAAGAGAATC AATGA-3ʹ; III, 5ʹ-GATCACGTCGTTTGCTCTGATTTTCACAG GTCGTGATATG-3ʹ; and IV, 5ʹ-GAAAATCAGAGCAA-ACGAC GTGATCTACATATATATTCCT-3ʹ. The final amiRNA PCR product was digested at the KpnI and BamHI sites flanking the sequence encoding the amiRNA hairpin. The resultant product was ligated into the pCHF3 vector (Ortiz-Masiá et al, 2007) using the KpnI and BamHI sites. The β3-directed amiRNA construct was purchased from Open Biosystems (AMR4844-99730584). Transformation of Arabidopsis was conducted according to the floral dip method (Clough and Bent, 1998). Transgenic plants were selected on half-strength MS medium containing appropriate antibiotics. Transgenic lines segregating 3:1 for antibiotic resistance were selected in the T2 generation of each transformation, and the T3 homozygous generation was used to characterize silencing by RT–PCR as above. Primer sequences for p24β2 were 5ʹ-AGGGTACGATCGTATTACTAG-3ʹ and 5ʹ-GACACGAGACA TGCCGAGTTTGCG-3ʹ and for p24β3 were 5ʹ-CGACAAGCGAA GATCCATG-3ʹ and 5ʹ-GACACAAGACCTCGCTCTGAGG-3ʹ. For further studies, the homozygous lines amiR-p24β2 and amiR-p24β3 that showed the best silencing for p24β2 and p24β3, respectively, were selected (Supplementary Fig. S6 at JXB online). RT–PCR analysis showed no silencing of p24β3 in the amiR-p24β2 line, while 20% p24β2 silencing was detected in the amiR-p24β3 line obtained from the amiRNA construct purchased from Open Biosystems (data not shown).
Results
Localization of endogenous p24 proteins of the delta and beta subfamilies
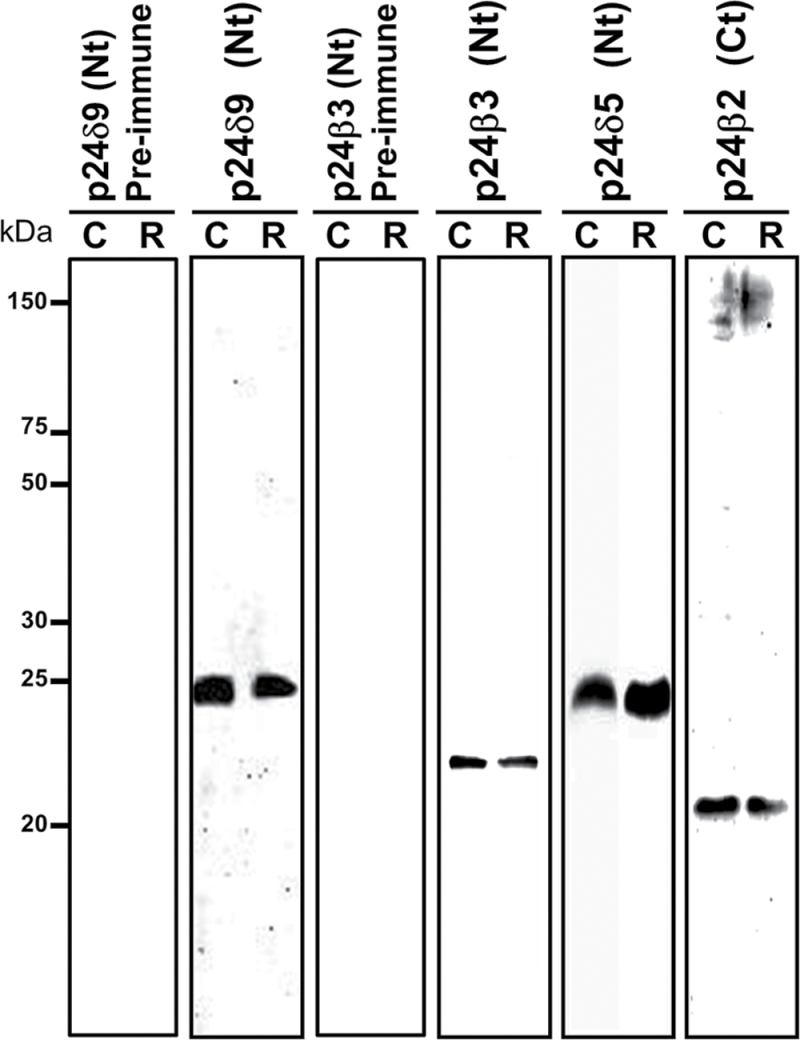
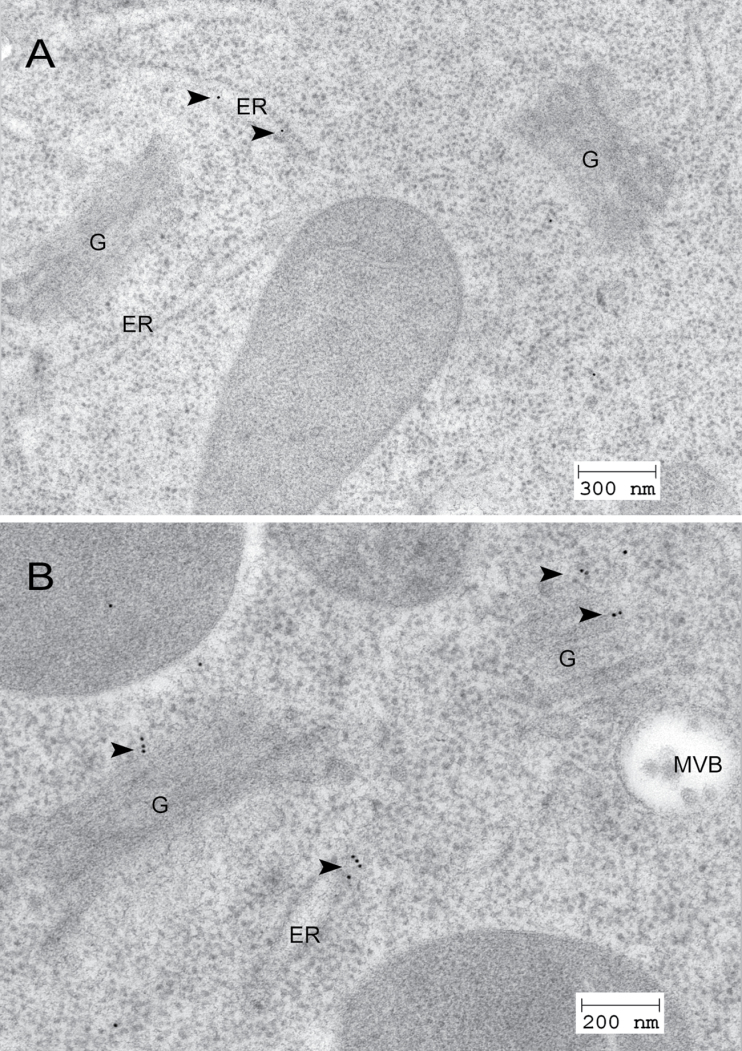
The localization of endogenous p24δ5 and p24δ4 (p24δ subfamily) and p24β2 (p24β subfamily) in Arabidopsis root cells was previously shown (Montesinos et al., 2012). The localization of p24δ9, in a branch of the p24δ subfamily different from that of p24δ5 or p24δ4, and of p24β3, the second member of the p24β subfamily in Arabidopsis, has now been investigated. To this end, peptide antibodies were generated against the N-terminus of both proteins, which, in contrast to the C-terminus, shows a high variability among different p24 proteins (Supplementary Fig. S1 at JXB online). p24 proteins were extracted from membranes of Arabidopsis cell suspension cultures or from Arabidopsis roots. As shown in Fig. 1, antibodies against the N-terminus of p24δ9 recognized a protein of the expected molecular weight (24kDa) in both membrane extracts, while antibodies against the N-terminus of p24β3 recognized a protein with an apparent molecular weight of ~22kDa. Interestingly, p24β2 and p24β3 showed a slightly different electrophoretic mobility, which in addition was also different from that of p24δ5 and p24δ9 (Fig. 1). These differences in electrophoretic mobility were also obvious when the luminal N-terminal portion of both p24δ5 and p24β2 with a C-terminal (His)6-tag was expressed in bacteria (Supplementary Fig. S2 at JXB online). Bacterial extracts were used to characterize further the specificity of the antibodies. As shown in Supplementary Fig. S2, antibodies against the N-terminus of p24δ9 did not recognize the N-terminus of p24δ5 (in contrast to Nt-p24δ5 or His antibodies), while antibodies against the N-terminus of p24β3 did not recognize the N-terminus of p24β2 (in contrast to Nt-p24β2 or His antibodies). These antibodies were used to localize p24δ9 and p24β3 by immunogold labelling on sections cut from cryofixed Arabidopsis roots. As shown in Fig. 2A, the N-terminal p24δ9 antibody mainly labelled ER membranes, as was found previously for endogenous p24δ5 and p24δ4 (Montesinos et al., 2012). Occasionally, some labelling was also seen on the cis-Golgi. In contrast, the N-terminal p24β3 antibody mainly labelled the Golgi apparatus, although some labelling could also be seen at ER membranes (Fig. 2B). This localization is very similar to that previously shown for endogenous p24β2 (Montesinos et al., 2012).
Fig. 1.
Characterization of antibodies against Arabidopsis p24 proteins. Protein extracts were obtained from membranes of Arabidopsis cell suspension cultures (C) or Arabidopsis roots (R), as described in the Materials and methods, and analysed by SDS–PAGE (14% acrylamide) and western blotting with antibodies against the p24δ9 N-terminus and p24β3 N-terminus, or with the corresponding pre-immune sera. Western blotting with antibodies against p24δ5 and p24β2 (Montesinos et al., 2012) is also shown. Note the slightly different electrophoretic mobility of p24δ5/p24δ9, p24β2, and p24β3.
Fig. 2.
Localization of p24δ9 and p24β3 by immunogold labelling on cryofixed Arabidopsis roots. (A) Labelling with antibodies against Nt-p24δ9 at the ER. (B) Labelling with antibodies against Nt-p24β3 at the Golgi apparatus and the ER. Arrowheads point to gold particles. ER, endoplasmic reticulum; G, Golgi apparatus; MVB, multivesicular body.
Trafficking properties of p24 proteins of the delta subfamily
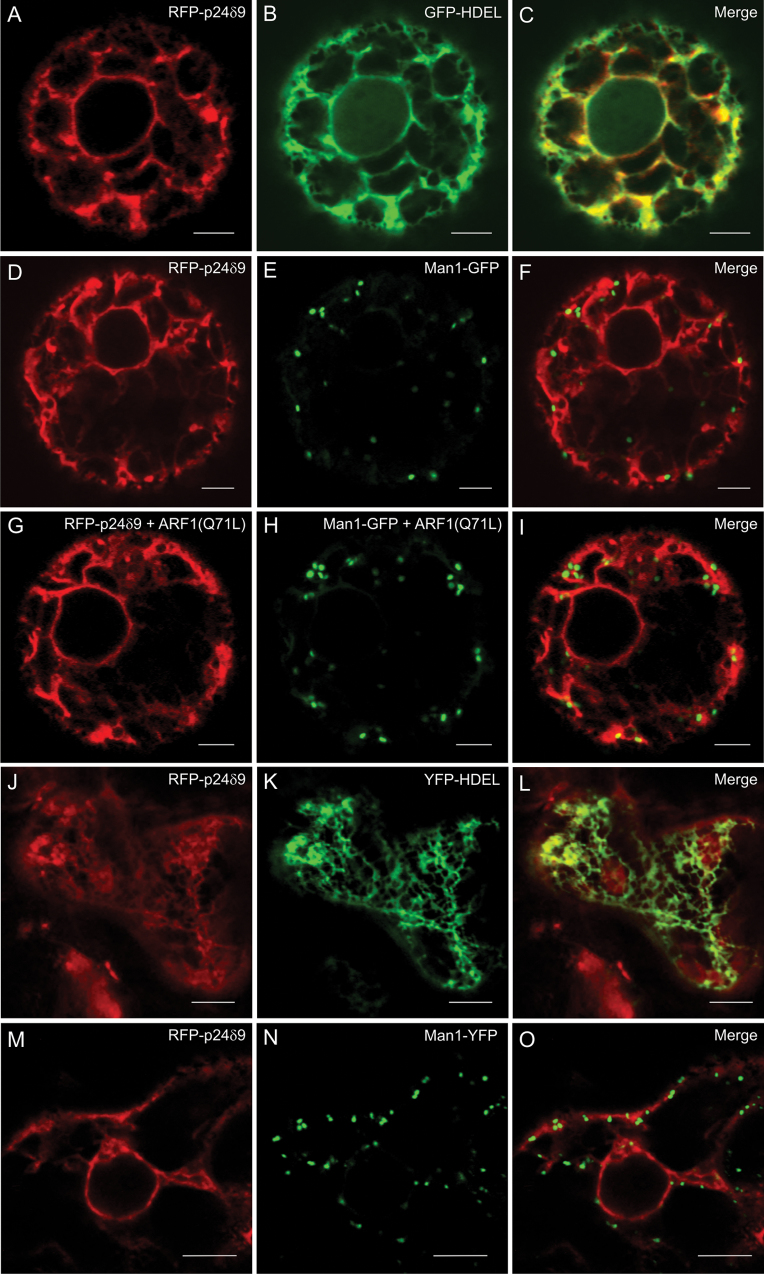
It was previously demonstrated that transiently expressed RFP–p24δ5 localizes to the ER at steady state but cycles between the ER and Golgi (Langhans et al., 2008; Montesinos et al., 2012). Arabidopsis p24 proteins of the delta subfamily have been suggested to belong to two different subclasses, p24δ1 and p24δ2, with different localization and trafficking properties (Chen and Zheng, 2012). In particular, members of the p24δ1 subclass (which comprise p24δ3–p24δ6) localized exclusively to the ER, while members of the p24δ2 subclass (which comprisee p24δ7–p24δ11) localized to both the ER and Golgi when transiently expressed in tobacco leaf epidermal cells (Chen and Zheng, 2012). Therefore, the trafficking properties of RFP–p24δ5 (p24δ1 subclass) and RFP–p24δ9 (p24δ2 subclass) were compared. In marked contrast to the data of Chen and Zheng (2012), it was found that transiently expressed RFP–p24δ9 localizes exclusively to the ER, both in tobacco protoplasts (Fig. 3A–F; Supplementary Fig. S3 at JXB online) and in tobacco leaf epidermal cells (Fig. 3J–O). In both cases, RFP–p24δ9 co-localized extensively with the ER markers GFP/YFP–HDEL but not with the Golgi markers ManI–GFP/YFP. At low expression levels, it showed a pattern nearly indistinguishable from that of RFP–p24δ5. At higher expression levels, it partially localized to dots, or even to ring-like structures, probably artefacts of overexpression. However, none of these structures was found to co-localize with the Golgi markers ManI–GFP/YFP (Supplementary Fig. S3). Therefore, transiently expressed RFP–p24δ9 seems to localize exclusively to the ER, like RFP–p24δ5 (Langhans et al., 2008; Montesinos et al., 2012).
Fig. 3.
Localization of RFP–p24δ9. (A–I) Transient gene expression in tobacco mesophyll protoplasts. (A–C) RFP–p24δ9 (A) co-localizes extensively with the ER marker GFP–HDEL (B) (merged image in C). (D–F) RFP–p24δ9 (D) does not co-localize with the Golgi marker Man1–GFP (E) (merged image in F). (G–I) RFP–p24δ9 (G) does not co-localize with the Golgi marker Man1–GFP (H) upon ARF1 (Q71L) expression (merged image in I). (J–O) Transient gene expression in tobacco leaf epidermal cells. (J–L) RFP–p24δ9 (J) co-localizes extensively with the ER marker YFP–HDEL (K) (merged image in L). (M–O) RFP–p24δ9 (M) does not co-localize with the Golgi marker Man1–YFP (N) (merged image in O). Scale bars=5 μm.
In order to investigate if transiently expressed RFP–p24δ9 also cycles between the ER and Golgi, as does RFP–p24δ5, RFP–p24δ9 was co-expressed with the GTP-restricted ARF1(Q71L) mutant, which prevents COPI-mediated Golgi–ER recycling (Pimpl et al., 2003). This treatment has been shown to redistribute RFP–p24δ5 partially to the Golgi apparatus (Langhans et al., 2008; Montesinos et al., 2012). Interestingly, this treatment did not change the ER localization of RFP–p24δ9 (Fig. 3G–I).
Trafficking properties and stability of p24 proteins of the beta subfamily
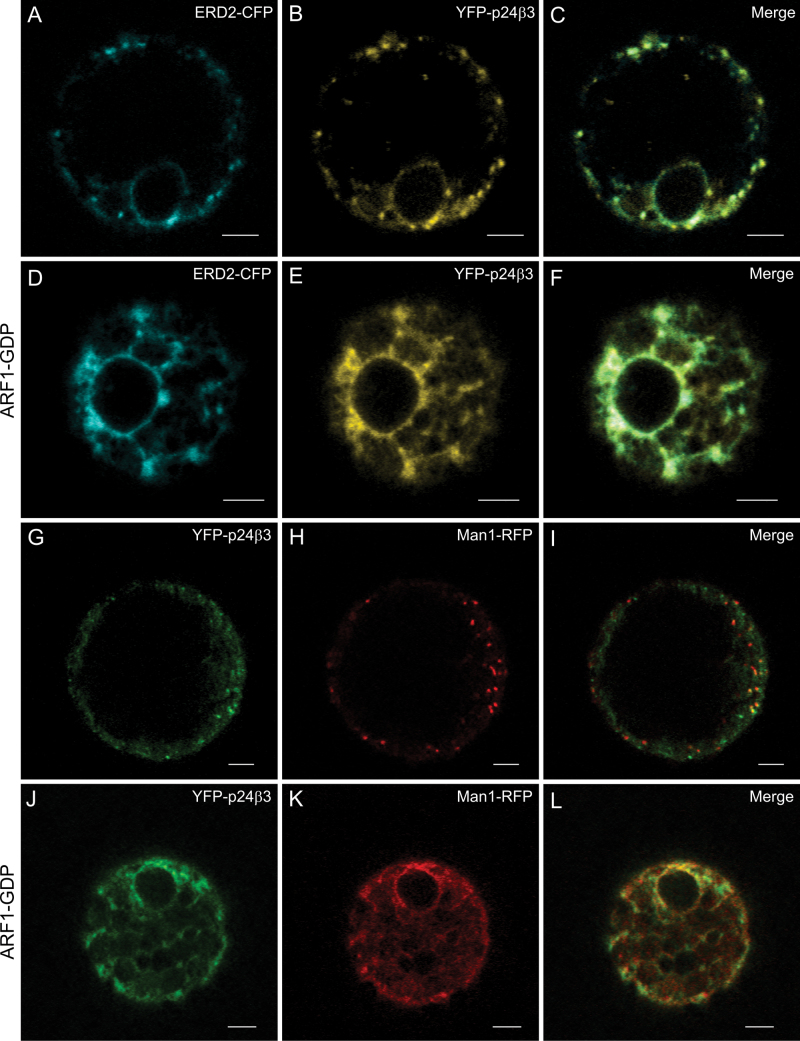
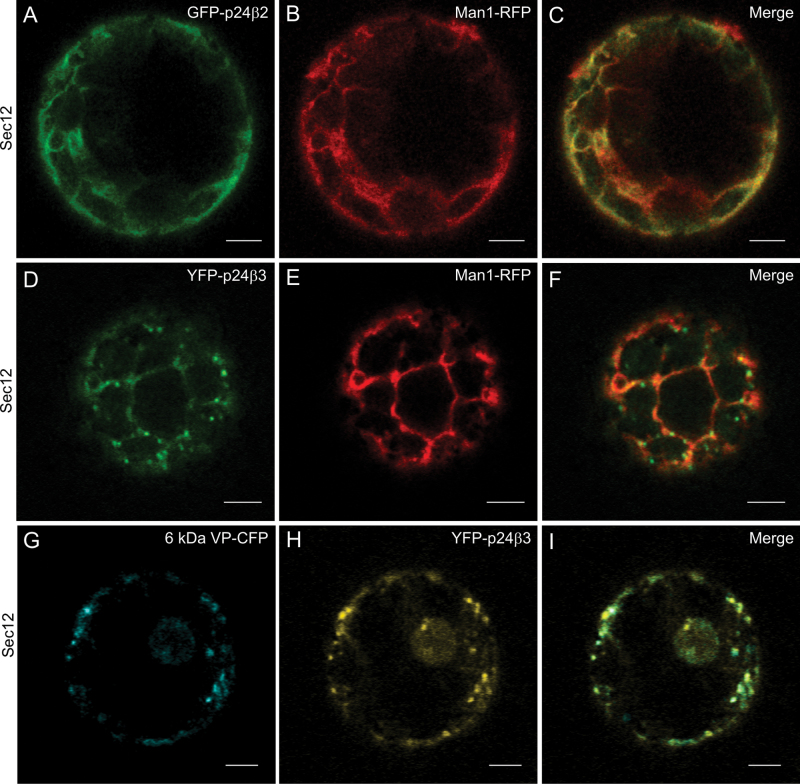
The trafficking properties of (X)FP–p24β3 were also investigated. Similar to GFP–p24β2, YFP–p24β3 showed a punctate pattern, which partially co-localized with the Golgi markers ERD2–CFP (Fig. 4A–C) or ManI–RFP (Fig. 4G–I). When co-expressed with the GDP-restricted ARF1(T31N) mutant, which causes relocalization of Golgi markers to the ER (Lee et al., 2002), YFP–p24β3 completely redistributed to the ER, where it co-localized with both ERD2–CFP (Fig. 4D-F) and Man I–RFP (Fig. 4J–L). This was also the case for YFP–p24β2 (Supplementary Fig. S4 at JXB online). These data suggest that transiently expressed p24β2 and p24β3 mainly localize to the Golgi apparatus, as had been observed for the endogenous proteins. When GFP–p24β2 or YFP–p24β3 was co-expressed with Sec12, to inhibit COPII-dependent ER export, both proteins were mainly localized to the ER, together with the Golgi marker Man I–RFP (Fig. 5A–F). This suggests that both proteins exit the ER in a COPII-dependent manner. In contrast to GFP–p24β2, co-expression of YFP–p24β3 and Sec12 not only led to a complete reticular pattern, but some dots were also very obvious (Fig. 5D–F). To test for the identity of these dots, YFP–p24β3 and Sec12 were co-expressed with 6kDa VP–CFP, a COPII/ERES marker (Lerich et al., 2011). As shown in Fig. 5G–I, many of the YFP–p24β3 punctae co-localized with 6kDa VP–CFP, suggesting that at least a fraction of YFP–p24β3 may accumulate at ERES under these conditions.
Fig. 4.
Localization of YFP–p24β3. (A–L) Transient gene expression in tobacco mesophyll protoplasts. (A–C) YFP–p24β3 (B) co-localizes with the Golgi marker ERD2–CFP (A) in punctate structures (merged image in C). (D–F) YFP–p24β3 (E) and ERD2–CFP (D) relocalized to the ER upon co-expression with the ARF1-GDP mutant (merged image in F). (G–I) YFP–p24β3 (G) co-localizes partially with the Golgi marker ManI–RFP (H) in punctate structures (merged image in I). (J–L) YFP–p24β3 (J) and ManI–RFP (K) relocalized to the ER upon co-expression with the ARF1-GDP mutant (merged image in L). Scale bars=5 μm.
Fig. 5.
ER export of p24 proteins of the beta subfamily is COPII dependent. (A–I) Transient gene expression in tobacco mesophyll protoplasts. (A–C) GFP–p24β2 (A) co-localizes with ManI–RFP (B) in the ER upon Sec12 overexpression (merged image in C). (D–F) YFP–p24β3 (D) co-localizes with ManI–RFP (E) in the ER upon Sec12 overexpression, but also in punctate structures (merged image in F). (G–I) YFP–p24β3 (H) co-localizes partially with the COPII/ERES marker 6kDa VP–CFP (G) in punctate structures upon Sec12 overexpression (merged image in I). Scale bars=5 μm.
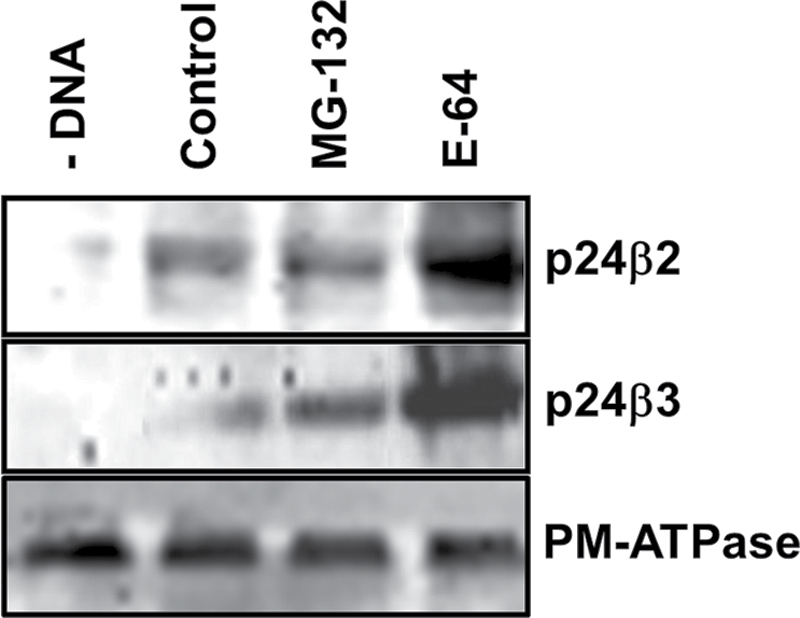
As had been observed with GFP–p24β2 (Montesinos et al., 2012), the signal obtained for YFP–p24β3 in the CLSM when expressed alone was relatively low. When the levels of GFP–p24β3 were analysed by western blotting, a relatively low signal was also detected (Fig. 6, lane 2). The stability of p24 proteins depends on their interactions with other family members (Montesinos et al., 2012). Therefore, transiently expressed individual proteins may be more susceptible to protein degradation. To investigate the mechanisms involved in the degradation of p24β2 and p24β3, both proteins were expressed in the presence of MG-132, a proteasome inhibitor, or E-64, an inhibitor of cysteine proteinases. Western blot analysis shows that the levels of both p24β2 and p24β3 significantly increased in the presence of E-64 (Fig. 6, lane 4), but not in the presence of MG-132 (Fig. 6, lane 3), suggesting that both proteins are mainly degraded by cysteine proteinases in acidic compartments.
Fig. 6.
Stability of p24 proteins of the beta subfamily. Tobacco mesophyll protoplasts were electroporated in the absence (–DNA) or the presence of 30 μg of plasmid DNAs corresponding to GFP–p24β2 (upper panel) or GFP–p24β3 (middle panel), in the absence (Control) or the presence of MG-132 or E-64. At 24h post-electroporation, protoplasts were washed and homogenized to obtain a post-nuclear supernatant, which was then centrifuged to obtain a total membrane fraction. Membranes were extracted in Laemmli sample buffer and analysed by SDS–PAGE (12% acrylamide) and western blot analysis with antibodies against Ct-p24β2 or GFP (to detect p24β3). A 30 μg aliquot of protein was loaded for each of the extracts. Western blotting with an antibody against the plasma membrane (PM) ATPase was used as a loading control (lower panel).
Interactions between different members of the p24 family in Arabidopsis
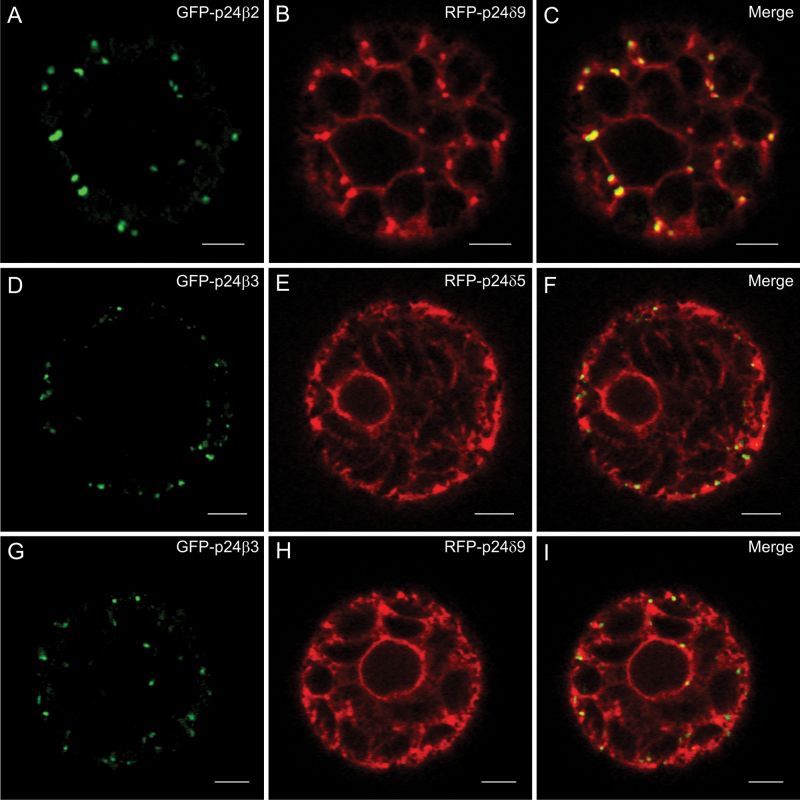
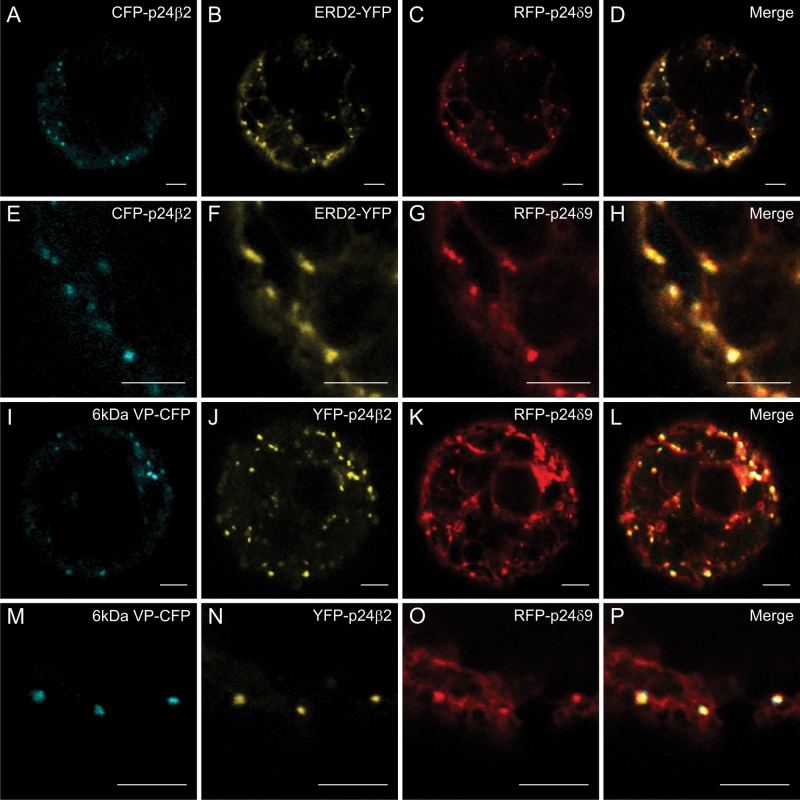
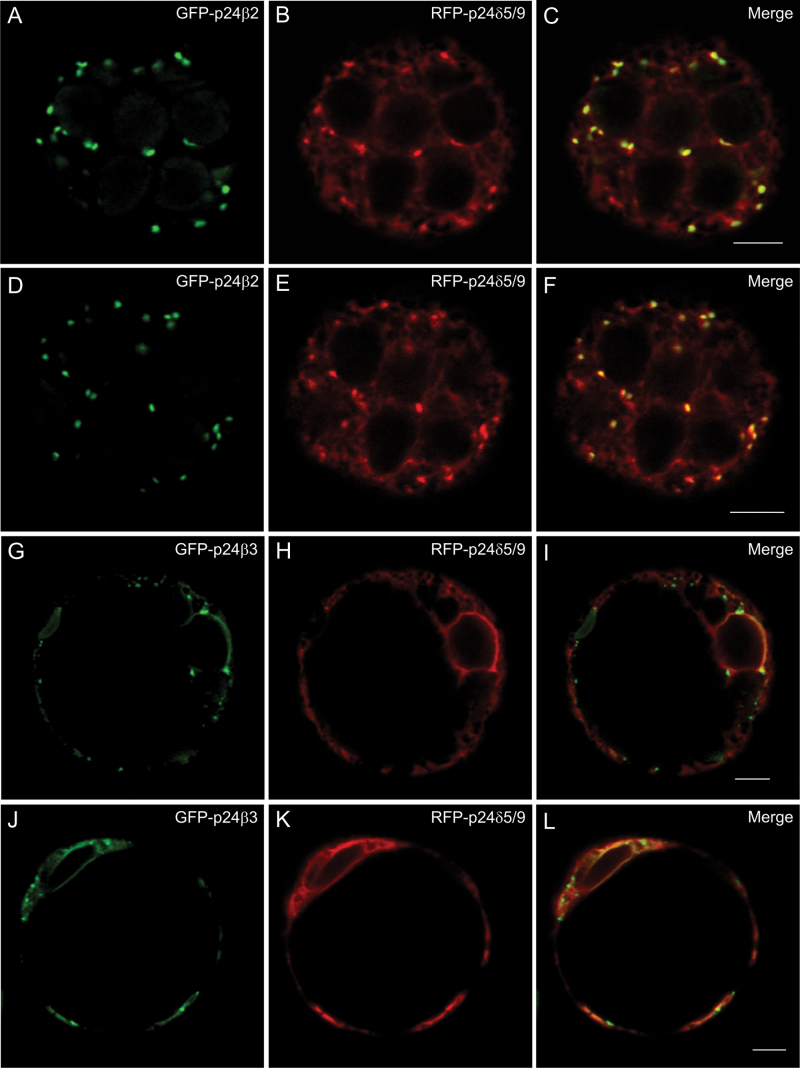
p24 proteins are thought to form hetero-oligomeric complexes, via their coiled-coil domains, which are essential for their trafficking and localization. It has been previously shown that p24δ5 and p24β2 interact with each other, probably at ERES, for their coupled transport to the Golgi apparatus (Montesinos et al., 2012). This analysis has now been extended to other members of the p24δ and p24β subfamilies. As shown in Fig. 7A–C, when GFP–p24β2 was co-expressed with RFP–p24δ9, the signal of GFP–p24β2 was clearly more intense than when expressed alone, but still localized to punctae. In addition, it was observed that RFP–p24δ9 showed its typical ER pattern but also localized to the same punctae under these conditions (see also Table 1A). The punctae where both proteins co-localize overlapped extensively with the Golgi marker ERD2–YFP (Fig. 8A–H), but also with the COPII/ERES marker 6kDa VP–CFP (Fig. 8I–P). This suggests that GFP–p24β2 is able to enhance the ER exit of RFP–p24δ9 and its transport to the Golgi apparatus, as happens with RFP–p24δ5 (Montesinos et al., 2012). This was also the case when GFP–p24β2 and RFP–p24δ9 were transiently co-expressed in tobacco leaf epidermal cells: while RFP–p24δ9 localized exclusively to the ER when expressed individually (Fig. 3J–O), it also localized to punctae when co-expressed with CFP–p24β2. Under these conditions, the punctae containing CFP–p24β2 showed an almost complete co-localization with RFP–p24δ9 (Supplementary Fig. S5 at JXB online). Whether GFP–p24β3 showed the same trafficking characteristics as GFP–p24β2 was next investigated in co-expression experiments. As shown in Fig. 7D–I, GFP–p24β3 punctae showed only a partial co-localization with either RFP–p24δ5 or RFP–p24δ9 (see also Table 1A). Strikingly, GFP–p24β3 did not significantly change the ER localization of RFP–p24δ5 or RFP–p24δ9, in contrast to GFP–p24β2 (Fig. 7D–I).
Fig. 7.
Co-expression of p24 proteins of the beta and delta subfamilies (I). (A–I) Transient gene expression in tobacco mesophyll protoplasts. (A–C) GFP–p24β2 (A) co-localizes extensively with RFP–p24δ9 (B) in punctate structures (merged image in C). (D–I) GFP–p24β3 (D, G) partially co-localizes with RFP–p24δ5 (E) or RFP–p24δ9 (H) (merged images in F and I). Scale bars=5 μm.
Table 1.
Co-localization of RFP–p24δ5/δ9 and GFP–p24β2/β3 in co-expression experiments
| A. | ||||
|---|---|---|---|---|
| Combination of proteins | Manders coefficient | |||
| A | B | M1 (A overlapping with B) | M2 (B overlapping with A) | |
| GFP–p24β2 | RFP–p24δ9 | 0.84±0.05 | 0.36±0.07 | |
| GFP–p24β3 | RFP–p24δ9 | 0.47±0.10 | 0.14±0.06 | |
| GFP–p24β3 | RFP–p24δ5 | 0.45±0.07 | 0.15±0.06 | |
| B. | ||||
| Combination of proteins | Co-localization (%) | |||
| RFP–p24δ5 | RFP–p24δ9 | YFP–p24β3 | CFP–p24β2 | 9.19±2.64 |
| RFP–p24δ5 | RFP–p24δ9 | YFP–p24β3 | – | 16.06±2.82 |
| RFP–p24δ5 | RFP–p24δ9 | – | CFP–p24β2 | 30.31±3.71 |
| – | – | YFP–p24β3 | CFP–p24β2 | 44.44±3.31 |
In A, measurements were made on 10 separate cells upon double co-expression (Fig. 7), and calculated with ImageJ 1.47i and the plugins JACoP (Bolte S, Cordelieres FP 2006) and PSC Colocalization (French et al., 2008).
Fig. 8.
(X)FP–p24β2 and RFP–p24δ9 localize partially to ERES and Golgi. (A–P) Transient gene expression in tobacco mesophyll protoplasts. (A–H) CFP–p24β2 (A, E) and RFP–p24δ9 (C, G) co-localize with the Golgi marker ERD2–YFP (B, F) in punctate structures (merged images in D and H). (I–P) YFP–p24β2 (J, N) and RFP–p24δ9 (K, O) co-localize with the ERES/COPII marker 6kDa VP–CFP (I, M) in punctate structures (merged images in L and P). Scale bars=5 μm.
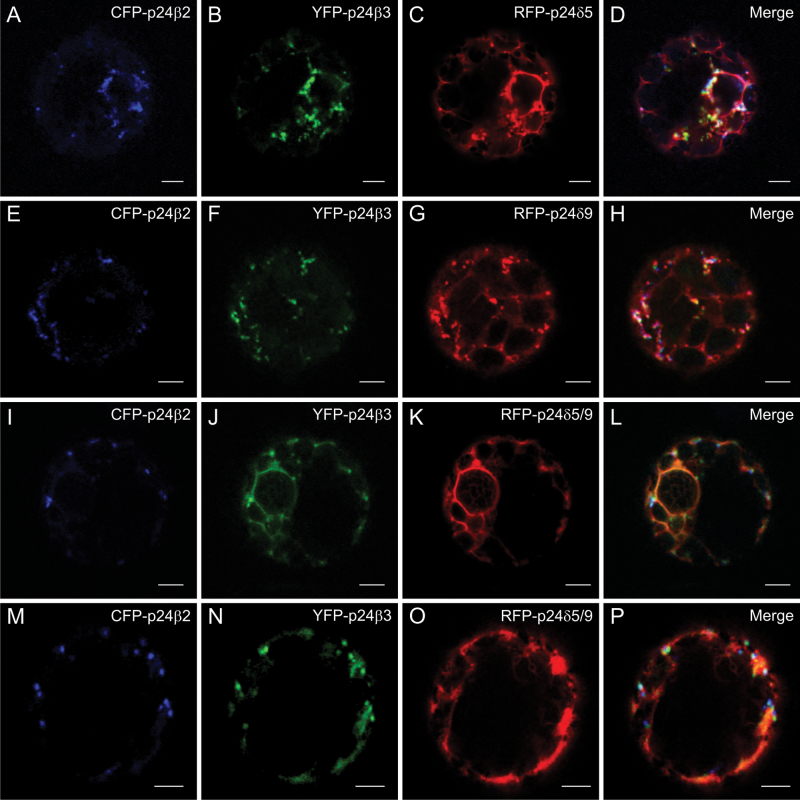
Triple or quadruple co-expression experiments were performed next. First, the two p24β proteins (CFP–p24β2 and YFP–p24β3) were co-expressed with either RFP–p24δ5 (Fig. 9A–D) or RFP–p24δ9 (Fig. 9E–H) (triple co-expression). In both cases, there was a partial co-localization of the three proteins in punctate structures. When CFP–p24β2, YFP–p24β3, RFP–p24δ5, and RFP–p24δ9 were expressed together (quadruple co-expression) (Fig. 9I–P), various degrees of co-localization between these proteins were obtained (see Table 1B). However, the most striking observation was that CFP–p24β2 remained mostly punctate, while in most of the protoplasts YFP–p24β3 was clearly less punctate and much more reticular. Under these conditions, RFP fluorescence (including both RFP–p24δ5 and RFP–p24δ9) was mostly reticular (Fig. 9I–P). Finally, the two p24δ proteins (RFP–p24δ5 and RFP–p24δ9) were co-expressed with either GFP–p24β2 or GFP–p24β3. Under these conditions, GFP–p24β2 and RFP–p24δ5/9 extensively co-localized in punctate structures (Fig. 10A–F). In contrast, GFP–p24β3 was only partially punctate and significantly redistributed to the ER, where it partially co-localized with RFP–p24δ5/9 (Fig. 10G–L).
Fig. 9.
Co-expression of p24 proteins of the beta and delta subfamilies (II). (A–P) Transient gene expression in tobacco mesophyll protoplasts. (A–D) CFP–p24β2 (A), YFP–p24β3 (B), and RFP–p24δ5 (C) co-localize partially in punctate structures (merged image in D). (E–H) CFP–p24β2 (E), YFP–p24β3 (F), and RFP–p24δ9 (G) co-localize partially in punctate structures (merged image in H). (I–P) Co-expression of CFP–p24β2 (I, M), YFP–p24β3 (J, N), and RFP–p24δ5/9 (K, O) (merged images in L and P) (see text for details). Scale bars=5 μm.
Fig. 10.
Co-expression of p24 proteins of the beta and delta subfamilies (III). (A–L) Transient gene expression in tobacco mesophyll protoplasts. (A–F) GFP–p24β2 (A, D) and RFP–p24δ5/δ9 (B, E) co-localize extensively in punctate structures (merged images in C and F). (G–L) GFP–p24β3 (G, J) relocalizes partially to the ER, where it co-localizes with RFP–p24δ5/δ9 (H, K) (merged images in I and L). Scale bars=5 μm.
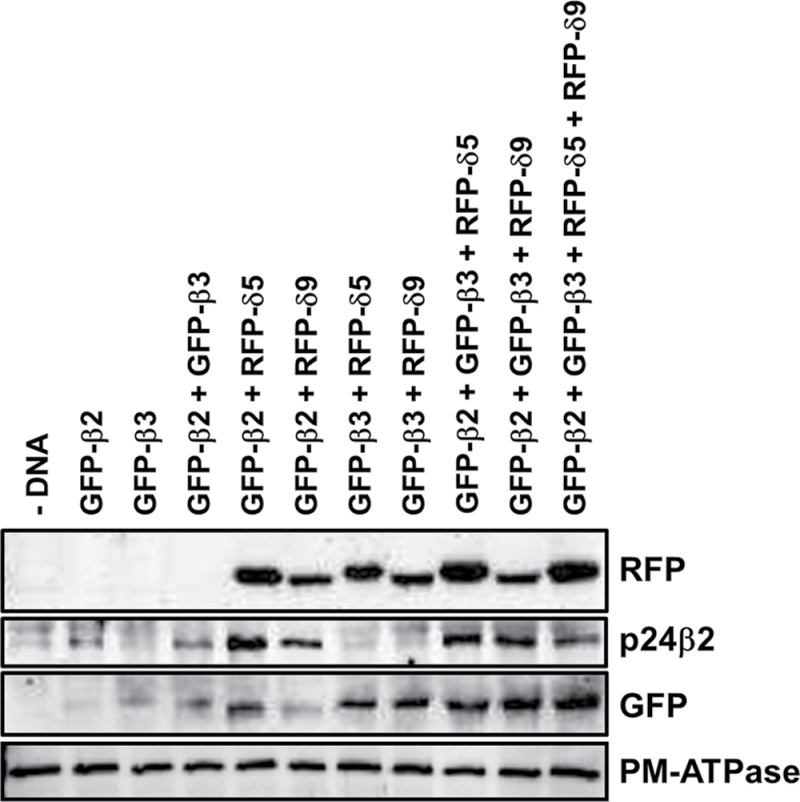
To quantify whether the levels of GFP–p24β proteins might depend on their trafficking properties, protoplasts were analysed by western blotting following the co-expression experiments shown above (Fig. 11). Since the N-terminal p24β3 antibodies could not detect transiently expressed GFP–p24β3 by western blotting, GFP antibodies, which recognized both GFP–p24β2 and GFP–p24β3, were used instead. The results are summarized in Table 2. As has been shown previously (Montesinos et al., 2012), the levels of GFP–p24β2 increased significantly upon co-expression with RFP–p24δ5. It has now been found that the levels of GFP–p24β2 also increased significantly when co-expressed with RFP–p24δ9. On the other hand, the levels of GFP–p24β2 or GFP–p24β3 were not significantly increased when both proteins were expressed together. In the case of GFP–p24β3, its levels increased upon co-expression with RFP–p24δ5 or RFP–p24δ9, but increased much more in the quadruple co-expression.
Fig. 11.
Biochemical analysis of co-expression experiments. Tobacco mesophyll protoplasts were electroporated in the absence (–DNA) or the presence of 25 μg of the indicated plasmid DNAs. At 24h post-electroporation, protoplasts were washed and homogenized to obtain a post-nuclear supernatant, which was then centrifuged to obtain a total membrane fraction. Membranes were extracted in Laemmli sample buffer and analysed by SDS–PAGE (12% acrylamide) and western blot analysis with antibodies against RFP (to detect RFP–p24δ5 and RFP–p24δ9), the p24β2 C-terminus, or GFP (to quantify the amount of both GFP–p24β2 and GFP–p24β3). A 30 μg aliquot of protein was loaded for each of the extracts. Western blot with an antibody against the plasma membrane (PM) ATPase was used as a loading control.
Table 2.
Levels of p24β2 and p24β3 in co-expression experiments
| Expression conditions | Intensity (arbitrary units) | ||
|---|---|---|---|
| p24β2 | GFP | p24β3 | |
| –DNA | 0.0 | 0.0 | 0.0 |
| p24β2 | 6.8 | 6.9 | 0.1 |
| p24β3 | 0.3 | 11.0 | 10.7 |
| p24β2+p24β3 | 7.0 | 17.0 | 10.0 |
| p24β2+p24δ5 | 27.0 | 27.2 | 0.2 |
| p24β2+p24δ9 | 12.0 | 12.1 | 0.1 |
| p24β3+p24δ5 | 0.5 | 17.0 | 16.5 |
| p24β3+p24δ9 | 0.3 | 24.0 | 23.7 |
| p24β2+p24β3+p24δ5 | 18.0 | 34.1 | 16.1 |
| p24β2+p24β3+p24δ9 | 16.0 | 36.2 | 20.2 |
| p24β2+p24β3+p24δ5+p24δ9 | 12.1 | 52.8 | 40.7 |
Quantification of western blots from two different co-expression experiments like the one shown in Fig. 11, using the Quantity One software (Bio-Rad Laboratories). The amount of p24β3 was calculated as the difference between the intensity of GFP (which includes the signal of both GFP–p24β2 and GFP–p24β3) and that of p24β2. Western blots in the linear range of detection that showed comparable intensities for p24β2 and GFP in the co-expression of p24β2+p24δ5 and p24β2+p24δ9 were selected.
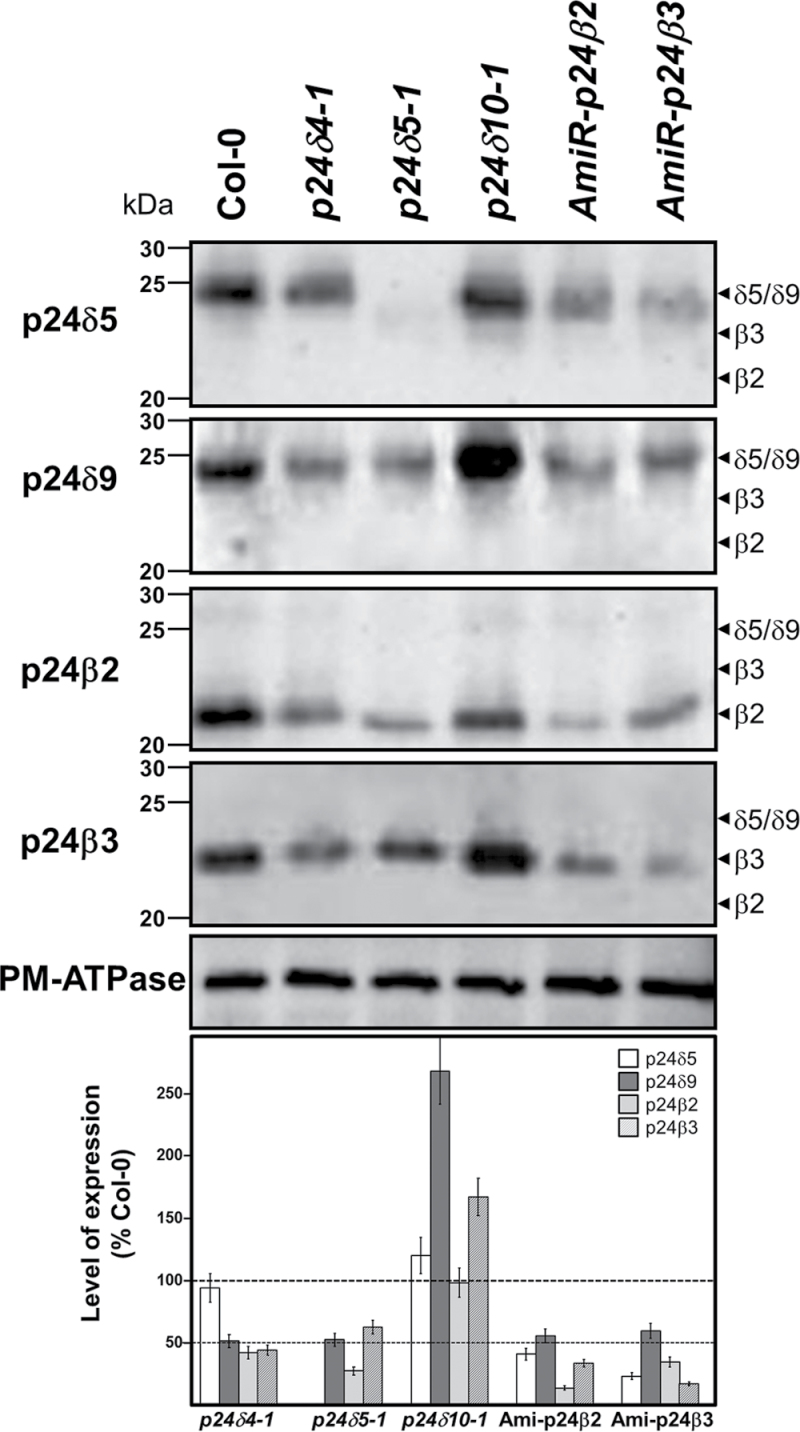
Previous studies where a single member of the p24 family had been deleted or knocked down showed that the protein level of other family members was reduced, which probably reflects the fact that these p24 proteins interact with each other in hetero-oligomeric complexes (Belden and Barlowe, 1996; Marzioch et al., 1999; Denzel et al., 2000; Vetrivel et al., 2007; Takida et al., 2008; Koegler et al., 2010; Jerome-Majewska et al., 2010; Zhang and Volchuk, 2010). Therefore, the protein levels of p24δ4, p24δ5, p24δ9, p24β2, or p24β3 were examined in knock-out/knock-down mutants. T-DNA insertion knock-out mutants lacking p24δ5 (p24δ5-1) and p24δ4 (p24δ4-1) had already been characterized (Montesinos et al., 2012). As no T-DNA insertion knock-out mutant for p24δ9 (δ2 subclass) was found in the Salk collection, a knock-out mutant for p24δ10 (δ2 subclass) was analysed (Supplementary Fig. S6 at JXB online). Plants from the three lines resembled wild-type plants under standard growth conditions (Supplementary Fig. S6). As T-DNA insertion mutants for p24β2 or p24β3 are not available in mutant collections, amiRNA was used to knock down p24β2 or p24β3 (Supplementary Fig. S6). No distinct phenotype was observed in amiR-p24β2 or amiR-p24β3 knock-down lines when compared with wild-type plants under standard growth conditions (Supplementary Fig. S6). Protein extracts from roots of wild-type (Col-0) plants, T-DNA knock-out insertion mutants, and amiR-p24β2 and amiR-p24β3 lines were analysed by western blotting with the corresponding antibodies (Fig. 12). It was previously shown that the T-DNA mutant lacking p24δ5 showed p24δ4 levels comparable with those of the wild type. The same was true for the levels of p24δ5 in the p24δ4 mutant (Montesinos et al., 2012). However, both mutants (p24δ4-1 and p24δ5-1) showed reduced levels of p24δ9, p24β2, and p24β3 (Fig. 12). In contrast, the T-DNA mutant lacking p24δ10 did not show reduced protein levels of p24δ5, p24β2, or p24β3, but showed a 2.5-fold increase in the protein levels of the highly related p24δ9 (Fig. 12). Finally, the lines expressing amiRNAs were analysed for p24β2 or p24β3. Expression of p24β2 and p24β3 was reduced by ~85% in the amiR-p24β2 and amiR-p24β3 lines, respectively (Fig. 12). In addition, the amiR-p24β2 line showed reduced protein levels of p24δ5, p24δ9, and p24β3, while the amiR-p24β3 line showed reduced protein levels of p24δ5, p24δ9, and p24β2 (Fig. 12). These results suggest that p24 proteins do interact with other family members to form heteromeric complexes.
Fig. 12.
Levels of p24 proteins in knock-out (KO) mutants or amiRNA lines. Western blot analysis with antibodies against the N-terminus of p24δ5, p24δ9, or p24β3, or the C-terminus of p24β2 in membranes from the wild type (Col-0) or the indicated KO mutants or amiRNA lines (see text for details). The expected positions for p24δ5/δ9, p24β3, and p24β2 (according to the western blot analysis shown in Fig. 1) are shown by arrowheads. Molecular weight markers are indicated on the left. A 25 μg aliquot of protein was loaded in each lane. Western blotting with an antibody against the plasma membrane (PM) ATPase was used as a loading control. Lower panel shows a quantification of the levels of each of the proteins in each mutant calculated as a percentage of the levels present in wild-type (Col-0) membranes (mean±SD from three independent experiments).
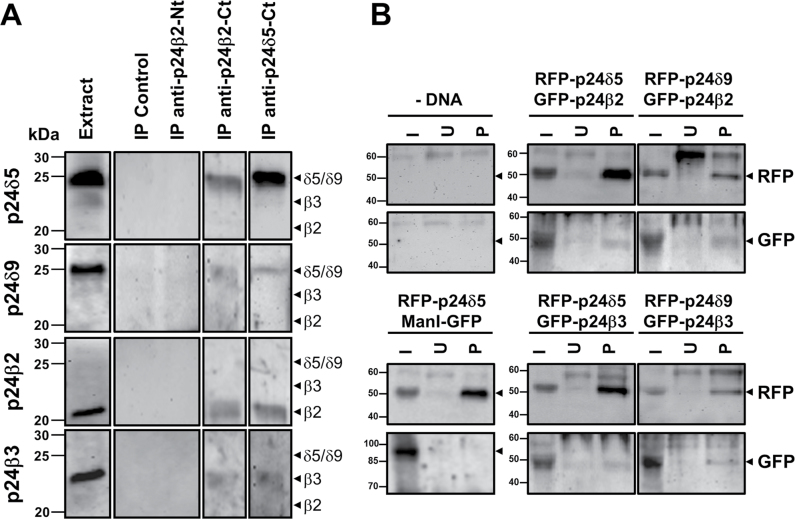
To test biochemically for interactions between endogenous p24 proteins, co-immunoprecipitation experiments were performed using C-terminal p24δ5 or p24β2 antibodies (Montesinos et al., 2012). As shown in Fig. 13A, antibodies against the C-terminus of p24β2 caused the co-immunoprecipitation of p24δ5, p24δ9, and p24β3, while antibodies against the C-terminus of p24δ5 caused the co-immunoprecipitation of p24δ9, p24β2, and p24β3. As a control, control beads or N-terminal p24β2 antibodies, which have previously beeen shown to be unable to immunoprecipitate p24β2 (Montesinos et al., 2012), were used. Pull-down experiments were also performed using membrane fractions from protoplasts co-expressing RFP–p24δ5 or RFP–p24δ9 and GFP–p24β2 or GFP–p24β3, using an RFP-trap for the pull-down of RFP-tagged proteins (p24δ5 or p24δ9) and GFP antibodies for the western blot analysis of the interacting proteins (p24β2 or p24β3). As shown in Fig. 13B, RFP–p24δ5 pulled-down GFP–p24β2 and, to a lesser extent, GFP–p24β3, but not ManI–GFP (used as a negative control). Similarly, RFP–p24δ9 pulled-down both GFP–p24β2 and GFP–p24β3 (Fig. 13B). This suggests that heterotypic interactions can occur between Arabidopsis p24 proteins from the beta and delta subfamilies.
Fig. 13.
Interactions between p24 proteins. (A) Co-immunoprecipitation experiments. Immunoprecipitation of endogenous p24 proteins was performed using affinity-purified antibodies against the C-terminus of p24β2 (IP anti-p24β2-Ct) or p24δ5 (IP anti-p24δ5-Ct). As a control, control beads (IP Control) or antibodies against the N-terminus of p24β2 (IP anti-p24β2-Nt) were used. Immunoprecipitates were analysed by SDS–PAGE and western blot with antibodies against the N-terminus of p24δ5, p24δ9, and p24β3, or the C-terminus of p24β2. Extract lane contains 20 μg of the membrane proteins used as input for the immunoprecipitation. The expected positions for p24δ5/δ9, p24β3, and p24β2 (according to the western blot analysis shown in Fig. 1) are shown by arrowheads. Molecular weight markers are indicated on the left. (B) Pull-down assays of RFP–p24δ5 or RFP–p24δ9 from membranes of protoplasts co-expressing the indicated proteins using an RFP-trap (see text). As a control, pull-downs were performed from membranes of untransfected protoplasts (–DNA) or protoplasts co-expressing RFP–p24δ5 and ManI–GFP. Bound proteins were analysed by SDS–PAGE and western blotting with antibodies against RFP (to detect RFP–p24δ5 or RFP–p24δ9) or GFP (to detect GFP–p24β2, GFP–p24β3, or ManI–GFP). In the case of protoplasts co-expressing RFP–p24δ5 and ManI–GFP, no RFP signal was detected in the 90kDa region (expected position for ManI–GFP) and no GFP signal was detected in the 50kDa region (expected position for RFP–p24δ5) (data not shown). I, input (5% of the membrane extracts used for the pull-down assay); U, unspecific binding (proteins bound to control blocked magnetic particles); P, pull-down.
p24δ5 may play a role in retrograde Golgi–ER transport of the KDEL-receptor ERD2
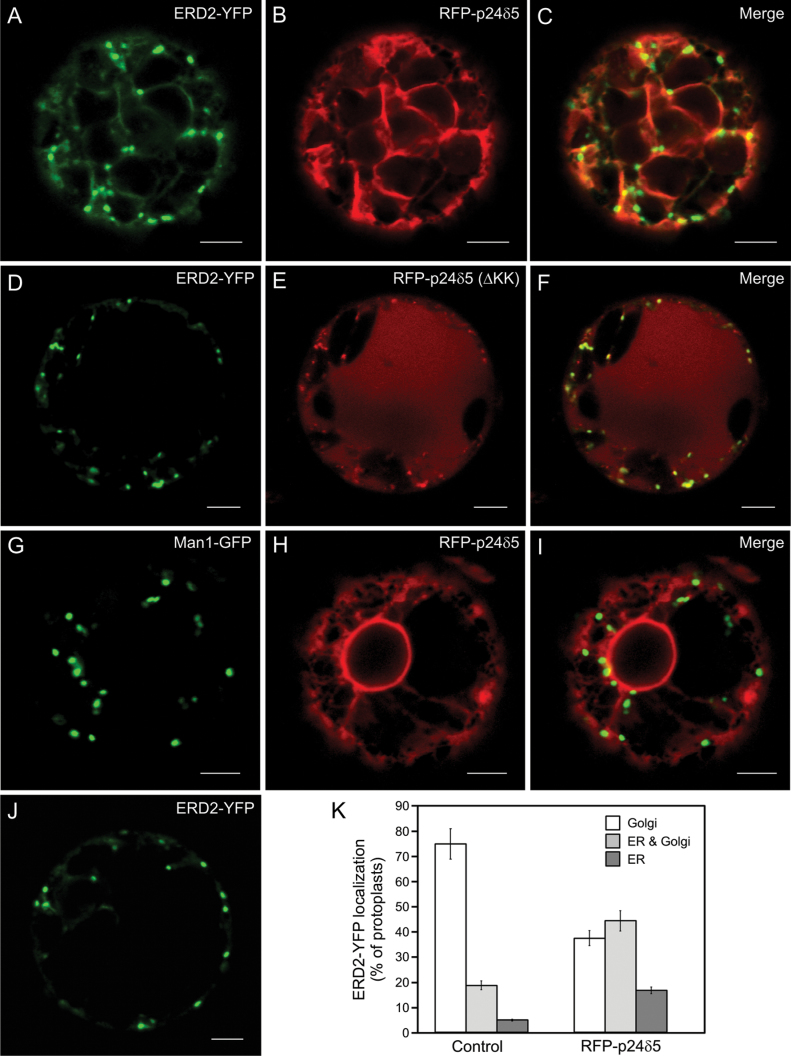
Finally, a functional analysis of p24 proteins in Arabidopsis was attempted. Since the single knock-down/knock-out mutants that were characterized did not show any obvious phenotypic alteration, a gain-of-function approach was tried, by overexpressing specific p24 proteins and testing for an influence on the trafficking of putative cargos. For these studies, the focus was on p24δ5, since its trafficking was previously characterized in the early secretory pathway and mutants were readily available (Langhans et al., 2008; Montesinos et al., 2012). However, the drawback of this strategy is the fact that specific p24 cargos have not yet been identified in plants. Since p24 proteins of the delta subfamily have been proposed to play a role in retrograde Golgi–ER transport (Majoul et al., 1998, 2001; Aguilera-Romero et al., 2008), it was thought that one good candidate might be ERD2, a receptor which retrieves KDEL/HDEL-containing cargo from the Golgi to the ER. Indeed, there is one previous report showing that mammalian p23 (p24δ subfamily) interacts with ERD2 and is involved in its retrograde Golgi–ER transport (Majoul et al., 1998, 2001). Therefore, ERD2–YFP was transiently co-expressed with increasing amounts of RFP–p24δ5. When expressed alone, ERD2–YFP localized mainly (76% of analysed protoplasts) to punctate Golgi structures, with a faint ER staining (Fig. 14J–K). Overexpression of RFP–p24δ5 produced a significant redistribution of ERD2–YFP from the Golgi to the ER. As quantified in Fig. 14K, Golgi localization of ERD2–YFP decreased to 38% in the presence of RFP–p24δ5. In the remaining protoplasts, ERD2–YFP localized either to the ER (17% of the protoplasts) or to both the ER and Golgi (45% of the protoplasts) (Fig. 14A–C). These experiments suggest that p24δ5 may play a role in retrograde Golgi–ER transport of ERD2. This was not a general effect on Golgi proteins, since the standard Golgi marker ManI–GFP was still Golgi localized under the same conditions (Fig. 14G–I). Interestingly, this effect was not observed when we used an RFP–p24δ5 mutant lacking the KK motif at the C-terminus, which is necessary for interaction with the COPI coat and therefore for retrograde Golgi–ER transport of p24δ5 (Langhans et al., 2008; Montesinos et al., 2012). Under these conditions, RFP–p24δ5(ΔKK) and ERD2–YFP extensively co-localized in punctate structures (Fig. 14D–F), which, based on previous results with this mutant, should correspond either to the Golgi or to the pre-vacuolar compartment (PVC) (Langhans et al., 2008; Montesinos et al., 2012). In addition, RFP–p24δ5(ΔKK) (but not ERD2–YFP) was also localized to the vacuole, as has been shown previously (Langhans et al., 2008).
Fig. 14.
RFP–p24δ5 [but not RFP–p24δ5(ΔKK)] partially relocalizes ERD2–YFP to the ER. (A–J) Transient gene expression in tobacco mesophyll protoplasts. (A–C) RFP–p24δ5 (B) caused a partial relocalization of ERD2–YFP (A) to the ER, although ERD2–YFP also showed a punctate localization (ER and Golgi localization) (merged image in C). (D–F) ERD2–YFP (D) and RFP–p24δ5(ΔKK) (E) (which also shows vacuolar localization) almost completely co-localized in punctate structures (merged image in F). (G–I) RFP–p24δ5 (H) did not significantly change the localization of ManI–GFP (G) (merged image in I). (J) ERD2–YFP showed mostly a Golgi localization when expressed alone. (K) Quantification of the localization of ERD2–YFP expressed alone (Control) or in the presence of RFP–p24δ5. Eighty protoplasts (from four independent experiments), showing comparable expression levels of ERD2–YFP, ManI–GFP, and RFP–p24δ5, were analysed per condition, using identical laser output levels and imaging conditions. The localization of ERD2–YFP was assigned as Golgi (punctae), ER and Golgi, or ER, and calculated as a percentage. Error bars represent the SEM. Images in the panels show the most representative pattern found for each condition. Scale bars=5 μm.
Discussion
p24 proteins have been known for quite some time, and numerous reports have been published on mammals and yeast concerning their trafficking and localization. However, only recently have their putative functions been addressed, and these appear to be highly dependent on their trafficking properties. In this respect, Hasegawa et al. (2010) highlighted the differential functional properties of p24 proteins from the alpha and delta subfamilies (both containing a dilysine motif in their cytoplasmic C-terminus) and the beta and gamma subfamilies. Comparatively, much less is known about these proteins in plants, which strikingly contain only members of the beta and delta subfamilies. Although Arabidopsis p24 proteins cycle in the early secretory pathway, their steady-state distribution appears to be different for members of the p24δ and p24β subfamilies. While endogenous p24δ5 or p24δ4 (Montesinos et al., 2012) and p24δ9 (Fig. 2) localize mainly to the ER, but also partially to the cis-Golgi, endogenous p24β2 (Montesinos et al., 2012) and p24β3 (Fig. 2) mainly localize to the Golgi, with only occasional ER labelling. This steady-state distribution may reflect the differential ability of their cytoplasmic tails to interact with COPI or COPII subunits (Contreras et al., 2004a , b ; Langhans et al., 2008) as well as their intrinsic ability to interact with other p24 family members (Montesinos et al., 2012).
Exit of p24 proteins from the ER appears to be COPII dependent (Fig. 5; Langhans et al., 2008; Chen et al., 2012) while Golgi–ER recycling of p24δ proteins is COPI dependent (Langhans et al., 2008; Montesinos et al., 2012). Interestingly, ER exit of p24β2 and p24β3 appears to show some striking differences. Upon Sec12 overexpression, which titrates cytosolic Sar1p and prevents COPII-coated vesicle formation (Philipson et al., 2001), both p24β2 and p24β3 showed a typical ER pattern and co-localized with a standard Golgi marker, suggesting that their ER exit is indeed COPII dependent. However, in the case of p24β3, an additional punctate pattern was also found. One possible explanation is that cycling of p24β3 occurs with slower kinetics. In this scenario, a population of the protein which has already reached the Golgi apparatus but has not been recycled would be insensitive to Sec12 treatment. However, most of the punctae where p24β3 was found under these conditions appear to correspond to ERES, as suggested by their co-localization with the ERES/COPII marker 6kDa VP–CFP. This differential behaviour has never been shown for p24 proteins. However, there is a previous report showing that the ER export of adrenergic and angiotensin II receptors is differentially regulated by Sar1 (Dong et al., 2008). In that study, the cell surface expression of the adrenergic receptors (ARs) α2B-AR or β2-AR and the angiotensin 1 receptor (AT1R) were significantly attenuated by the GTP-bound mutant Sar1H79G, suggesting that ER export of these receptors occurs via Sar1-dependent COPII-coated vesicles. Interestingly, subcellular distribution analyses showed that α2B-AR and AT1R receptor were highly concentrated at discrete locations near the nucleus in cells expressing Sar1H79G (presumably ERES), whereas β2-AR exhibited an ER distribution. These data indicate that Sar1-catalysed efficient GTP-hydrolysis differentially regulates ER export of ARs and AT1R and provided the first evidence indicating distinct mechanisms for the recruitment of different GPCRs into COPII vesicles on the ER membrane (Dong et al., 2008). Further work will be necessary to elucidate whether the differential behaviour of p24β2 and p24β3 in ER export may have any functional implications.
The stability of p24 proteins, which may be related to their trafficking, has also been investigated. While p24 proteins of the delta subfamily appear to be relatively stable, when expressed either alone or in combination with other p24 family members, p24 proteins of the beta subfamily seem to depend on the interaction with other p24 family members for stabilization. Therefore, the mechanism(s) involved in their degradation have been investigated. To the authors’ knowledge, there is only one previous study dealing with the mechanism of degradation of p24 proteins. In particular, TMP21 (also named p23, p24δ subfamily), a member of the presenilin complex, has been shown to have a short half-life of ~3h and to be degraded by the ubiquitin–proteasome pathway: while treatment with the proteasomal inhibitor MG-132 caused a significant increase in TMP21 protein levels, lysosomal inhibition was without effect (Liu et al., 2008). To the authors’ knowledge, there are no reports dealing with the degradation of p24 proteins of the beta subfamily. It has been shown that p24β2 localizes to the Golgi at steady state, but cycles between the ER and Golgi and may also be transported to the PVC and to the vacuole, which may result in an increased degradation by cysteine proteases present in these compartments (Montesinos et al., 2012). In this study, the protein levels of p24β2 and p24β3 were examined and they were found to be insensitive to MG-132 treatment, under conditions that have been shown to inhibit proteasome-mediated degradation in plant cells (Yanagawa et al., 2002). In contrast, treatment with the E-64, an inhibitor of cysteine proteases, caused a significant increase in the protein levels of both p24β2 and p24β3. This suggests that both proteins may be degraded by cysteine proteases upon transport to post-Golgi compartments (PVC, vacuole).
Proteins of the p24 family have been proposed to form functional heteromeric complexes, whereas it is still debatable whether they can exist as monomers, heterodimers, or heterotetramers, depending on their subcellular localization (Marzioch et al., 1999; Jenne et al., 2002). Recent data suggest that members of the four subfamilies in mammals (p25, p24, p28, and p23) can form hetero-oligomers, although the stoichiometry among them remains to be determined (Fujita et al., 2011). Plants contain only p24 proteins of the beta and delta subfamilies, the latter containing members of two different subclasses (δ1 and δ2) (Chen and Zheng, 2012). Interestingly, western blot analysis of the different mutants that have been analysed shows that the lack of a member of the δ1 subclass causes a reduction in the protein levels of members of the δ2 subclass but not of members of the δ1 subclass. In this respect, the p24δ 5–1 mutant shows no change in the levels of p24δ4 and vice versa (Montesinos et al., 2012), but reduced levels of p24δ9. In addition, p24δ1 mutants (p24δ 5–1 and p24δ 4–1) showed reduced protein levels of both p24β2 and p24β3. On the other hand, the p24δ 10–1 mutant that lacks p24δ10 (δ2 subclass) showed increased protein levels of the closely related p24δ9 (δ2 subclass), probably induced in the absence of p24δ10, and no decrease in the levels of p24δ5 (δ1 subclass) or p24β proteins. This indicates that the increased proteins levels of p24δ9 may compensate for the lack of p24δ10 in the p24δ10 mutant. On the other hand, amiRNA lines with reduced levels of p24β2 or p24β3 showed reduced protein levels of p24β3 or p24β2 (respectively) and of p24δ proteins from both subclasses. Altogether, these results suggest that p24 proteins may form heteromeric complexes containing members of the delta and beta subfamilies. This is consistent with the co-immunoprecipitation and pull-down experiments, which suggest that members of the beta and delta subfamilies interact with each other.
The co-expression experiments further support the existence of interactions between p24 proteins from both subfamilies and their coupled transport in the early secretory pathway. This can be deduced from the strong co-localization between p24β2 and p24δ5 or p24δ9, as well as by the fact that p24β2 changes the localization of p24δ5 and/or p24δ9 from a typical ER pattern to punctate structures corresponding to both pre-Golgi COPII (Langhans et al., 2012) and Golgi. This indicates that p24β2 is able to facilitate the transport of p24δ5 and p24δ9 from the ER to the Golgi. In addition, the stability of p24β2 increases significantly when co-expressed with p24δ5 or p24δ9 (Table 2). This is probably because p24δ proteins may hold back p24β proteins in the early secretory pathway (Montesinos et al., 2012). In the case of p24δ5 and p24β2, these effects require the coiled-coil domain, which suggests they are mediated by a direct interaction between both proteins (Montesinos et al., 2012). On the other hand, co-expression of p24β2 and p24β3 does not produce any significant stabilization of these proteins (Table 2). In contrast to p24β2, p24β3 shows only a partial co-localization with p24δ5 or p24δ9 and is also stabilized partially in the presence of these proteins. Interestingly, maximal stability of p24β3 was only achieved when co-expressed with both RFP–p24δ5 and RFP–p24δ9 (Table 2). The fact that p24β3 (but not p24β2) partially relocates to the ER under these conditions suggests that both RFP–p24δ5 and RFP–p24δ9 may cooperate in retrograde Golgi–ER transport of p24β3, contributing to its increased stability.
Altogether, the present experiments suggest that there are highly dynamic and complex interactions between p24 members of each subclass/subfamily which are needed for their correct localization and stability and therefore function. Although the stoichiometry and composition of these complexes remain to be established, the experiments described here suggest that ‘anterograde’ complexes should include p24β2, which facilitates transport of both p24δ5 (δ1 subclass) and p24δ9 (δ2 subclass) to the Golgi apparatus. On the other hand, ‘retrograde’ complexes should contain p24δ proteins (for sorting into COPI vesicles), probably including members from the δ1 and δ2 subclasses.
As a first attempt to elucidate putative functions for Arabidopsis p24 proteins, a gain-of-function approach has been used, given the lack of phenotypic alterations found in single knock-out mutants or knock-down lines. These experiments have convincingly shown that p24δ5 appears to play a role in the retrograde Golgi–ER transport of the KDEL-receptor ERD2, probably by facilitating its sorting into COPI vesicles. Indeed, ERD2-mediated retrograde transport of cholera toxin (a KDEL cargo) from the Golgi back to the ER has been shown to involve COPI, mammalian p23 (p24δ subfamily), and ERD2 (Majoul et al., 1998). In addition, it has been shown that p23 interacts with ERD2, suggesting that p23 participates directly in the retrograde transport of ERD2 (Majoul et al., 2001). The results are consistent with those observations. The fact that the C-terminus of p24δ5 has a high affinity for COPI (Contreras et al., 2004a , b ) makes this protein (and probably other members of the delta subfamily) ideal to perform a similar role in retrograde Golgi–ER transport in plants.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. The p24 family in Arabidopsis.
Figure S2. Western blot analysis with Nt-p24 antibodies of bacterial extracts expressing (His)6-tagged N-terminal p24 constructs.
Figure S3. Localization of RFP–p24δ9.
Figure S4. Localization of YFP–p24β2.
Figure S5. Co-expression of CFP–p24β2 and RFP–p24δ9 in tobacco leaf epidermal cells.
Figure S6. Characterization of the p24δ10 mutant and amiRNA-p24β2 and amiRNA-p24β3 lines.
Acknowledgements
We thank the Salk Institute Genomic Analysis Laboratory for providing the sequence-indexed Arabidopsis T-DNA insertion mutants, the greenhouse and electron microscopy sections of SCSIE (University of Valencia), and Pilar Selvi and Barbara Jesenofsky for excellent technical assistance. This work was supported by the Ministerio de Ciencia e Innovación (grant no. BFU2009-07039 to FA), Generalitat Valenciana (grant no. ISIC/2013/004 to FA) and the Deutsche Forschungsgemeinschaft (grant no. RO440/14-1 to DGR). JCM was the recipient of fellowships from Generalitat Valenciana (Geronimo Forteza Program) and from the Ministerio de Educación y Ciencia (FPU Program).
References
- Aguilera-Romero A, Kaminska J, Spang A, Riezman H, Muñiz M. 2008. The yeast p24 complex is required for the formation of COPI retrograde transport vesicles from the Golgi apparatus. Journal of Cell Biology 180, 713––720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anantharaman V, Aravind L. 2002. The GOLD domain, a novel protein module involved in Golgi function and secretion. Genome Biology 3, research0023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axelos M, Curie C, Bardet C, Lescure B. 1992. A protocol for transient expression in Arabidopsis thaliana protoplasts isolated from cell suspension cultures. Plant Physiology and Biochemistry 30, 123––128 [Google Scholar]
- Belden WJ, Barlowe C. 1996. Erv25p, a component of COPII-coated vesicles, forms a complex with Emp24p that is required for efficient endoplasmic reticulum to Golgi transport. Journal of Biological Chemistry 271, 26939––26946 [DOI] [PubMed] [Google Scholar]
- Belden WJ, Barlowe C. 2001. Distinct roles for the cytoplasmic tail sequences of Emp24p and Erv25p in transport between the endoplasmic reticulum and Golgi complex. Journal of Biological Chemistry 276, 43040––43048 [DOI] [PubMed] [Google Scholar]
- Brandizzi F, Frangne N, Marc-Martin S, Hawes C, Neuhaus JM, Paris N. 2002. The destination for single-pass membrane proteins is influenced markedly by the length of the hydrophobic domain. The Plant Cell 14, 1077––1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bubeck J, Scheuring D, Hummel E, Langhans M, Viotti C, Foresti O, Denecke J, Banfield DK, Robinson DG. 2008. The syntaxins SYP31 and SYP81 control ER–Golgi trafficking in the plant secretory pathway. Traffic 9, 1629––1652 [DOI] [PubMed] [Google Scholar]
- Carney GE, Bowen NJ. 2004. p24 proteins, intracellular trafficking, and behaviour: Drosophila melanogaster provides insights and opportunities. Biology of the Cell 96, 271––278 [DOI] [PubMed] [Google Scholar]
- Castillon GA, Aguilera-Romero A, Manzano-Lopez J, Epstein S, Kajiwara K, Funato K, Watanabe R, Riezman H, Muñiz M. 2011. The yeast p24 complex regulates GPI-anchored protein transport and quality control by monitoring anchor remodeling. Molecular Biology of the Cell 22, 2924––2936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Hasegawa H, Schmitt-Ulms G, et al. 2006. TMP21 is a presenilin complex component that modulates gamma-secretase but not epsilon-secretase activity. Nature 440, 1208––1212 [DOI] [PubMed] [Google Scholar]
- Chen J, Qi X, Zheng H. 2012. Subclass-specific localization and trafficking of Arabidopsis p24 proteins in the ER–Golgi interface. Traffic 13, 400––415 [DOI] [PubMed] [Google Scholar]
- Ciufo LF, Boyd A. 2000. Identification of a lumenal sequence specifying the assembly of Emp24p into p24 complexes in the yeast secretory pathway. Journal of Biological Chemistry 275, 8382––8388 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal 16, 735––743 [DOI] [PubMed] [Google Scholar]
- Contreras I, Ortiz-Zapater E, Aniento F. 2004a. Sorting signals in the cytosolic tail of membrane proteins involved in the interaction with plant ARF1 and coatomer. The Plant Journal 38, 685––698 [DOI] [PubMed] [Google Scholar]
- Contreras I, Yang Y, Robinson DG, Aniento F. 2004b. Plant COPI and COPII coat proteins show a differential affinity for p24 cytosolic tails. Plant and Cell Physiology 45, 1779––1786 [DOI] [PubMed] [Google Scholar]
- Contreras FX, Ernst AM, Haberkant P, et al. 2012. Molecular recognition of a single sphingolipid species by a protein’s transmembrane domain. Nature 481, 525––529 [DOI] [PubMed] [Google Scholar]
- Dancourt J, Barlowe C. 2010. Protein sorting receptors in the early secretory pathway. Annual Review of Biochemistry 79, 777––802 [DOI] [PubMed] [Google Scholar]
- Denzel A, Otto F, Girod A, Pepperkok R, Watson R, Rosewell I, Bergeron JJ, Solari RC, Owen MJ. 2000. The p24 family member p23 is required for early embryonic development. Current Biology 10, 55––58 [DOI] [PubMed] [Google Scholar]
- Dominguez M, Dejgaard K, Fullekrug J, Dahan S, Fazel A, Paccaud JP, Thomas DY, Bergeron JJ, Nilsson T. 1998. gp25L/emp24/p24 protein family members of the cis-Golgi network bind both COPI and COPII coatomer. Journal of Cell Biology 140, 751––765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong C, Zhou F, Fugetta EK, Filipeanu CM, Wu G. 2008. Endoplasmic reticulum export of adrenergic and angiotensin II receptors is differentially regulated by Sar1 GTPase. Cellular Signalling 20, 1035––1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery G, Rojo M, Gruenberg J. 2000. Coupled transport of p24 family members. Journal of Cell Science 113, 2507––2516 [DOI] [PubMed] [Google Scholar]
- French AP, Mills S, Swarup R, Bennett MJ, Pridmore TP. 2008. Colocalization of fluorescent markers in confocal microscope images of plant cells. Nature Protocols 3, 619––628 [DOI] [PubMed] [Google Scholar]
- Fujita M, Watanabe R, Jaensch N, Romanova-Michaelides M, Satoh T, Kato M, Riezman H, Yamaguchi Y, Maeda Y, Kinoshita T. 2011. Sorting of GPI-anchored proteins into ER exit sites by p24 proteins is dependent on remodeled GPI. Journal of Cell Biology 194, 61––75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Füllerkrug J, Suganuma T, Tang BL, Hong W, Storrie B, Nilsson T. 1999. Localization and recycling of gp27 (hp24g3): complex formation with other p24 family members. Molecular Biology of the Cell 10, 1939––1955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gommel D, Orci L, Emig EM, Hannah MJ, Ravazzola M, Nickel W, Helms JB, Wieland FT, Sohn K. 1999. p24 and p23, the major transmembrane proteins of COPI-coated vesicles, form hetero-oligomeric complexes and cycle between organelles of the early secretory pathway. FEBS Letters 447, 179––185 [DOI] [PubMed] [Google Scholar]
- Hasegawa H, Liu L, Nishimura M. 2010. Dilysine retrieval signal-containing p24 proteins collaborate in inhibiting γ-cleavage of amyloid precursor protein. Journal of Neurochemistry 115, 771––781 [DOI] [PubMed] [Google Scholar]
- Jenne N, Frey K, Brügger B, Wieland FT. 2002. Oligomeric state and stoichiometry of p24 proteins in the early secretory pathway. Journal of Biological Chemistry 277, 46504––46511 [DOI] [PubMed] [Google Scholar]
- Jerome-Majewska LA, Achkar T, Luo L, Lupu F, Lacy E. 2010. The trafficking protein Tmed2/p24beta(1) is required for morphogenesis of the mouse embryo and placenta. Developmental Biology 341, 154––166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koegler E, Bonnon C, Waldmeier L, Mitrovic S, Halbeisen R, Hauri HP. 2010. p28, a novel ERGIC/cis Golgi protein, required for Golgi ribbon formation. Traffic 11, 70––89 [DOI] [PubMed] [Google Scholar]
- Langhans M, Marcote MJ, Pimpl P, Virgili-López G, Robinson DG, Aniento F. 2008. In vivo trafficking and localization of p24 proteins in plant cells. Traffic 9, 770––785 [DOI] [PubMed] [Google Scholar]
- Langhans M, Meckel T, Kress A, Lerich A, Robinson DG. 2012. ERES (ER exit sites) and the ‘secretory unit concept’. Journal of Microscopy 247, 48––59 [DOI] [PubMed] [Google Scholar]
- Lavoie C, Paiement J, Dominguez M, Roy L, Dahan S, Gushue JN, Bergeron JJ. 1999. Roles for alpha(2)p24 and COPI in endoplasmic reticulum cargo exit site formation. Journal of Cell Biology 146, 285––300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MH, Min MK, Lee YJ, Jin JB, Shin DH, Kim DH, Lee KH, Hwang I. 2002. ADP-ribosylation factor 1 of Arabidopsis plays a critical role in intracellular trafficking and maintenance of endoplasmic reticulum morphology in Arabidopsis. Plant Physiology 129, 1507––20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerich A, Langhans M, Sturm S, Robinson DG. 2011. Is the 6kDa tobacco etch viral protein a bona fide ERES marker? Journal of Experimental Botany 62, 5013––5023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Bromley-Brits K, Xia K, Mittelholtz J, Wang R, Song W. 2008. TMP21 degradation is mediated by the ubiquitin–proteasome pathway. European Journal of Neuroscience 28, 1980––1988 [DOI] [PubMed] [Google Scholar]
- Luo W, Wang Y, Reiser G. 2007. p24A, a type I transmembrane protein, controls ARF1-dependent resensitization of protease-activated receptor-2 by influence on receptor trafficking. Journal of Biological Chemistry 282, 30246––30255 [DOI] [PubMed] [Google Scholar]
- Luo W, Wang Y, Reiser G. 2011. Proteinase-activated receptors, nucleotide P2Y receptors, and μ-opioid receptor-1B are under the control of the type I transmembrane proteins p23 and p24A in post-Golgi trafficking. Journal of Neurochemistry 117, 71––81 [DOI] [PubMed] [Google Scholar]
- Majoul I, Sohn K, Wieland FT, Pepperkok R, Pizza M, Hillemann J, Söling HD. 1998. KDEL receptor (Erd2p)-mediated retrograde transport of the cholera toxin A subunit from the Golgi involves COPI, p23, and the COOH terminus of Erd2p. Journal of Cell Biology 143, 601––612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majoul I, Straub M, Hell SW, Duden R, Söling HD. 2001. KDEL-cargo regulates interactions between proteins involved in COPI vesicle traffic: measurements in living cells using FRET. Developmental Cell 1, 139––153 [DOI] [PubMed] [Google Scholar]
- Marzioch M, Henthorn DC, Herrmann JM, Wilson R, Thomas DY, Bergeron JJ, Solari RC, Rowley A. 1999. Erp1p and Erp2p, partners for Emp24p and Erv25p in a yeast p24 complex. Molecular Biology of the Cell 10, 1923––1938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitrovic S, Ben-Tekaya H, Koegler E, Gruenberg J, Hauri HP. 2008. The cargo receptors Surf4, endoplasmic reticulum–Golgi intermediate compartment (ERGIC)-53, and p25 are required to maintain the architecture of ERGIC and Golgi. Molecular Biology of the Cell 19, 1976––1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montesinos JC, Sturm S, Langhans M, Hillmer S, Marcote MJ, Robinson DG, Aniento F. 2012. Coupled transport of Arabidopsis p24 proteins at the ER–Golgi interface. Journal of Experimental Botany 63, 4243––4261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñiz M, Nuoffer C, Hauri HP, Riezman H. 2000. The Emp24 complex recruits a specific cargo molecule into endoplasmic reticulum-derived vesicles. Journal of Cell Biology 148, 925––930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nebenführ A, Frohlick JA, Staehelin LA. 2000. Redistribution of Golgi stacks and other organelles during mitosis and cytokinesis in plant cells. Plant Physiology 124, 135––151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nebenführ A, Gallagher LA, Dunahay TG, Frohlick JA, Mazurkiewicz AM, Meehl JB, Staehelin LA. 1999. Stop-and-go movements of plant Golgi stacks are mediated by the acto-myosin system. Plant Physiology 121, 1127––1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz-Masia D, Perez-Amador MA, Carbonell J, Marcote MJ. 2007. Diverse stress signals activate the C1 subgroup MAP kinases of Arabidopsis. FEBS Letters 581, 1834––1840 [DOI] [PubMed] [Google Scholar]
- Ortiz-Zapater E, Soriano-Ortega E, Marcote MJ, Ortiz-Masiá D, Aniento F. 2006. Trafficking of the human transferrin receptor in plant cells: effects of tyrphostin A23 and brefeldin A. The Plant Journal 48, 757––770 [DOI] [PubMed] [Google Scholar]
- Ossowski S, Schwab R, Weigel D. 2008. Gene silencing in plants using artificial microRNAs and other small RNAs. The Plant Journal 53, 674––690 [DOI] [PubMed] [Google Scholar]
- Phillipson BA, Pimpl P, daSilva LL, Crofts AJ, Taylor JP, Movafeghi A, Robinson DG, Denecke J. 2001. Secretory bulk flow of soluble proteins is efficient and COPII dependent. The Plant Cell 13, 2005––2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimpl P, Hanton SL, Taylor JP, Pinto-DaSilva LL, Denecke J. 2003. The GTPase ARF1p controls the sequence-specific vacuolar sorting route to the lytic vacuole. The Plant Cell 15, 1242––1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojo M, Emery G, Marjomäki V, McDowall A, Parton RG, Gruenberg J. 2000. The transmembrane protein p23 contributes to the organization of the Golgi apparatus. Journal of Cell Science 113, 1043––1057 [DOI] [PubMed] [Google Scholar]
- Schimmöller F, Singer-Krüger B, Schröder S, Krüger U, Barlowe C, Riezman H. 1995. The absence of Em24p, a component of ER-derived COPII-coated vesicles, causes a defect in transport of selected proteins to the Golgi. EMBO Journal 14, 1329––1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab R, Ossowski S, Riester M, Warthmann N, Weigel D. 2006. Highly specific gene silencing by artificial microRNAs in Arabidopsis. The Plant Cell 18, 1121––1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strating JR, Martens GJ. 2009. The p24 family and selective transport processes at the ER–Golgi interface. Biology of the Cell 101, 495––509 [DOI] [PubMed] [Google Scholar]
- Strating JR, van Bakel NH, Leunissen JA, Martens GJ. 2009. A comprehensive overview of the vertebrate p24 family: identification of a novel tissue-specifically expressed member. Molecular Biology and Evolution 26, 1707––1714 [DOI] [PubMed] [Google Scholar]
- Takida S, Maeda Y, Kinoshita T. 2008. Mammalian GPI-anchored proteins require p24 proteins for their efficient transport from the ER to the plasma membrane. Biochemical Journal 409, 555––562 [DOI] [PubMed] [Google Scholar]
- Vetrivel KS, Gong P, Bowen JW, et al. 2007. Dual roles of the transmembrane protein p23/TMP21 in the modulation of amyloid precursor protein metabolism. Molecular Neurodegeneration 2, 4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei T, Wang A. 2008. Biogenesis of cytoplasmic membranous vesicles for plant potyvirus replication occurs at endoplasmic reticulum exit sites in a COPI- and COPII-dependent manner. Journal of Virology 82, 12252––12264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen C, Greenwald I. 1999. p24 proteins and quality control of LIN-12 and GLP-1 trafficking in Caenorhabditis elegans. Journal of Cell Biology 145, 1165––1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagawa Y, Hasezawa S, Kumagai F, et al. 2002. Cell-cycle dependent dynamic change of 26S proteasome distribution in tobacco BY-2 cells. Plant and Cell Physiology 43, 604––613 [DOI] [PubMed] [Google Scholar]
- Zhang L, Volchuk A. 2010. p24 family type 1 transmembrane proteins are required for insulin biosynthesis and secretion in pancreatic beta-cells. FEBS Letters 584, 2298––2304 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.