The actinomycetes family of bacteria is classically considered a rich source of natural products [1]. It is often suggested that such compounds have been selected by evolution to have biological activity [2]. For example, given the microbial-rich soil environment of the actinomycetes native habitat, their natural products have been suggested to serve in chemical warfare and/or signaling, which has been further connected to their leading role in providing pharmaceutically relevant antibiotic agents [3]. Nevertheless, surprisingly few of these molecules have been definitively assigned specific physiological roles in the native producing bacteria.
The vast majority of actinomycetes biosynthetic diversity is devoted to the production of either polyketides (PKs) and/or non-ribosomal peptides (NRPs), with relatively little production of terpenoids, which are commonly found in plants and fungi, and that form the largest class of natural products [4]. This dichotomy can perhaps be most easily seen in their genomes, which typically contain 25–30 gene clusters (i.e., operons) assigned to natural products biosynthesis [2], less than 5 of which produce terpenoids, with the vast majority of the remainder producing PKs and/or NRPs. For example, the genome of Streptomyces avermitilis MA-4680 contains 18 operons for the production of PKs and NRPs [5], but only four prototypical (i.e., class I) terpene synthases [6]. More generally, while there are many thousands of operons associated with bacterial PK and NRP biosynthesis [2], a recently reported bioinformatic search found only ~120 bacterial terpene synthases [6], demonstrating that bacterial terpenoid biosynthesis is relatively unusual.
The labdane-related diterpenoids form a natural products super-family that is characterized by protonation-initiated bicyclization of the general diterpenoid precursor (E,E,E)-geranylgeranyl diphosphate (GGPP) catalyzed by class II diterpene cyclases, most often to form a labdadienyl/copalyl diphosphate product from which the nomenclature was derived [7]. While largely associated with plants, where the requirement for gibberellin phytohormone production has provided a biosynthetic reservoir that has been repeatedly tapped to spawn the vast majority of the ~7,000 known such natural products [7], there are a few examples from actinomycetes. Those where the associated biosynthetic operon have been identified include that for terpentecin in Streptomyces griseolosporeus [8], phenalinolactone in Streptomyces sp. Tü6071 [9], and platensimycin/platencin in Streptomyces platensis [10], all of which are of some interest as antibiotics, although their native physiological function is unknown. However, the focus of this review is the labdane-related diterpenoid isotuberculosinol produced by Mycobacterium tuberculosis, which is unusual not only as an example of a bacterial (di)terpenoid, but even further as a bacterial natural product for which a physiologically relevant role can be assigned, and one with potential medical relevance at that.
While the vast majority of the actinomycetes do not cause human disease [11], the genus mycobacteria provides notable exceptions, containing a number of species that are pathogenic. Of particular medical relevance are those within the Mycobacterium tuberculosis complex, as many of these closely related species and sub-species are the causative agents of the widespread human disease Tuberculosis [12, 13]. It has been estimated that up to 30% of the global human population is infected with such bacteria, leading to over 1.4 million deaths annually [14]. The vast majority of human tuberculosis cases are thought to be due to the eponymous Mycobacterium tuberculosis (Mtb) because of its efficient infectivity in humans [15]. Moreover, the emerging multi- and extremely drug resistant variants of Mtb underline the continued relevance of this endemic human pathogen [16]. How the species within the Mtb complex cause tuberculosis is still not clear, as the pathogenic mechanism seemingly consists of an as yet not fully defined multipronged attack on the human immune system [17]. Nevertheless, while not pertinent to tuberculosis, the polyketide mycolactones produced by Mycobacterium ulcerans serve as important virulence factors in the resulting human disease Buruli ulcer [18], indicating the potential for a natural product(s) to play a role in the pathogenicity of species from the Mtb complex as well.
The ability of Mtb to infect its human host relies on circumvention of the human immune system, within which this pernicious pathogen actually carries out its lifecycle [19]. In particular, Mtb is thought to persist and replicate inside aveolar macrophage cells, enclosed in membrane-bound intracellular compartments derived from the phagosome into which the original infecting bacterium was first engulfed. This obviously requires subversion of this phagosome from its usual bacteriocidal purpose, which would normally be accomplished by fusion with pre-formed lysosomes in a complex process termed endocyctic/phagosomal maturation. However, Mtb containing phagosomes are arrested at an early stage of this process, wherein these compartments only undergo a relatively slight and transient decrease in pH, do not fuse with the protease containing lysosomes, and remain accessible to the early recycling endosomal system [19].
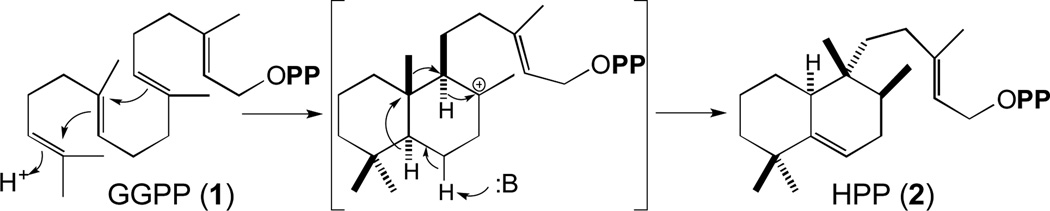
How Mtb blocks phagosomal maturation is unclear, although it has been attributed to multiple factors [19]. Nevertheless, while mycobacterial cell-surface lipids play a role, what the other effectors are remains less definitive, with different genetic screens indicating roles for non-overlapping sets of genes in the infection process [20–22]. Intriguingly, the only screen targeted at early stages of infection highlighted a role for two previously unstudied genes, Rv3377c and Rv3378c, found in a small operon that seemed to be involved in isoprenoid/terpenoid biosynthesis [23]. Almost immediately, Rv3377c was demonstrated to be a class II diterpene cyclase, catalyzing bicyclization and rearrangement of GGPP (1) to produce halimadienyl/tuberculosinyl diphosphate (HPP, 2)[24](Figure 1).
Figure 1.
Bicyclization and rearrangement of GGPP (1) to HPP (2) catalyzed by MtHPS/Rv3377c.
Given our long-standing interest in such labdane-related diterpenoid biosynthesis [7], we were interested in the unique activity of this cyclase, and were further intrigued by the similar phenotypic consequences for mutations in either Rv3377c or Rv3378c. This immediately suggests that Rv3378c encodes an enzyme that acts on HPP (2) to produce a bioactive natural product that contributes to the ability of Mtb to arrest phagosomal maturation. Although Rv3378c was not homologous to any known terpene synthases, we were able to show that it has (di)terpene synthase like activity, as well as produce small amounts of the resulting compound, finding that when this material was delivered to macrophages on beads the pH of the resulting phagosomes stabilized at a pH ~0.5 units above that of phagosome containing the control beads [25]. Furthermore, while this reflects a relatively minor portion of the overall phagosomal acidification (~25%), this seemed to be sufficient to block subsequent fusion with protease containing vesicles (Figure 2), while these beads containing phagosomes further remained accessible to the early endosomal recycling system, indicating that this compound has a role in the early stages of Mtb arrest of phagosomal maturation [25]. Spurred by the biological activity of this derived compound, we also investigated the HPP synthase encoded by Rv3377c (MtHPS), finding that it was susceptible to inhibition by an analog of the carbocation initially formed by protonation, 15-aza-14,15-dihydro-GGPP, which had previously been shown to be a sub-nanomolar inhibitor of such plant enzymes [26, 27], as well as a potentially more stable thiolo analog, suggesting that it may be possible to inhibit MtHPS/Rv3377c in vivo [28].
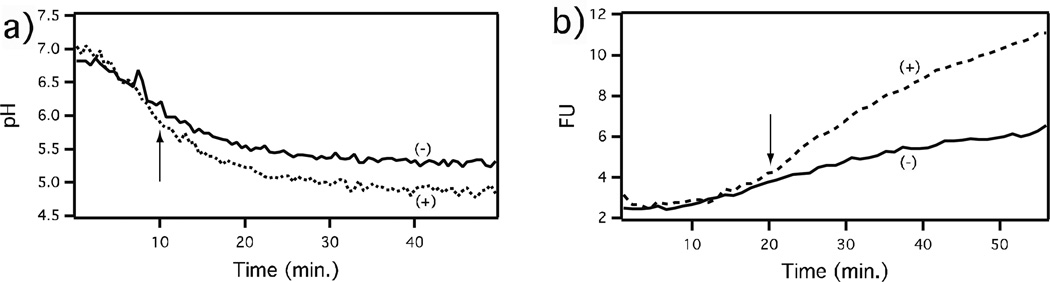
Figure 2.
Effect of isotuberculosinol on maturation of macrophage phagosomes containing control (−) or isotuberculosinol coated (+) beads, as indicated by solid and dotted lines, respectively. (a) Effect on pH. (b) Effect on proteolytic activity. Reprinted with permission from Ref. [25]. Copyright 2009 American Chemical Society. As indicated by the arrows, the effect on pH is observable within ~10 min., while that on proteolysis is only observed after ~20 min. The increase in proteolysis observed in the negative control phagosome presumably reflects its fusion with a protease containing lysosome. We hypothesize that, rather than reflecting a distinct effect, the latter occurring blockage of lysosomal fusion is a result of incomplete acidification.
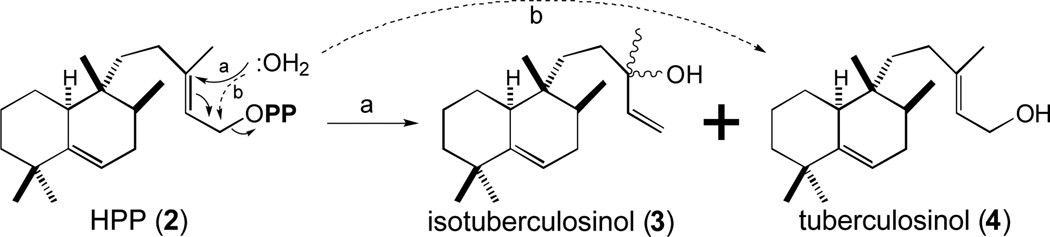
In our original report, we suggested that the Rv3378c encoded diterpene synthase (MtDTS) reacted with HPP (2) to produce a tricyclic diterpene olefin that we termed edaxadiene [25]. However, it was almost immediately suggested to us, by both Prof. Scott Rychnovsky (Univ. California, Irvine) and Prof. Barry Snider (Brandeis Univ.), that this compound in fact corresponded to the allylic tertiary alcohol resulting from nucleophilic addition of water after release of the diphosphate from HPP (2). This was quickly verified by comparison of our enzymatic product to an authentic standard synthesized by the Snider group [29], and also demonstrated via comparative synthesis by Prof. Eric Sorenson (Princeton Univ.) and co-workers [30]. While this structure previously had been identified as a sponge metabolite termed nosyberkol [31], there was a closely following Japanese language publication from the group of Prof. Tsutomu Hoshino (Nigata Univ.) indicating that MtDTS/Rv3378c produced an equal molar mixture of this and the corresponding primary alcohol [32], which they termed isotuberculosinol (3) and tuberculosinol (4), respectively, based on their earlier work with MtHPS/Rv3377c wherein its enzymatic product and the derived primary alcohol were termed tuberculosinyl diphosphate (i.e., HPP, 2) and tuberculosinol (4), respectively [24]. While our MtDTS/Rv3377c enzymatic preparations only produce the tertiary alcohol 3, we follow the precedent set by Prof. Hoshino, and refer to this compound here as isotuberculosinol (Figure 3).
Figure 3.
Hydrolysis of HPP (2) to isotuberculosinol (3) and tuberculosinol (4) catalyzed by MtDTS/Rv3378c.
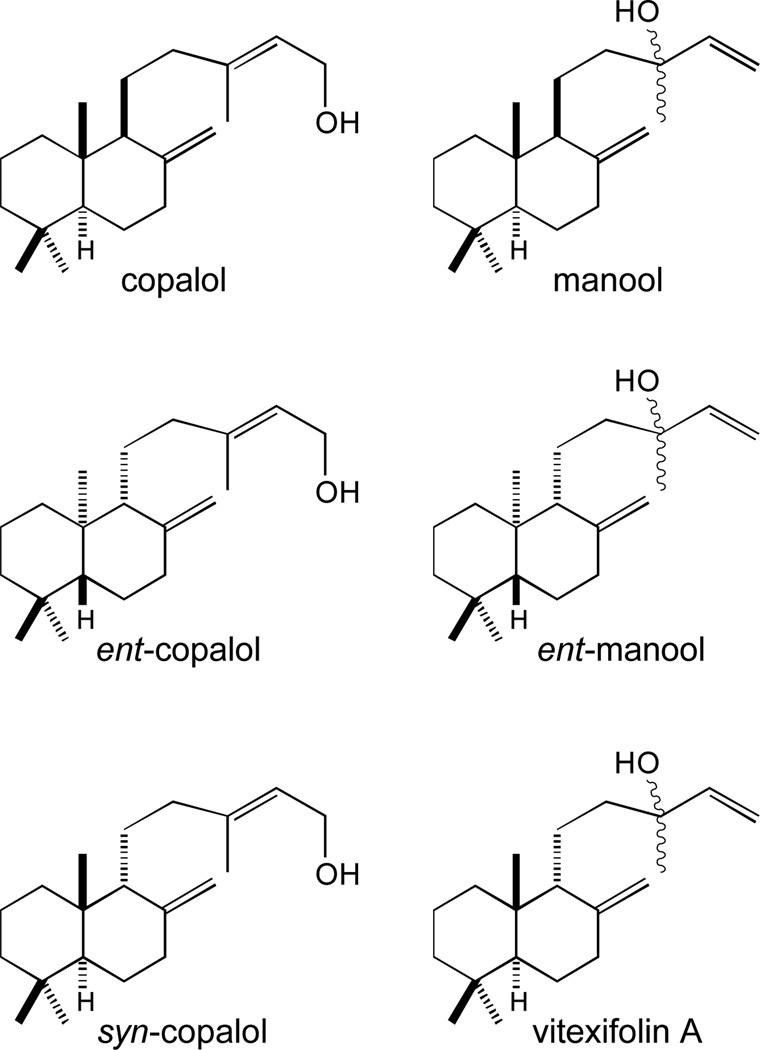
Later reported work by the Hoshino group further demonstrated the stereochemical conformation of the HPP (2) produced by MtHPS/Rv3377c, which is then conserved in the (iso)tuberculosinol (3/4) product of MtDTS/Rv3378c [33]. The Hoshino group also later reported a more detailed analysis of their MtDTS/Rv3378c enzymatic products, observing not only the equimolar production of tuberculosinol (4) and isotuberculosinol (3), but some stereoselectivity of the latter, with production of a 3:1 ratio of 13S- versus 13R- hydroxyl epimers [34]. In additionally reported work, the Hoshino group found that MtDTS/Rv3378c will react with the three known stereoisomers of copalyl diphosphate, which is the directly bicyclized product of class II cyclization of GGPP (1), to yield similar mixtures of derived primary and tertiary alcohols (Figure 4). Notably, this enabled a structure-activity relationship study of (iso)tuberculosinol (3/4) activity demonstrating the importance of the rearranged halimadienyl backbone. Specifically, isotuberculosinol (3) and tuberculosinol (4) individually and synergistically inhibited the phagocytosis of opsonized zymosan particles by human macrophage-like cells, but the analogous compounds derived from copalyl, rather than halimadienyl, diphosphate failed to do so [35].
Figure 4.
Products of MtDTS/Rv3378c from reaction with various stereoisomers of copalyl diphosphate.
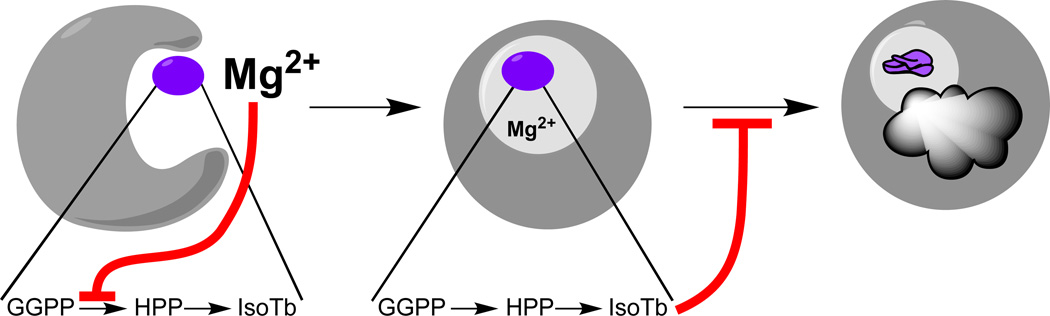
While the biochemical work described above strongly indicates a role for (iso)tuberculosinol in arresting phagosomal maturation, these compounds were only present at very low levels in Mtb grown in standard media [36], suggesting that such biosynthesis would need to be increased upon phagosomal engulfment. However, microarray analyses had not reported changes in the transcription of MtHPS/Rv3377c or MtDTS/Rv3378c during the infection process [37–39]. Our analysis of MtHPS/Rv3377c indicated it was susceptible to inhibition by its co-factor divalent magnesium ion [28], the levels of which are decreased in phagosomes [40, 41], approximately over the same range we observed affecting MtHPS/Rv3377c activity. We had previously observed similar enzymatic behavior in the distantly related class II diterpene cyclases specifically involved in gibberellin plant hormone biosynthesis [42], where it seems to be physiologically relevant [43], particularly given the matching variation in magnesium ion levels observed in response to light in the plastids where these enzymes are found [44]. Thus, we hypothesized that the decrease in magnesium ion level encountered upon phagosomal engulfment might act as a physiologically relevant biochemical trigger mechanism to induce (iso)tuberculosinol (3/4) production, and were able to provide evidence for this by demonstrating that shifting Mtb from the standard defined growth media, containing 0.43 mM magnesium, to media containing only 0.1 mM (but otherwise unchanged), was sufficient to induce a greater than 20-fold increase in (iso)tuberculosinol (3/4) levels, regardless of the promoter (native or recombinant) driving transcription of MtHPS/Rv3377c and MtDTS/Rv3378c [45](Figure 5).
Figure 5.
Effect of magnesium depletion on isotuberculosinol production by Mtb. Reprinted with permission from Ref. [45].
The original genetic screen only found mutants of MtHPS/Rv3377c and MtDTS/Rv3378c, with no hits found in the rest of the associated operon, which consists of Rv3377c – Rv3383c (although Rv3380 and Rv3381c seem to represent transposable elements)[23]. This presumably reflects functional redundancy, as suggested by the homology of these to other genes in Mtb, and/or loss of activity, as demonstrated already for Rv3379c [46], and highlights the requirement for Rv3377c and Rv3378c to produce (iso)tuberculosinol (3/4). Strikingly, this biosynthetic capacity is unevenly conserved across the Mtb complex – e.g., we earlier found that while MtHPS/Rv3377c is conserved in in all four of the Mtb strains whose genome sequences were available at that time, the M. bovis homolog contained an inactivating frameshift mutation that was conserved in the two sequenced strains, as well as another that we investigated directly [28]. Given the wider host range of M. bovis and its reduced infectivity in humans, we speculated that (iso)tuberculosinol (3/4) might play a role in the highly infectious nature of Mtb in humans [28]. The genomes of many more Mtb and other mycobacteria species have since been sequenced [47]. Notably, very few mutations are found in MtHPS/Rv3377c and MtDTS/Rv3378c in the sequenced Mtb diversity strains [48], with none in those from the highly infectious Beijing subgroup. By contrast, inactivating mutations in these genes were found in the sequences available for all other species within the Mtb complex, not only M. bovis, but also Mycobacterium canetti and Mycobacterium africanum, and these genes are completely missing in the genomes of other pathogenic mycobacteria (i.e., M. ulcerans and Mycobacterium marinum). These results then largely support the hypothesis that (iso)tuberculosinol (3/4) plays a role in the ability of Mtb to efficiently infect humans, which further may explain why MtHPS/Rv3377c and MtDTS/Rv3378c were not identified in genetic screens for pathogenesis that were carried out in mice [20, 22].
Arguably even more speculative is the origins of (iso)tuberculosinol (3/4) biosynthesis. While M. marinum produces tetraterpene carotenoid pigments [49], (iso)tuberculosinol (3/4) appears to have evolved from a separate gene pool. It has been noted that MtHPS/Rv3377c and MtDTS/Rv3378c exhibit lower guanine/cytosine (GC) content (54% and 48%, respectively) than the surrounding chromosome (65%), indicating that these were acquired by horizontal gene transfer [23]. Intriguingly, while MtHPS/Rv3377c shares homology with proteins from bacterial species in the actinomycetes genus Micromonospora, MtDTS has no homology to anything outside of Mycobacteria other than a hypothetical protein from social amoebae in the genus Dictyostelium, suggesting that MtHPS/Rv3377c and MtDTS/Rv3378c were originally obtained from disparate sources. The results with MtDTS/Rv3378c further highlight the unusual nature of this enzyme, which exhibits biochemical activity resembling that observed with certain terpene synthases, but does not seem to share homologous origins with these. On the other hand, the results with MtHPS/Rv3377c are consistent with the previously advanced hypothesis that class II diterpene cyclases from plants, bacteria, and fungi share homologous origins and seem to be derived from the bacterial squalene-hopene cyclases [50]. Nevertheless, these results also serve to demonstrate the unusual nature of labdane-related diterpenoid biosynthesis in bacteria, as only ~110 independent such sequences are found in BLAST searches with the MtHPS/Rv3377c amino acid sequence, and these include verified squalene-hopene cyclases along with the known bacterial class II diterpene cyclases [8–10, 51, 52].
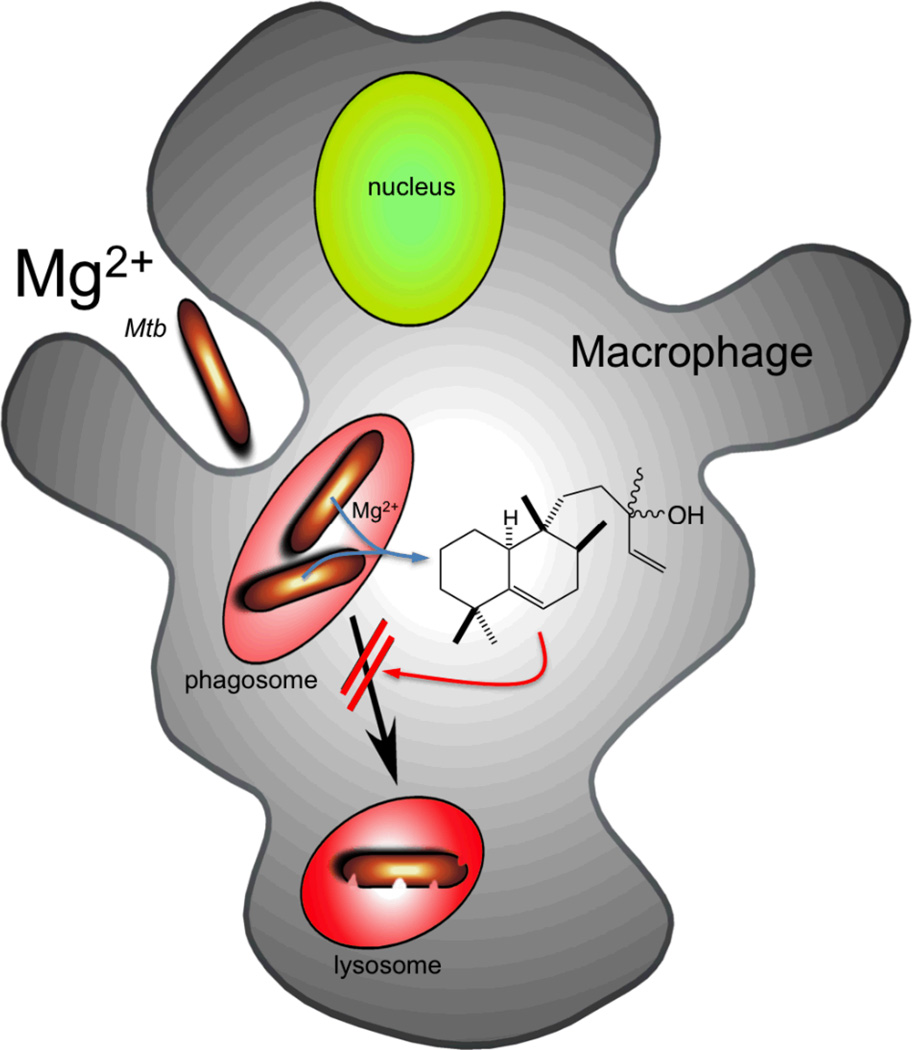
In conclusion, the production of (iso)tuberculosinol (3/4) by Mtb is unusual not only in being a rare example of a bacterial labdane-related diterpenoid, but also as an example of a bacterial natural product that has a plausible physiological role assigned to it. Specifically, serving as an immunomodulatory agent that seems likely to play a role in suppressing phagosomal maturation of the endocytic compartment into which Mtb is taken up by its aveolar macrophage host cell (Figure 6). To the extent that this contributes to infection and/or disease progress, such biosynthesis may serve as a potential target that seems likely to be druggable (i.e., based on the ability to inhibit the requisite MtHPS/Rv3377c with small molecules [28, 33]). Moreover, given the novel biological activity exhibited by (iso)tuberculosinol (3/4), it will be of significant interest to identify the molecular target through which the observed partial suppression of phagosomal/endocytic acidification is mediated.
Figure 6.
Role of isotuberculosinol in Mtb mediated block of phagosomal maturation.
Acknowledgements
The authors’ work described here was supported by a grant from the NIH (GM076324) to R.J.P.
Contributor Information
Francis M. Mann, Email: fmann@winona.edu.
Reuben J. Peters, Email: rjpeters@iastate.edu.
References
- 1.Baltz RH. Renaissance in antibacterial discovery from actinomycetes. Curr Opin Pharmacol. 2008;8(5):557–563. doi: 10.1016/j.coph.2008.04.008. [DOI] [PubMed] [Google Scholar]
- 2.Walsh CT, Fischbach MA. Natural products version 2.0: connecting genes to molecules. J Am Chem Soc. 2010;132(8):2469–2493. doi: 10.1021/ja909118a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mlot C. Microbiology. Antibiotics in nature: beyond biological warfare. Science. 2009;324(5935):1637–1639. doi: 10.1126/science.324_1637. [DOI] [PubMed] [Google Scholar]
- 4.Buckingham J. 1st ed. London ; New York: Chapman & Hall; 1994. Dictionary of natural products. v.<1–7, 10 >. [Google Scholar]
- 5.Omura S, et al. Genome sequence of an industrial microorganism Streptomyces avermitilis: deducing the ability of producing secondary metabolites. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(21):12215–12220. doi: 10.1073/pnas.211433198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cane DE, Ikeda H. Exploration and Mining of the Bacterial Terpenome. Accounts of chemical research. 2011 doi: 10.1021/ar200198d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peters RJ. Two rings in them all: The labdane-related diterpenoids. Nat. Prod. Rep. 2010;27(11):1521–1530. doi: 10.1039/c0np00019a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dairi T, et al. Eubacterial diterpene cyclase genes essential for production of the isoprenoid antibiotic terpentecin. J Bacteriol. 2001;183(20):6085–6094. doi: 10.1128/JB.183.20.6085-6094.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Durr C, et al. Biosynthesis of the terpene phenalinolactone in Streptomyces sp. Tu6071: analysis of the gene cluster and generation of derivatives. Chem. Biol. 2006;13(4):365–377. doi: 10.1016/j.chembiol.2006.01.011. [DOI] [PubMed] [Google Scholar]
- 10.Smanski MJ, et al. Dedicated ent-kaurene and ent-atiserene synthases for platensimycin and platencin biosynthesis. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(33):13498–13503. doi: 10.1073/pnas.1106919108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vos PD. Bergey's manual of systematic bacteriology. New York: Springer; 2009. [Google Scholar]
- 12.Brosch R, et al. A new evolutionary scenario for the Mycobacterium tuberculosis complex. Proc Natl Acad Sci U S A. 2002;99(6):3684–3689. doi: 10.1073/pnas.052548299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gutacker MM, et al. Genome-wide analysis of synonymous single nucleotide polymorphisms in Mycobacterium tuberculosis complex organisms: resolution of genetic relationships among closely related microbial strains. Genetics. 2002;162(4):1533–1543. doi: 10.1093/genetics/162.4.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Russell DG, Barry CE, 3rd, Flynn JL. Tuberculosis: what we don't know can, does, hurt us. Science. 2010;328(5980):852–856. doi: 10.1126/science.1184784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coscolla M, Gagneux S, Does M. tuberculosis genomic diversity explain disease diversity? Drug Discov Today Dis Mech. 7(1):e43–e59. doi: 10.1016/j.ddmec.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chiang CY, Centis R, Migliori GB. Drug-resistant tuberculosis: past, present, future. Respirology. 2010;15(3):413–432. doi: 10.1111/j.1440-1843.2010.01738.x. [DOI] [PubMed] [Google Scholar]
- 17.Deretic V, et al. Mycobacterium tuberculosis inhibition of phagolysosome biogenesis and autophagy as a host defence mechanism. Cell. Microbiol. 2006;8:719–727. doi: 10.1111/j.1462-5822.2006.00705.x. [DOI] [PubMed] [Google Scholar]
- 18.Stinear TP, et al. Giant plasmid-encoded polyketide synthases produce the macrolide toxin of Mycobacterium ulcerans. Proc Natl Acad Sci U S A. 2004;101(5):1345–1349. doi: 10.1073/pnas.0305877101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Russell DG. Who puts the tubercle in tuberculosis? Nat. Rev. Microbiol. 2007;5:39–47. doi: 10.1038/nrmicro1538. [DOI] [PubMed] [Google Scholar]
- 20.Joshi SM, et al. Characterization of mycobacterial virulence genes through genetic interaction mapping. Proc Natl Acad Sci U S A. 2006;103(31):11760–11765. doi: 10.1073/pnas.0603179103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rengarajan J, Bloom BR, Rubin EJ. Genome-wide requirements for Mycobacterium tuberculosis adaptation and survival in macrophages. Proc Natl Acad Sci U S A. 2005;102(23):8327–8332. doi: 10.1073/pnas.0503272102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sassetti CM, Rubin EJ. Genetic requirements for mycobacterial survival during infection. Proc Natl Acad Sci U S A. 2003;100(22):12989–12994. doi: 10.1073/pnas.2134250100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pethe K, et al. Isolation of Mycobacterium tuberculosis mutants defective in the arrest of phagosome maturation. Proc Natl Acad Sci U S A. 2004;101:13642–13647. doi: 10.1073/pnas.0401657101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakano C, et al. Mycobacterium tuberculosis H37Rv3377c encodes the diterpene cyclase for producing the halimane skeleton. Chem. Commun. 2005;2005:1016–1018. doi: 10.1039/b415346d. [DOI] [PubMed] [Google Scholar]
- 25.Mann FM, et al. Edaxadiene: a new bioactive diterpene from Mycobacterium tuberculosis. J. Am. Chem. Soc. 2009;131(48):15726–15727. doi: 10.1021/ja9019287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peters RJ, et al. Bifunctional abietadiene synthase: Free diffusive transfer of the (+)-copalyl diphosphate intermediate between two distinct active sites. J. Am. Chem. Soc. 2001;123(37):8974–8978. doi: 10.1021/ja010670k. [DOI] [PubMed] [Google Scholar]
- 27.Prisic S, et al. Probing the role of the DXDD motif in class II diterpene cyclases. ChemBioChem. 2007;8:869–874. doi: 10.1002/cbic.200700045. [DOI] [PubMed] [Google Scholar]
- 28.Mann FM, et al. Characterization and inhibition of a class II diterpene cyclase from Mycobacterium tuberculosis: implications for tuberculosis. J. Biol. Chem. 2009;284(35):23574–23579. doi: 10.1074/jbc.M109.023788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maugel N, et al. Synthesis of (+/−)-nosyberkol (isotuberculosinol, revised structure of edaxadiene) and (+/−)-tuberculosinol. Org. Lett. 2010;12(11):2626–2629. doi: 10.1021/ol100832h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spangler JE, Carson CA, Sorenson EJ. Synthesis enables a structural revision of the Mycobacterium tuberculosis-produced diterpene, edaxadiene. Chem. Sci. 2010;1:202–205. doi: 10.1039/C0SC00284D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rudi A, et al. Asmarines I, J, and K and nosyberkol: four new compounds from the marine sponge Raspailiac sp. J Nat Prod. 2004;67(11):1932–1935. doi: 10.1021/np049834b. [DOI] [PubMed] [Google Scholar]
- 32.Nakano C. Structure of Diterpene Produced by Rv3378c Gene Product from Mycobacterium tuberculosis. Koryo Terupen oyobi Seiyu Kagaku ni kansuru Toronkai Koen Yoshishu (Japan) 2005;49:247–249. [Google Scholar]
- 33.Nakano C, Hoshino T. Characterization of the Rv3377c gene product, a type-B diterpene cyclase, from the Mycobacterium tuberculosis H37 genome. Chembiochem. 2009;10(12):2060–2071. doi: 10.1002/cbic.200900248. [DOI] [PubMed] [Google Scholar]
- 34.Nakano C, et al. Characterization of the Rv3378c gene product, a new diterpene synthase for producing tuberculosinol and (13R, S)-isotuberculosinol (nosyberkol), from the Mycobacterium tuberculosis H37Rv genome. Bioscience, biotechnology, and biochemistry. 2011;75(1):75–81. doi: 10.1271/bbb.100570. [DOI] [PubMed] [Google Scholar]
- 35.Hoshino T, et al. Substrate specificity of Rv3378c, an enzyme from Mycobacterium tuberculosis, and the inhibitory activity of the bicyclic diterpenoids against macrophage phagocytosis. Organic & biomolecular chemistry. 2011;9(7):2156–2165. doi: 10.1039/c0ob00884b. [DOI] [PubMed] [Google Scholar]
- 36.Prach L, et al. Diterpene production in Mycobacterium tuberculosis. FEBS J. 2010;277(17):3588–3595. doi: 10.1111/j.1742-4658.2010.07767.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stewart GR, et al. Mycobacterial mutants with defective control of phagosomal acidification. PLoS Pathog. 2005;1(3):269–278. doi: 10.1371/journal.ppat.0010033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rohde K, et al. Mycobacterium tuberculosis and the environment with the phagosome. Immunol. Rev. 2007;291:37–54. doi: 10.1111/j.1600-065X.2007.00547.x. [DOI] [PubMed] [Google Scholar]
- 39.Waddell SJ, Butcher PD. Microarray analysis of whole genome expression of intracellular Mycobacterium tuberculosis. Curr Mol Med. 2007;7(3):287–296. doi: 10.2174/156652407780598548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Garcia-del Portillo F, et al. Characterization of the micro-environment of Salmonella typhimurium-containing vacuoles within MDCK epithelial cells. Mol Microbiol. 1992;6(22):3289–3297. doi: 10.1111/j.1365-2958.1992.tb02197.x. [DOI] [PubMed] [Google Scholar]
- 41.Lavigne JP, O'Callaghan D, Blanc-Potard AB. Requirement of MgtC for Brucella suis intramacrophage growth: a potential mechanism shared by Salmonella enterica and Mycobacterium tuberculosis for adaptation to a low-Mg2+ environment. Infect Immun. 2005;73(5):3160–3163. doi: 10.1128/IAI.73.5.3160-3163.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Prisic S, Peters RJ. Synergistic substrate inhibition of ent-copalyl diphosphate synthase: A potential feed-forward inhibition mechanism limiting gibberellin metabolism. Plant Physiol. 2007;144:445–454. doi: 10.1104/pp.106.095208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mann FM, et al. A single residue switch for Mg2+-dependent inhibition characterizes plant class II diterpene cyclases from primary and secondary metabolism. J. Biol. Chem. 2010;285(27):20558–20563. doi: 10.1074/jbc.M110.123307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ishijima S, et al. Light-induced increase in free Mg2+ concentration in spinach chloroplasts: Measurement of free Mg2+ by using a fluorescent probe and necessity of stromal alkalinization. Arch. Biochem. Biophys. 2003;412:126–132. doi: 10.1016/s0003-9861(03)00038-9. [DOI] [PubMed] [Google Scholar]
- 45.Mann FM, VanderVen BC, Peters RJ. Magnesium depletion triggers production of an immune modulating diterpenoid in Mycobacterium tuberculosis. Mol. Microbiol. 2011;79(6):1594–1601. doi: 10.1111/j.1365-2958.2011.07545.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bailey AM, et al. Identification, cloning, purification, and enzymatic characterization of Mycobacterium tuberculosis 1-deoxy-D-xylulose 5-phosphate synthase. Glycobiology. 2002;12(12):813–820. doi: 10.1093/glycob/cwf100. [DOI] [PubMed] [Google Scholar]
- 47.Reddy TB, et al. TB database: an integrated platform for tuberculosis research. Nucleic Acids Res. 2009;37(Database issue):D499–D508. doi: 10.1093/nar/gkn652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Comas I, et al. Human T cell epitopes of Mycobacterium tuberculosis are evolutionarily hyperconserved. Nature genetics. 2010;42(6):498–503. doi: 10.1038/ng.590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ichiyama S, Shimokata K, Tsukamura M. Relationship between mycobacterial species and their carotenoid pigments. Microbiology and immunology. 1988;32(5):473–479. doi: 10.1111/j.1348-0421.1988.tb01407.x. [DOI] [PubMed] [Google Scholar]
- 50.Cao R, et al. Diterpene cyclases and the nature of the isoprene fold. Proteins. 2010;78(11):2417–2432. doi: 10.1002/prot.22751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kawasaki T, et al. Presence of copalyl diphosphate synthase gene in an actinomycete possessing the mevalonate pathway. J. Antibiot. 2004;57(11):739–747. doi: 10.7164/antibiotics.57.739. [DOI] [PubMed] [Google Scholar]
- 52.Morrone D, et al. Gibberellin biosynthesis in bacteria: Separate ent-copalyl diphosphate and ent-kaurene synthases in Bradyrhizobium japonicum. FEBS Lett. 2009;583(2):475–480. doi: 10.1016/j.febslet.2008.12.052. [DOI] [PubMed] [Google Scholar]