Abstract
The perinuclear actin cap (or actin cap) is a recently characterized cytoskeletal organelle composed of thick, parallel, and highly contractile acto-myosin filaments that are specifically anchored to the apical surface of the interphase nucleus. The actin cap is present in a wide range of adherent eukaryotic cells, but is disrupted in several human diseases, including laminopathies and cancer. Through its large terminating focal adhesions and anchorage to the nuclear lamina and nuclear envelope through LINC complexes, the perinuclear actin cap plays a critical role both in mechanosensation and mechanotransduction, the ability of cells to sense changes in matrix compliance and to respond to mechanical forces, respectively.
What is the perinuclear actin cap?
Three distinct filamentous proteins - actin filaments, intermediate filaments, and microtubules - constitute the cytoskeleton, an expansive array of polymers that span the cytoplasm of all eukaryotic cells and mediate a multitude of important functions, including cell division, morphology, migration, and differentiation1-3. Globular actin (G-actin) polymerizes into semi-flexible polymers, actin filaments (F-actin)4-6 that bind myosin II to form contractile actomyosin stress fibers7-10. Bundles of cross-linked, rope-like intermediate filaments (keratins, vimentin, neurofilaments)11, 12 provide structural integrity to the cytoplasm and the nuclear lamina13-15, while thick, hollow, cylindrical-shaped rigid microtubules provide the guiding tracks for the intracellular transport of organelles, such as mitochondria and microvesicles16, 17, and set the polarization axis of migrating cells18, 19.
The perinuclear actin cap (or actin cap) is a recently characterized organelle composed of thick, parallel, and highly contractile acto-myosin filament bundles that are anchored to the apical surface of the interphase nucleus through highly conserved linker proteins in a wide range of adherent cells20 (Fig. 1). Actin-cap fibers are functionally, molecularly, and topologically distinct from conventional actin stress fibers, which include basal stress fibers, transverse arcs, and dorsal fibers21, 22. While conventional stress fibers are confined to the basal layer or cortex of the cell and are arranged in diverse directions, actin-cap fibers are typically aligned with the long axis of the cell and terminate at the leading and trailing edges of migratory cells. Critical to the distinct functions of the perinuclear actin cap is its specific anchorage to the nuclear envelope. In contrast, conventional stress fibers are directly or indirectly connected to the plasma membrane.
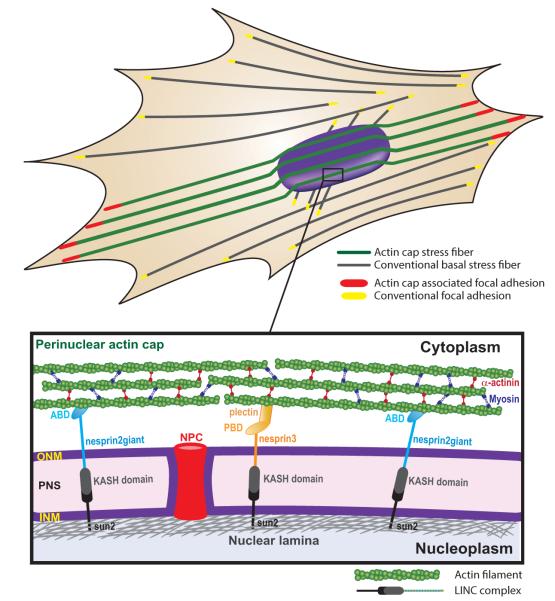
Fig. 1. Subcellular organization of the perinuclear actin cap and LINC complexes.
Perinuclear actin-cap fibers (green) are functionally and topologically distinct from conventional basal actin fibers (gray) due to tight coupling to the side and apical surfaces of the nuclear envelope through linkers of nucleoskeleton and cytoskeleton (LINC) complexes. LINC complexes consist of KASH-domain-containing nesprin isoforms (i.e., nesprin2giant and nesprin3) that are connected to the actin cap through actin binding domains (ABD) and plectin binding domains (PBD), respectively, localized in the outer nuclear membrane (ONM) and SUN proteins that interact with the nuclear lamina located underneath the inner nuclear membrane (INM). SUN proteins and Nesprins are connected through KASH-SUN interactions in the perinuclear space (PNS). (Reproduced from ref. 24, 28).
The actin cap has been confirmed in both fixed and live cells and has been so far identified and characterized in mouse embryonic fibroblasts, human lung and foreskin fibroblasts, mouse myoblasts, human umbilical vein endothelial cells, and human ovarian epithelial cells20, 23, 24. The actin cap is completely absent from undifferentiated embryonic stem cells and induced pluripotent stem cells, but starts to form and organize progressively during the differentiation23. The actin cap is also absent from epithelial sheets, but appears rapidly upon epithelial-to-mesenchymal (EMT) transition25.
Strikingly, the actin cap is disrupted or totally absent in cells from laminopathic mice and patients26, 27, a group of human diseases that stem from mutations in the LMNA gene that encodes the nuclear lamina protein lamin A/C (Fig. 1), an intermediate filament. The actin cap is also disrupted in human cancer cells such as the HeLa cervical cancer line28, U2OS osteosarcoma, MDA-MB-231 breast carcinoma, and MCF-7 breast adenocarcinoma 24 (Fig. 2). While a defective nuclear shape is a common trait of cancer cells, the biological significance of the lack of organized actin cap in cancer cells remains to be elucidated.
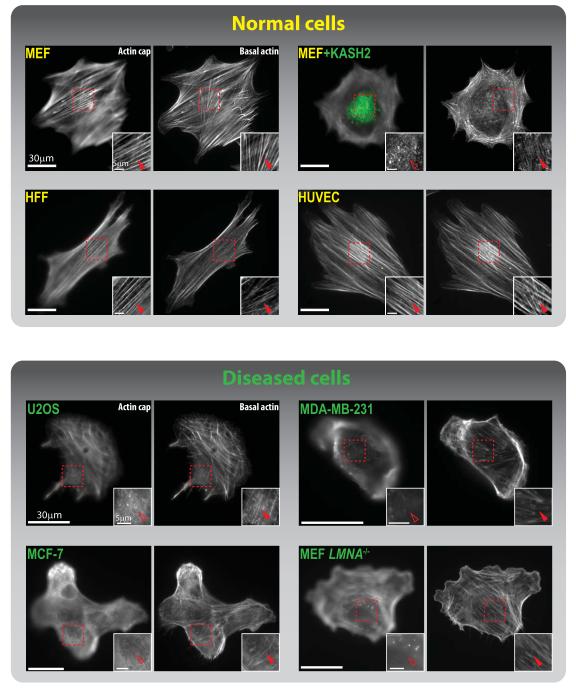
Fig. 2. Organization of the perinuclear actin cap in disease.
Actin cap (left panels) and basal actin filament organization (right panels) for healthy primary cells (MEFs: mouse embryonic fibroblasts, HFFs: human foreskin fibroblasts, HUVECs: human umbilical vein endothelial cells), compared to diseased cells (U2OS: osteosarcoma, MDA-MB-231: breast carcinoma, MCF-7: breast adenocarcinoma, and MEF LMNA−/−: a model for laminopathic diseases). Transfection of EGFP-KASH2 in MEFs (top right panels) displaces outer nuclear membrane-embedded LINC complex proteins nesprin2giant and nesprin3 from the nuclear envelope and disorganizes the actin cap. This in turn disrupts cellular mechanosensation and mechanotransduction. Insets magnify the red-boxed regions for detail. Filled red arrowheads point to organized, intact actin-cap fibers; empty arrowheads point to disrupted or absent actin cap. (Images of MEF, HFF, HUVEC, and U2OS taken from ref.24)
The actin cap is highly dynamic. Live-cell microscopy of cells transfected with GFP-lifeact readily reveals high contractility and dynamics, as the actin-cap fibers continuously undergo cycles of extension and retraction24, precisely positioning and actively deforming the nucleus20. Fluorescence recovery after photobleaching (FRAP) analysis also indicates that actin-cap fibers undergo much faster turnover than basal stress fibers24. In somatic cells, the actin cap does not appear for several hours following cell division20, indicating that the actin cap is not a permanent fixture of cells.
Molecular architecture of the actin cap and accessory proteins
The perinuclear actin cap is physically, yet dynamically, connected to the nuclear envelope through linkers of nucleoskeleton and cytoskeleton (LINC) complexes29 and is terminated by actin-cap associated focal adhesions (ACAFAs) at the basal surface of the adherent cell24 (Fig. 1). LINC-complex proteins nesprins bridge fibers of the actin cap to the nuclear envelope directly through the actin-binding domain of nesprin2giant30, 31 and indirectly through the multi-functional actin-binding protein plectin, which itself binds nesprin332, 33 (Fig. 1). Nesprins bind SUN (for Sad1p, UNC-84) proteins via KASH (for Klarsicht, ANC-1, Syne Homology) domains in the periplasmic space between the inner and outer membranes of the nuclear envelope 34, 35, and SUN proteins connect to major nuclear lamina protein lamin A/C35, which interacts directly and indirectly with chromosomal DNA36 (Fig. 1). Fibers of the actin cap contain more phosphorylated myosin II, the active form of the motor and F-actin-bundling protein myosin II, and the F-actin crosslinking/bundling protein α-actinin than basal stress fibers24. Fibers of the actin cap are terminated by particularly large focal adhesions, which are located at the edges of the cell and contain high density of phospho-FAK, the active form of focal adhesion kinase (FAK) 24.
Nuclear shaping by the perinuclear actin cap in health and disease
The first established function of the perinuclear actin cap was nuclear shaping20. When the actin cap of adherent cells is disrupted with a low-dose treatment of the F-actin depolymerizing drug latrunculin B or by disruption of the LINC complexes, nuclear lobulation ensues and the nucleus can bulge to almost twice its original height20 (Fig. 2). The actin cap serves to maintain the thin, disk-like morphology as well as the translocation and rotation of the interphase nucleus during the migration and shear-response of adherent cells37.
Defects in nuclear shape are a salient, commonly shared feature of nuclei in laminopathic cells26, 27. Embryonic fibroblasts collected from a mouse lacking LMNA (Lmna−/−), which displays an Emery-Dreyfus muscular dystrophy-like phenotype, do not show a significant difference in the organization of basal actin fibers from that of Lmna+/+ cells, but lamin A/C deficiency severely decreases the fraction of cells that contain an organized, intact actin cap (see the status of actin cap in healthy vs. diseased cells in Fig. 2). Additionally, cells with the homozygous mutation Lmnal530p/l530p, which causes an accelerated aging (progeria) phenotype in mice38, show even fewer actin caps than Lmna−/− cells20. Both Lmna−/− and Lmnal530p/l530p cells display disrupted LINC complexes at the nuclear envelope28, which could explain their lack of organized actin caps since specific disruption of LINC complexes causes specific disruption of the actin cap (Fig. 2) and abrogated nuclear-shaping ability20.
Cellular mechanosensation vs. cellular mechanotransduction
In addition to nuclear shaping, the perinuclear actin plays a central role in the ability of cells to sense changes in matrix compliance and to respond to mechanical forces. Here, we distinguish cellular mechanosensation from cellular mechanotransduction. The ability of cells to recognize, analyze, and respond to consistently changing extracellular substrate rigidity, termed mechanosensation, plays a crucial role in various physiological and pathological processes in vivo. For instance, the mechanical rigidity of the extracellular matrix seems to play an important regulatory factor during embryogenesis: mesenchymal stem cells grown on matrices of controlled stiffness mimicking brain tissue, striated muscle, and bone are differently differentiated towards neurogenic, myogenic, and osteogenic lineages, reflectively39, 40. Enhanced tissue stiffness by increased collagen cross-linking density also correlates with progression of malignant breast cancer41-43. Mechanosensation of extracellular physical cues indeed determines function and ultimate fate of cells44, while abnormality in mechanosensation is considered a distinct feature of diseased cells.
In contrast to mechanosensation, mechanotransduction is the mechanism by which cells respond to and convert externally applied mechanical forces into biochemical signals45. Cells in vivo are subjected to a variety of external forces, such as compressive and tensile forces caused by fluid shear stresses. These forces come into play in a multitude of physiologic and diseased conditions, such as blood flow through the vasculature46 and interstitial flow in connective tissues and tumor cells1. Shear stress applied to endothelial progenitor cells that freely circulate in blood or tissue and contribute to vascular regeneration, termed angiogenesis, promotes the formation of capillary-like tubule structures47. The absence of mechanical stimulation can be a major cause of bone loss in osteoporosis, another debilitating and progressive disease of aging48, and microtubule-based primary cilia regulate bone density by responding to fluid flow49, 50.
Two main cellular pathways are activated following mechanical stimulation. Ion channels open and close in response to changes in force and, in turn, alter ion flux across the cell membrane, activating signaling pathways and ultimately affecting gene transcription51. Additionally, focal adhesions are involved in transducing stress signals to the genome52, 53. Focal adhesions are large and dynamic protein clusters organized at the basal surface of the cell, through which cells are mechanically anchored to the extracellular substratum and biochemical signals are transmitted24, 54, 55. The dynamic assembly and disassembly of focal adhesion complexes regulate binding between transmembrane adhesion molecules (integrins) and the extracellular matrix, modulating cell migration and mechanosensation56-59. Both ion channel and focal adhesion-mediated signaling implicate the actin cytoskeleton. Cytoskeletal tension may regulate ion channel conductance in response to force51, 60, while actin stress fibers are anchored by focal adhesions, and actin-focal adhesion dynamics rely on transmission of force61.
Below we highlight recent experimental evidence showing how the perinuclear actin cap and the focal adhesions that terminate actin-cap fibers play a central role in both mechanosensation and mechanotransduction.
Cellular mechanosensation through actin-cap associated focal adhesions (ACAFAs)
Since actin-cap fibers are organized along the side and apical surfaces of the interphase nucleus and terminated by focal adhesions organized at the basal surface of adherent cells, the focal plane of a light microscope can be progressively lowered from the apical surface of the cell, identifying the actin cap, to the basal surface of the cell revealing ACAFAs, by following individual actin-cap fibers (Fig. 3A).
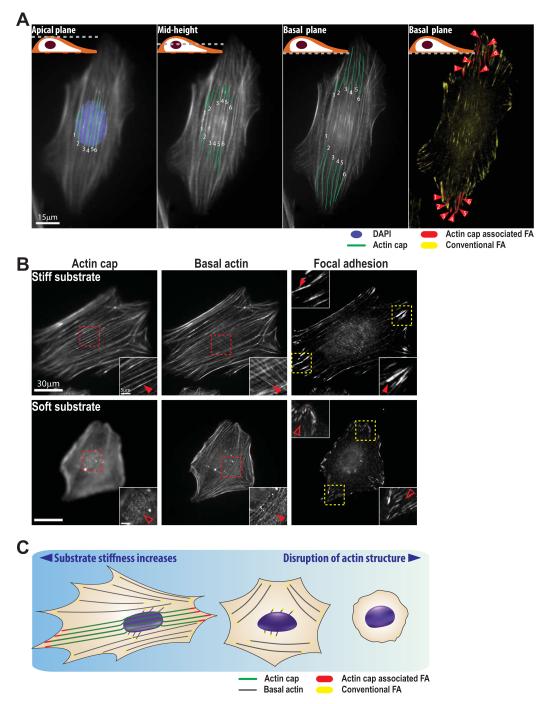
Fig. 3. Actin-cap associated focal adhesions mediate cellular mechanosensation.
A. Actin-cap associated focal adhesions (ACAFAs). Gradual lowering of the focal plane of the light microscope, from the apical surface of a cell, where actin-cap fibers are readily detected, to the basal surface, reveals that actin-cap fibers are draped over the nucleus (DAPI, blue) and terminated by a subset of focal adhesions termed actin-cap associated focal adhesions (ACAFAs, red), while all other focal adhesions that are not linked to the actin cap are termed conventional focal adhesions (CFAs, yellow). Segments of actin-cap fibers in the plane of focus are traced in green. Six individual actin-cap fibers and corresponding twelve actin-cap associated focal adhesions (at both ends of the actin-cap fibers) are visualized in a vinculin-stained mouse embryonic fibroblast (MEF). B. Mechanosensing response of the actin cap and associated ACAFAs to changes in substrate stiffness. On a stiff glass substrate, MEFs show highly ordered actin-cap fibers (top left panel) terminated by corresponding ACAFAs (top right panel) and organized basal actin fibers (top middle panel) terminated by CFAs. When these cells are placed on a soft polyacrylamide gel, actin-cap fibers are partially or completely disrupted (bottom left panel), and ACAFAs largely disappear (bottom right panel) without substantial changes in organization of basal actin fibers (bottom middle panel) and CFAs. Insets show a detail of the actin-cap fibers (red) and ACAFAs (yellow). Filled arrowheads indicate the presence of organized actin cap or organized ACAFAs; empty arrowheads indicate the absence of organized actin cap or CFAs. C. Schematic of cellular mechanosensation. Well-arranged actin-cap fibers (green) above the nucleus are terminated by large and elongated ACAFAs (red) at the periphery of the adherent cell. While substrate compliance increases, actin-cap fibers and corresponding ACAFAs disappear earlier than basal actin (gray) and CFAs (yellow), but on the extremely soft matrix, both types of actin structures and focal adhesions are largely absent. (Reprinted from ref. 24)
Quantitative microscopy analysis in human and mouse fibroblasts and endothelial cells reveals that around ~30% of all focal adhesions per cell are ACAFAs (labeled in red in Fig. 3A), which are significantly larger and more elongated than conventional focal adhesions (CFAs, labeled in yellow in Fig. 3A), which terminate basal stress fibers. Because actin-cap fibers are typically aligned with the long axis of the cell20, ACAFAs are typically located at the leading edge and trailing edges of motile cells, far from the nucleus24. ACAFAs are also more dynamic than CFAs, as measured by a significantly faster recovery of photobleached GFP-tagged focal-adhesion proteins (shorter halftime recovery, ~20s vs. ~30s) and much higher translocation speed than other focal adhesions24.
The morphological and dynamical differences between ACAFAs and CFAs are caused in part by the specific LINC-mediated connections of the actin-cap fibers to the nucleus. Due to the underlying nucleus and its internal pressure, actin-cap fibers and corresponding ACAFAs are under significantly higher tension than conventional actin structures and corresponding CFAs of the cell 24. This increased tension may promote the higher turnover dynamics of ACAFAs58, 62. Binding of actin-cap fibers to their corresponding focal adhesions not only distinguishes ACAFAs from CFAs, but also mediates their enhanced mechanosensing response to changes in mechanical properties of the microenvironment. For instance, variation of matrix compliance mimicking different stiffnesses of the microenvironment in vivo (i.e., bone, muscle, adipose tissue, and brain tissue) reveals a significantly more sensitive response of the actin cap and associated ACAFAs than conventional actin structures and CFAs (Fig. 3B). All actin structures and focal adhesions are disorganized on extremely compliant substrates (<1 kPa, close to the elasticity of brain tissue63), which confirms that CFAs also participate in mechanosensation64-71, but a larger amplitude of physical stimulation is required to match the level of ACAFA-mediated mechanosensation (Fig. 3C). Consequently, ACAFAs dominate mechanosensing response to changes in matrix compliance.
ACAFA-mediated cellular mechanosensation relies on the physical connections between the nucleus and the actin cap mediated by the LINC complexes (Fig. 1). When the LINC complex is disrupted by introducing KASH2 constructs35 that displace nesprin2-giant and nesprin3 from the nuclear envelope, actin-cap fibers are disrupted (top-right panels of Fig. 2) such that either no actin-cap associated focal adhesions are present, or any remaining actin-cap fibers and associated focal adhesions are less responsive to changes in matrix compliance. α-actinin24, which crosslinks and bundles actin filaments of the actin cap, synergistically with myosin II regulate cellular mechansensation by the actin cap 24. A recent theoretical model explains why ACAFAs, which are significantly larger than CFAs, are more mechanosensitive than CFAs, as it predicts that the level of mechanosensation scales with focal adhesion size58.
The actin cap mediates fast mechanotransduction
The actin cap, which along basal actin fibers is disrupted by serum starvation, is reformed and organized in response to fluid shear stresses as low as 0.01 dyn/cm2 within 5 min in mouse embryonic fibroblasts and myoblasts72 (left panel, Fig. 4). In contrast, basal actin fibers can only form past a threshold shear stress level 50-fold higher than required for the formation of the actin cap. Biochemical stimulation by addition of serum mediates actin-cap formation, but only at timescales orders-of-magnitude longer than required for fluid shear stresses. Similarly, the rate of actin cap disappearance following shear cessation is significantly faster than following the switch from serum-rich to serum-starved conditions72. Therefore, the response of the actin cap to mechanical forces is much faster and more dynamic than with biochemical stimulation, indicating that the actin cap could be part of an extremely fast and efficient, physical pathway of transmitting mechanical stimuli from the extracellular milieu all the way to the nucleus.
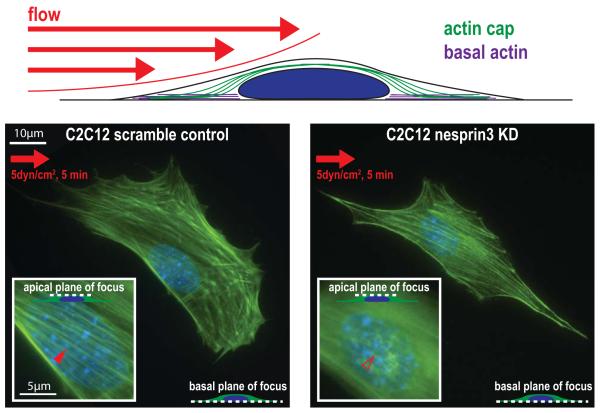
Fig. 4. LINC-mediated formation of the actin cap in cellular mechanotransduction.
Schematic of an adherent cell exposed to fluid flow shear force. Left, micrograph shows a serum-starved control C2C12 mouse myoblast cell (which contains no organized actin structures before stimulation) that forms an organized actin cap (inset) in response to exposure of a shear stress of 5 dyn/cm2 for 5 min. The right micrograph shows a serum-starved C2C12 cell that has been shRNA-depleted of LINC complex protein nesprin3. This cell is unable to form an organized actin cap after exposure to the same fluid shear stress.
Our recent studies have revealed two main regulators on either end of the actin-cap fibers that are critically required for shear-induced actin cap formation: focal adhesion protein zyxin (but not FAK and vinculin) in ACAFAs, lamin A/C, the LINC complex protein nesprin3, and to a lesser extent the LINC complex protein nesprin2giant (right panel, Fig. 4). The important role of these specific proteins in providing a cellular response to external shear stresses is consistent with the requirement of zyxin for chemokines interleukin-8 and CXCL1 to be upregulated after mechanical stretching of human primary endothelial cells53. Zyxin is mobilized from focal adhesions to actin filaments in response to cyclic stretching or shear stress, while FAK and other focal adhesion components remain at focal adhesion sites73. Nesprin3 is required for proper flow-induced MTOC re-positioning and polarization in human aortic endothelial cells74. Moreover, disruption of the LINC complex by way of nesprin-displacing dominant negative KASH2 or SUN1 constructs cause fibroblasts to lessen their displacement in response to microneedle manipulation of the cytoskeleton75. Together, these results suggest that the actin cap provides a completely continuous linkage between the extracellular milieu and the nucleus for effective and fast mechanotransduction.
Studies of the actin cap in mechanotransduction have predominantly been performed in isolated cells. For cardiovascular function and diseases, in particular, it remains to be determined how the actin cap organizes and functions in confluent monolayers of endothelial cells exposed to shear flow forces. Moreover, where actin-cap fibers are nucleated remains unknown: Actin-cap fibers could be formed at the nuclear surface or at focal adhesions. How the formation of the actin cap affects chromosomal organization, epigenetic modifications, and gene expression is currently under study.
ACKNOWLEDGEMENT
Authors thank Prof. Didier Hodzic and Prof. Gregory D. Longmore of the Washington University in St. Louis School of Medicine for useful discussions on the topics discussed in this review. Work in the Wirtz lab is funded by the NIH (grants R01GM084204 and U54CA143868). ABC is supported by an Achievement Rewards for College Scientists (ARCS) Foundation fellowship.
REFERENCES
- 1.Wirtz D, Konstantopoulos K, Searson PC. Nature reviews. Cancer. 2011;11:512–522. doi: 10.1038/nrc3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wirtz D. Annual review of biophysics. 2009;38:301–326. doi: 10.1146/annurev.biophys.050708.133724. [DOI] [PubMed] [Google Scholar]
- 3.Kole TP, Tseng Y, Jiang I, Katz JL, Wirtz D. Molecular biology of the cell. 2005;16:328–338. doi: 10.1091/mbc.E04-06-0485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tseng Y, Wirtz D. Biophysical journal. 2001;81:1643–1656. doi: 10.1016/S0006-3495(01)75818-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tseng Y, Schafer BW, Almo SC, Wirtz D. The Journal of biological chemistry. 2002;277:25609–25616. doi: 10.1074/jbc.M202609200. [DOI] [PubMed] [Google Scholar]
- 6.Tseng Y, Wirtz D. Physical review letters. 2004;93:258104. doi: 10.1103/PhysRevLett.93.258104. [DOI] [PubMed] [Google Scholar]
- 7.Kabsch W, Mannherz HG, Suck D, Pai EF, Holmes KC. Nature. 1990;347:37–44. doi: 10.1038/347037a0. [DOI] [PubMed] [Google Scholar]
- 8.Holmes KC, Popp D, Gebhard W, Kabsch W. Nature. 1990;347:44–49. doi: 10.1038/347044a0. [DOI] [PubMed] [Google Scholar]
- 9.Borisy GG, Svitkina TM. Current opinion in cell biology. 2000;12:104–112. doi: 10.1016/s0955-0674(99)00063-0. [DOI] [PubMed] [Google Scholar]
- 10.Pollard TD, Borisy GG. Cell. 2003;112:453–465. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- 11.Bousquet O, Ma L, Yamada S, Gu C, Idei T, Takahashi K, Wirtz D, Coulombe PA. The Journal of cell biology. 2001;155:747–754. doi: 10.1083/jcb.200104063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ma L, Yamada S, Wirtz D, Coulombe PA. Nature cell biology. 2001;3:503–506. doi: 10.1038/35074576. [DOI] [PubMed] [Google Scholar]
- 13.Fudge DS, Gardner KH, Forsyth VT, Riekel C, Gosline JM. Biophysical journal. 2003;85:2015–2027. doi: 10.1016/S0006-3495(03)74629-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kreplak L, Fudge D. BioEssays : news and reviews in molecular, cellular and developmental biology. 2007;29:26–35. doi: 10.1002/bies.20514. [DOI] [PubMed] [Google Scholar]
- 15.Goldman RD, Cleland MM, Murthy SNP, Mahammad S, Kuczmarski ER. J Struct Biol. 2012;177:14–23. doi: 10.1016/j.jsb.2011.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wacker I, Kaether C, Kromer A, Migala A, Almers W, Gerdes HH. Journal of cell science. 1997;110(Pt 13):1453–1463. doi: 10.1242/jcs.110.13.1453. [DOI] [PubMed] [Google Scholar]
- 17.Hamm-Alvarez SF, Sheetz MP. Physiological reviews. 1998;78:1109–1129. doi: 10.1152/physrev.1998.78.4.1109. [DOI] [PubMed] [Google Scholar]
- 18.Hale CM, Chen WC, Khatau SB, Daniels BR, Lee JS, Wirtz D. Journal of cell science. 2011;124:4267–4285. doi: 10.1242/jcs.091231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee JS, Chang MI, Tseng Y, Wirtz D. Molecular biology of the cell. 2005;16:871–880. doi: 10.1091/mbc.E03-12-0910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khatau SB, Hale CM, Stewart-Hutchinson PJ, Patel MS, Stewart CL, Searson PC, Hodzic D, Wirtz D. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:19017–19022. doi: 10.1073/pnas.0908686106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hotulainen P, Llano O, Smirnov S, Tanhuanpaa K, Faix J, Rivera C, Lappalainen P. Journal of Cell Biology. 2009;185:323–339. doi: 10.1083/jcb.200809046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tojkander S, Gateva G, Lappalainen P. Journal of cell science. 2012;125:1855–1864. doi: 10.1242/jcs.098087. [DOI] [PubMed] [Google Scholar]
- 23.Khatau SB, Kusuma S, Hanjaya-Putra D, Mali P, Cheng L, Lee JS, Gerecht S, Wirtz D. PloS one. 2012;7:e36689. doi: 10.1371/journal.pone.0036689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim DH, Khatau SB, Feng Y, Walcott S, Sun SX, Longmore GD, Wirtz D. Scientific reports. 2012;2:555. doi: 10.1038/srep00555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gay O, Gilquin B, Nakamura F, Jenkins ZA, McCartney R, Krakow D, Deshiere A, Assard N, Hartwig JH, Robertson SP, Baudier J. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:11464–11469. doi: 10.1073/pnas.1104211108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Capell BC, Collins FS. Nature reviews. Genetics. 2006;7:940–952. doi: 10.1038/nrg1906. [DOI] [PubMed] [Google Scholar]
- 27.Worman HJ. The Journal of pathology. 2012;226:316–325. doi: 10.1002/path.2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khatau SB, Kim DH, Hale CM, Bloom RJ, Wirtz D. Nucleus. 2010;1:337–342. doi: 10.4161/nucl.1.4.12331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rothballer A, Schwartz TU, Kutay U. Nucleus. 2013;4:29–36. doi: 10.4161/nucl.23387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Libotte T, Zaim H, Abraham S, Padmakumar VC, Schneider M, Lu W, Munck M, Hutchison C, Wehnert M, Fahrenkrog B, Sauder U, Aebi U, Noegel AA, Karakesisoglou I. Molecular biology of the cell. 2005;16:3411–3424. doi: 10.1091/mbc.E04-11-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhen YY, Libotte T, Munck M, Noegel AA, Korenbaum E. Journal of cell science. 2002;115:3207–3222. doi: 10.1242/jcs.115.15.3207. [DOI] [PubMed] [Google Scholar]
- 32.Ketema M, Wilhelmsen K, Kuikman I, Janssen H, Hodzic D, Sonnenberg A. Journal of cell science. 2007;120:3384–3394. doi: 10.1242/jcs.014191. [DOI] [PubMed] [Google Scholar]
- 33.Wilhelmsen K, Litjens SH, Kuikman I, Tshimbalanga N, Janssen H, van den Bout I, Raymond K, Sonnenberg A. The Journal of cell biology. 2005;171:799–810. doi: 10.1083/jcb.200506083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Crisp M, Liu Q, Roux K, Rattner JB, Shanahan C, Burke B, Stahl PD, Hodzic D. The Journal of cell biology. 2006;172:41–53. doi: 10.1083/jcb.200509124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stewart-Hutchinson PJ, Hale CM, Wirtz D, Hodzic D. Experimental cell research. 2008;314:1892–1905. doi: 10.1016/j.yexcr.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shoeman RL, Traub P. The Journal of biological chemistry. 1990;265:9055–9061. [PubMed] [Google Scholar]
- 37.Lee JSH, Chang MI, Tseng Y, Wirtz D. Molecular biology of the cell. 2005;16:871–880. doi: 10.1091/mbc.E03-12-0910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mounkes LC, Kozlov S, Hernandez L, Sullivan T, Stewart CL. Nature. 2003;423:298–301. doi: 10.1038/nature01631. [DOI] [PubMed] [Google Scholar]
- 39.Engler AJ, Sen S, Sweeney HL, Discher DE. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 40.Lanniel M, Huq E, Allen S, Buttery L, Williams PM, Alexander MR. Soft Matter. 2011;7:6501–6514. [Google Scholar]
- 41.Levental KR, Yu H, Kass L, Lakins JN, Egeblad M, Erler JT, Fong SF, Csiszar K, Giaccia A, Weninger W, Yamauchi M, Gasser DL, Weaver VM. Cell. 2009;139:891–906. doi: 10.1016/j.cell.2009.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gilkes DM, Bajpai S, Chaturvedi P, Wirtz D, Semenza GL. The Journal of biological chemistry. 2013 doi: 10.1074/jbc.M112.442939. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 43.Wong CC, Gilkes DM, Zhang H, Chen J, Wei H, Chaturvedi P, Fraley SI, Wong CM, Khoo US, Ng IO, Wirtz D, Semenza GL. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:16369–16374. doi: 10.1073/pnas.1113483108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Trappmann B, Gautrot JE, Connelly JT, Strange DGT, Li Y, Oyen ML, Stuart MAC, Boehm H, Li BJ, Vogel V, Spatz JP, Watt FM, Huck WTS. Nat Mater. 2012;11:642–649. doi: 10.1038/nmat3339. [DOI] [PubMed] [Google Scholar]
- 45.Ingber DE. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2006;20:811–827. doi: 10.1096/fj.05-5424rev. [DOI] [PubMed] [Google Scholar]
- 46.Malek AM, Alper SL, Izumo S. JAMA : the journal of the American Medical Association. 1999;282:2035–2042. doi: 10.1001/jama.282.21.2035. [DOI] [PubMed] [Google Scholar]
- 47.Obi S, Yamamoto K, Shimizu N, Kumagaya S, Masumura T, Sokabe T, Asahara T, Ando J. J Appl Physiol. 2009;106:203–211. doi: 10.1152/japplphysiol.00197.2008. [DOI] [PubMed] [Google Scholar]
- 48.Burge R, Dawson-Hughes B, Solomon DH, Wong JB, King A, Tosteson A. J Bone Miner Res. 2007;22:465–475. doi: 10.1359/jbmr.061113. [DOI] [PubMed] [Google Scholar]
- 49.Malone AM, Anderson CT, Tummala P, Kwon RY, Johnston TR, Stearns T, Jacobs CR. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:13325–13330. doi: 10.1073/pnas.0700636104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Temiyasathit S, Tang WJ, Leucht P, Anderson CT, Monica SD, Castillo AB, Helms JA, Stearns T, Jacobs CR. PloS one. 2012;7 doi: 10.1371/journal.pone.0033368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Morris CE. The Journal of membrane biology. 1990;113:93–107. doi: 10.1007/BF01872883. [DOI] [PubMed] [Google Scholar]
- 52.Cattaruzza M, Lattrich C, Hecker M. Hypertension. 2004;43:726–730. doi: 10.1161/01.HYP.0000119189.82659.52. [DOI] [PubMed] [Google Scholar]
- 53.Wojtowicz A, Babu SS, Li L, Gretz N, Hecker M, Cattaruzza M. Circulation research. 2010;107:898–902. doi: 10.1161/CIRCRESAHA.110.227850. [DOI] [PubMed] [Google Scholar]
- 54.Geiger B, Spatz JP, Bershadsky AD. Nat Rev Mol Cell Bio. 2009;10:21–33. doi: 10.1038/nrm2593. [DOI] [PubMed] [Google Scholar]
- 55.Fraley SI, Feng Y, Krishnamurthy R, Kim DH, Celedon A, Longmore GD, Wirtz D. Nature cell biology. 2010;12:598–604. doi: 10.1038/ncb2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kanchanawong P, Shtengel G, Pasapera AM, Ramko EB, Davidson MW, Hess HF, Waterman CM. Nature. 2010;468:580–584. doi: 10.1038/nature09621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kim DH, Wirtz D. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2013;27:1351–1361. doi: 10.1096/fj.12-220160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Walcott S, Kim DH, Wirtz D, Sun SX. Biophysical journal. 2011;101:2919–2928. doi: 10.1016/j.bpj.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Patla I, Volberg T, Elad N, Hirschfeld-Warneken V, Grashoff C, Fassler R, Spatz JP, Geiger B, Medalia O. Nature cell biology. 2010;12:909–915. doi: 10.1038/ncb2095. [DOI] [PubMed] [Google Scholar]
- 60.Janmey PA, McCulloch CA. Annual review of biomedical engineering. 2007;9:1–34. doi: 10.1146/annurev.bioeng.9.060906.151927. [DOI] [PubMed] [Google Scholar]
- 61.Schwarz US, Gardel ML. Journal of cell science. 2012;125:3051–3060. doi: 10.1242/jcs.093716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Riveline D, Zamir E, Balaban NQ, Schwarz US, Ishizaki T, Narumiya S, Kam Z, Geiger B, Bershadsky AD. Journal of Cell Biology. 2001;153:1175–1185. doi: 10.1083/jcb.153.6.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Buxboim A, Ivanovska IL, Discher DE. Journal of cell science. 2010;123:297–308. doi: 10.1242/jcs.041186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang HB, Dembo M, Hanks SK, Wang Y. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:11295–11300. doi: 10.1073/pnas.201201198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Peyton SR, Kim PD, Ghajar CM, Seliktar D, Putnam AJ. Biomaterials. 2008;29:2597–2607. doi: 10.1016/j.biomaterials.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Alexander NR, Branch KM, Parekh A, Clark ES, Iwueke IC, Guelcher SA, Weaver AM. Current biology : CB. 2008;18:1295–1299. doi: 10.1016/j.cub.2008.07.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Colombelli J, Besser A, Kress H, Reynaud EG, Girard P, Caussinus E, Haselmann U, Small JV, Schwarz US, Stelzer EH. Journal of cell science. 2009;122:1665–1679. doi: 10.1242/jcs.042986. [DOI] [PubMed] [Google Scholar]
- 68.Michael KE, Dumbauld DW, Burns KL, Hanks SK, Garcia AJ. Molecular biology of the cell. 2009;20:2508–2519. doi: 10.1091/mbc.E08-01-0076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pasapera AM, Schneider IC, Rericha E, Schlaepfer DD, Waterman CM. The Journal of cell biology. 2010;188:877–890. doi: 10.1083/jcb.200906012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Prager-Khoutorsky M, Lichtenstein A, Krishnan R, Rajendran K, Mayo A, Kam Z, Geiger B, Bershadsky AD. Nature cell biology. 2011;13:1457–1465. doi: 10.1038/ncb2370. [DOI] [PubMed] [Google Scholar]
- 71.Trichet L, Le Digabel J, Hawkins RJ, Vedula SR, Gupta M, Ribrault C, Hersen P, Voituriez R, Ladoux B. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:6933–6938. doi: 10.1073/pnas.1117810109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chambliss AB, Khatau SB, Erdenberger N, Robinson DK, Hodzic D, Longmore GD, Wirtz D. Scientific reports. 2013;3:1087. doi: 10.1038/srep01087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yoshigi M, Hoffman LM, Jensen CC, Yost HJ, Beckerle MC. The Journal of cell biology. 2005;171:209–215. doi: 10.1083/jcb.200505018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Morgan JT, Pfeiffer ER, Thirkill TL, Kumar P, Peng G, Fridolfsson HN, Douglas GC, Starr DA, Barakat AI. Molecular biology of the cell. 2011;22:4324–4334. doi: 10.1091/mbc.E11-04-0287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lombardi ML, Jaalouk DE, Shanahan CM, Burke B, Roux KJ, Lammerding J. The Journal of biological chemistry. 2011;286:26743–26753. doi: 10.1074/jbc.M111.233700. [DOI] [PMC free article] [PubMed] [Google Scholar]