Abstract
We present a simple, yet flexible microfluidic mixer with a demonstrated mixing time as short as 80 µs that is widely accessible because it is made of commercially available parts. To simplify the study of fast protein dynamics, we have developed an inexpensive continuous-flow microfluidic mixer, requiring no specialized equipment or techniques. The mixer uses three-dimensional, hydrodynamic focusing of a protein sample stream by a surrounding sheath solution to achieve rapid diffusional mixing between the sample and sheath. Mixing initiates the reaction of interest. Reactions can be spatially observed by fluorescence or absorbance spectroscopy. We characterized the pixel-to-time calibration and diffusional mixing experimentally. We achieved a mixing time as short as 80 µs. We studied the kinetics of horse apomyoglobin (apoMb) unfolding from the intermediate (I) state to its completely unfolded (U) state, induced by a pH jump from the initial pH of 4.5 in the sample stream to a final pH of 2.0 in the sheath solution. The reaction time was probed using the fluorescence of 1-anilinonapthalene-8-sulfonate (1,8-ANS) bound to the folded protein. We observed unfolding of apoMb within 760 µs, without populating additional intermediate states under these conditions. We also studied the reaction kinetics of the conversion of pyruvate to lactate catalyzed by lactate dehydrogenase using the intrinsic tryptophan emission of the enzyme. We observe sub-millisecond kinetics that we attribute to Michaelis complex formation and loop domain closure. These results demonstrate the utility of the three-dimensional focusing mixer for biophysical studies of protein dynamics.
Introduction
The study of protein dynamics in folding and enzyme reactions has been hindered by the time limit of conventional mixing methods for initiating these processes, some of which are intrinsically fast. For instance, the development of secondary and tertiary structure during protein folding may take place in microseconds or even nanoseconds.1 Fast processes in enzymatic reactions, which include proton or hydride transfer and conformational rearrangements, occur on the microsecond to millisecond timescale under common experimental conditions.2 In order to observe such rapid events, new experimental techniques capable of initiating and monitoring protein dynamics on the same or even shorter timescales have been developed. Some of these techniques include temperature jump, photochemical triggering, and ultra-rapid mixing of fluid samples.1, 3 Fast mixers have become particularly useful in the study of protein dynamics because they can quickly introduce a large perturbation in the solution environment (e.g. pH or denaturant concentration) to trigger folding or an enzymatic reaction. Furthermore, relaxation methods that modulate intensive properties such as temperature or pressure usually do not perturb the system very far from equilibrium. In contrast, with mixing methods it is possible to initiate a reaction very far from equilibrium, for example refolding from a completely unfolded state under conditions that strongly favor folding. Such experiments are useful because they access different regions of the protein energy landscape.4, 5
Several groups have studied protein folding dynamics using fast mixers in conjunction with different optical techniques, and they report folding rates in agreement with those determined using other experimental techniques.6–8 Unfortunately, these methods have not been widely adopted despite their demonstrated utility, because of the complexity and expense of the mixing devices and difficulty in applying these devices to the study of protein dynamics. Improving the performance of fast mixers remains an active area of research. 8–15 The three most important criteria for evaluating the performance of fast mixers are: 1) mixing time, 2) functionality and 3) flexibility (compatibility with multiple spectroscopic probes). Mixing time is important because it limits the timescale on which events can be observed. Therefore it has been a major focus of new fast mixer development with a push towards accessing much shorter times than is possible with conventional stop-flow systems. However, it is also important that mixers are developed so that they are functional, which we define as reliable and widely accessible in terms of cost, and simplicity of construction. This concern is particularly relevant as the size of mixers shrinks and clogging becomes more frequent, necessitating replacement of the mixer. Cost and accessibility are significant drawbacks to mixers constructed by lithography or with expensive materials like silicon. Finally, mixers should be flexible in design, such that they are compatible with a range of spectroscopic techniques and wavelengths, since not all phenomena can be well studied by visible fluorescence excitation and emission. Mixer material and geometry must be considered from this perspective as not all common materials (e.g. polydimethylsiloxane) are UV transparent and not all designs allow for probe light to transverse the entire sample region. We review below the state of the field in terms of these criteria to establish context for our mixer’s design and performance.
Microfluidic devices with observed mixing times on the order of 50 µs10, 16, 17 and theoretical mixing times as low as 1 µs12 have been reported. These short mixing times have been achieved with laminar-flow devices employing hydrodynamic focusing. An emphasis of current efforts to improve mixer performance is to use three-dimensional (3D), rather than two-dimensional (2D), hydrodynamic focusing.8–10, 18–21 Diffusional mixing in a 3D focused sample stream (cylindrical) is inherently more efficient than a 2D one (slab), because the ratio of sample volume to surface area in contact with the sheath is a factor of two greater for the case of the 2D focused slab compared to the 3D focused cylinder. Furthermore, the 2D focused slab geometry leaves part of the sample stream in contact with the walls of the mixing device, causing this part to travel more slowly than the region of the sample not contacting the walls. This creates a gradient of flow velocities within the sample stream, leading to blurring of the measurement and uncertainty in the mixing time. Confocal fluorescence detection of the central part of the sample stream circumvents this problem, but it is unavoidable with any technique such as absorbance that probes the full width of the sample stream. In contrast, 3D-focusing devices surround the sample on all sides with the sheath solution so that the entire sample stream flows with uniform velocity and faster mixing is achieved. The full potential of 3D focusing has not yet been realized in existing devices, which have mixing times of hundreds of µs. Silicon-based 3D mixers are further limited in their optical transparency since only one side of the mixer admits light; therefore, it is difficult to monitor reaction progress using absorbance spectroscopy.22 In addition, many 3D mixers are often complicated to construct, and not all of them have been tested for compatibility with a biological system.
We have designed a 3D focusing microfluidic mixer with an emphasis on improving the functionality and spectroscopic flexibility while maintaining a biophysically relevant mixing time. Most notably, the mixer design is highly accessible to the biophysical chemistry community due to its simple construction from commercially available components. The mixer is assembled by hand with fused silica capillaries, standard plastic fittings, and plastic tubing, all of which are inexpensive and readily available. Because the capillaries are made of fused silica, the mixer is transparent to probe wavelengths from the ultraviolet to the near-infrared. This makes the mixer compatible with a variety of different optical spectroscopy techniques. We emphasize fluorescence detection, but Raman and UV-visible absorption spectroscopy are also compatible with this device.14 Furthermore, the simple construction of the mixer makes clogs easy to eliminate, since the mixer parts can be quickly disassembled and reassembled to remove any obstruction to solution flow. Finally, the mixer is compatible with deep UV excitation and therefore with direct excitation of intrinsic fluorophores such as tryptophan, eliminating the need for labeling with extrinsic probes. Other laminar flow mixers have not been able to access this important spectral region, although there is one report of a unique turbulent flow design that uses Trp fluorescence as a probe.23
We have demonstrated the capability of this fast mixer by addressing two important biophysical problems. First, we studied the unfolding kinetics of horse apomyoglobin (apoMb), an important model system for fundamental protein folding studies because of its simple structure and fast folding kinetics. It is postulated to fold through a series of progressively more structured intermediates: the unfolded form (U), the extended form (E), the intermediate form (I), and the native form (N).1, 24–31 A major issue in the study of protein folding mechanisms is the question of whether such intermediate states are obligatory or if they represent unproductive states that can lead to misfolding and pathology. For this study, we induced unfolding of apoMb from its I to U form at low salt concentration, via a rapid pH drop within our mixer. We observe direct conversion of I to U without population of intermediate states under these conditions. This result is in contrast to T-jump measurements and turbulent flow mixing experiments at higher pH (4.2) that show population of intermediates on the unfolding pathway.31, 32 We conclude that the rapid pH jump achieved in the continuous flow mixing experiment accesses part of the unfolding energy landscape not previously observed. Second, we studied the enzyme dynamics in the conversion of pyruvate to lactate catalyzed by lactate dehydrogenase (LDH). The unique optical properties of the fast mixer and imaging system allow us to follow the submillisecond kinetics of this reaction using the intrinsic tryptophan fluorescence of LDH. Thus we have observed the conformational changes of the enzyme associated with catalysis, including loop closure, structural changes within the Michaelis complex and the chemistry step. These results clearly demonstrate the effectiveness of the mixer for the study of fast protein dynamics.
EXPERIMENTAL
Mixer Construction
The mixer is constructed from two concentric cylindrical capillaries and achieves uniform 3D focusing by completely surrounding a sample stream held in the inner capillary with a flowing sheath solution held in the outer capillary. This sheath solution focuses the sample hydrodynamically into a narrow jet so that rapid diffusional mixing between the solutions is initiated. Since the protein diffusion is slow, it remains in the central sample stream, whereas substrates within the sheath solution diffuse into the protein stream, initiating the reaction. Our design is related to that developed by Pabit and Hagen for fluorescence kinetics studies.9 However, our mixer produces better focusing of the sample stream and therefore shortens mixing times through the use of a tapered inner capillary and a cylindrical, rather than rectangular, outer capillary. While Pabit and Hagen report a mixing time of approximately 425 µs, we observed complete mixing in under 100 µs.
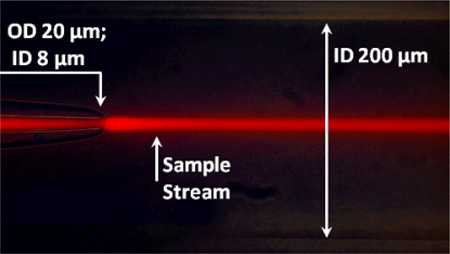
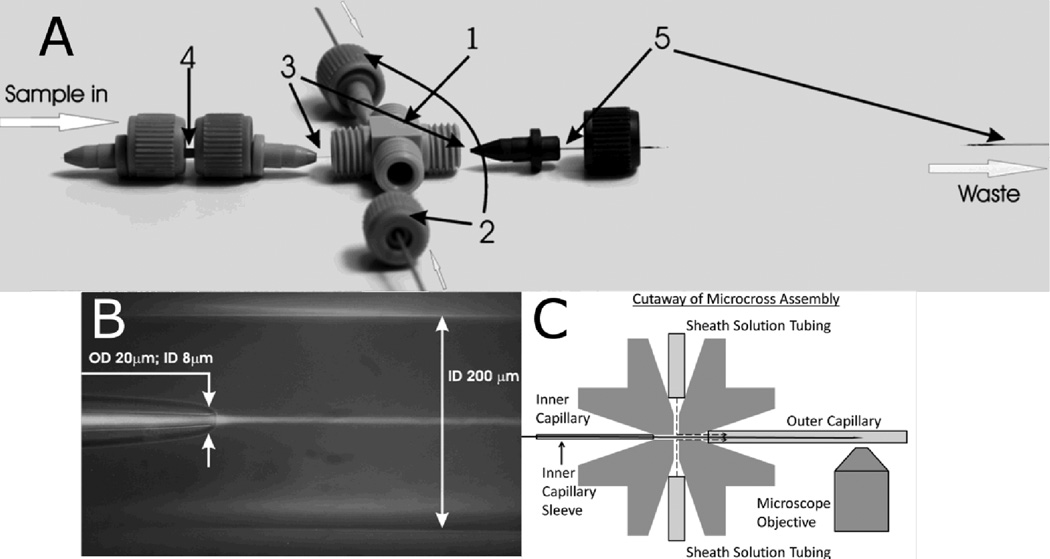
The mixer is made with standard plastic parts and PEEK tubing supplied by Idex (Oak Harbor, WA) and fused silica capillaries supplied by Polymicro Technologies (Phoenix, AZ) (Figure 1A). The mixer contains an inner and outer capillary that carry the sample and sheath solutions, respectively. The outer capillary has a 350-µm outer diameter (OD) and 200-µm inner diameter (ID), and the inner capillary has a 90-µm OD and 20-µm ID before its narrowed tip. The outer and inner diameters at the tip of the inner capillary were reduced at the outlet to 20 µm and 8 µm, respectively (Figure 1B). This tapered tip was produced by a simple flame-pulling method. First, the capillary was attached to a plastic 50-mL cylindrical tube filled with 45 mL of water, approximately 57 g total mass. Second, the capillary was suspended from a support stand with the tube hanging off the stand to provide a pulling force on the capillary. Finally, the suspended capillary was melted with a propane pencil flame burner torch until the force of the suspended tube separated the capillary into two segments, each having a narrowed tip where the flame had been applied. The characteristics of the tapered outlet are easily tuned by adjusting the weight of the “puller”, i.e. by changing the volume of water in the suspended tube; decreasing the volume of water produces smaller outlet diameters and vice versa.
Figure 1.
(A) Mixer design. The components include: (1) MicroCross that holds 360 µm outer diameter tubing and has a 150-µm through-hole (2) 1/32” PEEK tubing that has a 250 µm inner diameter and carries sheath flow from HPLC pumps (3) Fused silica capillary for sample flow: outer diameter = 90µm; inner diameter = 20µm (4) MicroTight tubing sleeve (5) Fused silica capillary for sheath flow: outer diameter = 350 µm; inner diameter = 200 µm. All parts are connected via fitting nuts and MicroFerrules with 0.025” diameters. (B) Overlay of bright field and fluorescence images of capillary flow system. The sample flows from left to right through the inner capillary, and the sheath solution flows from left to right through the outer capillary, with dimensions as indicated. The tapered capillary tip and 3D focusing by the sheath solution create a narrow sample jet that is 2–5 µm in diameter. A laminar flow of the sample is established and rapid mixing occurs via diffusion. The sample appears as a bright stream due to fluorescence of Eu beads. The sample flow rate is 0.025 µL/min, and the sheath flow rate is 100 µL/min. (C) Cutaway of MicroCross assembly showing how solutions are kept separate (not to scale). Ferrules (not shown) hold each tube or capillary in place. The sheath solution can only flow out through the outer capillary, whereas the inner capillary outlet is outside of the cross and therefore the mixing region is outside of the cross, in the vicinity of the microscope objective.
This simple flame-pulling method produces tapered tips of highly reproducible dimensions. We confirmed the uniformity of the tips by measuring the average tip diameter using the calibrated imaging microscope (1 pixel = 0.37 µm). A typical batch of 12 tips gave an average tip size of 7.4 µm with a standard deviation of 0.74 µm (approaching the uncertainty of the measurement itself). Furthermore, we determined the error in the pixel to time calibration introduced by different tips using the Eu nanosphere method described below (Figure 2). We found that the uncertainty in this calibration introduced by different tips is less than the overall uncertainty of the calibration (see Table 2 of Supporting Information).
Figure 2.
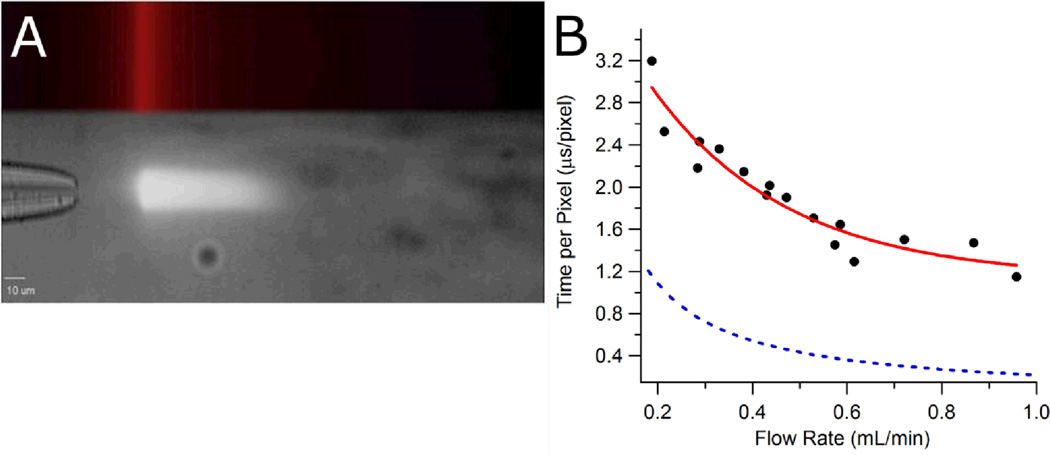
(A) Single-point laser excitation of Eu nanospheres flowing through the mixer. The top part of the image shows the excitation laser position and profile. The laser is focused to a narrow line perpendicular to the axis of sample flow. The bottom part of the image shows the nanosphere fluorescence as a white streak downstream of the line-focused laser. The nanospheres exit the inner capillary and are excited at the position of the focused laser, then as they move away from the excitation point, the intensity of their fluorescence decays at a known rate. (B) Mixer time resolution at various sheath flow rates. Experimental data (black circles) were fit to a single exponential function (red line): τ = 1.1 + 3.5e −3.5v. Also shown is the theoretical time resolution (blue dashes) based on Equation 2.
The pulled inner capillary was placed in an Idex MicroTight tubing sleeve to maintain its position within the mixing system. The inner capillary was then inserted into a MicroCross, which directed the inner capillary through the center of the outer capillary (Figure 1C). Both capillaries were secured to the MicroCross via 0.025” -diameter MicroFerrules and fitting nuts.
The sample and sheath solutions were delivered separately to the inner and outer capillaries, respectively. The sample solution was held in a 1-mL Tuberculin (Franklin Lakes, NJ) plastic syringe and was flowed by a KD Scientific (Holliston, MA) continuous cycle syringe pump through 1/32” PEEK tubing to reach the inner capillary. The sample was filtered with a 0.5-µm stainless steel frit in-line syringe filter from Idex. The sheath solution was flowed by two HPLC pumps, Rainin SD-200 (Oakland, CA), through 1/32” PEEK tubing channels that were directed into opposite sides of the MicroCross to bring the sheath solution into the outer capillary. The two pumps were operated in asynchronous cycles to minimize pulsing of the flow due to the nonuniform pumping cycles of each pump.
The mixer was mounted onto an Applied Scientific Instrumentation (Eugene, OR) MS-2000 automated stage using Newport (Irvine, CA) miniature optical rails and lens mounts. A 3-in rail and a 6-in rail were attached at their ends. The 3-in rail was fixed to the stage, while the 6-in rail was placed over the objectives. Two fixed-position lens mounts were attached to the 6-in rail, and the capillary system was secured within these lens mounts. The stage was attached to an Olympus (Center Valley, PA) IX81 microscope, and the capillary system was rotated around the connection between the 3-in and 6-in rails to place the mixer directly into the microscope objective’s image plane. Microscope images were collected on a Hamamatsu (Bridgewater, NJ) C9100-14 ImageM-1k EM-CCD Camera. Integration time of the camera was varied from 100 to 1000 ms depending on the intensity of the sample fluorescence. The microscope and camera were controlled by Slidebook 5.0 software from Intelligent Imaging Innovations, Inc. (Denver, CO). Images were exported as TIFF files into Igor Pro software from WaveMetrics (Portland, OR) for analysis.
Sample Preparation
Three different sample solutions were used in the mixer: Eu nanospheres, fluorescein, and apoMb. Before use, all sample solutions were filtered with 0.2-µm PTFE filters supplied by Nalgene (Rochester, NY). All sheath solutions were filtered with 0.22-µm PES filters supplied by Corning (Lowell, MA). Details of each sample preparation are given below.
Europium nanospheres
Europium (Eu) carboxylate-modified nanospheres 40 nm in size were purchased from Invitrogen (Grand Island, NY) and used without further modification.
Fluorescein
Fluorescein sodium salt was purchased from Sigma-Aldrich (St. Louis, MO). A solution of fluorescein was prepared by dissolving it in deionized (DI) water and then adding NaOH to the solution to reach a pH of approximately 8. The concentration of fluorescein was determined using the extinction coefficient of 80,000 M−1cm−1 at 488 nm.33
Apomyoglobin
Equine skeletal muscle myoglobin was purchased from Sigma-Aldrich. A solution containing 5 mg of protein/mL of deionized water was prepared. Myoglobin was precipitated from this solution by adding ammonium sulfate to 68% saturation at room temperature. The sample was dialyzed overnight against deionized water using regenerated cellulose dialysis tubing by Fisherbrand (Pittsburg, PA). ApoMb was prepared from this sample according to a modification of the procedure of Adams.34 In this modification, the diethyl ketone extraction and dialysis were performed twice to improve sample purity. In addition, apoMb was dialyzed against deionized water only. The concentration of apoMb was determined using the extinction coefficient of 15,570 M−1cm−1 at 280 nm.34
Fluorescence detection of apoMb unfolding was accomplished using the fluoresent probe 1,8-ANS, which was also purchased from Sigma-Aldrich. A solution of 1,8-ANS was prepared by dissolving the compound in DI water. The 1,8-ANS solution was added to apoMb to make samples with a 1,8-ANS: apoMb concentration ratio of approximately 1:1. The concentration of 1,8-ANS was determined using the extinction coefficient of 5000 M−1cm−1 at 350 nm.35
Lactate Dehydrogenase
Porcine heart LDH was purchased from EMD Chemicals (San Diego, CA). NADH and pyruvate were purchased from Sigma-Aldrich (St. Louis, MO). Excitation of tryptophan was from a frequency tripled Ti:Sapphire laser centered at 850 nm. Excitation light was filtered with a band pass filter centered at 280 nm. Emission was filtered with a band-pass centered at 357 nm.
Experimental Design and Procedures
All filters, lenses, and mirrors described in the optical set-ups of the experiments below were purchased from Semrock (Rochester, NY). Objectives used in these studies are from Olympus (Center Valley, PA). The objectives were 40X water immersion (UApoN340), 60X water immersion (UPlanSApo), and 100X oil immersion (UPlanSApo).
Pixel-to-Time Calibration
The amount of time represented by each pixel in fluorescence images of the sample flow was determined by observing the fluorescence decay of Eu nanospheres within the mixer. The fluorescence lifetime of the Eu nanospheres was measured separately using a time-resolved fluorescence instrument. Dividing the measured lifetime of the Eu nanosphere fluorescence by the observed decay in pixels along an image collected under the microscope gave a pixel-to-time conversion factor for the mixer at a given sample and sheath flow rate. This conversion factor represents the time resolution of the mixer. The fluorescence decay time of the Eu nanospheres was measured using a pulsed laser based fluorescence lifetime instrument. A sample of Eu nanospheres in a 1-cm pathlength quartz fluorescence cuvette, Model 29F-Q-10, from Starna Cells (Atascadero, CA) was excited with a 2 ns pulse at 351 nm from a diode-pumped, Q-switched Nd:YLF laser, Model: QUV-351-100, from CrystaLaser (Reno, NV) with a repetition rate of 1 kHz. The Eu fluorescence was collected perpendicular to the incident beam and focused onto a SPEX 1681A monochromator (Spex Industries, Edison, NJ) set to pass 610 nm light with an input slit width of 0.58 mm and output slit width of 0.62 mm. A Hamamatsu R7518 photomultiplier tube (PMT) was attached to the monochromator output slit for detection of the fluorescence signal. The PMT signal was terminated at 50 Ω into a Lecroy (Chesnut Ridge, NY) WaveSurfer 62 Xs-A digital oscilloscope. The incident light that passed through the sample was collected on a ThorLabs (Newton, NJ) DET10A photodiode, terminated at 50 Ω into the same oscilloscope, and used as the trigger source for data collection. The recorded transients had a time resolution of 2 ns. Signal averaging of 1000 transients produced the fluorescence decay used for the lifetime analysis.
The mixer was calibrated by flowing a sample of Eu nanospheres through a sheath solution of DI water. The Eu nanospheres were flowed at a constant rate of 0.1 µL/min, while the DI water sheath was flowed at rates ranging from 0.2 to 1.0 mL/min. A cw UV diode laser emitting at 375 nm (Newport, Irvine, CA Model LQC375-16C) was used to excite the sample of flowing Eu nanospheres with less than 1 mW average power. The beam was shaped into a narrow line with a cylindrical lens and a slit and then directed into a side port of the microscope to ultimately illuminate the objective. The laser beam is focused by the objective into the central sample stream after it exits the inner capillary, and the laser excites the Eu nanospheres flowing through the mixer at a single point along their trajectory (Figure 2A). The fluorescence of the excited nanospheres is collected back through the objective, filtered for Eu emission at 610 nm, and directed to the camera. All calibration experiments were performed using the 100X oil objective.
Diffusional Mixing Time Calibration
The time required for diffusional mixing of the sheath solution with the sample stream was determined by observing the quenching of fluorescein fluorescence by potassium iodide (KI). The mixing time was determined by flowing a fluorescein sample stream through a sheath solution of KI and measuring the time required for the iodide to diffuse into the sample stream and quench the fluorescence. Fluorescein was excited at 488 nm using the filtered output of a HBO mercury short-arc lamp from Osram (Danvers, MA). Emission from the sample was transmitted through a long-pass filter and directed to the camera. All fluorescein and KI quenching experiments were performed using the 60X water objective.
ApoMb Unfolding
Unfolding of apoMb was induced by a jump to lower pH, accomplished by flowing the folded protein sample stream through a low pH sheath solution. Thus, folded apoMb at a pH of 4.5 was flowed through a sheath solution of HCl at a pH of 2 to initiate the unfolding reaction. A control study was also performed by flowing apoMb through DI water. The sample flow rate remained at 0.1 µL/min, and the sheath flow rate was varied from 0.5 to 1.4 mL/min.
The apoMb unfolding kinetics was observed by monitoring the fluorescent probe bound to the folded protein. The 1,8-ANS fluorescence was excited using light from the mercury lamp filtered through a band-pass filter centered at 405 nm. Fluorescence from the 1,8-ANS probe was filtered with a long-pass filter to block stray light and was directed to the camera. All apoMb experiments were performed using the 60X water objective.
Data Analysis
For every image collected in one of the experimental procedures above, the sample fluorescence intensity was plotted against pixels along the length of the image. To make this plot, the sample fluorescence intensity and the background signal intensity were integrated over a specific number of pixels across the width of the image at each pixel along the length of the image. The background signal was subtracted from the sample signal, which was then plotted against pixels along the length of the image. The data were fit to appropriate functions in Igor Pro (Wavemetrics).
RESULTS AND DISCUSSION
Flow Rate Calibration
The flow rate calibration was performed to determine the time resolution of the mixer for specific sheath flow rates. The fluorescence decay time of the Eu nanospheres was measured independently using a time-resolved fluorescence instrument. Eu nanospheres were excited by a pulsed 351-nm laser, and their fluorescence intensity was recorded over time. This signal was fit to a single exponential function, which yielded a lifetime of 548 µs (Figure S-1 in the Supporting Information) that is comparable to the approximate 600-µs lifetime reported by the manufacturer.
The measured fluorescence lifetime was applied to the flow cell calibration experiment in order to determine the linear flow velocity of the sample stream within the mixer. Eu nanospheres were flowed through a sheath solution of DI water and were excited at a single point along their trajectory, thereby impulsively exciting the Eu fluorescence. Downstream of this excitation point, the fluorescence of the Eu nanospheres decays with the established lifetime of 548 µs (Figure 2A). Images of this decay were collected for 16 different sheath volumetric flow rates. For each image, a plot of Eu fluorescence intensity against pixels along the length of that image was fit with a single-exponential function from the point of laser excitation to the point at which fluorescence signal was no longer detected above background. The decay in pixels of each single exponential fit was then determined. Dividing the Eu fluorescence decay time of 548 µs by the Eu fluorescence decay in pixels for an image collected at a specific sheath flow rate gave the time resolution (time per pixel in µs/pixel) at that sheath volumetric flow rate. The linear flow velocity of the sample stream is then given by the pixel size (1.15 × 10−5 cm/pixel at 100X magnification) divided by the time per pixel. Table S-1 of the Supporting Information gives the time resolution values for each sheath volumetric flow rate studied. These time resolution values were plotted against their corresponding sheath volumetric flow rates in order to model the dependence of the linear flow velocity on the volumetric flow rate (Figure 2B). The calibration data were fit to a single-exponential function (τ = 1.1 + 3.5e−3.5v) where τ is the time per pixel in µs/pixel and v is the sheath flow rate in mL/min. All fit values and errors are given in Table S-2 of the Supporting Information. Using this calibration, we calculate for the flow rate of 1.17 ml/min, used in the apoMb experiment described below, a time/pixel ratio of 1.2 µs/pixel. This value is near the limit of 1.1 µs/pixel predicted for high flow rates.
All time calibration values used to analyze the following results are derived from this equation. Note that time calibration fit function is calculated for when the mixer is viewed at 100X magnification. For data collected at 40X and 60X magnification, τ is multiplied by 2.5 and 1.5, respectively, to adjust for the difference in magnification. These different magnification factors were verified using the resolution target RES-1 from Newport Corporation (Irvine, CA). The magnification factors were determined by comparing the number of image pixels spanning a given distance on the target and then calculating the pixel count ratio for each objective relative to the 100X.
The theoretical time calibration was calculated by assuming that the fluid flow in the mixer equals the rate of the sheath solution flow, since the sheath comprises greater than 99.9% of the total volumetric flow. The volumetric sheath flow rate is converted into a linear flow velocity using the dimensions of the flow cell and assuming a uniform flow velocity in the center of the cell. Equation 1 below gives the explicit expression for the time calibration, TC, in terms of the volumetric flow rate, FR (mL/min), the cross-sectional area of the outer capillary, A (cm2), and the distance/pixel ratio, D/P (cm/pixel), which is a function of the objective being used. Equation 2 gives the numerical evaluation of this expression for the 200-µm diameter capillary and the 100X objective. Figure 2B also displays this theoretical time calibration as a dashed line.
| (1) |
| (2) |
This theoretical expression predicts that the µs/pixel resolution of the mixer is enhanced by using higher sheath flow rates, as the resolution should be inversely proportional to sheath flow rate. The experimentally observed behavior of the mixer deviates from this prediction in two ways. First, the time calibration data are best fit to an exponential function of FR and not 1/FR. In addition, the theoretical time calibration values are lower than the experimental ones.
The differences between the calculated and experimentally observed behaviors are likely due to theoretical assumptions that were not realized experimentally. The calculation makes the simplifying assumption that the linear flow velocity is constant along the cross section of the mixer, but we expect that there is actually a gradient of flow velocities due to drag on the capillary walls. At the interface between the sheath solution and the wall of the outer capillary, the sheath flow rate should be at a minimum due to the drag of the wall. The sheath flow rate should increase towards the center of the capillary, where drag is minimal. The linear flow velocity used in the theoretical calculation represents a weighted average of all velocities in the flow gradient and is realized somewhere between the center and edges of the outer capillary. Therefore, the position of the inner capillary within the outer capillary will determine the sheath solution linear flow velocity the sample experiences and may not be the same as the average linear flow velocity used in the calculation. Since the cross-sectional flow gradient varies nonlinearly with the volumetric flow rate, the observed linear flow velocity should not follow the expected 1/FR relationship, which explains the observed exponential relationship shown in Figure 2. The calculations also assumed that the sample is flowing at the same rate as the sheath solution. Initially, the sample flow rate is significantly slower, but it is rapidly accelerated by contact with the sheath flow. It is possible that the two streams are not yet in equilibrium within the time of the measurements, thereby making the observed time per pixel greater than the theoretical prediction. These results emphasize the importance of experimental determination of the actual flow rate, although this is not common practice for microfluidic mixing studies.
Diffusional Mixing Time
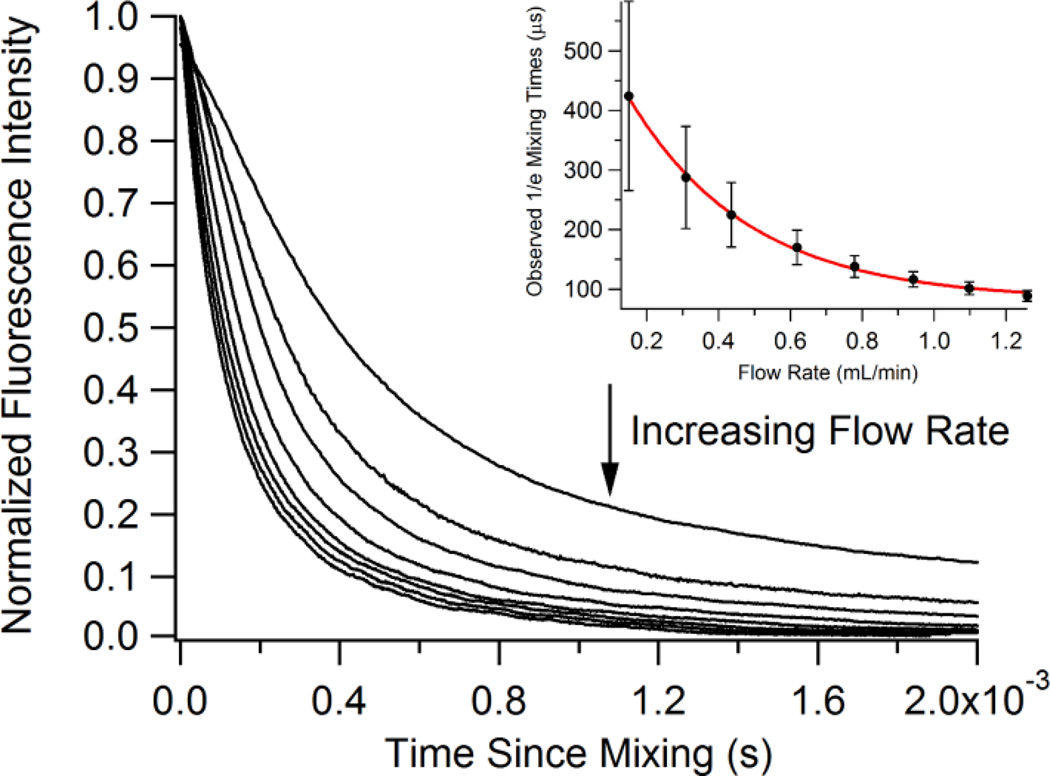
The diffusional mixing time indicates the time required for the sheath solution to diffuse through the cross-sectional area of the sample stream. We observe this diffusion as an exponential rate and therefore report the 1/e time of the fluorescence decay as the mixing time. A sample of 20-µM fluorescein was flowed through a sheath solution of 2.5-M KI, which quenches the fluorescence of fluorescein in a diffusion controlled reaction. Images of the fluorescence quenching were collected for eight different flow rates. The fluorescein fluorescence signal across each image was fit to a single exponential function, and the decay in pixels of each function was determined. These decays were multiplied by the appropriate time resolution value, based on the sheath flow rate that was used, to obtain a mixing time. We considered time zero as the point where the sample and sheath solutions were first able to mix, so in this setup this is the end of the inner capillary. The reported mixing times are times from the end of the inner capillary. Figure 3 illustrates the mixing times plotted against their corresponding sheath flow rates. The data demonstrate a decrease in mixing time as sheath flow rate increased. The data were fit to a single exponential function (τmix = 82 + 530e−3.0v) where τmix is the 1/e mixing time and v is the sheath flow rate in mL/min. All fit values and error are given in Table S-3 of the Supporting Information. The fit indicates a minimal diffusional mixing time of 82 µs for iodide that can be achieved at the highest sheath flow rate. Iodide has a relatively slow rate of diffusion, so we expect that shorter mixing times may be achieved with smaller, faster diffusing solutes. A mixing time of 98 µs is calculated for the flow rate of 1.17 mL/min used in the pH jump experiments with apoMb described below. The decrease in mixing time as flow rate increases shown in Figure 3 is likely due to increased hydrodynamic focusing of the sample at higher sheath flow rates and constant sample flow rate. As hydrodynamic focusing increases, the width of the sample stream decreases. The distance through which a solute in the sheath solution must diffuse in order to mix with the sample effectively decreases as the sheath flow rate increases, allowing diffusion to occur more rapidly.
Figure 3.
Fluorescence transients from the quenching of fluorecein fluorescence by mixing with potassium iodide at different sheath flow rates. Inset shows the observed mixing times as determined by exponential fits. Inset data were fit to a single exponential function: τmix = 82 + 530e −3.0·v.
The mixing times determined for our mixer are faster than many protein structural dynamics, thus making continuous flow mixing a viable approach to initiate fast protein events, including enzymatic catalysis and protein folding. It is also reasonable to expect that mixing experiments that use a pH change might have a faster mixing time than we report here because protons diffuse faster due to their small mass and additional transport mechanisms like the Grotthuss mechanism.36 However, for the pH jump experiments described here, similar mixing times are observed (verified by pH quenching of fluorescein under the same conditions as the protein experiment) despite the two orders of magnitude increase in diffusion rate of protons compared to iodide. The reason for similar mixing times is that the increase in diffusion rate is negated by the comparatively low concentration of protons available at pH 2, more than two orders of magnitude lower than the iodide concentration in the calibration experiment.
ApoMb Unfolding Kinetics
ApoMb is an important model system for the study of protein folding mechanisms. The protein consists of only 153 amino acids organized into 8 alpha helices labeled A-H.37 Several studies have demonstrated that the protein adopts four unique conformational states, depending on solution conditions: the native form (N), the intermediate form (I), the extended form (E), and the unfolded form (U).1, 24–26 These distinct equilibrium conformations of apoMb have also been observed as kinetic intermediates in T-jump and fast mixing studies.25, 27–31 All of these states can be accessed in equilibrium by changing the pH and ionic strength of the protein solution. During acid denaturation, apoMb has been shown to transition between its structural states in two distinct folding events. At pH 7, apoMb exists in its N form. Between pH 6 and 4, the protein unfolds from its N form to its I form due to the protonation of the imidazole side chains of three histidine residues (His24, His64, and His113).38 An additional expansion of apoMb’s structure from its I form to E form can occur at pH 3 when the salt concentration of the protein solution is low.39 Below pH 3, the protein completely unfolds to its U form as the carboxyl groups on the side chains of a glutamate (Glu6) and an aspartate (Asp122) are protonated.38
We used the fast mixer to study the unfolding kinetics of apoMb initiated by a rapid pH drop. The pH drop was induced by flowing apoMb through a sheath solution of HCl and the reaction was probed using the fluorescence of 1,8-ANS bound in the heme pocket of the protein in a 1:1 molar ratio.40 The apoMb sample was at a pH of 4.5, and the HCl sheath was at a pH of 2.0. The diffusion of protons from the flowing sheath solution into the apoMb sample stream lowered the pH of the protein solution to pH 2.0, causing the apoMb to unfold and therefore the 1,8-ANS fluorescence to be quenched. The unfolding lifetime of apoMb is determined from the time dependence of the 1,8-ANS fluorescence quenching. Equilibrium measurements have previously shown that under the initial conditions employed in the present study of minimal salt and pH 4.5, apoMb is in the I form, a partially folded intermediate state with a native-like AGH core. When the pH drops to 2.0, apoMb unfolds to its U form, having very little residual structure.39 Hence we expect the pH jump to initiate the unfolding transition from I to U under conditions that strongly favor the completely unfolded state U.
The unfolding lifetime was measured at five different sheath flow rates. In principle, the observed lifetime should be independent of the sheath flow rate provided that it is significantly longer than the effective mixing time. Control experiments compared the apoMb-1,8-ANS fluorescence images for a sample stream flowed through DI water (no pH change) at these five flow rates. We found that the fluorescence signal for the control experiment in DI water decreases along the sample stream as it is hydrodynamically focused by the sheath flow. When the sample stream is focused, there are fewer molecules in the excitation volume of the imaging system, resulting in an apparent decrease in the total fluorescence signal. This effect is removed from the pH jump data by dividing the fluorescence image of an HCl experiment by the image of the reference DI water experiment at the same flow rate. This method yields a corrected 1,8-ANS fluorescence quenching profile. A similar approach was applied by Hertzog and coworkers to analyze fluorescence decays in a different microfluidic flow system.17 Each fluorescence quenching profile was fit to a single exponential to determine the profile’s decay in pixels. Each decay was then converted to an unfolding lifetime by multiplying by the appropriate flow dependent time resolution. The dependence of the observed unfolding lifetime on the sheath flow rate (Figure 4) can be understood in terms of the inverse relationship between sheath flow rate and mixing time due to hydrodynamic focusing of the sample stream. At slow sheath flow rates, the mixing time is slow and it overlaps with the unfolding time, yielding an apparent lifetime that is slower. As the sheath flow rate is increased, the apparent lifetime increases until it reaches the limiting value of 760 ± 8 µs and becomes independent of flow rate. At this point the mixing time is less than 100 µs, meaning it no longer perturbs the observed unfolding time. We conclude that the observed decay time of 760 ± 8 µs at the fastest sheath flow rates represents the actual lifetime of the pH induced transition from the I to U form of apoMb.
Figure 4.
Fluorescence transients from the rapid change in pH of apoMb from 4.5 to 2.0. Inset shows apoMb unfolding times as determined by exponential fits. The unfolding time decreases according to an exponential function: τunf = 760 + 80000e −8.8·v. The baseline indicates a flow rate independent unfolding time (I ↔ U) of 760 ± 8 µs.
The unfolding mechanism of apoMb has been studied by many different methods, including T-jump, rapid mixing and H/D exchange.1, 24, 31, 32, 41–44 The consensus of previous work is a multi-state unfolding mechanism:
The earliest intermediate in the folding reaction, the E state, consists of a hydrophobically collapsed chain stabilized by packing of the nearly fully formed A, G and H helices to make a core structure. The formation of this state was not resolved in early stopped flow and continuous flow experiments, but rather was observed as a burst phase.42, 44 T-jump IR measurements, however, observed sub-millisecond relaxation lifetimes for the E state.32 The I state has been observed both as a kinetic intermediate in refolding experiments at pH 6,43 and also as an equilibrium intermediate at pH 4.2–4.5 and high ionic strength (20 mM NaCl).39 It resembles the E state, having a folded AGH core, but it also has the fully folded B and E helices. A recent study measured the rate of urea induced unfolding of sperm whale apoMb starting from the I state at pH 4.2 (labeled “M” by the Roder group), using a turbulent flow mixer with similar time resolution to the present work31 This study postulated an additional state “L” between I and E, to account for the observed kinetics and observed an overall unfolding lifetime (I ↔ L ↔ E ↔ U) at pH 4.2 extrapolated to zero urea concentration of 1.5 ms. We find a significantly shorter unfolding lifetime (760 µs) at pH 2 and no urea. More importantly, we find that the unfolding reaction under these conditions does not populate any intermediates, but rather can be modeled as a simple two-state process (I ↔ U).
We emphasize that the rapid pH jump achievable in our continuous flow mixer enables the investigation of the unfolding kinetics of apoMb on a previously unexplored part of the energy landscape. At pH 2 and low salt, apoMb unfolds in a highly cooperative, all or nothing transition that does not populate intermediate states. This result means that the “E” and “L” intermediates are not obligatory and that it is possible for the protein to unfold through a more direct route under highly destabilizing conditions that impose a large driving force for the unfolding reaction. Alternatively, it is possible that the protein still unfolds through the same intermediates observed at pH 4.2, but they are not populated to any great extent under these conditions due to differences in the activation barriers. One other difference worth noting is that the while the horse protein used in the present study has considerable sequence identity (88%) to the sperm whale protein, and while overall folding behavior of the two forms is similar, some subtle differences have been observed.45 These differences are small, however, and unlikely to cause a change in the unfolding mechanism by themselves. Furthermore, the observation of two state unfolding of I to U is not unprecedented, since Baryshnikova et al. found that unfolding of apoMb starting in the native state N proceeds through a single intermediate state I (N ↔ I ↔ U) under strongly denaturing conditions.46 Thus it is possible to effectively bypass the L and E intermediate states under conditions that strongly destabilize the folded structure.
Lactate dehydrogenase kinetics
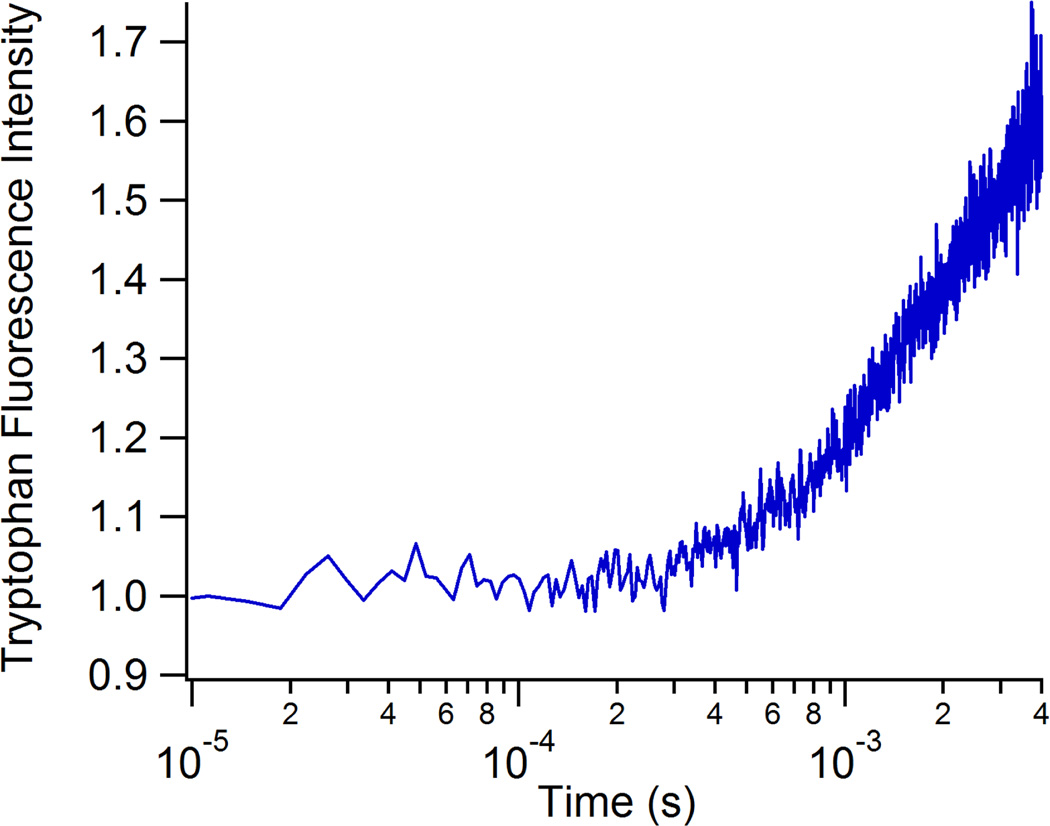
We also demonstrated the application of the fast mixer to the study of enzyme reaction kinetics by studying the conversion of pyruvate to lactate catalyzed by the enzyme lactate dehydrogenase (LDH). This reaction involves the direct transfer of a hydride ion from C4 of the reduced nicotinamide group of NADH to the C2 carbon of pyruvate accompanied by the protonation of the keto oxygen of pyruvate, the proton being supplied by His195. The on-enzyme substrate turnover occurs in about 1 ms, but faster processes are expected due to loop closure and structural changes of the Michaelis complex prior to the chemistry step.47 Previous studies have used tryptophan fluorescence and T-jump perturbation to study the enzyme dynamics faster than 1 ms.47, 48 Trp fluorescence has not been widely used as a probe in fast mixing studies either because of optical limitations of the mixers, or of the confocal fluorescence microscopes required for these studies. Our fast mixer overcomes these limitations because the high quality fused silica capillaries are transparent down to 200 nm and minimally fluorescent, allowing direct excitation at 280 nm and detection of the Trp emission at 350 nm. Furthermore, confocal detection is not required due to the 3D focusing of the sample jet and minimal background fluorescence from the capillary walls. Figure 5 shows the Trp fluorescence transient recorded from the reaction of the binary complex of lactate dehydrogenase and NADH with its substrate, pyruvate. The observed change in Trp emission is not well fit by a single exponential, showing at least two components including a submillisecond event. We attribute these multiple rates to a combination of events including Michaelis complex formation, loop domain closure and the chemistry step, by comparison to kinetics measurements using T-jump relaxation methods.47, 48 The full elucidation of these processes will require further study, but the present results demonstrate the viability of the mixer coupled with native Trp fluorescence to study fast enzyme dynamics.
Figure 5.
Transient tryptophan fluorescence profile obtained from the rapid mixing of 200 µM pig heart lactate dehydrogenase/350 µM NADH with 1000 mM pyruvate. The observed increase in fluorescence intensity is due to a combination of enzyme conformational change and enzymatic turnover.
Conclusions
We have demonstrated the utility of a new continuous flow rapid mixer for the study of protein dynamics. The mixer has a robust design that is easy and inexpensive to implement and allows rapid recovery from clogs. The mixer utilizes 3D, hydrodynamic focusing of a sample solution by an all-encompassing sheath solution in order to achieve rapid diffusive mixing between the sample and sheath. Characterization of the mixer’s performance was conducted completely via experimentation, rather than simulation. The mixer achieves a minimum time resolution of 1.2 µs/pixel at sheath flow rates above 1 mL/min. Flowing the sample at a rate of 0.1 µL/min minimizes its consumption. At a sheath flow rate of 1 mL/min, diffusive mixing of small molecules from the sheath into the sample is accomplished in under 100 µs. This fast mixing time is sufficient to resolve the unfolding kinetics of apoMb from its I form (pH 4.5) to its U form (pH 2.0) with an unfolding time of 760 ± 8 µs. This reaction occurs as a single cooperative transition, without populating intermediates, unlike its behavior in the presence of denaturants at higher pH. The rapid pH jump made possible by the continuous flow mixer allows access to a part of the folding energy landscape of apoMb not previously explored. These results demonstrate the applicability of this rapid continuous flow mixer to the study of protein dynamics in general, including enzymatic catalysis. Complete optical access is available to the entire mixing region, and it is possible to use UV excitation of intrinsic fluorophores (280 nm excitation of tryptophan) or extrinsic labels. The extension of possible probe wavelengths deeper into the UV substantially expands the applicability of fast mixers. Moreover, the mixer is simple enough to construct and operate in any biophysical chemistry laboratory. This ease of access and usage is a substantial advantage over similar devices, which have comparable experimental mixing times but are more complicated to operate.5,8,25 We expect these advances to lead to more widespread application of this methodology to the study of biomolecular reactions.
Supplementary Material
Acknowledgements
This work was supported by NIH grants GM068036 and GM053640 (RBD) and by an NSF graduate fellowship DGE-0940903 (MJR).
Footnotes
Electronic Supplementary Information (ESI) available: Fluorescence decay measurement of Eu nanospheres and fit coefficients with error as indicated in the text. See DOI:10.1039/b000000x/
Notes and references
- 1.Callender RH, Dyer RB. R. Gilmanshin and W. H. Woodruff. Annu Rev Phys Chem. 1998;49:173–202. doi: 10.1146/annurev.physchem.49.1.173. [DOI] [PubMed] [Google Scholar]
- 2.Klinman JP. Chem Phys Lett. 2009;471:179–193. doi: 10.1016/j.cplett.2009.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eaton WA, Thompson PA, Chan CK. S. J. Hagen and J. Hofrichter. Structure. 1996;4:1133–1139. doi: 10.1016/s0969-2126(96)00121-9. [DOI] [PubMed] [Google Scholar]
- 4.Dyer RB. Curr Opin Struct Biol. 2007;17:38–47. doi: 10.1016/j.sbi.2007.01.001. [DOI] [PubMed] [Google Scholar]
- 5.Wang C, Ye D-K, Wang Y-Y. T. Lu and X.-H. Xia. Lab Chip. 2013;13:1546–1553. doi: 10.1039/c3lc41319e. [DOI] [PubMed] [Google Scholar]
- 6.Lapidus LJ, Yao SH, McGarrity KS, Hertzog DE. E. Tubman and O. Bakajin. Biophys J. 2007;93:218–224. doi: 10.1529/biophysj.106.103077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pollack L, Tate MW, Darnton NC, Knight JB, Gruner SM. W. A. Eaton and R. H. Austin. Proc Natl Acad Sci USA. 1999;96:10115–10117. doi: 10.1073/pnas.96.18.10115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hamadani KM, Weiss S. Biophys J. 2008;95:352–365. doi: 10.1529/biophysj.107.127431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pabit SA, Hagen SJ. Biophys J. 2002;83:2872–2878. doi: 10.1016/S0006-3495(02)75296-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gambin Y, Simonnet C, VanDelinder V. A. Deniz and A. Groisman. Lab Chip. 2010;10:598–609. doi: 10.1039/b914174j. [DOI] [PubMed] [Google Scholar]
- 11.Yao S, Bakajin O. Anal Chem. 2007;79:5753–5759. doi: 10.1021/ac070528n. [DOI] [PubMed] [Google Scholar]
- 12.Park HY, Qiu XY, Rhoades E, Korlach J, Kwok LW, Zipfel WR. W. W. Webb and L. Pollack. Anal Chem. 2006;78:4465–4473. doi: 10.1021/ac060572n. [DOI] [PubMed] [Google Scholar]
- 13.Hertzog DE, Ivorra B, Mohammadi B. O. Bakajin and J. G. Santiago. Anal Chem. 2006;78:4299–4306. doi: 10.1021/ac051903j. [DOI] [PubMed] [Google Scholar]
- 14.Toepke MW, Brewer SH, Vu DM, Rector KD, Morgan JE, Gennis RB. P. J. A. Kenis and R. B. Dyer. Anal Chem. 2007;79:122–128. doi: 10.1021/ac061193x. [DOI] [PubMed] [Google Scholar]
- 15.Abonnenc M. J. Josserand and H. H. Girault. Lab Chip. 2009;9:440–448. doi: 10.1039/b815581j. [DOI] [PubMed] [Google Scholar]
- 16.Knight JB, Vishwanath A. J. P. Brody and R. H. Austin. Phys Rev Lett. 1998;80:3863–3866. [Google Scholar]
- 17.Hertzog DE, Michalet X, Jager M, Kong XX, Santiago JG. S. Weiss and O. Bakajin. Anal Chem. 2004;76:7169–7178. doi: 10.1021/ac048661s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lim TW, Son Y, Jeong YJ, Yang D-Y, Kong H-J. K.-S. Lee and D.-P. Kim. Lab Chip. 2011;11:100–103. doi: 10.1039/c005325m. [DOI] [PubMed] [Google Scholar]
- 19.Chiu Y-J, Cho SH, Mei Z, Lien V. T.-F. Wu and Y.-H. Lo. Lab Chip. 2013;13:1803–1809. doi: 10.1039/c3lc41202d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mao X. J. R. Waldeisen and T. J. Huang. Lab Chip. 2007;7:1260–1262. doi: 10.1039/b711155j. [DOI] [PubMed] [Google Scholar]
- 21.Shi J, Yazdi S, Steven Lin S-C, Ding X, Chiang IK. K. Sharp and T. J. Huang. Lab Chip. 2011;11:2319–2324. doi: 10.1039/c1lc20042a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beebe DJ. G. A. Mensing and G. M. Walker. Annu Rev Biomed Eng. 2002;4:261–286. doi: 10.1146/annurev.bioeng.4.112601.125916. [DOI] [PubMed] [Google Scholar]
- 23.Shastry MCR. S. D. Luck and H. Roder. Biophys J. 1998;74:2714–2721. doi: 10.1016/S0006-3495(98)77977-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Uzawa T, Akiyama S, Kimura T, Takahashi S, Ishimori K. I. Morishima and T. Fujisawa. Proc Natl Acad Sci USA. 2004;101:1171–1176. doi: 10.1073/pnas.0305376101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meinhold DW, Wright PE. Proc Natl Acad Sci USA. 2011;108:9078–9083. doi: 10.1073/pnas.1105682108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weisbuch S, Gerard F, Pasdeloup M, Cappadoro J. Y. Dupont and M. Jamin. Biochemistry. 2005;44:7013–7023. doi: 10.1021/bi047372v. [DOI] [PubMed] [Google Scholar]
- 27.Gilmanshin R, Williams S, Callender RH, Woodruff WH, Dyer RB. Proc Natl Acad Sci USA. 1997:3709–3713. doi: 10.1073/pnas.94.8.3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gilmanshin R, Callender RH, Dyer RB. Nat Struct Biol. 1998:363–365. doi: 10.1038/nsb0598-363. [DOI] [PubMed] [Google Scholar]
- 29.Gulotta M, Gilmanshin R, Buscher T. R. Callender and R. Dyer. Biochemistry. 2001;40:5137–5143. doi: 10.1021/bi002256n. [DOI] [PubMed] [Google Scholar]
- 30.Gulotta M, Rogatsky E. R. H. Callender and R. B. Dyer. Biophys. J. 2003;84:1909–1918. doi: 10.1016/S0006-3495(03)74999-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu M, Beresneva O. R. Rosario and H. Roder. J Phys Chem B. 2012;116:7014–7025. doi: 10.1021/jp3012365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gilmanshin R. R. H. Callender and R. B. Dyer. Nat Struct Biol. 1998;5:363–365. doi: 10.1038/nsb0598-363. [DOI] [PubMed] [Google Scholar]
- 33.Tsien RY, Ernst L, Waggoner A. In: Handbook of Biological Confocal Microscopy. 3rd edn. Pawley JB, editor. ch. 16. New York: Springer Science+Business Media, LLC; 2006. pp. 338–351. [Google Scholar]
- 34.Adams PA. Biochem J. 1977;163:153–158. doi: 10.1042/bj1630153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weber G, Young LB. J Biol Chem. 1964;239 1415-&. [PubMed] [Google Scholar]
- 36.Cukierman S. Biochim Biophys Acta Bioeng. 2006;1757:876–885. doi: 10.1016/j.bbabio.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 37.Evans SV, Brayer GD. J Biol Chem. 1988;263:4263–4268. [PubMed] [Google Scholar]
- 38.Yang AS, Honig B. J Mol Biol. 1994;237:602–614. doi: 10.1006/jmbi.1994.1258. [DOI] [PubMed] [Google Scholar]
- 39.Gilmanshin R. R. B. Dyer and R. H. Callender. Prot Sci. 1997;6:2134–2142. doi: 10.1002/pro.5560061008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stryer L. J Mol Biol. 1965;13:482–495. doi: 10.1016/s0022-2836(65)80111-5. [DOI] [PubMed] [Google Scholar]
- 41.Jamin M, Baldwin RL. J Mol Biol. 1998;276:491–504. doi: 10.1006/jmbi.1997.1543. [DOI] [PubMed] [Google Scholar]
- 42.Uzawa T, Nishimura C, Akiyama S, Ishimori K, Takahashi S. H. J. Dyson and P. E. Wright. Proc Natl Acad Sci USA. 2008;105:13859–13864. doi: 10.1073/pnas.0804033105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jennings PA, Wright PE. Science. 1993;262:892–896. doi: 10.1126/science.8235610. [DOI] [PubMed] [Google Scholar]
- 44.Jamin M, Yeh SR. D. L. Rousseau and R. L. Baldwin. J Mol Biol. 1999;292:731–740. doi: 10.1006/jmbi.1999.3074. [DOI] [PubMed] [Google Scholar]
- 45.Haruta N, Kitagawa T. Biochemistry. 2002;41:6595–6604. doi: 10.1021/bi012027g. [DOI] [PubMed] [Google Scholar]
- 46.Baryshnikova EN, Melnik BS. G. V. Semisotnov and V. E. Bychkova. Mol Biol. 2005;39:1008–1016. [PubMed] [Google Scholar]
- 47.Ghanem M, Saen-Oon S, Zhadin N, Wing C, Cahill S, Schwartz SD. R. H. Callender and V. Schramm. Biochemistry. 2008;47:3202–3215. doi: 10.1021/bi702491d. [DOI] [PubMed] [Google Scholar]
- 48.Deng H, Brewer SH, Vu DV, Clinch K. R. Callender and R. B. Dyer. Biophys. J. 2008;95:804–813. doi: 10.1529/biophysj.108.128884. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.