Abstract
Telomeres are the physical ends of eukaryotic chromosomes. They are important for maintaining the integrity of chromosomes and this function is mediated through a number of protein factors. In Saccharomyces cerevisiae, Cdc13p binds to telomeres and affects telomere maintenance, telomere position effects and cell cycle progression through G2/M phase. We identified four genes encoding Pol1p, Sir4p, Zds2p and Imp4p that interact with amino acids 1–252 of Cdc13p using a yeast two-hybrid screening system. Interactions of these four proteins with Cdc13p were through direct protein–protein interactions as judged by in vitro pull-down assays. Direct protein–protein interactions were also observed between Pol1p–Imp4p, Pol1p–Sir4p and Sir4p–Zds2p, whereas no interaction was detected between Imp4p–Sir4p and Zds2p–Imp4p, suggesting that protein interactions were specific in the complex. Pol1p was shown to interact with Cdc13p. Here we show that Zds2p and Imp4p also form a stable complex with Cdc13p in yeast cells, because Zds2p and Imp4p co-immunoprecipitate with Cdc13p, whereas Sir4p does not. The function of the N-terminal 1–252 region of Cdc13p was also analyzed. Expressing Cdc13(252–924)p, which lacks amino acids 1–252 of Cdc13p, causes defects in progressive cell growth and eventually arrested in the G2/M phase of the cell cycle. These growth defects were not caused by progressive shortening of telomeres because telomeres in these cells were long. Point mutants in the amino acids 1–252 region of Cdc13p that reduced the interaction between Cdc13p and its binding proteins resulted in varying level of defects in cell growth and telomeres. These results indicate that the interactions between Cdc13(1–252)p and its binding proteins are important for the function of Cdc13p in telomere regulation and cell growth. Together, our results provide evidence for the formation of a Cdc13p-mediated telosome complex through its N-terminal region that is involved in telomere maintenance, telomere length regulation and cell growth control.
INTRODUCTION
Cdc13p is a single-stranded TG1–3 binding protein that interacts with telomeres in Saccharomyces cerevisiae (1–5). It binds specifically to single-stranded TG1–3 DNA, a tailed duplex DNA molecule with single-stranded TG1–3 tail, or a quadruplex DNA formed by TG1–3 sequences (1,2,6,7). CDC13 is an essential gene (8). It is involved in cell cycle control (8), since a temperature-sensitive allele, cdc13-1 (a Pro371→Ser change), causes cell arrest in the G2/M phase of the cell cycle at non-permissive temperatures (8). This cell cycle control appears to be dependent on the presence of a single-stranded TG1–3 tail since masking its presence rescues cell cycle arrest (9). The gene product of RAD9, which is involved in surveying the integrity of chromosomes, also plays an essential role in cell cycle control (10). Cdc13p may enable Rad9p to differentiate whether the ends of a linear DNA are telomeres or broken ends by binding to telomeres. At least part of the cell cycle control function contributed by Cdc13p is through the interaction with Stn1p and Ten1p (11–13). It appears that these two genes act together in maintaining telomere function because telomere defects caused by STN1 or TEN1 mutation are similar to CDC13 mutation (11,13,14).
Cdc13p is also involved in regulating the length of telomeres (2,15). A mutation allele of CDC13, cdc13-2 (a Glu252→Lys change), brings about a gradual loss of telomere length whereas a truncated form of CDC13, cdc13-5 (an N-terminal amino acids 1–694 fragment of Cdc13p), has long telomeres. Genetic and physical interactions were also observed between CDC13 and EST1, and mutation in EST1 caused a gradual loss of telomere length (2,14,16,17). Est1p is associated with the RNA component of telomerase and is bound to single-stranded telomeric DNA in vitro (18–21). Cdc13p might cooperate with Est1p to recruit telomerase to telomeres or activate telomere-bound telomerase for its replication (5,22). Moreover, it might promote C1–3A DNA synthesis through its interaction with the catalytic subunit of polymerase α (17).
CDC13 encodes a 924 amino acid protein with a predicted molecular mass of 104 895 (8). The DNA-binding region of Cdc13p is within the polypeptide, comprising amino acids 497–693 (6,12,23,24). A region comprising amino acids 190–340 has been reported to be involved in telomerase recruitment, probably through interaction with Est1p (14). The Cdc13p fragment from amino acid 252 to 924, which contains both the telomere-binding region and Stn1p-interacting region, are required to maintain cell viability (12). In the present work we show that the N-terminal region comprising amino acids 1–252 of Cdc13p interacts with Pol1p, Sir4p, Zds2p and Imp4p. Moreover, CDC13-deleted yeast cells expressing Cdc13p lacking the N-terminal 1–252 amino acids region or Cdc13p with point mutations in this region caused defects in progressive cell growth and in cell cycle control. These cells also have defects in telomere length regulation. Thus, we conclude that the N-terminal region of Cdc13p is involved in telomere maintenance, telomere length regulation and cell growth control through its interaction with its binding proteins.
MATERIALS AND METHODS
Plasmids
Escherichia coli DH5α was used as the host for plasmid construction and propagation. To obtain bait plasmids expressing amino acids 1–252 of Cdc13p, plasmid pVZ-CDC13 (17) was first digested with SalI, followed by EcoRI partial digestion. CDC13 fragments encoding amino acids 1–924 were then isolated and ligated to EcoRI and SalI digested pEG202 (Ampr HIS3 2µ) to generate plasmids pEG-CDC13. Plasmid pEG-CDC13(1–252) was constructed by ligating the EcoRI–EcoRI DNA fragment of pEG-CDC13 containing CDC13(1–252) to EcoRI digested pEG202.
To map the minimal POL1 interacting region on CDC13, different fragments of CDC13 were generated by PCR. Primers cdc1w (5′-AACCATGGATGGATACCTTAGA AGA-3′, NcoI site) and cdc163c (5′-AAGTCGACTGTC GATTTTGTAGATAT-3′, SalI site) were used to amplify CDC13(1–163); primers cdc78w (5′-AACCATGGATAGG GGCTCGGAGAGG-3′, NcoI site) and cdc163c were used to amplify CDC13(78–163); primers cdc78w and cdc252c (5′-AAGTCGACGAATTCGTTTTGGGATTC-3′, SalI site) were used to amplify CDC13(78–252). The amplified products were digested with NcoI and SalI and ligated with pEG202, which was previously treated with the same two enzymes.
To map the minimal region of POL1 interacting with CDC13, different fragments of POL1 were generated by PCR. Primers pol47w (5′-TAGAATTCGCCAGAAAGCGCCA GGAA-3′, EcoRI site) and pol412c (5′-TTCTCGAGCGG AATTATTTCCTCGTG-3′, XhoI site) were used to amplify POL1(47–412); primers pol227w (5′-TAGAATTCGCAAA TGATGTACAGGAT-3′, EcoRI site) and pol412c were used to amplify POL1(227–412). The PCR products were treated with EcoRI and XhoI and ligated with pJG4-5 (Ampr TRP1 2µ) which had been previously treated with EcoRI and XhoI.
Plasmid pGST-CDC13(1–252) was constructed by digesting pGST-CDC13 (12) with EcoRI and followed by self-ligation. The resulting plasmid was used to express glutathione S-transferase (GST) fused with Cdc13(1–252)p under control of the tac promoter. Plasmid pMAL-POL1(47–412) was constructed by ligating the EcoRI fragment of pJG-POL1(47–412) with EcoRI digested pMAL™-p2X (New England Biolabs). To construct a plasmid that expresses full-length Imp4p, IMP4 was first PCR amplified from plasmid p415GPD-IMP2-HA using primers imp1w (5′-AACCATG GTAAGAAGACAAGCCCGTG-3′) and imp290c (5′-AAACTAGTTCACAAATAGTCTTTTTATTGGC-3′) (25). Plasmid pET6H-IMP4 was constructed by ligating the NcoI and SpeI digested PCR fragment with pET-6H, which was digested with the same enzymes. To construct a plasmid that expresses amino acids 1143–1358 of Sir4p, DNA encoding this fragment of SIR4 was PCR amplified from two-hybrid plasmid pJG-SIR4(1143–1358) using primers JGSIR4-5 (5′-TACCATGGAGCTGAGCACAATGGAAACGAA-3′) and SIR43 (5′-CGGATCCTCAATACGGTTTTATCTCCTT-3′). The PCR amplified DNA was digested with NcoI and BamHI and ligated with pET-6H that had been digested with the same enzymes. Plasmid pMAL-SIR4 that expresses amino acids 1143–1358 of Sir4p fused to maltose-binding protein (MBP) was constructed by ligating the EcoRI fragment of pJG-SIR4(1143–1358) to EcoRI digested pMAL-p2 (New England Biolabs). To construct a plasmid that expresses amino acids 553–786 of Zds2p, DNA encoding this fragment of ZDS2 was PCR amplified from two-hybrid pJG-ZDS2(553–786) using primers ZDS25 (5′-ACCA TGGAGCTTACAGATGATGAAGAC-3′) and ZDS23 (5′-AGGATCCACTAGAGTGCACTTCGACGT-3′). The PCR amplified DNA was digested with NcoI and BamHI and ligated with pET-6H that had been digested with the same enzymes.
Two-hybrid screening
For two-hybrid screening, the DNA fragment encoding the N-terminal 1–252 amino acids of Cdc13p was first ligated downstream of a LexA DNA-binding domain in plasmid pEG202 (26). The resulting bait plasmid, pEG-CDC13(1–252), was transformed into yeast YEM1α (MATa his3 trp1 leu2::pLexAop6-LEU2 URA3-pLexAop8-LacZ) (17). To screen for interacting proteins, a plasmid library with yeast DNA cloned downstream of a B42 activation domain was transformed into YEM1α/pEG-CDC13(1–252) and selected for Leu+ cells in the presence of 3% galactose. Leu+ cells were then subjected to a colony-lift filter β-galactosidase assay. For confirmation, prey plasmids of Leu+ and LacZ+ colonies were recovered from E.coli and transformed back into YEM1α/pEG-CDC13(1–252) and YEM1α/pEG202. Subsequently, plasmids of the positives were sequenced using Sequenase (US Biochemical).
Complementation of cdc13Δ by CDC13 fragments
Plasmid loss experiments were carried out to test if Cdc13(252–924)p is sufficient to complement the essential functions of a cdc13Δ mutation (12). Briefly, haploid strains YJL501 (cdc13Δ::HIS3/YEP24-CDC13) and YJL501rad52 (cdc13Δ::HIS3 rad52Δ::TRP1/YEP24-CDC13) were constructed, which require a plasmid carrying CDC13 (YEP24-CDC13) for viability. Plasmids pTHA-NLS, pTHA-NLS-CDC13 and pTHA-NLS-CDC13(252–924) were separately transformed into YJL501 or YJL501rad52 cells. The resulting transformants were spotted on plates containing 0.5 mg/ml 5-fluoroorotic acid (5-FOA) and incubated at 30°C until colonies formed.
In vitro association analysis
To prepare soluble fusion proteins GST–Cdc13(1–252)p (12), a 20 ml culture of E.coli cells was grown in LB medium containing 50 µg/ml ampicillin at 30°C to OD600 = 0.5. After induction by 1 mM IPTG for 3 h, cells were collected and sonicated in 1 ml of extraction buffer [1× phosphate-buffered saline (PBS) pH 7.4, 5% glycerol, 0.5% Triton X-100, 1 mM EDTA, 1 mM DTT, 1 mM phenylmethylsulfonyl fluoride, 1× protease inhibitors cocktail (Calbiochem)]. The supernatant with soluble fusion proteins was collected by centrifugation (10 000 g) at 4°C for 5 min. The soluble fusion proteins MBP–Pol1(47–412)p, 6×His–Imp4, 6×His–Sir4(1143–1358)p, 6×His–Zds2(553–786)p and MBP–Sir4(1143–1358)p were obtained from 50 ml E.coli cultures by the same procedures.
Three different types of resins, amylose resin, glutathione–agarose and Ni–NTA–agarose, were used in our association assays depending on the fusion protein tested. For example, the bacterial extract with soluble MBP or MBP–Pol1(47–412)p was incubated with amylose resin at 4°C for 1 h. The resin was collected by centrifugation, washed once with extraction buffer and then mixed with the supernatant containing GST or GST–Cdc13(1–252)p. The incubation was allowed to continue at 4°C for 1 h. Subsequently, the resin was collected, washed and eluted with the extraction buffer containing 10 mM maltose. The GST–Cdc13(1–252)p bound was detected by western blotting using polyclonal anti-GST–Cdc13(1–252)p antibodies. Similar experiments were conducted with various combinations of proteins tested.
Co-immunoprecipitation
Yeast cells [YPH258 Δimp4::HIS3 pGAL::IMP4 (25)] harboring HA-tagged Imp4p were used to analyze the interaction between Cdc13p and Imp4p. Polyclonal antibodies against Cdc13(451–693)p or control preimmune sera was added to the total yeast extract (∼500 µg) in buffer A [50 mM Tris–HCl pH 7.5, 1 mM EDTA, 50 mM NaOAc, 1 mM DTT, 1× protease inhibitor cocktail (Calbiochem), 0.1% Tween 20, 20% glycerol] and incubated by mixing at 4°C for 1 h. An aliquot of 50 µl of protein A–Sepharose 4B beads was added to the mixture followed by continued incubation for another 1 h. The beads were then washed three times with buffer A. The immunoprecipitates were eluted with 0.1 M citric acid (pH 3.0) and then subjected to SDS–PAGE analysis. The monoclonal antibody 12CA5 against the HA tag was used in western blotting analysis to detect the presence of HA-tagged Imp4p. BJ2168 yeast cells were used to determine the interaction between Cdc13p and Zds2p. Polyclonal antibodies against Cdc13(451–693)p were used to precipitate the Cdc13p–protein complex and polyclonal antibodies against 6×His–Zds2(553–786)p (Chen and Lin, unpublished results) were used to detect Zds2p in western blotting analysis.
Fluorescence-activated cell sorting (FACS) and budding index analysis
To examine the cell cycle defect of cdc13Δ/Cdc13(252–924)p cells, the cells were grown in liquid medium to OD600 = 1. The cells were harvested by centrifugation, rinsed with water and fixed in 70% ethanol at 4°C for 16 h. The cells were then washed with PBS (10 mM Na phosphate pH 7.4, 0.9% NaCl) and incubated in the same buffer containing 1 mg/ml RNase A at 37°C for another 16 h. Thereafter, cells were stained with 50 µg/ml propidium iodide for 3 h, diluted to ∼106 cells/ml and sonicated before being subjected to FACS analysis.
To determine the budding index, cdc13Δ/Cdc13(252–924)p cells were grown as described. The cells were then harvested, washed, fixed with 3.7% formaldehyde, sonicated and examined under a light microscope. Single cells (G1 phase), small bud cells (S phase) and large bud or two equal-sized cells (G2/M phase) were scored.
Telomere length determination
To determine telomere length, yeast DNA was prepared, digested with XhoI and separated on 1% agarose gels. The DNA fragments were transferred to a Hybond N+ paper (Amersham) for hybridization and a random primed C1–3A/TG1–3 DNA fragment was used as probe (27).
Identification of Cdc13(1–252)p mutants that fail to bind to its binding proteins
We used PCR mutagenesis to generate mutations in Cdc13(1–252)p that failed to interact with its binding proteins. Primers EG5 (5′-GAAATCGCGCAGCGTTTGGG-3′) and EG3 (5′-CGTGCTGCTAGCGCTATATGC-3′) were used to amplify a DNA fragment that covered 500 bp from 5′ and 3′ of Cdc13(1–252)p in plasmid pEG-CDC13(1–252)p. Nucleotide dITP was added to increase the mutation frequency. The PCR amplified DNA fragment was mixed with EcoRI and XhoI digested pEG202 and transformed into YEM1α/pJG-IMP4(38–290), YEM1α/pJG-SIR4(1143–1358), YEM1α/pJG-ZDS2(553–786) or YEM1α/pJG-POL1(47–560). The transformants were grown on YC plates lacking histidine and tryptophan. Mutations of Cdc13(1–252)p that prevented cells from growing on YC plates lacking leucine in the presence of 3% galactose were selected. Total cell extracts from these mutants were analyzed by western blotting using anti-Cdc13(1–252)p antibodies to confirm the presence of mutant proteins. Plasmids from these mutants were then isolated, sequenced and transformed back into YEM1α/pJG4-5, YEM1α/pJG-IMP4(38–290), YEM1α/pJG-SIR4(1143–1358), YEM1α/pJG-ZDS2(553–786) or YEM1α/pJG-POL1(47–560) to examine the interactions with other binding proteins using two-hybrid assays.
RESULTS
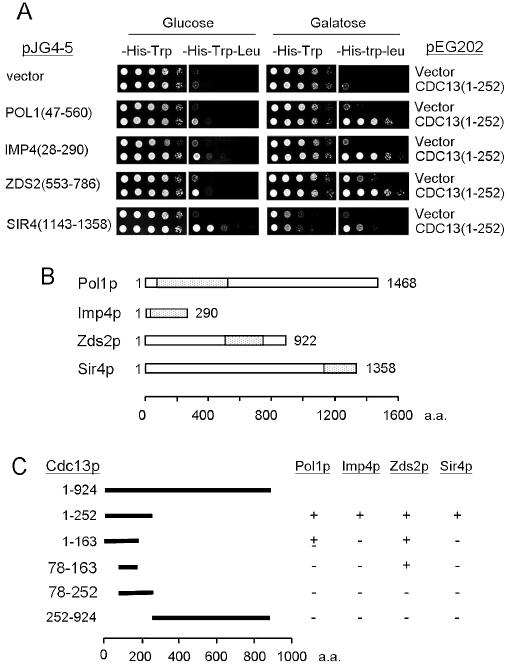
Cdc13(1–252)p interacts with Pol1p, Imp4p, Zds2p and Sir4p
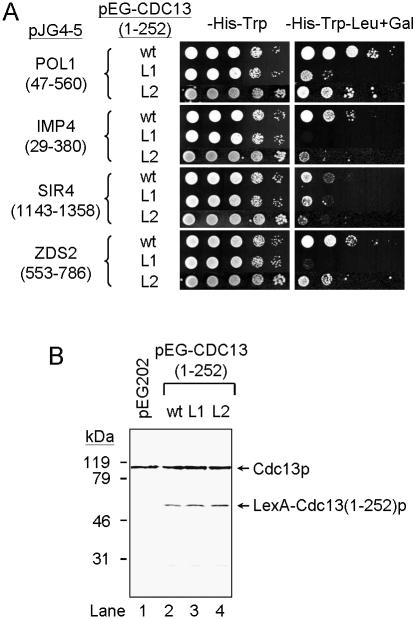
Even though sequence analysis did not reveal any functional domain of Cdc13p, several studies on Cdc13p have revealed structure–function relationships. The telomeric DNA-binding region was mapped to a region comprising amino acids 497–693 (6,12,23,24). Moreover, a region, comprising amino acids 190–340 has been reported to be involved in telomerase recruitment (14). The function of the N-terminal region of Cdc13p is less clear. To understand the function of this region in Cdc13p, we conducted yeast two-hybrid screening using Cdc13(1–252)p as the bait to search for proteins that interact with it. Cdc13(1–252)p was fused to the LexA DNA-binding domain and used as the bait in a yeast two-hybrid system to identify its interacting proteins. After screening a total of ∼650 000 transformants, we identified four clones that interacted strongly with Cdc13(1–252)p (Fig. 1A). DNA sequence analysis revealed that these four clones encoded Pol1(47–560)p, Imp4(28–290)p, Zds2(553–786)p and Sir4(1143–1358)p fused in-frame to the B42 activation domain (Fig. 1B). Pol1p is the catalytic subunit of DNA polymerase α. Previous genetic studies and yeast two-hybrid analyses have indicated an interaction between Cdc13p and Pol1p (17). Imp4p is a component of the U3 small nucleolar ribonucleoprotein (snoRNP) that is essential for pre-18S rRNA processing (25,28). Zds2p has roles in a variety of cellular functions, including signal transduction, translation, chromatin organization, cell cycle regulation, mating type cassettes and rDNA silencing, and may also affect the lifespan of yeast cells (29–34). Sir4p forms a complex with Sir3p that participates in mating type cassettes and telomere silencing (35–37). Two-hybrid analyses indicate that Zds2p also interacts with Sir4p (33). It is interesting to note that in the absence of galactose induction, the Sir4p construct gave Leu+-positive cells in the presence of Cdc13(1–252)p (Fig. 1A). This is probably due to leaky repression of the GAL1 promoter by glucose. Expression of a B42 activation domain–Sir4(1143–1358)p fusion protein appears to have a negative effect on cell growth (Fig. 1A). It is unclear how this fusion protein causes such an effect. However, since the C-terminus of Sir4p also interacts with other proteins that are important for cell growth, expression of excess amount of the C-terminal portion of Sir4p might titrate away these proteins from their normal functions.
Figure 1.
Interaction of Cdc13(1–252)p with Pol1(47–560)p, Imp4(28–290)p, Zds2(553–786)p and Sir4(1143–1358)p. (A) Two-hybrid analysis of Cdc13(1–252)p and its interacting proteins. Yeast cells YEM1α/pEG202 or YEM1α/pEG-CDC13(1–252) carrying plasmid pJG4-5, pJG-POL1(47–560), pJG-IMP4(28–290), pJG-ZDS2(553–786) or pJG-SIR4(1143–1358) were grown on YC medium lacking histidine and tryptophan at 30°C. Cells were then analyzed by their ability to grow in the absence of leucine. Ten-fold serial dilution of cells were spotted on glucose or galactose plates either without histidine and tryptophan or without leucine, tryptophan and histidine and incubated at 30°C until colonies formed. Photographs of the plates are shown. (B) Polypeptides that interact with Cdc13(1–252)p in two-hybrid analysis. Dotted regions indicate the portion of proteins expressed from the two-hybrid vector pJG4-5. (C) Defining the region of Cdc13p that interacted with Pol1(47–560)p, Imp4(28–290)p, Zds2(553–786)p and Sir4(1143–1358)p. Yeast cells YEM1α/pEG202, YEM1α/pEG-CDC13(1–252), YEM1α/pEG-CDC13(1–163), YEM1α/pEG-CDC13(78–163) or YEM1α/pEG-CDC13(78–252) carrying plasmid pJG4-5, pJG-POL1(47–560), pJG-IMP4(28–290), pJG-ZDS2(553–786) or pJG-SIR4(1143–1358) were analyzed in two-hybrid system. The results are summarized. The plus sign (+) indicates cells that are able to grow on medium lacking leucine whereas the minus sign (–) indicates the opposite.
To further narrow down the region within Cdc13p interacting with these proteins, we generated a series of deletion mutants of Cdc13(1–252)p. Two-hybrid assays with various fragments of Cdc13p were employed to test if these proteins interact with smaller regions of Cdc13(1–252)p. As shown in Figure 1C, only the fragment(s) containing Cdc13(1–252)p interacted with Imp4(28–290)p, Sir4(1143–1358)p and Pol1(47–560)p, whereas fragment Cdc13(78–163)p is sufficient to interact with Zds2(553–786)p. Cdc13p lacking the N-terminal 252 amino acid region of the protein, Cdc13(252–924)p, did not interact with these four proteins (Fig. 1C). These results indicate that the region of interaction of Cdc13p with these four proteins is limited to the N-terminal sequence.
Interactions among Cdc13p, Pol1p, Imp4p, Sir4p and Zds2p in vitro
Yeast two-hybrid analyses detect not only direct protein–protein interactions but also interactions mediated through other factors. Therefore, pull-down assays were carried out to further analyze the nature of the interaction between Cdc13p and its binding proteins. We first fused Pol1(47–412)p to MBP and the recombinant protein was allowed to bind amylose resin. Then, bacterial extracts containing Cdc13(1–252)p fused to GST were passed through the resin. Retention of Cdc13(1–252)p fusion protein was detected by western blotting analysis using polyclonal antibodies against GST–Cdc13(1–252)p (12). The results shown in Figure 2A indicate that Cdc13(1–252)p was associated with Pol1(47–412)p. Similar experiments were conducted to test the interactions between Cdc13(1–252)p and 6×His-tagged Imp4p, Sir4(1143–1358)p and Zds2(553–786)p (see Materials and Methods for details). The results shown in Figure 2B–D indicate that these three proteins were able to interact physically with Cdc13(1–252)p in the absence of other yeast proteins. Faint bands were also detected in Figure 2B (lanes 2 and 4) in samples with GST–CDC13(1–252)p; these bands are non-specific cross-hybridization with the anti-6×His antibody. It appears that 6×His–Sir4p and 6×His–Zds2p have weak affinity for GST protein under our experimental conditions because we also detected weak signals corresponding to the positions of 6×His–Sir4p and 6×His–Zds2p in the GST controls in Figure 2C and D (lane 2). However, since samples with Cdc13(1–252)p reproducibly gave stronger signals than that of GST controls (Fig. 2C and D, lanes 2 and 4), we concluded that Cdc13(1–252)p interacted with Sir4p and Zds2p. Together, our results indicate that Cdc13(1–252)p interacts directly with Pol1(47–412)p, Imp4p, Sir4(1143–1358)p and Zds2(553–786)p.
Figure 2.
Interactions between Cdc13p and its binding proteins in vitro. (A) Bacterial extracts containing MBP or MBP-Pol1(47–412)p were incubated with 50 µl of amylose resin at 4°C for 1 h. The resins were then washed and mixed with the extracts containing GST or GST–Cdc13(1–252)p. The incubation was allowed to continue for 1 h and proteins that bound to resins were eluted with 200 µl of 10 mM maltose. Aliquots of 10 µl of the eluates were analyzed by 10% SDS–PAGE. The results of immunoblotting analysis using an antibody against Cdc13(1–252)p are presented. The bands were visualized by chemiluminescence using an ECL kit (Amersham-Pharmacia). The molecular standard and the position of GST–Cdc13(1–252)p are indicated. Similar experiments were conducted in (B)–(D) with the exception of use of GST–Sepharose resin and detection using anti-6×His antibody. The non-specific band in (B) was detected by the anti-6×His antibody.
We next asked whether these Cdc13-binding proteins can interact with each other using similar in vitro pull-down assays. As shown in Figure 3, interactions of Pol1(47–412)p and Imp4p, Pol1(47–412)p and Sir4(1143–1358)p and Sir4(1143–1358)p and Zds2(553–786)p are apparent, indicating that these proteins interact directly. So far, we have been unable to detect an interaction in vitro between Pol1(47–412)p and Zds2(553–786)p and Imp4p and Sir4(1143–1358)p. Together, these results suggest the formation of a Cdc13p-mediated complex at telomeres.
Figure 3.
Interactions between Pol1p, Imp4p, Sir4p and Zds2p in vitro. The experimental procedures were similar to those described in Figure 5. Bacterial extracts containing MBP, MBP-Pol1(47–412)p (A and B) or MBP-Sir4(1143–1358)p (C) were incubated with 50 µl of amylose resin at 4°C for 1 h. The resins were then washed and mixed with the extracts containing 6×His or 6×His–Imp4p (A), 6×His–Sir4(1143–1358)p (B) or 6×His–Zds2(553–786)p (C). The incubation was allowed to continue for 1 h and proteins that bound to resins were eluted with 200 µl of 10 mM maltose. Aliquots of 10 µl of the eluates were analyzed by 10% SDS–PAGE. The result of immunoblotting analysis using an antibody against 6×His is presented. The bands were visualized by chemiluminescence using an ECL kit (Amersham-Pharmacia).
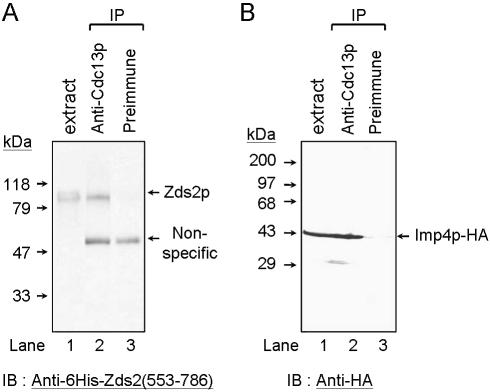
Cdc13p interacts with Zds2p and Imp4p in vivo
It was shown that Cdc13p co-immunoprecipitated with Pol1p (17). Here we apply a similar approach to determine if the interactions between fragments of Cdc13p and Imp4p, Zds2p and Sir4p reflect an in vivo association of full-length proteins. To determine the interaction between Cdc13p and Zds2p, total cell extracts were prepared from cells carrying endogenous copies of both genes expressed from their own promoters. Polyclonal anti-GST–Cdc13(451–693)p antibodies were used to immunoprecipitate Cdc13p. The precipitate was analyzed by western blotting using polyclonal anti-6×His–Zds2(553–786)p antibodies (Chen and Lin, unpublished results). Zds2p, with a predicted mass of 103 732, was readily detected in the anti-Cdc13p immunoprecipitate (Fig. 4A). In the experiment to determine the interaction between Cdc13p and Imp4p, cell extracts were prepared from imp4Δ cells that expressed Imp4p with a triple HA tag on the C-terminus (25). Polyclonal anti-GST–Cdc13(451–693)p antibodies were used to immunoprecipitate Cdc13p. The precipitate was analyzed by western blotting using monoclonal anti-HA antibody. The results shown in Figure 4B showed that Imp4p was present in the anti-Cdc13p immunoprecipitate. Together, we conclude that full-length Cdc13p interacts with Zds2p and Imp4p in vivo. So far, we have been unable to detect Sir4p in the anti-Cdc13p immunoprecipitate (Chen and Lin, data not shown). Consequently, we do not know whether the interaction detected between Cdc13p and Sir4p fragments reflects the interaction of full-length proteins in vivo.
Figure 4.
Interactions between Cdc13p and Zds2p and Imp4p in vivo. (A) Yeast extracts were incubated with polyclonal anti-Cdc13(451–693)p antibodies. Protein A resin was then added to the mixtures to immunoprecipitate the Cdc13–protein complexes. The immunoprecipitates were analyzed by immunoblotting analysis using anti-6×His–Zds2(553–786)p antibodies. The bands were visualized by chemiluminescence using an ECL kit (Amersham-Pharmacia). The molecular standard and the position of Zds2p are indicated. The non-specific bands detected by the anti-Zds2p serum in lanes 2 and 3 are rabbit IgG used in immunoprecipitation. Similar experiments were conducted in (B) with the exceptions that the cell extracts were prepared from yeast cells carrying a HA tag on the C-terminus of Imp4p (Imp4p–HA) and detected using anti-HA antibody. In both blots, one tenth of the cell extracts used in the experiments were loaded as a control (lane 1).
Loss of Cdc13(1–252)p affects cell growth and telomere maintenance
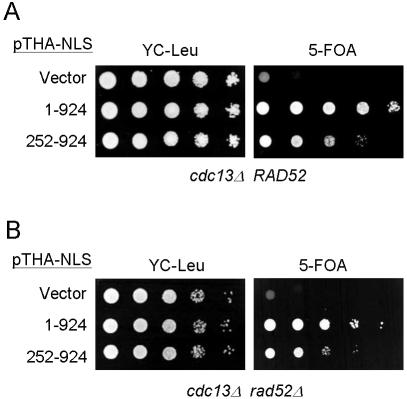
It is known that Cdc13p is essential for cell viability. We have shown that Cdc13(451–693)p containing the telomere-binding region of Cdc13p could not complement the growth arrest caused by cdc13-1 or cdc13-null mutations and, therefore, that the telomere-binding activity alone was not sufficient to account for the essentiality of Cdc13p (12). Instead, we showed that Cdc13(252–924)p complemented the null allele of cdc13 using a plasmid loss experiment. The effect on yeast cells of a lack of Cdc13(1–252)p was analyzed further. Here, the cdc13Δ::HIS3 strain YJL501 requires a CDC13-bearing plasmid (YEP24-CDC13, with a URA3 marker) to grow. If a second plasmid introduced into yeast expresses functional CDC13, then YJL501 no longer requires plasmid YEP24-CDC13 for viability. Growth on 5-FOA is used to monitor loss of YEP24-CDC13. As shown in Figure 5A, 5-FOA-resistant cells were observed in YJL501 transformed with a plasmid expressing Cdc13p. Similarly, 5-FOA-resistant cells were observed in YJL501 cells expressing Cdc13(252–924)p, although this rescue was ∼10–100 times less efficient than that of Cdc13p. However, transformation of YJL501 with plasmid vector pTHA-NLS or plasmids expressing other fragments of CDC13 did not yield any 5-FOA-resistant cells (see also 12). To eliminate the possibility that this effect was due to recombination, the same experiments were carried out in rad52Δ cells that were defective in recombination. Identical results were obtained (Fig. 5B), indicating that survival was not due to a recombination effect.
Figure 5.
Complementation of a cdc13Δ mutant by CDC13(252–924). Yeast strain YJL501 (cdc13Δ::HIS3/YEP24-CDC13) (A) or YJL501rad52 (cdc13Δ::HIS3 rad52Δ::TRP1/YEP24-CDC13) (B) carrying plasmid pTHA-NLS (vector), pTHA-NLS-CDC13 (1–924) or pTHA-NLS-CDC13(252–924) was spotted in 10-fold serial dilutions on YC-Leu plates or plates containing 5-FOA and incubated at 30°C until colonies were formed. The plates are shown here.
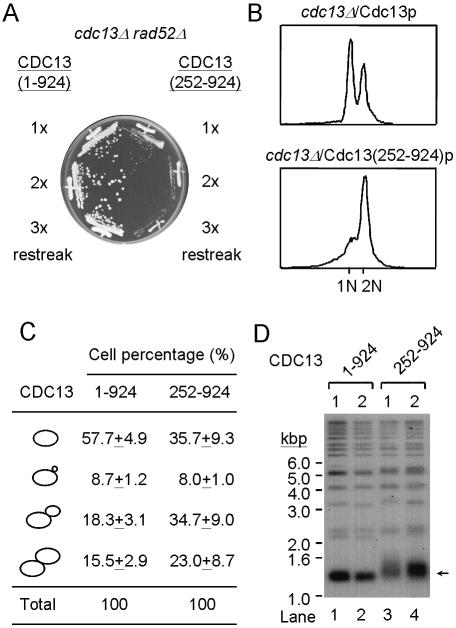
We further analyzed the cell growth and telomere phenotypes. cdc13Δ/Cdc13(252–924)p colonies in 5-FOA plates were generally smaller than cdc13Δ/Cdc13p cells, cdc13Δ/Cdc13(252–924)p cells produced smaller colonies in the first re-streak and did not form colonies at the third re-streak (Fig. 6A). The results suggest that there is a growth defect in these cells. Therefore, FACS analysis was conducted to investigate whether this phenotype is related to the cell cycle defect. The results shown in Figure 6B indicate that the proportion of cdc13Δ/Cdc13(252–924)p cells in G2/M stage was significantly higher than that of cdc13Δ/Cdc13p cells. The budding index was also determined. As shown in Figure 6C, the proportion of large bud and two equal-sized cells (G2/M) increased from 33% in cdc13Δ/Cdc13p cells to 58% in cdc13Δ/Cdc13(252–924)p cells. Thus, cdc13Δ/Cdc13(252–924)p cells have a cell cycle defect in G2/M phase. This cell cycle defect was dependent on RAD9, since cdc13Δ/Cdc13(252–924)p cells failed to arrest at G2/M phase in the absence of RAD9 (data not shown).
Figure 6.
Yeast cdc13Δ/Cdc13(252–924)p cells cause a cell cycle defect and telomere lengthening. (A) 5-FOA-resistant cells from Figure 5 were restreaked on YEPD plates. A photograph of cells that had been grown for the indicated cell divisions is given. (B) A 5 ml overnight culture of cdc13Δ/Cdc13p or cdc13Δ/Cdc13(252–924)p cells was stained with propidium iodide and subjected to FACS analysis. FACS profiles of these cultures are presented. (C) A 5 ml overnight culture of cdc13Δ/Cdc13p or cdc13Δ/Cdc13(252–924)p cells was fixed and sonicated before scoring under a light microscope. The morphologies and the percentage of cells in a specific stage are presented. In each experiment, a total of ∼300–400 cells were scored. The values presented are the averages of three independent determinations. (D) Telomere phenotype of cdc13Δ/Cdc13(252–924)p cells. Yeast DNA was isolated from two different cultures of the indicated strain, digested with XhoI, separated on a 1% agarose gel and analyzed by Southern blotting. The blot was hybridized with probes prepared from 270 bp TG1–3/C1–3A. Arrow indicates the Y′ bearing telomeres.
In S.cerevisiae, middle repetitive sequences known as Y′ elements are found in the subtelomeric regions of most chromosomes (38). As shown in Figure 6D (lanes 1 and 2), in cdc13Δ/Cdc13p cells XhoI digestion produces an ∼1.3 kb fragment from the ends of Y′-bearing chromosomes that contains ∼950 bp of Y′ and a terminal tract of ∼350 bp of TG1–3/C1–3A DNA. However, the length of Y′-bearing telomeres in cdc13Δ/Cdc13(252–924)p cells appeared to be longer and more heterogeneous than those in cdc13Δ/Cdc13p cells (Fig. 6D, lanes 3 and 4). This telomere length phenotype was not progressive, as increased cell divisions did not further affect its length (data not shown). The growth defect observed in cdc13Δ/Cdc13(252–924)p cells does not appear to be caused by gradual telomere shortening. The single-stranded telomeric tail did not appear to be altered in cdc13Δ/Cdc13(252–924)p cells, as judged by Southern hybridization under non-denaturing conditions (data not shown). Yeast CDC13/Cdc13(252–924)p cells that expressed Cdc13(252–924)p with wild-type chromosomal CDC13 did not have a growth defect, a cell cycle delay and long telomeres, suggesting that the phenotype observed in cdc13Δ/Cdc13(252–924)p cells is recessive (data not shown). Therefore, it is likely that the N-terminal region of Cdc13p plays a role in causing the phenotype and that the phenotype we observed in cdc13Δ/Cdc13(252–924)p cells is caused by loss of interactions between Cdc13(1–252)p and its binding proteins.
Identification of Cdc13p mutations that reduce the interaction with its binding proteins
To identify point mutations in Cdc13p that disrupted the interaction between Cdc13(1–252)p and Pol1(47–412)p, Imp4p, Sir4(1143–1358)p or Zds2(553–786)p, we used PCR to mutagenize Cdc13(1–252)p (39). The mutagenized fragments were then assessed for their ability to interact with Pol1(47–412)p, Imp4p, Sir4(1143–1358)p and Zds2(553–786)p in the two-hybrid assay. While screening a total of ∼1200 colonies from each pair of interactions, we isolated a total of 23 mutants that failed to interact with all four proteins and two mutants (cdc13-L1 and cdc13-L2) that failed to interact with some of these four proteins. Since we are interested in analyzing the role of these interactions in telomere function, only cdc13-L1 and cdc13-L2 cells were picked for further analysis (Fig. 7A). The cdc13-L1 mutant failed to interact with Pol1p, Imp4p or Zds2p. Sequence analysis revealed three point mutations that generated three amino acid changes, D2G, L53Q and S134R, in cdc13-L1. Mutant cdc13-L2 failed to interact with Imp4p and Sir4p while it maintained the interactions with Pol1p and Zds2p (Fig. 7A). There were two point mutations that generated two amino acid differences, V18G and K129E, in cdc13-L2. These two mutants showed similar expression levels to that of wild-type protein by western blotting, suggesting that their defects were not due to a loss of protein stability (Fig. 7B).
Figure 7.
Identification of mutant Cdc13p that failed to interact with Pol1(47–560)p, Imp4(28–290)p, Zds2(553–786)p or Sir4(1143–1358)p. (A) Two-hybrid analysis of cdc13 mutants, L1 and L2, and their interacting proteins. Yeast cells YEM1α/pEG-CDC13(1–252), YEM1α/pEG-CDC13(1–252)-L1 or YEM1α/pEG-CDC13(1–252)-L2 carrying plasmid pJG4-5, pJG-POL1(47–560), pJG-IMP4(28–290), pJG-ZDS2(553–786) or pJG-SIR4(1143–1358) were grown on YC medium lacking histidine and tryptophan at 30°C. Cells were then analyzed by their abilities to grow in the absence of leucine. Ten-fold serial dilution of cells were spotted on glucose or galactose plates either without histidine and tryptophan or without leucine, tryptophan and histidine and incubated at 30°C until colonies formed. Photographs of the plates are shown. (B) Western analysis of cdc13 mutants that disrupt interactions with its binding proteins. Extracts prepared from the indicated strains were analyzed by SDS–PAGE and western blotting using an anti-Cdc13(1–252)p antiserum. The positions of endogenous Cdc13p and LexA-Cdc13(1–252)p are indicated.
Similar to cdc13(252–924), a plasmid loss assay indicated that mutant cdc13-L1 could only partially complement a cdc13 null mutation (Fig. 8A). The 5-FOA-resistant cdc13-L1 cells also lost viability after two or three re-streaks, similarly to 5-FOA-resistant cdc13(252–924) cells (data not shown). Telomere length analysis indicates that 5-FOA-resistant cdc13-L1 cells have lengthened telomeres, although the telomere lengthening is not as great as in 5-FOA-resistant cdc13(252–924) cells (Fig. 8B). Mutant cdc13-L2 had different properties. It complemented the cdc13 null mutation and 5-FOA-resistant cells grew almost as well as wild-type cells (Fig. 8A and data not shown). The telomeres of 5-FOA-resistant cells were ∼50 bp shorter than in full-length CDC13 cells (Fig. 8B). Together, several conclusions can be drawn from these analyses. First, the interaction between Cdc13p and Sir4p does not affect cell growth. Second, the interaction between Cdc13p and Pol1p and/or Cdc13p and Zds2p affects the essential function of CDC13 and negatively modulates telomere length. Finally, the interaction of Cdc13p with Imp4p or Sir4p is not required for the essential function of Cdc13p, but positively modulates telomere length.
Figure 8.
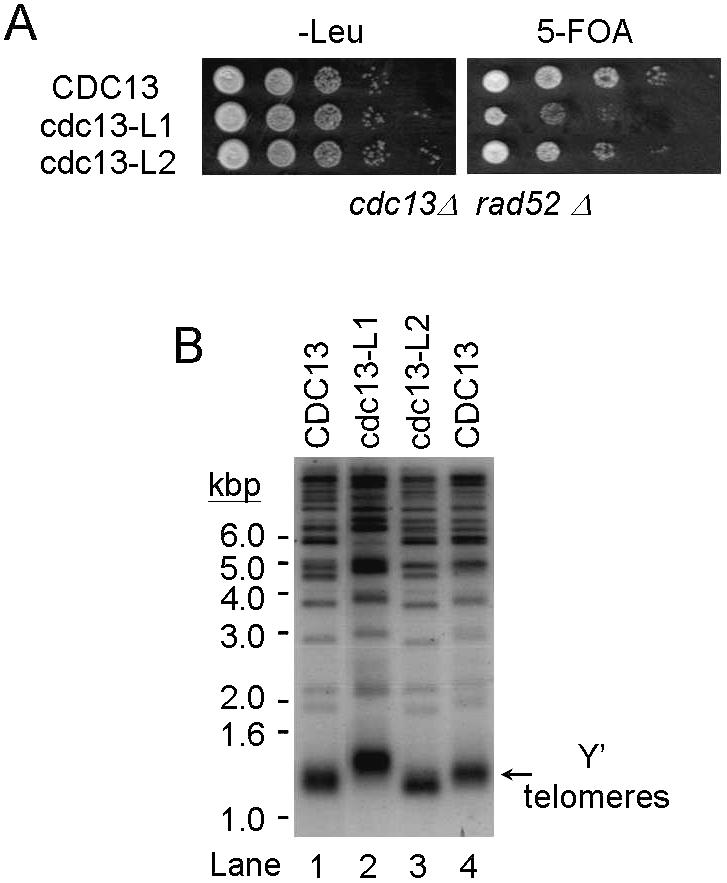
The cdc13 mutants cause telomere maintenance defects. (A) Complementation of a cdc13Δ mutant by cdc13-L1 or cdc13-L2. Yeast strain YJL501rad52 (cdc13Δ::HIS3 rad52Δ::TRP1/YEP24-CDC13) carrying plasmid pTHA-NLS-CDC13(1–924), pTHA-NLS-CDC13-L1 or pTHA-NLS-CDC13-L2 was spotted in 10-fold serial dilutions on YC-Leu plates or plates containing 5-FOA and incubated at 30°C until colonies were formed. The plates are shown here. (B) Telomere phenotype of cdc13Δ/Cdc13-L1p and cdc13Δ/Cdc13-L2p cells. Yeast DNA was isolated from two different cultures of the indicated strain, digested with XhoI, separated on a 1% agarose gel and analyzed by Southern blotting. The blot was hybridized with probes prepared from 270 bp TG1–3/C1–3A. Arrow indicates the Y′ bearing telomeres.
DISCUSSION
Our analyses of cdc13Δ/Cdc13(252–924)p cells revealed several interesting features of Cdc13(1–252)p in telomere function. Even though Cdc13(252–924)p is sufficient to maintain immediate cell growth, it cannot support the long-term survival of cells. Although this type of growth defect is a signature of mutants that show a gradual loss of telomeres (16), the telomeres in cdc13Δ/Cdc13(252–924)p cells are significantly longer than in wild-type cells and remain long when cell growth is affected. It appears that telomere shortening is not the reason for the growth defect in cdc13Δ/Cdc13(252–924)p cells. A similar conclusion was drawn for mammalian cells. In human cells which overexpress TRF2, a telomere-binding protein, the rate of telomere shortening is increased without an acceleration in the rate of senescence (40). It was suggested that the alteration of telomere state is responsible for triggering cell growth arrest.
It is still unclear to us what the primary cause of the cell cycle arrest in cdc13Δ/Cdc13(252–924)p cells is. In pol1 mutants that failed to interact with Cdc13p, cells do not show detectable growth defects (17). Thus, failure of the interaction between Cdc13p and Pol1p might not contribute to the growth phenotype. Even though similar lengthening phenotypes were observed in stn1 and ten1 mutants (11), it is unlikely that failed interactions between Cdc13p and these two proteins causes such telomere changes. The region of interaction between Cdc13p and Stn1p was mapped to amino acids 252–924 of Cdc13p, since interactions between Cdc13p and these two proteins were normal in cells lacking the N-terminal 252 amino acids of Cd1c3p (12). Since both Sir4p and Zds2p are not essential for cell survival, failed interactions between Cdc13p and these two proteins could not contribute to the defect in the cell cycle. We show in the cdc13-L2 mutant that a failed interaction between Cdc13p and Imp4p does not cause the cell cycle defect, indicating that their interaction is not the cause of the cell cycle defect. Thus, any failed interaction between Cdc13p and individual interacting proteins cannot completely explain the cell cycle defect in cdc13Δ/Cdc13(252–924)p cells. Since all the mutants we isolated caused at least two failed interactions, we propose that the growth defect observed was the combined effect of failed interactions between Cdc13p and multiple proteins. Even though the mechanism of how Cdc13(1–252)p affects telomere maintenance and the cell cycle remains to be tested, our results indicate that the N-terminal 1–252 amino acids of Cdc13p affect multiple aspects of telomere function, including replication, cell cycle control and length maintenance.
During telomere replication, telomerase is involved in extending the G-rich strand of the telomere (41). Subsequently, the C-rich strand should be synthesized by lagging strand synthesis (42–44). At least two DNA polymerases, α and δ, were reported to be involved in telomere replication (43,44). However, the molecular mechanism of how these polymerases mediate telomere replication is unclear. The telomeres in several pol1 mutants are longer than in wild-type cells (45). In cdc13 mutants, in which the interaction with Pol1p failed, the telomeres were long (17). Thus, the long telomeres observed in cdc13Δ/Cdc13(252–924)p and cdc13Δ/Cdc13-L1p cells are probably due to loss of the interaction between Cdc13p and Pol1p. These results provide a clue as to how DNA polymerase α might mediate telomere replication; Cdc13p, a telomere-binding protein, might recruit DNA polymerase α to the telomere for C-rich strand synthesis and at the same time inhibit telomere extension by telomerase.
We have shown that the telomere-binding domain maps within amino acids 451–693 of Cdc13p both in vitro and in vivo (6,12). We also showed that the N-terminal 1–251 amino acids of Cdc13p did not bind to single-stranded TG1–3 DNA in vitro (12). However, using a one-hybrid system, Bourns et al. (3) showed that the N-terminal 251 amino acids of Cdc13p interacts with the telomere. Given that Cdc13(1–252)p interacts with Sir4p, binding of Cdc13(1–252)p to the telomere depends on the presence of Sir3p (3) and Sir4p interacts with both Sir3p and Rap1p (46,47), it is likely that this N-terminal 251 amino acid fragment of Cdc13p interacts indirectly with the telomere through its interaction with Sir4p in the one-hybrid system. The interaction between Cdc13p and Sir4p is probably transient or unstable, because we could not detect the interaction by co-immunoprecipitation analysis. It is also evident from the observation that the C-terminal region of Sir4p which interacts with Cdc13p and Zds2p also interacts with several other proteins, including Sir2p, Sir3p, Dot4p, Ubp3p, Zds1p, Rap1p, Hdf1, Dis1p and itself (33,34,46–52). Sir4p may utilize its C-terminal region, which contains a lamin-like coiled-coil domain, to mediate its various interactions in yeast cells (53). These proteins might form different complexes through the combination of different proteins in telomeres and work together for telomere maintenance. The function of the interaction between Cdc13p and Sir4p is still unclear to us. However, since Sir4p is required for telomere silencing and overexpressing Cdc13-1p affects telomere silencing at non-permissive temperatures, we speculate that the interaction might be important for telomere silencing (1).
It has been shown that ZDS1 and ZDS2 have pleiotropic effects in a large number of assays, suggesting roles of these two genes in a variety of cell functions (29–34). Deletion analysis indicates that the ZDS1 mutation affects telomere silencing, whereas the ZDS2 mutation does not (34). Zds2p is implicated in telomere function because it interacts with Sir2p, Sir3p, Sir4p and Rap1p and its overexpression slightly affects telomere silencing (33,34). Here we show an interaction between Cdc13p and Zds2p by both two-hybrid and biochemical criteria. Our results provide evidence that Zds2p is indeed a component of telomeres. It will be interesting to learn how the interaction between these two proteins affects telomere function.
snoRNP particles are known to function in the modification and processing of pre-rRNA. However, additional roles of these complexes have been reported. For example, the class of box H/ACA snoRNPs has been implicated in telomerase function in that the protein components bind the 3′-terminal domain of telomerase RNA (54–56). Mutations in the human H/ACA snoRNP dyskerin and telomerase RNA affect telomerase RNA accumulation and subsequently affect telomerease activity (57,58). Clearly, box H/ACA snoRNPs have a role in telomere maintenance. Here we report another example of a different class of snoRNP, the box C/D snoRNPs, that might also be involved in telomere function. U3 snoRNP is a member of the box C/D class of snoRNAs which is essential for pre-18S rRNA processing (59). U3 snoRNP forms a large ribonucleoprotein complex containing over 20 proteins, including Imp4p (25,28). The interaction between Cdc13p and Imp4p might imply a role of U3 snoRNP in telomere function. Imp4p contains a Brix domain that might be involved in binding to RNA (60). The Brix domain has also been identified in several yeast proteins involved in RNA metabolism (60). Nucleic acids binding analysis also demonstrates that Imp4p is capable of binding to RNA and single-stranded DNA (61). Imp4p might also utilize the Brix domain to interact with the single-stranded telomeric tail or to bind telomerase RNA. It will be interesting to learn how Imp4p affects telomere function.
From our results and genetic evidence provided by others we propose a model for formation of the telosome complex mediated by Cdc13p. Here, Cdc13p binds to the single-stranded tail of the telomere (1,2) and cooperates with Est1p and telomerase for G-rich strand extension (17,22). Cdc13p might also interact and recruit DNA polymerase α to the telomere for C-strand synthesis. The interaction between Cdc13p and Pol1p may negatively regulate telomerase extension so that the telomere would not be extended further, and DNA polymerase α will fill the single-stranded, G-rich DNA by initiating lagging strand synthesis. Genetic and biochemical evidence indicates that Cdc13p interacts with Stn1p and Ten1p for telomere length maintenance (11,13). We identify three additional protein factors that interact with Cdc13p, Imp4p, Sir4p and Zds2p. The interaction between Cdc13p and Sir4p and/or Zds2 might contribute to telomere silencing mediated by Cdc13p. Moreover, interaction of Cdc13p with Imp4p might modulate telomere length, even though the molecular mechanism is unclear. Nevertheless, Cdc13p appears to play a central role in the nucleation of telosome structures.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Dr S.J. Baserga (Yale University, USA) for providing IMP4 reagents, Dr K.W. Runge (Lerner Research Institute, Cleveland Clinic Foundation, USA) for providing ZDS2 reagents and Dr T. Formosa (University of Utah, USA) for providing polymerase α antibodies. We also thank Drs C. Wang and S.C. Teng for critical reading of the manuscript. This work was supported by grants from the NSC (92-2311-B-010-005 and 92-3112-B-010-012).
REFERENCES
- 1.Lin J.-J. and Zakian,V.A. (1996) The Saccharomyces CDC13 protein is a single-strand TG1–3 telomeric DNA-binding protein in vitro that affects telomere behavior in vivo. Proc. Natl Acad. Sci. USA, 93, 13760–13765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nugent C.I., Hughes,T.R., Lue,N.F. and Lundblad,V. (1996) Cdc13p: a single-strand telomeric DNA-binding protein with a dual role in yeast telomere maintenance. Science, 274, 249–252. [DOI] [PubMed] [Google Scholar]
- 3.Bourns B.D., Alexander,M.K., Smith,A.M. and Zakian,V.A. (1998) Sir proteins, rif proteins and Cdc13p bind Saccharomyces telomeres in vivo. Mol. Cell. Biol., 18, 5600–5608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tsukamoto Y., Taggart,A.K.P. and Zakian,V.A. (2001) The role of the Mre11-Rad50-Xrs2 complex in telomerase-mediated lengthening of Saccharomyces cerevisiae telomeres. Curr. Biol., 11, 1328–1335. [DOI] [PubMed] [Google Scholar]
- 5.Taggart A.K.P., Teng,S.-C. and Zakian,V.A. (2002) Est1p as a cell cycle-regulated activator of telomere-bound telomerase. Science, 297, 1023–1026. [DOI] [PubMed] [Google Scholar]
- 6.Lin Y.-C., Hsu,C.-L., Shih,J.-W. and Lin,J.-J. (2001) Specific binding of single-stranded telomeric DNA by Cdc13p of Saccharomyces cerevisiae. J. Biol. Chem., 276, 24588–24593. [DOI] [PubMed] [Google Scholar]
- 7.Lin Y.-C., Shih,J.-W., Hsu,C.-L. and Lin,J.-J. (2001) Binding and partial denaturing of G-quartet DNA by Cdc13p of Saccharomyces cerevisiae. J. Biol. Chem., 276, 47671–47674. [DOI] [PubMed] [Google Scholar]
- 8.Garvik B., Carson,M. and Hartwell,L. (1995) Single-stranded DNA arising at telomeres in cdc13 mutants may constitute a specific signal for the RAD9 checkpoint. Mol. Cell. Biol., 15, 6128–6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pang T.-L., Wang,C.Y., Hsu,C.-L., Chen,M.Y. and Lin,J.-J. (2003) Exposure of single-stranded telomeric DNA causes G2/M cell cycle arrest in Saccharomyces cerevisiae. J. Biol. Chem., 278, 9318–9321. [DOI] [PubMed] [Google Scholar]
- 10.Weinert T.A. and Hartwell,L.H. (1988) The RAD9 gene controls the cell cycle response to DNA damage in Saccharomyces cerevisiae. Science, 241, 317–322. [DOI] [PubMed] [Google Scholar]
- 11.Grandin N., Reed,S.I. and Charbonneau,M. (1997) Stn1, a new Saccharomyces cerevisiae protein, is implicated in telomere size regulation in association with Cdc13. Genes Dev., 11, 512–527. [DOI] [PubMed] [Google Scholar]
- 12.Wang M.-J., Lin,Y.-C., Pang,T.-L., Lee,J.-M., Chou,C.-C. and Lin,J.-J. (2000) Telomere-binding and Stn1p-interacting activities are required for the essential function of Saccharomyces cerevisiae Cdc13p. Nucleic Acids Res., 28, 4733–4741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grandin N., Damon,C. and Charbonneau,M. (2001) Ten1 functions in telomere end protection and length regulation in association with Stn1 and Cdc13. EMBO J., 20, 1173–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pennock E., Buckley,K. and Lundblad,V. (2001) Cdc13 delivers separate complexes to the telomere for end protection and replication. Cell, 104, 387–396. [DOI] [PubMed] [Google Scholar]
- 15.Chandra A., Hughes,T.R., Nugent,C.I. and Lundblad,V. (2001) Cdc13 both positively and negatively regulates telomere replication. Genes Dev., 15, 404–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lundblad V. and Szostak,J.W. (1989) A mutant with a defect in telomere elongation leads to senescence in yeast. Cell, 57, 633–643. [DOI] [PubMed] [Google Scholar]
- 17.Qi H. and Zakian,V.A. (2000) The Saccharomyces telomere-binding protein Cdc13p interacts with both the catalytic subunit of DNA polymerase α and the telomerase-associated Est1 protein. Genes Dev., 14, 1777–1788. [PMC free article] [PubMed] [Google Scholar]
- 18.Lin J.-J. and Zakian,V.A. (1995) An in vitro assay for Saccharomyces telomerase required EST1. Cell, 81, 1127–1135. [DOI] [PubMed] [Google Scholar]
- 19.Steiner B.R., Hidaka,K. and Futcher,B. (1996) Association of the EST1 protein with telomerase activity in yeast. Proc. Natl Acad. Sci. USA, 93, 2817–2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Virta-Pearlman V., Morris,D.K. and Lundblad,V. (1996) Est1 has the properties of a single-stranded telomere end-binding protein. Genes Dev., 10, 3094–3104. [DOI] [PubMed] [Google Scholar]
- 21.Zhou J., Hidaka,K. and Futcher,B. (2000) The Est1 subunit of yeast telomerase binds the Tlc1 telomerase RNA. Mol. Cell. Biol., 20, 1947–1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Evans S.K. and Lundblad,V. (1999) Est1 and Cdc13 as comediators of telomerase access. Science, 286, 117–120. [DOI] [PubMed] [Google Scholar]
- 23.Hughes T.R., Weilbaecher,R.G., Walterscheid,M. and Lundblad,V. (2000) Identification of the single-strand telomeric DNA binding domain of the Saccharomyces cerevisiae Cdc13 protein. Proc. Natl Acad. Sci. USA, 97, 6457–6462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Anderson E.M., Halsey,W.A. and Wuttke,D.S. (2002) Delineation of the high-affinity single-stranded telomeric DNA-binding domain of Saccharomyces cerevisiae Cdc13. Nucleic Acids Res., 30, 4305–4313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee S.J. and Baserga,S.J. (1999) Imp3p and Imp4p, two specific components of the U3 smallnucleolar ribonucleoprotein that are essential for pre-18S rRNA processing. Mol. Cell. Biol., 19, 5441–5452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gyuris J., Golemis,E., Chertkov,H. and Brent,R. (1993) Cdi1, a human G1 and S phase protein phosphatase that associates with Cdk2. Cell, 75, 791–803. [DOI] [PubMed] [Google Scholar]
- 27.Lin J.-J. and Zakian,V.A. (1994) Isolation and characterization of two Saccharomyces cerevisiae genes that encode proteins that bind to (TG1–3)n single strand telomeric DNA in vitro. Nucleic Acids Res., 22, 4906–4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dragon F., Gallagher,J.E.G., Compagnone-Post,P.A., Mitchell,B.M., Porwancher,K.A., Wehner,K.A., Wormsley,S., Settlage,R.E., Shabanowitz,J., Beyer,A.L. et al. (2002) A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature, 417, 967–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bi E. and Pringle,J.R. (1996) ZDS1 and ZDS2, genes whose products may regulate Cdc42p in Saccharomyces cerevisiae. Mol. Cell. Biol., 16, 5264–5275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schwer B., Linder,P. and Shuman,S. (1998) Effects of deletion mutations in the yeast Ces1 protein on cell growth and morphology and on high copy suppression of mutations in mRNA capping enzyme and translation initiation factor 4A. Nucleic Acids Res., 26, 803–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ma X.J., Lu,Q. and Grunstein,M. (1996) A search for proteins that interact genetically with histone H3 and H4 amino termini uncovers novel regulatorsof the Swe1 kinase in Saccharomyces cerevisiae. Genes Dev., 10, 1327–1340. [DOI] [PubMed] [Google Scholar]
- 32.Yu Y., Wellinger,R.J., Carlson,K., Roberts,J.M. and Stillman,D.J. (1996) Mutations in the homologous ZDS1 and ZDS2 genes affect cell cycle progression. Mol. Cell. Biol., 16, 5254–5263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roy N. and Runge,K.W. (1999) The ZDS1 and ZDS2 proteins require the Sir3p component of yeast silent chromatin to enhance the stability of short linear centromeric plasmids. Chromosoma, 108, 146–161. [DOI] [PubMed] [Google Scholar]
- 34.Roy N. and Runge,K.W. (2000) Two paralogs involved in transcriptional silencing that antagonistically control yeast life span. Curr. Biol., 10, 111–114. [DOI] [PubMed] [Google Scholar]
- 35.Ivy J.M., Klar,A.J. and Hicks,J.B. (1986) Cloning and characterization of four SIR genes of Saccharomyces cerevisiae. Mol. Cell. Biol., 6, 688–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schnell R. and Rine,J. (1986) A position effect on the expression of a tRNA gene mediated by the SIR genes in Saccharomyces cerevisiae. Mol. Cell. Biol., 6, 494–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aparicio O.M., Billington,B.L. and Gottschling,D.E. (1991) Modifiers of position effect are shared between telomeric and silent mating-type loci in S. cerevisiae. Cell, 66, 1279–1287. [DOI] [PubMed] [Google Scholar]
- 38.Chan C.S.M. and Tye,B.-K. (1983) A family of Saccharomyces cerevisiae repetitive autonomously replicating sequences that have very similar genomic environments. J. Mol. Biol., 168, 505–523. [DOI] [PubMed] [Google Scholar]
- 39.Spee J.H., de Vos,W.M. and Kuipers,O.P. (1993) Efficient random mutagenesis method with adjustable mutation frequency by use of PCR and dITP. Nucleic Acids Res., 21, 777–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Karlseder J., Smogorzewska,A. and de Lange,T. (2002) Senescence induced by altered telomere state, not telomere loss. Science, 295, 2446–2449. [DOI] [PubMed] [Google Scholar]
- 41.Blackburn E.H. (1992) Telomerases. Annu. Rev. Biochem., 61, 113–129. [DOI] [PubMed] [Google Scholar]
- 42.Zakian V.A. (1995) Telomeres: beginning to understand the end. Science, 270, 1601–1607. [DOI] [PubMed] [Google Scholar]
- 43.Carson M. and Hartwell,L. (1985) CDC17: an essential gene that prevents telomere elongation in yeast. Cell, 42, 249–257. [DOI] [PubMed] [Google Scholar]
- 44.Diede S.J. and Gottschling,D.E. (1999) Telomerase-mediated telomere addition in vivo requires DNA primase and DNA polymerases α and δ. Cell, 99, 723–733. [DOI] [PubMed] [Google Scholar]
- 45.Adams A.K. and Holm,C. (1996) Specific DNA replication mutations affect telomere length in Saccharomyces cerevisiae. Mol. Cell. Biol., 16, 4614–4620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moazed D., Kistler,A., Axelrod,A., Rine,J. and Johnson,A.D. (1997) Silent information regulator protein complexes in Saccharomyces cerevisiae: a SIR2/SIR4 complex and evidence for a regulatory domain in SIR4 that inhibits its interaction with SIR3. Proc. Natl Acad. Sci. USA, 94, 2186–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moretti P., Freeman,K., Coodly,L. and Shore,D. (1994) Evidence that a complex of SIR proteins interacts with the silencer and telomere-binding protein RAP1. Genes Dev., 8, 2257–2269. [DOI] [PubMed] [Google Scholar]
- 48.Park Y., Hanish,J. and Lustig,A.J. (1998) Sir3p domains involved in the initiation of telomeric silencing in Saccharomyces cerevisiae. Genetics, 150, 977–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kahana A. and Gottschling,D.E. (2002) DOT4 links silencing and cell growth in Saccharomyces cerevisiae. Mol. Cell. Biol., 19, 6608–6620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moazed D. and Johnson,A.D. (1996) A deubiquitinating enzyme interacts with SIR4 and regulates silencing in S. cerevisiae. Cell, 86, 667–677. [DOI] [PubMed] [Google Scholar]
- 51.Tsukamoto Y., Kato,J. and Ikeda,H. (1997) Silencing factors participate in DNA repair and recombination in Saccharomyces cerevisiae. Nature, 388, 900–903. [DOI] [PubMed] [Google Scholar]
- 52.Zhang Z. and Buchman,A.R. (1997) Identification of a member of a DNA-dependent ATPase family that causes interference with silencing. Mol. Cell. Biol., 17, 5461–5472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Diffley J.F.X. and Stillman,B. (1989) Transcriptional silencing and lamins. Nature, 342, 24. [DOI] [PubMed] [Google Scholar]
- 54.Mitchell J.R., Cheng,J. and Collins,K. (1999) A box H/ACA small nucleolar RNA-like domain at the human telomerase RNA 3′ end. Mol. Cell. Biol., 19, 567–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dragon F., Pogacic,V. and Filipowicz,W. (2000) In vitro assembly of human H/ACA small nucleolar RNPs reveals unique features of U17 and telomerase RNAs. Mol. Cell. Biol., 20, 3037–3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pogacic V., Dragon,F. and Filipowicz,W. (2000) Human H/ACA small nucleolar RNPs and telomerase share evolutionarily conserved proteins NHP2 and NOP10. Mol. Cell. Biol., 20, 9028–9040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mitchell J.R., Wood,E. and Collins,K. (1999) A telomerase component is defective in the human disease dyskeratosis congenita. Nature, 402, 551–555. [DOI] [PubMed] [Google Scholar]
- 58.Vulliamy T., Marrone,A., Goldman,F., Dearlove,A., Bessler,M., Mason,P.J. and Dokal,I. (2001) The RNA component of telomerase is mutated in autosomal dominant dyskeratosis congenita. Nature, 413, 432–435. [DOI] [PubMed] [Google Scholar]
- 59.Filipowicz W. and Pogacic,V. (2002) Biogenesis of small nucleolar ribonucleoproteins. Curr. Opin. Cell Biol., 14, 319–327. [DOI] [PubMed] [Google Scholar]
- 60.Eisenhaber F., Wechsellberger,C. and Kreil,G. (2001) The Brix domain protein family—a key to the ribosomal biogenesis pathway? Trends Biochem. Sci., 26, 345–347. [DOI] [PubMed] [Google Scholar]
- 61.Wehner K.A. and Baserga,S.J. (2002) The σ70-like motif: a eukaryotic RNA binding domain unique to a superfamily of proteins required for ribosome biogenesis. Mol. Cell, 9, 329–339. [DOI] [PubMed] [Google Scholar]