Summary
The basic helix-loop-helix transcription factors oligodendrocyte transcription factor 1 (OLIG1) and OLIG2 are structurally similar and, to a first approximation, coordinately expressed in the developing CNS and postnatal brain. Notwithstanding these similarities, it was apparent from early on after their discovery that OLIG1 and OLIG2 have non-overlapping developmental functions in patterning, neuron subtype specification and the formation of oligodendrocytes. Here, we summarize more recent insights into the separate functions of these transcription factors in the postnatal brain during repair processes and in neurological disease states, including multiple sclerosis and malignant glioma. We discuss how the unique biological functions of OLIG1 and OLIG2 may reflect their distinct genetic targets, co-regulator proteins and/or post-translational modifications.
Introduction
The appearance of myelinating oligodendrocytes represented a major step forward in the evolution of the vertebrate CNS, as these cells enabled more efficient axon insulation and conduction of action potentials and, consequently, much greater brain complexity. The essential roles of oligodendrocytes in myelin production, establishing the nodal architecture of the axon, and saltatory conductivity were recognized by the late 1960s. The biological relevance of the structures and functions enabled by oligodendrocytes came from the gradual realization that damage to the myelin sheath, subsequent demyelination and focal depletion of myelinating oligodendrocytes constitute the cellular basis of multiple sclerosis (MS)1-4. The typical relapsing–remitting course of this disorder highlighted the repair potential of oligodendrocytes and focused attention on two important unresolved issues in myelin development, namely the anatomical origins of oligodendrocytes and the lineage relationships between oligodendrocytes and other neural cell types.
The progress towards answering these questions was initially slow and the findings from early studies were contentious. For example, on the subject of anatomical origins, tissue explant studies suggested that oligodendrocytes arise exclusively from the ventral neural tube5-8. However, quail–chick grafting experiments suggested a more complex picture, with some oligodendrocytes originating from dorsal regions of the neural tube9. With respect to lineage relationships, tissue explant studies identified glial-restricted progenitor cells from the optic nerve and the spinal cord, suggesting a lineage relationship between oligodendrocytes and astrocytes10-12. However, grafting experiments and analysis of pattern formation in the developing spine highlighted the role of anatomy in glial specification and suggested that oligodendrocytes and astrocytes arise from separate, bipotent progenitor cells (for a review see REF. 13).
A major lacuna in the field was a lack of genetic tools. This void was filled with the cloning and characterization of the basic helix–loop–helix (bHLH) transcription factors oligodendrocyte transcription factor 1 (OLIG1) and OLIG2 — so named because early in situ hybridization experiments revealed that these transcription factors showed pronounced expression in myelinating oligodendrocytes and oligodendrocyte progenitors14-16. Gain-of-function and loss-of-function genetic assays involving Olig1 or Olig2, together with Cre-lox fate-mapping experiments, resolved long-running polemics on the cellular and anatomical origins of oligodendrocytes in the developing CNS14-18. With respect to anatomical origins, it is now clear that oligodendrocyte progenitors arise from both the ventral and dorsal regions of the developing CNS. Most oligodendrocytes in the adult spinal cord are derived from ventral progenitors. By contrast, oligodendrocytes in the mature forebrain are derived from dorsal progenitors 19-21. Regarding lineage relationships, the notion that oligodendrocytes and astrocytes arise from a common, bi-potent glial-restricted progenitor proved to be incompatible with genetic analysis and fate mapping studies focusing on Olig1 and Olig217,18. It is now evident that oligodendrocyte progenitor origins are more closely aligned to neuron subtype progenitors than to astrocytes — at least in the developing CNS.
More recent studies have revealed biological functions for Olig1 and Olig2 in the postnatal brain during neurogenesis22 and reactive gliosis23, 24, and in repair functions in experimental models of MS25. Preliminary, but nevertheless provocative, studies have also suggested that these genes have links to schizophrenia26-28 and to the cognitive deficiencies associated with Down's syndrome29. Overshadowing these links to development and disease is a broad body of literature linking one of these genes — OLIG2 — to a cohort of primary brain cancers known collectively as the diffuse gliomas30-34.
Against initial expectations based on expression patterns and striking similarities in their DNA-targeting bHLH motifs, the biological functions of OLIG1 and OLIG2 are for the most part separate and non-overlapping. In this Review, we examine facets of OLIG1 and OLIG2 molecular biology that may account for their diverse functional repertoires. In the fullness of time, genetic targets, co-regulator proteins and post-translational modifications unique to OLIG1 or OLIG2 may lend themselves to development of targeted drugs for CNS injuries and for a variety of neurological disease states, including cerebral palsy, MS and malignant glioma.
OLIG structure and expression
The human genome encodes approximately 125 transcription factors that are defined by a canonical bHLH motif35,36. OLIG1 and OLIG2, together with a third transcription factor, OLIG3, form a recognizable subset of proteins in the bHLH family by virtue of their amino acid sequence homology — especially within the bHLH regions, which mediate transcription factor dimerization and DNA targeting (BOX 1). Olig1 and Olig2 are expressed exclusively within the CNS14-16 (TABLE 1). To a first approximation, these genes are coordinately expressed in both space and time, although there are some important distinctions in their patterns of expression — especially at the level of subcellular localization of the two OLIG proteins (see below).
Table 1.
Developmental and postnatal expression of Olig genes
| Olig1 | Olig2 | Olig3 | References | |
|---|---|---|---|---|
| Developmental | ||||
| Ventral spinal cord (pMN domain) | ++ | +++ | − | 14, 15 |
| Dorsal spinal cord | − | − | +++ | 42, 123 |
| Early oligodendrocyte progenitors | +++ | +++ | − | 14, 15 |
| Postnatal | ||||
| SVZ transit-amplifying cells | +++ | +++ | NT | 22 |
| NG2-positive glia | +++ | +++ | NT | 124 |
| Myelinating oligodendrocytes | +++ | +++ | NT | 14, 15 |
| External to CNS | − | − | ++ | 16 |
Expression levels (arbitrary units) ranging from no expression (−) to strong expression (+++). NT not tested.
Olig2 expression is detected at very early time points in CNS development within the radial glia of the neural tube that ultimately give rise to motor neurons and oligodendrocytes37,38. Expression of Olig2 in multipotent progenitor cells of developing forebrain is recapitulated in vitro in cultures of multipotent neurospheres, in which most mitotic cells are OLIG2-positive39.
Anatomically refined studies of Olig1 and Olig2 expression have been carried out in developing spinal cord. Here, both Olig1 and Olig2 have been shown to be downstream targets of the ventralizing signal Sonic hedgehog14, 15. Olig1 is first expressed in the dorsal portion of the ‘p3’ progenitor domain of the ventral neural tube and then becomes confined to the pMN (progenitors of motor neurons) domain by embryonic day (E) 10.5. Olig2 expression is also observed at early development stages prior to neural tube closure in the ventral-most p3 domain and then becomes confined to the pMN domain by approximately E9–E9.5. By the time that Hb9 (homeobox protein Hb9) is expressed (E9-E9.5), Olig1 and Olig2 expression is down-regulated in motor neuron lineage cells. Studies by Chen et al. show that a microRNA (mir-17-3p) may be the effector of this down-regulation — at least for Olig2 40. Both Olig1 and Olig2 show sustained expression in oligodendrocyte precursor cells as these cells progress to become mature oligodendrocytes (FIG. 1)14-16. In contrast to Olig1 and Olig2, Olig3 is expressed in the dorsal-most domains of the developing spinal cord, patterned by bone morphogenetic proteins (BMPs), and is also expressed in multiple tissues outside of the CNS (TABLE 1)16, 41, 42.
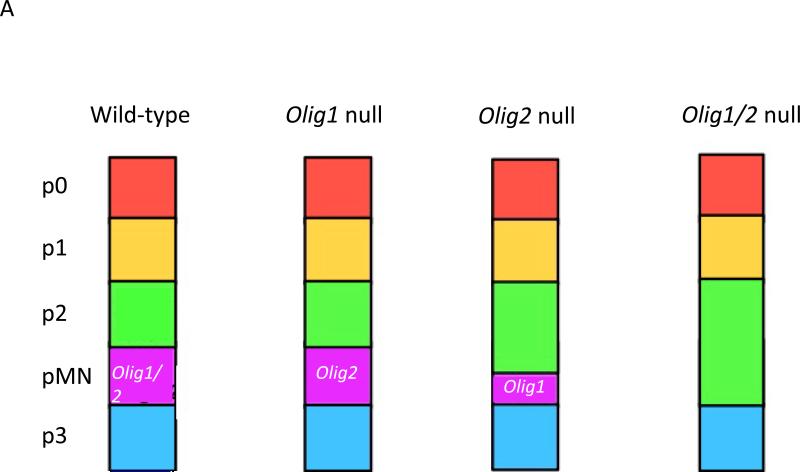
FIG. 1. The developmental roles of OLIG1 and OLIG2.
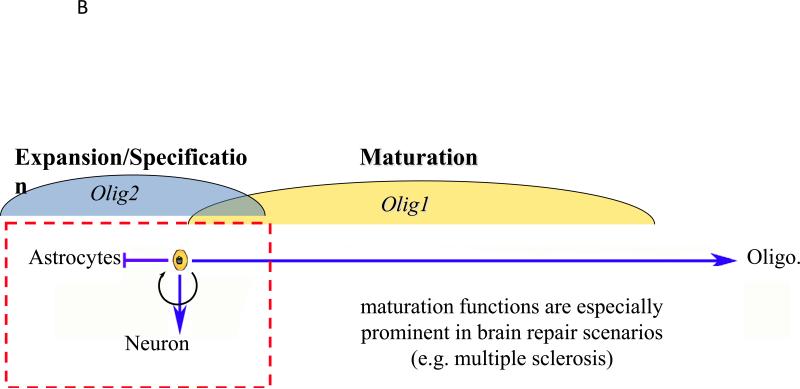
a | Impact of Olig1 and/or Olig2 ablation on spinal cord patterning in mice. The combinatorial interactions of homeodomain proteins organize the ventral spinal cord of developing mouse embryos into five distinct regions, namely the p0, p1, p2, pMN and p3 domains. Different classes of ventral interneurons arise from the p0–p3 domains, whereas motor neurons and oligodendrocytes are derived from progenitors in the pMN domain (see REF. 13 for a review). Knockout of Olig1 has little affect on maintenance of the pMN domain, whereas knockout of Olig2 results in ventral expansion of the p2 domain. Combinatorial knockout leads to the complete disappearance of the pMN domain (see REF. 122 for a review). b | Non-overlapping roles for Olig1 and Olig2 in proliferation and differentiation of neural progenitors. OLIG1 promotes the differentiation of committed oligodendrocyte progenitors, a function that may be even more readily apparent in repair scenarios than in development. By contrast, OLIG2 functions at earlier developmental stages. Initially, OLIG2 acts to oppose cell differentiation and sustains the replication competent state so as to expand the pool of progenitors. At later stages of development, OLIG2 promotes the fate choice decision to form early oligodendrocyte progenitors and certain types of neurons, notably motor neurons in the developing spinal cord. Generally speaking, OLIG2 suppresses the formation of astrocytes, although there may be regional exceptions to this rule and it has been suggested that OLIG2 has a role in reactive gliosis.
Roles in development
Although structurally related and, to a large extent, coordinately expressed in developing embryos, the biological functions of OLIG1 and OLIG2 are only partially redundant (TABLE 2). Olig1 and Olig2 each contribute to patterning of the spinal cord, although Olig2 plays the dominant role (FIG. 1A). Indeed, in the spinal cord, OLIG2 excusively promotes the specification of motor neurons and early oligodendrocyte progenitors that express platelet-derived growth factor receptor α (PDGFRα) (FIG. 1B). The switch from maintaining immature pMN progenitors to the production of motor neurons involves regulation of OLIG2 levels as well as phosphorylation of this protein at a site within its bHLH domain (see below)43, 44. Loss-of-function studies also indicate roles for Olig2 in neuron development within the ventral forebrain, particularly in cholinergic neuron populations45.
Table 2.
Functions of OLIG proteins
| OLIG1 | OLIG2 | OLIG1 + 2 | OLIG3 | References | |
|---|---|---|---|---|---|
| Developmental | |||||
| Spinal cord patterning | − | + | +++ | − | 14, 15 |
| Expansion of progenitor pool | − | +++ | +++ | − | 39 |
| Specification of motor neurons | − | +++ | +++ | − | 14, 15 |
| Specification of other neurons | − | + | + | +++ | 29, 42, 123, 125 |
| Specification of NG2-positive glia | − | +++ | NT | NT | 124 |
| Specification of oligodendrocytes | +/− | ++ | +++ | − | 14, 15 |
| Maturation of oligodendrocytes | + | NT | NT | − | 14 |
| Postnatal | |||||
| Malignant glioma | +/− | +++ | +++ | NT | 39, 57 |
| Myelin repair | +++ | NT | NT | NT | 25 |
| Reactive gliosis | − | +++ | NT | NT | 49 |
| Down syndrome | NT | NT | +++ | NT | 29 |
| Alzheimer's / Schizophrenia* | NT | + | NT | NT | 26-28 |
| Rheumatoid arthritis* | NT | NT | NT | + | 126, 127 |
Genome wide association studies correlate certain OLIG2 single nucleotide polymorphisms to these disorders. These observations have not progressed beyond the correlative level.
Function (arbitrary units) ranging from no involvement (−) to essential for the specific process (+++). NT not tested.
Ablation of Olig1 has no impact on the formation of motor neurons and early oligodendrocyte progenitors (FIG. 1B). However, subsequent maturation of oligodendrocyte progenitors within the spinal cord is delayed in Olig1 knockout mice17, 46.However, occasional foci of PDGFRα-positive oligodendrocyte progenitors are seen in the forebrain of Olig2 knockout mouse embryos and these PDGFRα-expressing cells are even more abundant in the hindbrain17. Combinatorial knockout of Olig1 and Olig2 is required to ablate these last vestiges of oligodendrocyte formation completely18, suggesting that the role of OLIG1 in the specification phase of oligodendrocyte formation is partially redundant in the presence of OLIG2.
In rodents and humans, expression of OLIG2 in the parenchyma is generally considered to identify a cell of the oligodendrocyte lineage, but there are several exceptions to this characterization. First, Olig2 expression in putative astrocyte precursor cells has been reported in the postnatal day (P) 7 neonatal rodent brain47. Second, Cahoy et al. reported that a small percentage (~3%) cells in the adult mouse brain that were positive for the astrocytic marker ALDH1L1, also expressed Olig2 48. Third, Olig2 expression has been reported in proliferating reactive astrocyte precursor cells 49. However, Olig2 expression is down-regulated upon terminal differentiation of astrocytes, an observation confirmed in vitro (see below).
One critical neurogenic function seems to be unique to OLIG2. Generally, the bHLH transcription factors that control neurogenesis can be classified as being either anti-neurogenic (pro-mitotic) or neurogenic (anti-mitotic). At early stages of development, expression of anti-neurogenic (pro-mitotic) transcription factors prevents cell cycle exit and thereby expands the pool of neural progenitors. At later stages of neural development, expression of neurogenic (anti-mitotic) factors promotes cell cycle exit, subtype specification and differentiation50-52. In this context, OLIG2 stands apart, showing functional characteristics of both sets of transcription factors in multipotent neural progenitor cells. In the embryonic spinal cord, for example, OLIG2 is required for specification and the ultimate differentiation of motor neurons and oligodendrocytes17. However, at early time points in development in the pMN domain, OLIG2 functions in pattern formation as an anti-neurogenic factor (pro-mitotic factor), to sustain the replication competent state of some pMN progenitors that are destined for the second wave of gliogenesis43. An emerging body of literature suggests that this early anti-neurogenic (pro-mitotic) function of OLIG2 may be co-opted by the stem-like ‘tumor-initiating cells’ of malignant glioma (see below).
The developmental functions (TABLE 1) and expression (TABLE 2) of OLIG3 are divergent from those of OLIG1 and OLIG2. Accordingly, OLIG3 will not be discussed beyond this point.
Roles in disease and neural repair
A broadening body of literature documents non-overlapping postnatal roles for OLIG1 and OLIG2 in neurological diseases and in response to injury (TABLE 2). For example, both proteins are expressed in fresh surgical isolates of human diffuse gliomas30-34. However, human gliomas have been reported to contain a subpopulation of highly tumorigenic cells53 and OLIG2, but not OLIG1, is selectively expressed in such cells39. Beyond merely marking glioma progenitors, OLIG2 is required for intracranial tumor formation in genetically relevant murine models of human glioma39, 54-56 and for the proliferation of authentic human glioma cells implanted into the brains of severe combined immunodeficiency (SCID) mice57. These findings do not rule out a role for OLIG1 in gliomas, but show that it is dispensable — unlike OLIG2 — in certain subtypes of these tumors.
As indicated in TABLE 2, a postnatal role for OLIG1 is demonstrated in the repair of white matter injury. Although Xin et al. have described an Olig1 knockout mouse with a relatively severe block in myelination, leading to early postnatal lethality, the originally reported Olig1 knockout mouse strain exhibits only a mild developmental delay in oligodendrocyte maturation, even when both copies of Olig1 are ablated, and survives to develop fully myelinated axons in the brain and spinal cord17,46. The two mouse strains differ in terms of the targeting of the Olig1 locus and genetic background, and these differences might underlie the observed differential of the strong versus mild knockout phenotypes. In any case, the relatively benign developmental phenotype of the original Olig1 knockout strain allows scrutiny of Olig1 functions in murine models of MS, and Arnett et al. have showed that the functions of Olig1 in response to demyelinating injury are more readily apparent than the developmental functions. Indeed, Olig1 null mice were severely limited in their ability to repair demyelinated lesions that were induced by various gliotoxins (cuprizone, lysolecithin and ethidium bromide). The loss of Olig1 had no effect on the genesis or recruitment of early oligodendrocyte progenitors expressing NG2, OLIG2 or homeobox protein NKX2.2 into the lesion. Rather, the Olig1 null progenitors were markedly impaired in their ability to differentiate into myelinating oligodendrocytes and wild-type levels of OLIG2 could not compensate for the absence of OLIG1 in this regard25.
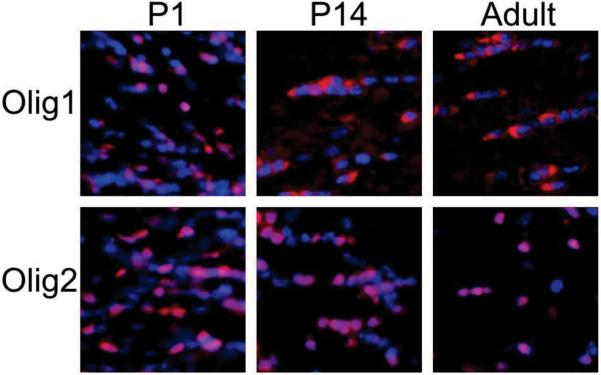
One distinctive feature of OLIG1 in the developing CNS is recapitulated in the repair of demyelinating insults in murine models and also in postmortem brain tissues from patients with MS. During mouse embryonic development, OLIG1 is localized to the nucleus of oligodendrocyte progenitors (FIG. 2). However, in mature oligodendrocytes in the CNS from 2 weeks after birth, OLIG1 is mostly located in the cytoplasm. Translocation of OLIG1 from the nucleus to the cytoplasm is a precise marker of the terminal differentiation of oligodendrocytes58. By contrast, OLIG2 is localized to the nucleus at all stages examined and in all regions of the CNS. The differential localization of OLIG1 and OLIG2 is also seen in the adult human brain. Arnett et al. showed that demyelinating injuries to the adult mouse CNS create an environment that recruits immature oligodendrocyte progenitors with nuclear localization of OLIG1 25. The relocalization of OLIG1 observed in murine models of MS is recapitulated in postmortem brain tissue from patients with MS25. Cells containing cytosolic OLIG1 are present in normal-appearing white matter of the human brains but nuclear OLIG1 is present at the edges of active MS lesions. Collectively, these findings fit into an emerging theme in human white matter injuries as diverse as adult MS and neonatal brain injury leading to cerebral palsy, whereby various cell-intrinsic and environmental influences may limit the repair response by arresting maturation of oligodendrocyte progenitors, resulting in fixed demyelinated lesions59-63.
Figure 2. OLIG1 and OLIG2 localization.
OLIG1 and OLIG2 (both visualized in red) are both present in the nuclei (blue) of oligodendrocytes and their progenitors at postnatal day (P) 1 in the mouse brain. OLIG2 continues to have a nuclear localization at all developmental stages, whereas OLIG1 is found almost completely in the cytoplasm in the adult mouse brain.
Two other facets of the response to demyelination in both rodents and humans are the proliferation of microglia and astrocytes (known as reactive gliosis; reviewed in REF. 64). Neither of these reactive responses to demyelination was impaired in the Olig1 null mice17, 25. However, a series of mouse studies supports a functional role for Olig2 in reactive gliosis. Within several days following a cortical stab-wound injury, the number of Olig2-expressing cells increased in the lesioned area, but there was no rise in the number of Olig1-expressing cells24. Going beyond the correlative level, Chen et al. showed that targeted ablation of Olig2fl/fl with a GFAP-Cre driver, reduced the number of reactive astrocytes following injury, whereas ablation of Olig2 in neuronal cells or oligodendroglial cells had no impact on this phenotype 49.
The OLIG2-positive cell type that gives rise to reactive astrocytes is somewhat of a mystery. Early studies showed that neither Olig1 nor Olig2 is expressed in mature astrocytes14, 15 and that OLIG2 developmentally acts as a repressor of astrogenesis65-67. Moreover, fate-mapping studies in the developing CNS failed to reveal any lineage relationship between OLIG protein-positive cells and astrocytes17, 68. In the postnatal brain, Olig expression is confined almost exclusively to transit-amplifying cells of the subventricular zone, NG2-positive glia and mature myelinating oligodendrocytes. NG2-positive glia do have progenitor-like qualities. However, fate-mapping studies indicate that these cells are probably not the progenitors of reactive astrocytes69-71. It is conceivable that injury scenarios trigger transient re-expression of Olig2 in an as yet poorly characterized type of progenitor of reactive astrocytes, which appears to express excitatory amino acid transporter 1 (EAAT1; also known as GLAST), as well as other astroglial markers 72. A transient non-lineage-restricted period of Olig2 expression would be consistent with several reports on reactive gliosis that document the early export of OLIG2 from the nucleus of proliferating progenitors followed by apparent degradation of the cytosolic protein45, 73-75.
The most common genetic cause of intellectual disability is triplication of chromosome 21, giving rise to Down syndrome. OLIG1 and OLIG2 are co-localized to chromosome 21 within or near a region on the long (q) arm (the so-called Down syndrome critical region) that is thought to be most tightly associated with the cognitive facets of the Down syndrome phenotype. Does overexpression of the two OLIG genes contribute to the neurological facets of Down syndrome? Chakrabarti et al. have generated support for this view in their studies with a well-characterized murine model of trisomy 21, the Ts65dn mouse29. The parental Ts65dn mouse shows many of the cognitive and neuro-anatomical defects associated with Down syndrome. In careful anatomical studies, Chakrabarti et al. first noted that the Ts65dn mice have a substantial increase in the number of forebrain inhibitory neurons. This observed increase in forebrain inhibitory neurons resonated with earlier studies suggesting that a major functional defect underlying the behavioral abnormalities in Ts65dn mice is an imbalance between excitation and inhibition76, 77. A total of 128 genes are triplicated in the Ts65dn mouse. Chakrabarti intercrossed these mice with a heterozygous Olig1/2 double knockout mouse (Olig1/2+/−), so as to selectively reduce the dosage of Olig1 and Olig2 from three copies to two29, 78, 79. This genetically precise reduction in gene dosage rescued the overproduction of interneurons. Cognitive tests were not performed on the Olig1/Olig2-rescued animals; however, the published observations show a plausible molecular and cellular route towards the neurological facets of Down syndrome wherein OLIG1 and OLIG2 have pivotal roles.
Reports that have yet to undergo functional validation link OLIG2, but not OLIG1, to schizophrenia and Alzheimer's disease. Genome-wide association work identified several single nucleotide polymorphisms (SNPs) in OLIG2 that are associated with schizophrenia in a UK population26. One of the SNPs identified by the UK team (SNP rs762178) has been confirmed in a study of schizophrenia in a Chinese Han population28. Two of the above-mentioned OLIG2 SNPs (SNP rs762237 and rs2834072) have also been linked to a cohort of Alzheimer's disease patients with psychotic symptoms27.
Genetic targets
As discussed above and summarized in TABLE 2, data from multiple studies indicate that OLIG1 and OLIG2 have largely non-overlapping roles in development, tissue repair and disease. Paradoxically, these two genes are co-localized within 40 kb of each other on human chromosome 21 and their expression patterns are largely overlapping. Moreover, OLIG1 and OLIG2 have highly homologous DNA-targeting bHLH motifs. In this, and subsequent sections, we explore ways in which their non-overlapping biological functions may reflect separate genetic targets, co-regulator proteins and post-translational modifications. We begin with discussion of their genetic targets.
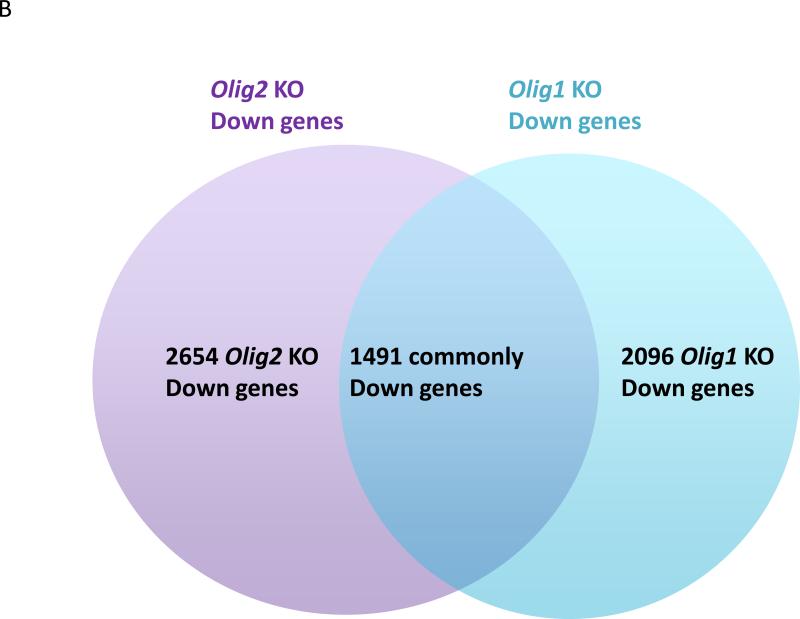
Expression profiling studies of Olig1 and Olig2 knockout mice versus their wild-type counterparts show that various non-overlapping sets of genes are up-regulated or down-regulated in the absence of Olig1 or Olig2 39, 80 (S. Mehta, H. Liu, J. Alberta, E. Huillard, D. Rowitch, C. Stiles, unpublished observations) (FIG. 3). However, expression profiling cannot discriminate between direct genetic targets of OLIG1 and OLIG2 and downstream sequelae of their deletion. The basic domain of bHLH transcription factors mediates the interaction between these proteins and DNA sequences that contain the core hexanucleotide motif CANNTG, known as an E-box (reviewed in REF. 81). For OLIG2, specification of motor neurons following E-box binding seems to be channeled largely through its functions as a transcription repressor. In chick embryo electroporation assays, involving ectopic motor neuron formation as a biological readout, the neurogenic effect of OLIG2 is mimicked when the OLIG2 DNA-binding domain is fused to the transcription repressor domain of engrailed. Equivalent fusions to the transcription activator VP-16 lack neurogenic activity82-84. One important downstream effector gene of the OLIG2 repressor function seems to be Hb9 43. Lee et al. have shown that OLIG2 binds to an E-box element in the Hb9 promoter. The OLIG2–Hb9 promoter interaction results in repression of Hb9 transcription, preventing differentiation into post-mitotic motor neurons and sustaining the replication competent state of pMN neural progenitors during neural tube development43. OLIG2 also binds to promoter elements of the cell cycle repressor gene p21 and suppresses its expression39. Suppression of p21 transcription by OLIG2 may contribute in part to the growth of normal and malignant neural progenitors and to the notorious resistance of p53-positive human gliomas to radiation and genotoxic drugs57.
Figure 3. OLIG1 and OLIG2 downstream gene targets.
The gene sets regulated by OLIG1 and OLIG2 only partially overlap. a | The number of genes that show upregulated expression in Olig1 and/or Olig2 null neural progenitor cells compared with wild-type neural progenitor cells. b | The number of genes that show down-regulated expression in Olig1 and/or Olig2 null neural progenitor cells compared with wild-type counterparts. The Olig2 data set is taken from REF. 39, whereas the Olig1 expression profiling data sets (S. Mehta, H. Liu, J. Alberta, E. Huillard, D. Rowitch, C. Stiles, unpublished observations) are available from the NCBI GEO database (GSE39706).
All that being said, it is unclear if OLIG2 solely acts as a transcriptional repressor. Expression profiling reveals multiple genes that are down-regulated in Olig2 null neural progenitors compared with wild-type counterparts, consistent with the view that OLIG2 might stimulate expression of such genes39. Recent data show that, in mice, OLIG2 binds to an enhancer site upstream of Sox10, thereby inducing its expression and increasing oligodendroglial activity85. More recently, two groups have used ChIP-seq protocols to identify new OLIG2 target genes86, 87. Weng et al. identified Sip1 as a direct, inducible target gene of OLIG2. Specifically, they showed that OLIG2 stimulates expression of Sip1, which goes on to promote the maturation of oligodendrocyte progenitors via inhibition of SMAD7 signalling 87.
What about genetic targets of OLIG1? As noted in the section on disease and neural repair, the truly distinctive feature of OLIG1 is that it is localized to the cytosol in the postnatal brain where it cannot possibly have any direct genetic targets25. Indeed, as nuclear localization is essential for transcription factor function, it can be surmised that the critical phase of OLIG1 transcriptional activity in oligodendrocyte differentiation has been completed by the time it becomes localized to the cytoplasm. Genetic targets of OLIG1 have been inferred from gene expression profiles of wild-type versus Olig1 null tissues. Compared with their wild-type counterparts, optic nerve, spinal cord and brain tissue from Olig1 null mice show reduced mRNA and protein levels for several genes involved in oligodendrocyte maturation, including myelin basic protein (Mbp), myelin oligodendrocyte glycoprotein (Mog) and myelin proteolipid protein (Plp1) 25, 46, 80, 88, 89. OLIG1 directly binds to the promoter region of Mbp, thereby inducing its expression46, 90. The Mbp promoter contains one E-box that is entirely conserved between human, mouse, rat, chick and zebrafish90. Mutation of this E-box reduces the binding affinity of OLIG1 for the Mbp promoter in luciferase promoter assays and gel mobility shift assays90. OLIG1 also induces expression of zinc finger protein 488 (Zfp488), as shown in a luciferase reporter assay. There are 20 conserved E-box sequences in the Zfp488 promoter region, but it has not been tested whether OLIG1 directly or indirectly activates Zfp488 expression80.
Co-regulator proteins
All bHLH transcription factors function in a dimeric state as homodimers or as heterodimers with another bHLH protein. Once in contact with promoter or enhancer elements of a target, bHLH homodimers and heterodimers serve as scaffolding upon which a multimeric complex of transcriptional co-regulator proteins can be assembled91-94. Transcriptional co-regulators serve many functions, but broadly speaking they can be assigned into two groups — those that are components of the basal transcriptional machinery and those that modify the structure of chromatin92. Against this backdrop, the non-overlapping biological functions of OLIG1 and OLIG2 could reflect differential dimeric partners for the two transcription factors and/or differential interactions with co-regulator proteins.
As a general rule, tissue-specific bHLH transcription factors (termed class B factors) form heterodimers with ubiquitously expressed class A bHLH transcription factors such as E12, E47 (E2A immunoglobulin enhancer binding factors E12/E47, also known as transcription factor 3, TCF3) or TCF12 (transcription factor 12)81. However, OLIG2, which is highly tissue-specific, seems to favour homodimerization over heterodimerization with class A bHLH transcription factors. Indeed, although OLIG2–E12 and OLIG2–E47 heterodimers have been detected 43,95, Lee et al. found that OLIG2 homodimers are the preferred product when OLIG2 and E47 are co-expressed in yeast43. In addition, OLIG2 will form heterodimers with OLIG1 and, under certain conditions, HLH transcription factors such as DNA-binding protein inhibitor ID-2 (ID2) and ID4. These OLIG–ID interactions are detected when differentiation of progenitors towards the oligodendrocyte lineage is suppressed by BMPs 95. Lacking a basic domain, HLH factors cannot interact with DNA. Accordingly, HLH transcription factors are thought to function as natural dominant-negative agents for bHLH transcription factors by forming heterodimers that cannot bind to E-box elements of their target genes96.
Physical interactions between bHLH proteins and members of the homeodomain protein family regulate the development of several other tissues, including the pancreas, the pituitary gland and muscle97-99. OLIG2 has been shown to interact with Nkx2.2, a homeodomain protein that defines the p3 progenitor domain of developing spinal cord and is specifically required for production of V3 interneurons and maturation of oligodendrocytes100.
Another co-regulator protein that has been linked to OLIG2 is histone acetyltransferase p300 (p300)94, 101, which, via its associated histone acetyl transferase activity, functions to decondense the structure of chromatin and thus promotes transcription94, 101, 102. The presence of p300 as a co-regulator resonates with data suggesting that OLIG2 can stimulate the expression of some genes85. Considering the known function of OLIG2 as a transcription repressor in the genesis of motor neurons 43, it is somewhat surprising that studies to date have not identified any known members of the general transcription co-repressor complex in association with OLIG2. Conceivably, directed antibody pull down experiments and unbiased yeast-two-hybrid screens lack sensitivity or cell-type specificity to display these interactions.
Most HLH-containing proteins that have been identified as OLIG2-binding partners show some binding affinity for OLIG1 as well (the exception is cyclin-D1-binding protein 1 (CCNDBP1)). This is not surprising since the bHLH domains of OLIG1 and OLIG2 show more than 80% amino acid identity (BOX 1). CCNDBP1 is a helix–loop–helix protein that lacks a DNA-binding region. Ikushima et al. showed that this protein inhibits transcription of transforming growth factor β (TGF-β)-induced genes that require the SMAD complex for their activation103. They demonstrated in NmuMG and U373MG cells (normal murine mammary gland and glioma cell lines, respectively) that CCNDBP1 recruits OLIG1, thereby interfering with the OLIG1–SMAD interaction. A comprehensive list of all co-regulator proteins that have been associated with OLIG1 and OLIG2 can be found in Supplementary information S1.
Post-translational modifications
How are the proliferative and developmental functions of OLIG1 and OLIG2 regulated at different developmental stages and in different cell types? Most neurogenic bHLH transcription factors (for example, mammalian achaete-scute homologue 1 (MASH1), mammalian atonal homologue MATH, neurogenin 1 (NGN1) and NGN2) are only expressed transiently in progenitor cells at times when their functions are required. Notably, neither neurogenic nor anti-neurogenic bHLH transcription factors are generally expressed in fully formed, terminally differentiated neurons50-52. However, expression of OLIG1 and OLIG2, initiated in oligodendrocyte progenitors, is sustained throughout development and occurs in the postnatal brain, where initial in situ hybridization images indicate that the two genes are expressed in white matter tracts of the corpus callosum, the optic nerve and the cerebellum14-16. Thus, for OLIG1 and OLIG2, post-translational modifications, rather than the timing of gene expression per se, might be the key to the developmental control of their functions. Obviously, factor-specific post-translational modifications could contribute to differential interactions with co-regulator proteins and genetic targets as discussed above.
Phosphorylation regulates OLIG2 function
Computer algorithms reveal a number of conserved potential phosphorylation sites in OLIG2 and recent studies indicate that several of these sites are functional (FIG. 4A and Supplementary information S2). Developmentally regulated phosphorylation events may account for the functional versatility of OLIG2 in cell cycle regulation and differentiation.
Figure 4. Post-translational modification motifs in OLIG1 and OLIG2.
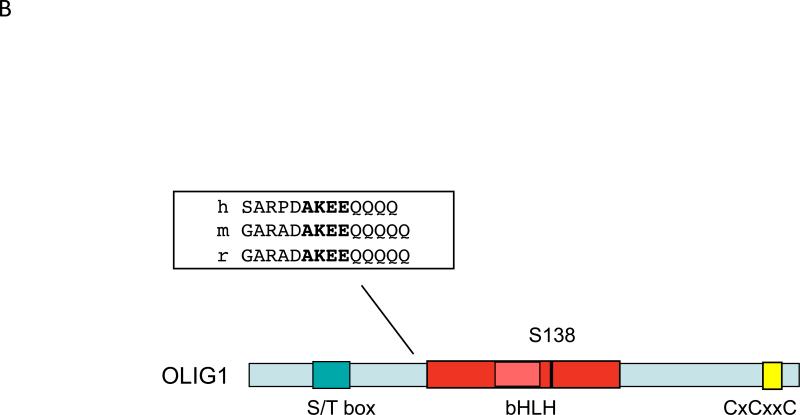
a | Human OLIG2 contains a number of serine phosphorylation sites, which are conserved in mouse OLIG2. The triple serine motif in N terminus of human OLIG2 and Ser147 are entirely conserved in chicken, Xenopus laevis and zebrafish. b | Human OLIG1 contains a serine residue at position 138 that seems to correspond to Ser147 in OLIG2. Protein alignment of OLIG1 orthologues from human, mouse and rat reveals a conserved putative SUMOylation motif close to the basic helix–loop–helix (bHLH) domain (inset).
A striking structural feature of OLIG2 is a string of 12 contiguous serine and threonine residues at position Ser77-Ser88 in its N-terminal region (known as the ST box). Huillard et al. showed that a murine OLIG2 protein fragment containing the ST box, fused to glutathione S-transferase, was a substrate for the serine–threonine protein kinase CK2104. Tryptic peptide digests revealed phosphorylation of the OLIG2 fragment at residues Ser85 and Ser87 within the ST box. Interestingly, targeted disruption of the gene encoding CK2β (an essential regulatory subunit of CK2) results in impaired oligodendrocyte differentiation in vivo and in vitro. These observations largely resemble the differentiation phenotype of the Olig2 knockout mice. Moreover, an Olig2 deletion mutant that lacks the entire ST box is unable to rescue the formation of oligodendrocyte progenitor cells when it is transduced in Olig2 null neural progenitor cells. Collectively, these observations provide circumstantial evidence that the phosphorylation state of the ST box could regulate separate functions of OLIG2 in proliferation and differentiation.
One concern, however, is that phosphorylation of the ST box has never been detected in living cells. Sun et al. isolated endogenous OLIG2 from mouse neural stem cells, human malignant gliomas and ectopic OLIG2 transfected into COS7 cells and screened these isolates for post-translational phosphorylation events by mass spectrometry. Phosphorylation of the ST box in the OLIG2 extracts was not detected105. Conceivably, phosphorylation of the ST box in vivo is a spatially and temporally restricted event that was not duplicated in the cell types assessed by Sun and colleagues. However, it is also possible that misfolding of the OLIG2 fragment in the in vitro experiment described above exposed CK2 substrates that are normally occluded in native OLIG2. It may be noteworthy that the ST box itself is not especially well conserved through phylogeny, especially when compared with several other phosphorylation motifs interesting OLIG2 (see FIG. 4A and Supplementary information S2). Thus, the biological functions of the ST region have yet to be fully resolved in vivo.
Cortical progenitor cultures that have been expanded by incubation with basic fibroblast growth factor (FGF) and epidermal growth factor (EGF) for two weeks contain 90% OLIG2-positive cells39. Following withdrawal of FGF and EGF and exposure to ciliary neurotrophic factor (CNTF), OLIG2 is exported from the nucleus to the cytosol where it is rapidly degraded. In an elegant series of studies, Setoguchi and Kondo showed that this CNTF-induced relocalization of OLIG2 coincides with activation of the serine–threonine protein kinase Akt66. Moreover, they showed that Ser30 in the N-terminus of OLIG2 is phosphorylated by Akt in vitro. A phosphomutant version of OLIG2 containing a Ser30Ala mutation, is retained in the nucleus of neural progenitor cells. This coincides with impaired CNTF-induced astrocytic differentiation after FGF and EGF withdrawal, compared to cells overexpressing vector control. Together, their observations are consistent with a model wherein the phosphorylation state of OLIG2 at Ser30 dictates a fate choice decision for cortical progenitor cells to differentiate into astrocytes or remain as uncommitted neuronal progenitors.
Setoguchi et al. did not demonstrate Ser30 phosphorylation of endogenous OLIG2 in CNTF-treated cortical progenitor cells. In addition, Sun et al. did not detect Ser30 phosphorylation in OLIG2 that was isolated from proliferating mouse neurospheres or glioma progenitor cells105. However, OLIG2 in cycling neurosphere cultures is localized strictly to the nucleus, and Sun et al. did detect some Ser30 phosphorylation in OLIG2 that was isolated from COS7 cells, in which an appreciable amount of the protein is found in the cytosol105, 106. These observations would be consistent with the nuclear export and degradation function of Ser30 phosphorylation, as was suggested by Setoguchi and Kondo66. One caveat regarding the functional relevance of the Ser30 site is that this site is not well conserved in OLIG2 from different animal species (FIG. 4A).
As noted above in our comments on development, one important function of OLIG2 in the early stages of development is to sustain the replication-competent state of neural progenitors43. At later stages of development however, OLIG2 is pivotal for specification of motor neurons and oligodendrocytes (FIG. 1B and TABLE 2). Studies by Sun et al. show that the proliferative functions of OLIG2 are largely controlled by developmentally regulated phosphorylation of a triple serine motif comprising Ser10, Ser13 and Ser14 105. When phosphorylated at these positions, OLIG2 maintains pro-mitotic functions in normal neural progenitors. Using a phosphorylation state-specific antibody to the triple serine motif, Sun et al. showed that endogenous OLIG2 is phosphorylated at these residues during early stages of embryonic development when oligodendrocyte progenitors are proliferating. In postnatal white matter, the same serine residues are in a non-phosphorylated state. Strikingly, cells expressing a phosphomimetic mutant version of OLIG2 (wherein the negatively charged amino acids aspartate or glutamate were substituted for Ser10, Ser13 and Ser14) were more tumorigenic in a murine model of high-grade glioma. Conversely, cells expressing a phosphonull mutant of OLIG2 (with the neutral amino acids glycine or alanine were substituted for the three serine residues) were less tumorigenic. On a final note, OLIG2 expressed in p53-positive human gliomas is phosphorylated at the triple serine motif. The data presented by Sun et al. and in a related paper by Mehta et al. suggest that phosphorylated OLIG2 inhibits the genetic and biological responses to p53 57. Collectively, these data suggest that phosphorylated OLIG2 may contribute to the notorious resistance of human gliomas to radiation and genotoxic drugs. This OLIG2 triple serine motif is stringently conserved throughout evolution (FIG. 4A). In fact, these serines and their flanking amino acids are nearly as well conserved as the bHLH motif itself. To date, the protein kinases and phosphatases that regulate the phosphorylation state of this triple serine motif have not been identified.
As described above (TABLE 2), OLIG2 is essential for specification of motor neurons as well as oligodendrocytes in the pMN domain of the developing spinal cord. Li et al. identified a phosphorylation site in the bHLH domain of OLIG2 that regulates the motor neuron to oligodendrocyte transition44. A bio-informatic search for predicted phosphorylation sites and conserved amino acids in OLIG2 drew the authors attention to a predicted protein kinase A (PKA)-phosphorylated serine at position 147 in the second helix of OLIG2. A phospho-specific antibody showed that Ser147 is phosphorylated in the developing mouse spinal cord during the window of time wherein OLIG2 specifies the formation of motor neurons. The phosphorylation state of Ser147 reaches a maximum at day E9.5 during the peak of neurogenesis in the pMN and then decreases over time, disappearing entirely after E12, when the switch towards oligodendrocyte production takes place. Li et al. also generated mice that expressed Olig2 encoding a serine-to-alanine substitution at position 147 (Ser147Ala OLIG2) to study the biological function of Ser147 phosphorylation in vivo. Ser147Ala OLIG2 mice have a diminished pMN domain (revealed by Pax6 and Nkx2.2 expression), as do Olig2 null mice. Also, as noted with Olig2 null mice, Ser147Ala OLIG2 mice die at birth, because they do not generate motor neurons.
At a biochemical level, Li et al. showed that Ser147Ala OLIG2 less readily forms OLIG2 homodimers and OLIG2–OLIG1 heterodimers relative to wild-type OLIG2 44. Conversely, Ser147Ala OLIG2 binds NGN2 with higher affinity than does wild-type OLIG2. The authors speculate that dephosphorylation of OLIG2 in the developing pMN domain sequesters NGN2, thereby preventing motor neuron development. In a perfect world, this sequestration of NGN2 would be antagonized by substitution of a negatively charged amino acid (aspartic acid or glutamic acid) at Ser147, as per rescue experiments with the phosphomimetic mutant for the triple serine motif, described by Sun and colleagues105. However, rescue experiments of this sort are not always successful and Ser147Asp and Ser147Glu OLIG2 seem to be phenotypically equivalent at a biochemical level to the Ser147Ala OLIG2 — at least with respect to their diminished capacity to form OLIG2 homodimers 44. Interestingly, the Ser147 phosphorylation motif is conserved in the bHLH domain of OLIG1 (Ser138), which is not involved in motor neuron development (FIG. 4B and TABLE 2). It would be interesting to see if the bHLH domain of OLIG1 could be substituted for the OLIG2 bHLH domain in ‘domain swap’ experiments focusing on motor neuron formation.
Is OLIG1 protein post-translationally regulated?
Regardless of the structural similarities in OLIG1 and OLIG2, neither the OLIG2 triple serine motif nor the AKT phosphorylation site at Ser30 have equivalents in OLIG1. However, as mentioned above, the predicted PKA-phosphorylated serine at Ser147 in the bHLH domain of OLIG2 is represented at an equivalent position in OLIG1 (Ser138). As described earlier, one of the features of OLIG1 is its nuclear to cytoplasm relocalization during oligodendrocyte development25. Niu et al. transduced Olig1 null rat oligodendrocyte progenitor cells with a OLIG2 phosphomutant construct (Ser138Ala) or with its rescue construct (Ser138Asp)107. They showed that Ser138Ala OLIG1 is expressed primarily in the nucleus of oligodendrocyte progenitor cells, whereas Ser138Asp OLIG2 has a cytoplasmic localization. Future work should clarify whether this serine has a functional role in vivo, as this study did not include any direct analysis of the phosphorylation state of this serine with either a phosphorylation state-specific antibody or mass spectrometry, or any in vivo mouse work.
One other intriguing protein modifier that is relevant to transcription factor biology is SUMO (small ubiquitin-like modifier). SUMOylation has been implicated in numerous biological functions within the nucleus, including transcription factor activity, protein–protein interactions, promyelocytic leukemia (PML) nuclear body integrity, DNA repair and sub-nuclear localization 108-111. SUMOylation of NF-κB essential modulator (NEMO), which regulates NF-κB, is sufficient for nuclear import of NEMO 112,113. By contrast, several studies show an increase in substrate SUMOylation concurrent with substrate nuclear export114-116. It is unclear whether SUMOylation initiates export or occurs in the cytoplasm, perhaps as a mechanism to retain proteins in the cytoplasm through masking of a nuclear localization signal or context-dependent protein–protein interactions. As shown in FIG. 4B, OLIG1 does contain a conserved SUMOylation motif, and it was predicted to be a target of SUMOylation in a genome-wide screen for this modification117. Accordingly, it is at least conceivable that a SUMOylation event dictates the developmentally regulated localization of OLIG1 observed in remyelinating white matter (FIG. 2) 25.
Conclusions and future directions
This Review has highlighted disparate aspects of the biology of OLIG1 and OLIG2 during development and in human disease. Despite much progress in elucidating this biology, many important questions remain regarding OLIG protein functions and activity. Indeed, given their multiple stage-specific roles, we have argued that insight into the post-translational regulation of OLIG1 and OLIG2 activity is crucial for understanding the functional roles of these proteins.
The phenotype of Olig2 knockout mice is severe and the biological functions of the OLIG2 are readily apparent (TABLE 2). By contrast, the phenotype of Olig1 null mice is relatively nuanced 17. Investigators naturally gravitate to systems that are ‘black and white’ rather than ‘shades of gray’ and an OLIG2 bias is readily apparent in the literature. Indeed, the PubMed database at the National Library of Medicine (USA) currently shows a four to one ratio of papers with OLIG2 in their title to those with OLIG1. OLIG1 and OLIG2 lie in close proximity to each other in humans, rodents and numerous other species in a region that is genetically well conserved118. In our view, the biological functions of OLIG1 in development and disease are understudied and deserving of more attention. Indeed, many questions regarding its function remain. For example, are there early roles for OLIG1 in forebrain neurogenesis? Can OLIG1 compensate in part for OLIG2 during gliomagenesis? What are the important factors regulating OLIG1 at the post-translational level?
Overshadowing these technical challenges and underserved areas of inquiry are a series of therapeutic opportunities. Small molecule activators of OLIG1 and OLIG2 could have practical applications in MS and spinal cord injury respectively. Conversely, recent data suggest that small molecule antagonists of OLIG2 might serve as highly targeted therapeutics for malignant glioma. Against these therapeutic opportunities lies one formidable challenge. Transcription factors are generally considered to be unattractive targets for drug development because their interactions with DNA and heterodimeric partner proteins involve large and complex surface area contacts. Surrogate targets for OLIG drug development may be embedded within enzymatically active gene targets, partner proteins or post-translational modifying enzymes.
Supplementary Material
Box 1 | The oligodendrocyte transcription factor family: a closer look at structure.
In humans, the genes encoding oligodendrocyte transcription factor 1 (OLIG1) and OLIG2 are localized within 40 kb of each other on chromosome 21 (syntenic to mouse chromosome 16). Co-localization of these genes is also observed in numerous other species and the chromosomal region in question is well conserved118. By contrast, OLIG3 maps to human chromosome 6 (mouse chromosome 10)16.
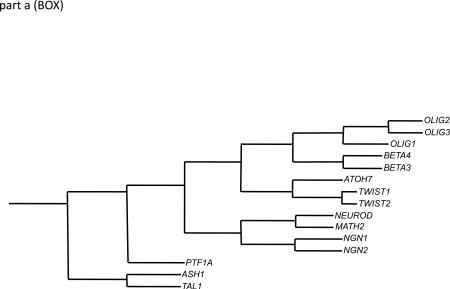
OLIG1, OLIG2 and OLIG3 are recognizable as a core subset of genes in the family of basic helix–loop–helix (bHLH) transcription factors by nucleotide sequence homologies in regions encoding to the bHLH domain and amino and carboxy termini of the corresponding proteins (see part a; the phylogenetic tree was generated utilizing ClustalW2).
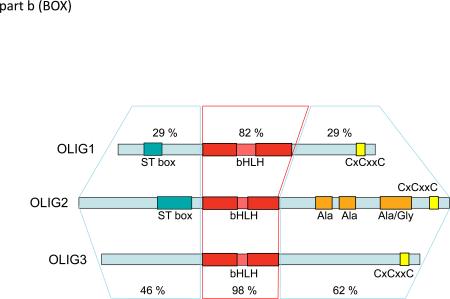
Standard amino acid sequence alignment algorithms utilizing Clustal Omega and further adjusted to “best-fit” by eye, indicate that at the protein level, OLIG2 is more closely related to OLIG3 than to OLIG1. Minimal conservation of domains outside the bHLH domains of OLIG1 and OLIG2 is suggested (see FIG. B, percentages reflect conserved substitution weighting following ClustalW2, Gonnet Pam250). However, the close relationship between OLIG1 and OLIG2 becomes more compelling when the alignment is adjusted to highlight short regions of homology, multiple insertions and deletions, and to utilize the conservation of the spacing of presumed conserved structural elements, such as proline (Supplementary information S2).
The amino terminal domain of OLIG1 is smaller than that of OLIG2 (89 amino acids versus 108 amino acids in humans). Also, the overall amino acid identity between OLIG1 and OLIG2 in this region is low, despite the presence of a distinctive serine–threonine-rich ‘ST box’ in both proteins (see part b). The OLIG3 amino terminus is even smaller (83 amino acids in humans) than that of OLIG1 and lacks the ST box; however, the amino termini of OLIG2 and OLIG3 are rather similar. Importantly, both contain a critical triple serine motif (see main text). It should be noted that the ST box common to OLIG1 and OLIG2 in humans and rodents is actually not well-conserved in OLIG1 and OLIG2 in other species (see Supplementary information S2).
The bHLH region is very highly conserved across all homologues in the OLIG family. However, the bHLH domain of OLIG1 is distinctive even within the broader family of bHLH transcription factors in several respects. In particular, the loop region of this domain has several unusual features in OLIG1. First, it is nearly twice the length of the loops from the other members of the OLIG family. Second, it has an extremely rare shift in the position of the helix 1-breaking proline. Third, the proline that precedes helix 2 is displaced. Last, it lacks the serine that directly precedes helix 2 in OLIG2 and which is a predicted candidate for phosphorylation.
Identity of the carboxy terminal domain in OLIG1 and OLIG2 is very low. By contrast, OLIG2 and OLIG3 are homologous in this region even though OLIG3 lacks a set of distinctive alanine and glycine-rich domains that is seen in OLIG2. The OLIG2–OLIG3 similarities extend well past the end of helix 2 of the bHLH domain and include a distinctive multi-proline motif directly next to the cysteine domain. The only domain common to all three OLIGs and conserved in all orthologues is a cysteine motif (CxCxxC) close to the C terminus. Cysteine residues and domains are implicated in multiple roles, including disulphide-bond formation, post-translational modifications such as S-palmitoylation, and recruitment of histone-modifying activities to chromatin119-121.
Acknowledgements
The authors gratefully acknowledge helpful conversations with W. Richardson (University College London, UK), R. Miller (Case Western Reserve University, USA) and Y. Sun (Dana-Farber Cancer Institute, USA). Work from the authors’ laboratories that is cited here was supported by grants from the National Institutes of Health (NS047572 and NS057727 to C. D. S. and NS040511 to D. H. R.) and from the Pediatric Low-Grade Astrocytoma Foundation (grant to C. D. S.). D. H. R. is supported by the Howard Hughes Medical Institute.
Glossary
- Cre-lox fate-mapping
This procedure installs a stable marker protein (usually colorimetric, such as LacZ) into a genetically defined cell type and all of its daughter cells.
- Tumor-initiating cells
In tumors with a heterogeneous cell population (such as glioblastoma multiforme) these undifferentiated “stemlike” cells are thought to be responsible for propagating the tumors in serial animal transplantation protocols.
- SCID mice
This “severe combined immunodeficiency” mouse strain is used as a host animal for transplantation experiments with human tumors.
- Transit-amplifying cells
Also known as “type C” cells, these rapidly dividing neural progenitor cells in the subventricular zone of postnatal brain are the immediate progeny of the more slowly replicating multipotent adult neural stem cells.
- SNP
Single nucleotide polymorphisms (“SNPs”) are DNA sequence variations that differ between individual members of a biological species or between paired chromosomes in a single individual.
- Expression profiling
This procedure identifies the gene types that are expressed in a particular cell type by processing mRNA into cDNA and then annealing the cDNA to gene sequences arrayed on a solid surface.
- Sumoylation
This post-translational modification event involves covalent ligation of Small Ubiquitin-like Modifier (SUMO) proteins to regulate various cellular processes.
References
- 1.Charcot JM. Histologie de la sclerose en plaques. Gazette des hopitaux Paris. 1868;41:554–555. [Google Scholar]
- 2.Marburg O. Die sogenannte akute multiple Sklerose. Jahrb Psychiatre. 1906;27:211–312. [Google Scholar]
- 3.Prineas JW, Connell F. Remyelination in multiple sclerosis. Annals of neurology. 1979;5:22–31. doi: 10.1002/ana.410050105. [DOI] [PubMed] [Google Scholar]
- 4.Lucchinetti C, et al. A quantitative analysis of oligodendrocytes in multiple sclerosis lesions. A study of 113 cases. Brain : a journal of neurology. 1999;122(Pt 12):2279–95. doi: 10.1093/brain/122.12.2279. [DOI] [PubMed] [Google Scholar]
- 5.Warf BC, Fok-Seang J, Miller RH. Evidence for the ventral origin of oligodendrocyte precursors in the rat spinal cord. J Neurosci. 1991;11:2477–88. doi: 10.1523/JNEUROSCI.11-08-02477.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pringle NP, Richardson WD. A singularity of PDGF alpha-receptor expression in the dorsoventral axis of the neural tube may define the origin of the oligodendrocyte lineage. Development. 1993;117:525–33. doi: 10.1242/dev.117.2.525. [DOI] [PubMed] [Google Scholar]
- 7.Yu WP, Collarini EJ, Pringle NP, Richardson WD. Embryonic expression of myelin genes: evidence for a focal source of oligodendrocyte precursors in the ventricular zone of the neural tube. Neuron. 1994;12:1353–62. doi: 10.1016/0896-6273(94)90450-2. [DOI] [PubMed] [Google Scholar]
- 8.Timsit S, et al. Oligodendrocytes originate in a restricted zone of the embryonic ventral neural tube defined by DM-20 mRNA expression. J Neurosci. 1995;15:1012–24. doi: 10.1523/JNEUROSCI.15-02-01012.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cameron-Curry P, Le Douarin NM. Oligodendrocyte precursors originate from both the dorsal and the ventral parts of the spinal cord. Neuron. 1995;15:1299–310. doi: 10.1016/0896-6273(95)90009-8. [DOI] [PubMed] [Google Scholar]
- 10.Raff MC, Miller RH, Noble M. A glial progenitor cell that develops in vitro into an astrocyte or an oligodendrocyte depending on culture medium. Nature. 1983;303:390–6. doi: 10.1038/303390a0. [DOI] [PubMed] [Google Scholar]
- 11.Rao MS, Noble M, Mayer-Proschel M. A tripotential glial precursor cell is present in the developing spinal cord. Proc Natl Acad Sci U S A. 1998;95:3996–4001. doi: 10.1073/pnas.95.7.3996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee JC, Mayer-Proschel M, Rao MS. Gliogenesis in the central nervous system. Glia. 2000;30:105–21. doi: 10.1002/(sici)1098-1136(200004)30:2<105::aid-glia1>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 13.Jessell TM. Neuronal specification in the spinal cord: inductive signals and transcriptional codes. Nature reviews. Genetics. 2000;1:20–9. doi: 10.1038/35049541. [DOI] [PubMed] [Google Scholar]
- 14.Lu QR, et al. Sonic hedgehog--regulated oligodendrocyte lineage genes encoding bHLH proteins in the mammalian central nervous system. Neuron. 2000;25:317–29. doi: 10.1016/s0896-6273(00)80897-1. [DOI] [PubMed] [Google Scholar]
- 15.Zhou Q, Wang S, Anderson DJ. Identification of a novel family of oligodendrocyte lineage-specific basic helix-loop-helix transcription factors. Neuron. 2000;25:331–43. doi: 10.1016/s0896-6273(00)80898-3. [DOI] [PubMed] [Google Scholar]
- 16.Takebayashi H, et al. Dynamic expression of basic helix-loop-helix Olig family members: implication of Olig2 in neuron and oligodendrocyte differentiation and identification of a new member, Olig3. Mech Dev. 2000;99:143–8. doi: 10.1016/s0925-4773(00)00466-4. [DOI] [PubMed] [Google Scholar]
- 17.Lu QR, et al. Common developmental requirement for Olig function indicates a motor neuron/oligodendrocyte connection. Cell. 2002;109:75–86. doi: 10.1016/s0092-8674(02)00678-5. [DOI] [PubMed] [Google Scholar]
- 18.Zhou Q, Anderson DJ. The bHLH transcription factors OLIG2 and OLIG1 couple neuronal and glial subtype specification. Cell. 2002;109:61–73. doi: 10.1016/s0092-8674(02)00677-3. [DOI] [PubMed] [Google Scholar]
- 19.Cai J, et al. Generation of oligodendrocyte precursor cells from mouse dorsal spinal cord independent of Nkx6 regulation and Shh signaling. Neuron. 2005;45:41–53. doi: 10.1016/j.neuron.2004.12.028. [DOI] [PubMed] [Google Scholar]
- 20.Vallstedt A, Klos JM, Ericson J. Multiple dorsoventral origins of oligodendrocyte generation in the spinal cord and hindbrain. Neuron. 2005;45:55–67. doi: 10.1016/j.neuron.2004.12.026. [DOI] [PubMed] [Google Scholar]
- 21.Fogarty M, Richardson WD, Kessaris N. A subset of oligodendrocytes generated from radial glia in the dorsal spinal cord. Development. 2005;132:1951–9. doi: 10.1242/dev.01777. [DOI] [PubMed] [Google Scholar]
- 22.Menn B, et al. Origin of oligodendrocytes in the subventricular zone of the adult brain. J Neurosci. 2006;26:7907–18. doi: 10.1523/JNEUROSCI.1299-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fancy SP, Zhao C, Franklin RJ. Increased expression of Nkx2.2 and Olig2 identifies reactive oligodendrocyte progenitor cells responding to demyelination in the adult CNS. Mol Cell Neurosci. 2004;27:247–54. doi: 10.1016/j.mcn.2004.06.015. [DOI] [PubMed] [Google Scholar]
- 24.Buffo A, et al. Expression pattern of the transcription factor Olig2 in response to brain injuries: implications for neuronal repair. Proc Natl Acad Sci U S A. 2005;102:18183–8. doi: 10.1073/pnas.0506535102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arnett HA, et al. bHLH transcription factor Olig1 is required to repair demyelinated lesions in the CNS. Science. 2004;306:2111–5. doi: 10.1126/science.1103709. [DOI] [PubMed] [Google Scholar]
- 26.Georgieva L, et al. Convergent evidence that oligodendrocyte lineage transcription factor 2 (OLIG2) and interacting genes influence susceptibility to schizophrenia. Proc Natl Acad Sci U S A. 2006;103:12469–74. doi: 10.1073/pnas.0603029103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sims R, et al. Evidence that variation in the oligodendrocyte lineage transcription factor 2 (OLIG2) gene is associated with psychosis in Alzheimer's disease. Neurosci Lett. 2009;461:54–9. doi: 10.1016/j.neulet.2009.05.051. [DOI] [PubMed] [Google Scholar]
- 28.Huang K, et al. Positive association between OLIG2 and schizophrenia in the Chinese Han population. Hum Genet. 2008;122:659–60. doi: 10.1007/s00439-007-0434-z. [DOI] [PubMed] [Google Scholar]
- 29.Chakrabarti L, et al. Olig1 and Olig2 triplication causes developmental brain defects in Down syndrome. Nat Neurosci. 2010;13:927–34. doi: 10.1038/nn.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bouvier C, et al. Shared oligodendrocyte lineage gene expression in gliomas and oligodendrocyte progenitor cells. J Neurosurg. 2003;99:344–50. doi: 10.3171/jns.2003.99.2.0344. [DOI] [PubMed] [Google Scholar]
- 31.Ligon KL, et al. The oligodendroglial lineage marker OLIG2 is universally expressed in diffuse gliomas. J Neuropathol Exp Neurol. 2004;63:499–509. doi: 10.1093/jnen/63.5.499. [DOI] [PubMed] [Google Scholar]
- 32.Lu QR, et al. Oligodendrocyte lineage genes (OLIG) as molecular markers for human glial brain tumors. Proc Natl Acad Sci U S A. 2001;98:10851–6. doi: 10.1073/pnas.181340798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marie Y, et al. OLIG2 as a specific marker of oligodendroglial tumour cells. Lancet. 2001;358:298–300. doi: 10.1016/S0140-6736(01)05499-X. [DOI] [PubMed] [Google Scholar]
- 34.Ohnishi A, et al. Expression of the oligodendroglial lineage-associated markers Olig1 and Olig2 in different types of human gliomas. J Neuropathol Exp Neurol. 2003;62:1052–9. doi: 10.1093/jnen/62.10.1052. [DOI] [PubMed] [Google Scholar]
- 35.Ledent V, Paquet O, Vervoort M. Phylogenetic analysis of the human basic helix-loop-helix proteins. Genome Biol. 2002;3:RESEARCH0030. doi: 10.1186/gb-2002-3-6-research0030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gray PA, et al. Mouse brain organization revealed through direct genome-scale TF expression analysis. Science. 2004;306:2255–7. doi: 10.1126/science.1104935. [DOI] [PubMed] [Google Scholar]
- 37.Malatesta P, et al. Neuronal or glial progeny: regional differences in radial glia fate. Neuron. 2003;37:751–64. doi: 10.1016/s0896-6273(03)00116-8. [DOI] [PubMed] [Google Scholar]
- 38.Tsai HH, et al. Regional astrocyte allocation regulates CNS synaptogenesis and repair. Science. 2012;337:358–62. doi: 10.1126/science.1222381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ligon KL, et al. Olig2-regulated lineage-restricted pathway controls replication competence in neural stem cells and malignant glioma. Neuron. 2007;53:503–17. doi: 10.1016/j.neuron.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen JA, et al. Mir-17-3p controls spinal neural progenitor patterning by regulating Olig2/Irx3 cross-repressive loop. Neuron. 2011;69:721–35. doi: 10.1016/j.neuron.2011.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takebayashi H, et al. Non-overlapping expression of Olig3 and Olig2 in the embryonic neural tube. Mech Dev. 2002;113:169–74. doi: 10.1016/s0925-4773(02)00021-7. [DOI] [PubMed] [Google Scholar]
- 42.Liu Z, et al. Control of precerebellar neuron development by Olig3 bHLH transcription factor. J Neurosci. 2008;28:10124–33. doi: 10.1523/JNEUROSCI.3769-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee SK, Lee B, Ruiz EC, Pfaff SL. Olig2 and Ngn2 function in opposition to modulate gene expression in motor neuron progenitor cells. Genes Dev. 2005;19:282–94. doi: 10.1101/gad.1257105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li H, de Faria JP, Andrew P, Nitarska J, Richardson WD. Phosphorylation regulates OLIG2 cofactor choice and the motor neuronoligodendrocyte fate switch. Neuron. 2011;69:918–29. doi: 10.1016/j.neuron.2011.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Furusho M, et al. Involvement of the Olig2 transcription factor in cholinergic neuron development of the basal forebrain. Dev Biol. 2006;293:348–57. doi: 10.1016/j.ydbio.2006.01.031. [DOI] [PubMed] [Google Scholar]
- 46.Xin M, et al. Myelinogenesis and axonal recognition by oligodendrocytes in brain are uncoupled in Olig1-null mice. J Neurosci. 2005;25:1354–65. doi: 10.1523/JNEUROSCI.3034-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marshall CA, Novitch BG, Goldman JE. Olig2 directs astrocyte and oligodendrocyte formation in postnatal subventricular zone cells. J Neurosci. 2005;25:7289–98. doi: 10.1523/JNEUROSCI.1924-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cahoy JD, et al. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J Neurosci. 2008;28:264–78. doi: 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen Y, et al. The basic helix-loop-helix transcription factor olig2 is critical for reactive astrocyte proliferation after cortical injury. J Neurosci. 2008;28:10983–9. doi: 10.1523/JNEUROSCI.3545-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kageyama R, Nakanishi S. Helix-loop-helix factors in growth and differentiation of the vertebrate nervous system. Current opinion in genetics & development. 1997;7:659–65. doi: 10.1016/s0959-437x(97)80014-7. [DOI] [PubMed] [Google Scholar]
- 51.Lee JE. Basic helix-loop-helix genes in neural development. Current opinion in neurobiology. 1997;7:13–20. doi: 10.1016/s0959-4388(97)80115-8. [DOI] [PubMed] [Google Scholar]
- 52.Parras CM, et al. Mash1 specifies neurons and oligodendrocytes in the postnatal brain. The EMBO journal. 2004;23:4495–505. doi: 10.1038/sj.emboj.7600447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Singh SK, et al. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 54.Bao S, et al. Targeting cancer stem cells through L1CAM suppresses glioma growth. Cancer Res. 2008;68:6043–8. doi: 10.1158/0008-5472.CAN-08-1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Barrett LE, et al. Self-renewal does not predict tumor growth potential in mouse models of high-grade glioma. Cancer Cell. 2012;21:11–24. doi: 10.1016/j.ccr.2011.11.025. [DOI] [PubMed] [Google Scholar]
- 56.Appolloni I, et al. Antagonistic modulation of gliomagenesis by Pax6 and Olig2 in PDGF-induced oligodendroglioma. Int J Cancer. 2012 doi: 10.1002/ijc.27606. [DOI] [PubMed] [Google Scholar]
- 57.Mehta S, et al. The central nervous system-restricted transcription factor Olig2 opposes p53 responses to genotoxic damage in neural progenitors and malignant glioma. Cancer Cell. 2011;19:359–71. doi: 10.1016/j.ccr.2011.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kitada M, Rowitch DH. Transcription factor co-expression patterns indicate heterogeneity of oligodendroglial subpopulations in adult spinal cord. Glia. 2006;54:35–46. doi: 10.1002/glia.20354. [DOI] [PubMed] [Google Scholar]
- 59.Chang A, Tourtellotte WW, Rudick R, Trapp BD. Premyelinating oligodendrocytes in chronic lesions of multiple sclerosis. N Engl J Med. 2002;346:165–73. doi: 10.1056/NEJMoa010994. [DOI] [PubMed] [Google Scholar]
- 60.Kuhlmann T, et al. Differentiation block of oligodendroglial progenitor cells as a cause for remyelination failure in chronic multiple sclerosis. Brain : a journal of neurology. 2008;131:1749–58. doi: 10.1093/brain/awn096. [DOI] [PubMed] [Google Scholar]
- 61.Billiards SS, et al. Myelin abnormalities without oligodendrocyte loss in periventricular leukomalacia. Brain Pathol. 2008;18:153–63. doi: 10.1111/j.1750-3639.2007.00107.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Verney C, et al. Microglial reaction in axonal crossroads is a hallmark of noncystic periventricular white matter injury in very preterm infants. J Neuropathol Exp Neurol. 2012;71:251–64. doi: 10.1097/NEN.0b013e3182496429. [DOI] [PubMed] [Google Scholar]
- 63.Fancy SP, et al. Axin2 as regulatory and therapeutic target in newborn brain injury and remyelination. Nat Neurosci. 2011 doi: 10.1038/nn.2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pekny M, Nilsson M. Astrocyte activation and reactive gliosis. Glia. 2005;50:427–34. doi: 10.1002/glia.20207. [DOI] [PubMed] [Google Scholar]
- 65.Gabay L, Lowell S, Rubin LL, Anderson DJ. Deregulation of dorsoventral patterning by FGF confers trilineage differentiation capacity on CNS stem cells in vitro. Neuron. 2003;40:485–99. doi: 10.1016/s0896-6273(03)00637-8. [DOI] [PubMed] [Google Scholar]
- 66.Setoguchi T, Kondo T. Nuclear export of OLIG2 in neural stem cells is essential for ciliary neurotrophic factor-induced astrocyte differentiation. J Cell Biol. 2004;166:963–8. doi: 10.1083/jcb.200404104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Muroyama Y, Fujiwara Y, Orkin SH, Rowitch DH. Specification of astrocytes by bHLH protein SCL in a restricted region of the neural tube. Nature. 2005;438:360–3. doi: 10.1038/nature04139. [DOI] [PubMed] [Google Scholar]
- 68.Dimou L, Simon C, Kirchhoff F, Takebayashi H, Gotz M. Progeny of Olig2-expressing progenitors in the gray and white matter of the adult mouse cerebral cortex. J Neurosci. 2008;28:10434–42. doi: 10.1523/JNEUROSCI.2831-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Komitova M, Serwanski DR, Lu QR, Nishiyama A. NG2 cells are not a major source of reactive astrocytes after neocortical stab wound injury. Glia. 2011;59:800–9. doi: 10.1002/glia.21152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zawadzka M, et al. CNS-resident glial progenitor/stem cells produce Schwann cells as well as oligodendrocytes during repair of CNS demyelination. Cell Stem Cell. 2010;6:578–90. doi: 10.1016/j.stem.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tripathi RB, Rivers LE, Young KM, Jamen F, Richardson WD. NG2 glia generate new oligodendrocytes but few astrocytes in a murine experimental autoimmune encephalomyelitis model of demyelinating disease. J Neurosci. 2010;30:16383–90. doi: 10.1523/JNEUROSCI.3411-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Buffo A, et al. Origin and progeny of reactive gliosis: A source of multipotent cells in the injured brain. Proc Natl Acad Sci U S A. 2008;105:3581–6. doi: 10.1073/pnas.0709002105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Magnus T, et al. Evidence that nucleocytoplasmic Olig2 translocation mediates brain-injury-induced differentiation of glial precursors to astrocytes. J Neurosci Res. 2007;85:2126–37. doi: 10.1002/jnr.21368. [DOI] [PubMed] [Google Scholar]
- 74.Zhao JW, Raha-Chowdhury R, Fawcett JW, Watts C. Astrocytes and oligodendrocytes can be generated from NG2+ progenitors after acute brain injury: intracellular localization of oligodendrocyte transcription factor 2 is associated with their fate choice. Eur J Neurosci. 2009;29:1853–69. doi: 10.1111/j.1460-9568.2009.06736.x. [DOI] [PubMed] [Google Scholar]
- 75.Cassiani-Ingoni R, et al. Cytoplasmic translocation of Olig2 in adult glial progenitors marks the generation of reactive astrocytes following autoimmune inflammation. Exp Neurol. 2006;201:349–58. doi: 10.1016/j.expneurol.2006.04.030. [DOI] [PubMed] [Google Scholar]
- 76.Fernandez F, Garner CC. Over-inhibition: a model for developmental intellectual disability. Trends Neurosci. 2007;30:497–503. doi: 10.1016/j.tins.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 77.Belichenko PV, et al. Synaptic structural abnormalities in the Ts65Dn mouse model of Down Syndrome. J Comp Neurol. 2004;480:281–98. doi: 10.1002/cne.20337. [DOI] [PubMed] [Google Scholar]
- 78.Haydar TF, Reeves RH. Trisomy 21 and early brain development. Trends in neurosciences. 2012;35:81–91. doi: 10.1016/j.tins.2011.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lu J, et al. OLIG2 over-expression impairs proliferation of human Down syndrome neural progenitors. Hum Mol Genet. 2012 doi: 10.1093/hmg/dds052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang SZ, et al. An oligodendrocyte-specific zinc-finger transcription regulator cooperates with Olig2 to promote oligodendrocyte differentiation. Development. 2006;133:3389–98. doi: 10.1242/dev.02522. [DOI] [PubMed] [Google Scholar]
- 81.Massari ME, Murre C. Helix-loop-helix proteins: regulators of transcription in eucaryotic organisms. Molecular and cellular biology. 2000;20:429–40. doi: 10.1128/mcb.20.2.429-440.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhou Q, Choi G, Anderson DJ. The bHLH transcription factor Olig2 promotes oligodendrocyte differentiation in collaboration with Nkx2.2. Neuron. 2001;31:791–807. doi: 10.1016/s0896-6273(01)00414-7. [DOI] [PubMed] [Google Scholar]
- 83.Novitch BG, Chen AI, Jessell TM. Coordinate regulation of motor neuron subtype identity and pan-neuronal properties by the bHLH repressor Olig2. Neuron. 2001;31:773–89. doi: 10.1016/s0896-6273(01)00407-x. [DOI] [PubMed] [Google Scholar]
- 84.Mizuguchi R, et al. Combinatorial roles of olig2 and neurogenin2 in the coordinated induction of pan-neuronal and subtype-specific properties of motoneurons. Neuron. 2001;31:757–71. doi: 10.1016/s0896-6273(01)00413-5. [DOI] [PubMed] [Google Scholar]
- 85.Kuspert M, Hammer A, Bosl MR, Wegner M. Olig2 regulates Sox10 expression in oligodendrocyte precursors through an evolutionary conserved distal enhancer. Nucleic Acids Res. 2011;39:1280–93. doi: 10.1093/nar/gkq951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mazzoni EO, et al. Embryonic stem cell-based mapping of developmental transcriptional programs. Nature methods. 2011;8:1056–8. doi: 10.1038/nmeth.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Weng Q, et al. Dual-mode modulation of smad signaling by smad-interacting protein sip1 is required for myelination in the central nervous system. Neuron. 2012;73:713–28. doi: 10.1016/j.neuron.2011.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Guo X, et al. Delayed onset of experimental autoimmune encephalomyelitis in Olig1 deficient mice. PLoS One. 2010;5 doi: 10.1371/journal.pone.0013083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chen Y, et al. The oligodendrocyte-specific G protein-coupled receptor GPR17 is a cell-intrinsic timer of myelination. Nature neuroscience. 2009;12:1398–406. doi: 10.1038/nn.2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Li H, Lu Y, Smith HK, Richardson WD. Olig1 and Sox10 interact synergistically to drive myelin basic protein transcription in oligodendrocytes. J Neurosci. 2007;27:14375–82. doi: 10.1523/JNEUROSCI.4456-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Beckett D. Regulated assembly of transcription factors and control of transcription initiation. Journal of molecular biology. 2001;314:335–52. doi: 10.1006/jmbi.2001.5134. [DOI] [PubMed] [Google Scholar]
- 92.Featherstone M. Coactivators in transcription initiation: here are your orders. Current opinion in genetics & development. 2002;12:149–55. doi: 10.1016/s0959-437x(02)00280-0. [DOI] [PubMed] [Google Scholar]
- 93.Torchia J, Glass C, Rosenfeld MG. Co-activators and co-repressors in the integration of transcriptional responses. Current opinion in cell biology. 1998;10:373–83. doi: 10.1016/s0955-0674(98)80014-8. [DOI] [PubMed] [Google Scholar]
- 94.Ravasi T, et al. An atlas of combinatorial transcriptional regulation in mouse and man. Cell. 2010;140:744–52. doi: 10.1016/j.cell.2010.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Samanta J, Kessler JA. Interactions between ID and OLIG proteins mediate the inhibitory effects of BMP4 on oligodendroglial differentiation. Development. 2004;131:4131–42. doi: 10.1242/dev.01273. [DOI] [PubMed] [Google Scholar]
- 96.Benezra R, Davis RL, Lockshon D, Turner DL, Weintraub H. The protein Id: a negative regulator of helix-loop-helix DNA binding proteins. Cell. 1990;61:49–59. doi: 10.1016/0092-8674(90)90214-y. [DOI] [PubMed] [Google Scholar]
- 97.Poulin G, Lebel M, Chamberland M, Paradis FW, Drouin J. Specific protein-protein interaction between basic helix-loop-helix transcription factors and homeoproteins of the Pitx family. Molecular and cellular biology. 2000;20:4826–37. doi: 10.1128/mcb.20.13.4826-4837.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Babu DA, Chakrabarti SK, Garmey JC, Mirmira RG. Pdx1 and BETA2/NeuroD1 participate in a transcriptional complex that mediates short-range DNA looping at the insulin gene. The Journal of biological chemistry. 2008;283:8164–72. doi: 10.1074/jbc.M800336200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Makarenkova HP, Gonzalez KN, Kiosses WB, Meech R. Barx2 controls myoblast fusion and promotes MyoD-mediated activation of the smooth muscle alpha-actin gene. The Journal of biological chemistry. 2009;284:14866–74. doi: 10.1074/jbc.M807208200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sun T, et al. Olig bHLH proteins interact with homeodomain proteins to regulate cell fate acquisition in progenitors of the ventral neural tube. Curr Biol. 2001;11:1413–20. doi: 10.1016/s0960-9822(01)00441-9. [DOI] [PubMed] [Google Scholar]
- 101.Fukuda S, Kondo T, Takebayashi H, Taga T. Negative regulatory effect of an oligodendrocytic bHLH factor OLIG2 on the astrocytic differentiation pathway. Cell Death Differ. 2004;11:196–202. doi: 10.1038/sj.cdd.4401332. [DOI] [PubMed] [Google Scholar]
- 102.Ogryzko VV, Schiltz RL, Russanova V, Howard BH, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–9. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 103.Ikushima H, et al. An Id-like molecule, HHM, is a synexpression grouprestricted regulator of TGF-beta signalling. The EMBO journal. 2008;27:2955–65. doi: 10.1038/emboj.2008.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Huillard E, et al. Disruption of CK2beta in embryonic neural stem cells compromises proliferation and oligodendrogenesis in the mouse telencephalon. Mol Cell Biol. 2010;30:2737–49. doi: 10.1128/MCB.01566-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Sun Y, et al. Phosphorylation state of Olig2 regulates proliferation of neural progenitors. Neuron. 2011;69:906–17. doi: 10.1016/j.neuron.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sun T, et al. Cross-repressive interaction of the Olig2 and Nkx2.2 transcription factors in developing neural tube associated with formation of a specific physical complex. J Neurosci. 2003;23:9547–56. doi: 10.1523/JNEUROSCI.23-29-09547.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Niu J, et al. Phosphorylated olig1 localizes to the cytosol of oligodendrocytes and promotes membrane expansion and maturation. Glia. 2012 doi: 10.1002/glia.22364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gill G. SUMO and ubiquitin in the nucleus: different functions, similar mechanisms? Genes & development. 2004;18:2046–59. doi: 10.1101/gad.1214604. [DOI] [PubMed] [Google Scholar]
- 109.Hay RT. SUMO: a history of modification. Molecular cell. 2005;18:1–12. doi: 10.1016/j.molcel.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 110.Johnson ES. Protein modification by SUMO. Annual review of biochemistry. 2004;73:355–82. doi: 10.1146/annurev.biochem.73.011303.074118. [DOI] [PubMed] [Google Scholar]
- 111.Verger A, Perdomo J, Crossley M. Modification with SUMO. A role in transcriptional regulation. EMBO reports. 2003;4:137–42. doi: 10.1038/sj.embor.embor738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Huang TT, Wuerzberger-Davis SM, Wu ZH, Miyamoto S. Sequential modification of NEMO/IKKgamma by SUMO-1 and ubiquitin mediates NF-kappaB activation by genotoxic stress. Cell. 2003;115:565–76. doi: 10.1016/s0092-8674(03)00895-x. [DOI] [PubMed] [Google Scholar]
- 113.Hay RT. Modifying NEMO. Nature cell biology. 2004;6:89–91. doi: 10.1038/ncb0204-89. [DOI] [PubMed] [Google Scholar]
- 114.Imoto S, et al. Sumoylation of Smad3 stimulates its nuclear export during PIASy-mediated suppression of TGF-beta signaling. Biochemical and biophysical research communications. 2008;370:359–65. doi: 10.1016/j.bbrc.2008.03.116. [DOI] [PubMed] [Google Scholar]
- 115.Martin S, Wilkinson KA, Nishimune A, Henley JM. Emerging extranuclear roles of protein SUMOylation in neuronal function and dysfunction. Nat Rev Neurosci. 2007;8:948–59. doi: 10.1038/nrn2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Imoto S, et al. Sumoylation of Smad3 stimulates its nuclear export during PIASy-mediated suppression of TGF-beta signaling. Biochem Biophys Res Commun. 2008;370:359–65. doi: 10.1016/j.bbrc.2008.03.116. [DOI] [PubMed] [Google Scholar]
- 117.Zhou F, Xue Y, Lu H, Chen G, Yao X. A genome-wide analysis of sumoylation-related biological processes and functions in human nucleus. FEBS letters. 2005;579:3369–75. doi: 10.1016/j.febslet.2005.04.076. [DOI] [PubMed] [Google Scholar]
- 118.Frazer KA, et al. Evolutionarily conserved sequences on human chromosome 21. Genome Res. 2001;11:1651–9. doi: 10.1101/gr.198201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Muskal SM, Holbrook SR, Kim SH. Prediction of the disulfide-bonding state of cysteine in proteins. Protein engineering. 1990;3:667–72. doi: 10.1093/protein/3.8.667. [DOI] [PubMed] [Google Scholar]
- 120.Smith E, Shilatifard A. The chromatin signaling pathway: diverse mechanisms of recruitment of histone-modifying enzymes and varied biological outcomes. Molecular cell. 2010;40:689–701. doi: 10.1016/j.molcel.2010.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.el-Husseini Ael D, Bredt DS. Protein palmitoylation: a regulator of neuronal development and function. Nature reviews. Neuroscience. 2002;3:791–802. doi: 10.1038/nrn940. [DOI] [PubMed] [Google Scholar]
- 122.Rowitch DH, Lu QR, Kessaris N, Richardson WD. An ‘oligarchy’ rules neural development. Trends Neurosci. 2002;25:417–22. doi: 10.1016/s0166-2236(02)02201-4. [DOI] [PubMed] [Google Scholar]
- 123.Muller T, et al. The bHLH factor Olig3 coordinates the specification of dorsal neurons in the spinal cord. Genes Dev. 2005;19:733–43. doi: 10.1101/gad.326105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ligon KL, et al. Development of NG2 neural progenitor cells requires Olig gene function. Proc Natl Acad Sci U S A. 2006;103:7853–8. doi: 10.1073/pnas.0511001103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Miyoshi G, Butt SJ, Takebayashi H, Fishell G. Physiologically distinct temporal cohorts of cortical interneurons arise from telencephalic Olig2-expressing precursors. J Neurosci. 2007;27:7786–98. doi: 10.1523/JNEUROSCI.1807-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Plenge RM, et al. Two independent alleles at 6q23 associated with risk of rheumatoid arthritis. Nature genetics. 2007;39:1477–82. doi: 10.1038/ng.2007.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Thomson W, et al. Rheumatoid arthritis association at 6q23. Nature genetics. 2007;39:1431–3. doi: 10.1038/ng.2007.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.