Abstract
The composition of RNase H2 has been a long-standing problem. Whereas bacterial and archaeal RNases H2 are active as single polypeptides, the Saccharomyces cerevisiae homolog, Rnh2Ap, when expressed in Escherichia coli, fails to produce an active RNase H2. By affinity chromatography purification and identification of polypeptides associated with a tagged S.cerevisiae Rnh2Ap, we obtained a complex of three proteins (Rnh2Ap, Ydr279p and Ylr154p) that together are necessary and sufficient for RNase H2 activity. Deletion of the gene encoding any one of the proteins or mutations in the catalytic site in Rnh2A led to loss of RNase H2 activity. Even when S.cerevisiae RNase H2 is catalytically compromised, it still exhibits a preference for cleavage of the phosphodiester bond on the 5′ side of a ribonucleotide–deoxyribonucleotide sequence in substrates mimicking RNA-primed Okazaki fragments or a single ribonucleotide embedded in a duplex DNA. Interestingly, Ydr279p and Ylr154p have homologous proteins only in closely related species. The multisubunit nature of S.cerevisiae RNase H2 may be important both for structural purposes and to provide a means of interacting with other proteins involved in DNA replication/repair and transcription.
INTRODUCTION
RNases H are enzymes that specifically hydrolyze RNA when annealed to a complementary DNA and are present in all living organisms (1). Most organisms have more than one type, leading to the classification into two distinct groups, RNase HI/H1 and RNase HII/H2, based on amino acid sequence similarity (2). Despite the lack of amino acid sequence homology between the two classes of RNases H, they have similar three-dimensional structures and a common arrangement of carboxylic acid residues that form the catalytic site and presumably cleave their substrates via similar mechanisms (3,4). Some bacteria and archaea have only one type of RNase H, while other bacteria such as Escherichia coli have both RNases HI and HII (5). Eukaryotic RNases H also are of both type I and II. Within a given class, there is a wide variation in specific activity, divalent metal ion preference and sites at which the enzymes are capable of cleaving RNA–DNA hybrids (2). In addition to having a similar catalytic mechanism, both enzymes seem to have overlapping functions in cells, since over-expression of E.coli RNase HII can complement an RNase HI deletion phenotype in E.coli (6) and RNase H2 exhibits an increased activity in rnh1Δ deletion strains of yeast, probably to partially compensate for loss of RNase H1 activity (7).
In single-celled eukaryotes, both RNase H-encoding genes can be deleted with only minor defects on the cell’s ability to respond to DNA-damaging agents (7,8). In contrast, deletion of the RNase H1-encoding gene results in embryonic lethality in mouse (9) and Drosophila melanogaster (10), the former being due to an inability to amplify mitochondrial DNA (9). Unlike RNases H1, the cellular function(s) of eukaryotic RNases H2 is still not well defined but is thought to be involved in removal of Okazaki fragment primers (7,11) and for removal of single ribonucleotides in DNA–DNA duplexes (12–14). For many years, the composition of this class of RNase H remained elusive, with some studies suggesting multiple subunits and others indicating a single polypeptide (15). Although this enzyme is readily detected in enzymatic assays, it is present in low amounts and proved to be difficult to purify by classical techniques.
Highly purified RNases H2 from human tissue culture cells (12,16) and calf thymus (17) exhibit biochemical properties distinct from RNase H1 but share many common properties with archaeal and bacterial RNase HII. Genes encoding clear homologs of bacterial RNases HII are found in eukaryotes and, when the gene is deleted in Saccharomyces cerevisiae, a 70% reduction in RNase H activity in crude extracts is observed (7,18). Overproduction of this polypeptide results in only a modest increase in RNase H activity and expression of this polypeptide in E.coli yields an RNase H with little (11) or no enzymatic activity (19). In contrast, RNases HII from bacteria and archaea are active when expressed in E.coli (3,4,6,20,21). Therefore, it seems likely that eukaryotic RNases H2 may require post-translational modification or an additional subunit(s). Many reports using highly purified RNases H2 derived from mammalian sources suggest the possibility of at least one additional component (15). To determine the composition and enzymatic properties of eukaryotic RNases H2, we employed the well-studied eukaryotic organism, S.cerevisiae.
MATERIALS AND METHODS
Yeast strains and plasmids
The X2180-A (wt), rnh1Δ (HIRO29), rnh2Δ (HIRO16A) and rnh1Δrnh2Δ (HIRO27AF) strains used in this work have been described (7). The BY4741 (wt), ydr279wΔ and ylr154cΔ isogenic strains were obtained from the Yeast Deletion Consortium, and the ydr279wΔylr154cΔ strain was constructed by introducing a HIS3 cassette into the YDR279W allele of the ylr154cΔ strain. The genotypes of all yeast strains used in this work are listed in Table 1S of the Supplementary Material available at NAR Online.
For expression in yeast, we used the previously described pVANPH2 plasmid (7) and added FLAG and hemagglutinin (HA) tags (two each) at the C-terminus of the protein with transcription driven by the native RNH2A promoter. Transfer of the NdeI–XhoI fragment to the pYX242 vector results in expression of Rnh2Ap with triple affinity tags (2× FLAG, 2× HA and six-His) at the C-terminus where transcription from the TPI promoter directs the expression of the cloned gene. Details of the construction of the plasmids used in this work are provided in the Supplementary Material.
Oligonucleotide substrates
Oligonucleotide substrates used for RNase H2 assays were synthesized by Integrated DNA Technologies (Coralville, IA). Each oligonucleotide was labeled using T4 polynucleotide kinase (NEB) following the manufacturer’s recommendation. Labeled oligonucleotides were separated by 15% TBE–urea PAGE, excised from the gel, and 32P-labeled oligonucleotides were eluted from gel fragments by passive diffusion in 50 mM Tris–HCl pH 8.0, 1 mM EDTA, 0.5 M NaCl with gentle agitation at room temperature overnight. Oligonucleotides in the supernatant were recovered by ethanol precipitation and dissolved in 100 µl of diethylpyrocarbonate (DEPC)-treated water and stored at –20°C. The sequences of oligonucleotide substrates used for this work are listed in the Supplementary Material.
Purification of recombinant Rnh2Ap from S.cerevisiae
An rnh1Δrnh2Δ (HIRO27AF) strain harboring either pYESH2HF or pYESH2HFH was used for expression of recombinant Rnh2Ap containing multiple affinity tags. Cells were cultured in YPD broth using a 10 l fermentor and harvested in mid-log phase. Cells (∼30 g wet weight) were washed with sterile water and resuspended in 30 ml of lysis buffer [50 mM Tris–HCl, pH 7.9, 150 mM NaCl, 1 mM EDTA, 10 mM MgCl2, 1 mM dithiothreitol (DTT)], and one tablet of ‘Complete protease inhibitor’ (Roche Biochemicals). Lysates were made by passing the cell suspension at least three times through a French press (Thermo, Inc.) followed by two centrifugations at 12 000 r.p.m. at 4°C for 30 min (Sorvall) and at 35 000 r.p.m. at 4°C for 2 h (60Ti rotor, Beckman).
The cleared lysate was loaded onto 2 ml of FLAG affinity resin (Sigma) and the column was washed with 25 ml of wash buffer (same as lysis buffer except 250 mM NaCl and 0.1% NP-40) at 4°C. Elutions were made by applying three bed volumes of elution buffer (lysis buffer plus 200 µg/ml FLAG peptide).
For the triple-tagged protein, an additional metal affinity column (Talon, Clontech) was performed for further purification of Rnh2Ap. Eluates from FLAG affinity purification were applied to the metal affinity column (1 ml bed volume) and washed with 25 ml of His wash buffer (50 mM Tris–HCl pH 7.9, 250 mM NaCl, 10 mM imidazole, 10% glycerol). Final elution was made by applying three bed volumes of His elution buffer (His wash buffer containing 250 mM imidazole) onto the column. Purification of the protein was monitored by SDS–PAGE and immunoblotting using monoclonal anti-HA antibody (1:2000 dilution) (Tropix) for detection of recombinant Rnh2Ap (7).
Glycerol gradient sedimentation
A 100 µl aliquot of the RNase H2 preparation (∼50 µg of protein) from the FLAG affinity resin was applied onto a 4 ml glycerol gradient (15–30%) in lysis buffer. Centrifugation was in a SW60.1 rotor (Beckman) for 16 h at 45 000 r.p.m. at 4°C, with fractions collected by pipetting out 200 µl aliquots starting from the top. Protein standards consisting of thyroglobulin (669 kDa), ferritin (440 kDa), catalase (232 kDa), γ-globulin (158 kDa) and ovalbumin (44 kDa) (Amersham Bioscience) were also applied onto a parallel gradient.
RNase H assays
RNase H activity was measured using 32P-labeled poly(rA)–poly(dT) (22). Assays using 5′-32P-labeled RNA–DNA hybrids as substrate were performed in the same buffer condition as liquid assays [50 mM Tris–HCl pH 7.9, 10 mM MgCl2, 1 mM DTT, 5% glycerol, 100 µg/ml bovine serum albumin (BSA)] and contained ∼100 fmol of labeled RNA oligonucleotide. To form RNA–DNA hybrids, 5 µl of 200 nM RNA was mixed with 1 µl of 100 µM solution of the corresponding complementary DNA oligonuclotide and incubated at room temperature for 30 min. The substrate was then added to a reaction mixture of 100 µl and divided into 10 µl aliquots for assays. Reactions were pre-incubated for 5 min at 37°C and initiated by adding 1 µl of protein preparation. Reactions were stopped at the indicated times by adding 10 µl of 90% formamide (v/v), 15 mM EDTA, 0.01% bromophenol blue and xylene cyanol-ff. Cleavage products were separated by 15% TBE–urea PAGE and analyzed using a PhosphoImager (Molecular Dynamics).
Peptide identification
Peptides were identified by SELDI ProteinChip® and tandem LC-MS/MS after the affinity-purified Rnh2Ap preparation was separated by SDS–PAGE (8–16%) and visualized by staining in Coomassie solution (7.5% methanol, 0.2% Coomassie blue R-350). After a brief incubation with destaining solution (7.5% methanol), gel segments containing polypeptides were excised and digested by sequencing grade trypsin (Roche). The digested polypeptides were further identified as indicated in the Supplementary Material. For N-terminal amino acid sequencing, the sample was separated by SDS–PAGE and transferred to a PVDF membrane by electroblotting. After visualization by brief Coomassie staining, polypeptide bands were cut out and subjected to Edman degradation microsequencing using an Applied Biosystems model 474A protein sequencer.
RESULTS
Purification of RNase H2 from S.cerevisiae
Using a solution-based assay, a gel renaturation assay and examination of complementation of a temperature-sensitive E.coli strain in which the bacterial RNase HI can be replaced by many different RNases H (19), we failed to detect any RNase H activity from expression of Rnh2Ap in E.coli (data not shown). To determine the differences between the Rnh2Ap expressed in E.coli and the enzymatically active enzyme, we purified RNase H2 from S.cerevisiae. We expressed an epitope-tagged Rnh2Ap from a low copy plasmid with transcription driven by the native RNH2A promoter or a constitutive TPI promoter. Recombinant Rnh2Ap, carrying two copies of the HA and FLAG epitopes or two copies of the HA and FLAG epitopes with a six-His affinity tag at the C-terminus of the protein to facilitate further purification, was generally expressed in a rnh1Δrnh2Δ strain (HIRO27AF) to avoid contamination with any residual RNase H activity. Tagged Rnh2Ap was purified by FLAG affinity chromatography and glycerol gradient ultracentrifugation as described in Materials and Methods.
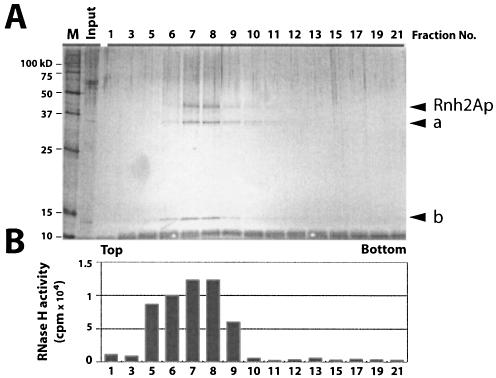
Glycerol gradient fractions were examined for polypeptide composition by silver staining gels after an 8–16% SDS–PAGE as shown in Figure 1A. Fractions 7 and 8 contained three polypeptides detectable by silver staining and showed the highest RNase H activity using poly(rA)–poly(dT) as substrate (Fig. 1B). In a separate western blot analysis, the band noted as Rnh2Ap was detected using anti-HA antibodies (data not shown). The coincidence of the RNase H activity and Rnh2Ap suggests that Rnh2Ap is at least partially responsible for hydrolysis of the RNA–DNA.
Figure 1.
Purification of S.cerevisiae RNase H2 by glycerol gradient ultracentrifugation following FLAG immunopurification. (A) A low copy plasmid carrying the tagged RNH2A gene driven by the RNH2A promoter was used to prepare extracts. Aliquots of glycerol gradient fractions were analyzed by silver staining after SDS–PAGE. Rnh2Ap (marked by an arrow) with two other polypetides (a and b). In several preparations, additional bands were identified as Ydr279p and Ylr154p by mass spectrometry of tryptic peptides (data not shown). (B) RNase H assays of the glycerol gradient. A 1 µl aliquot of each fraction was assayed for RNase H activity using 32P-labeled poly(rA)–poly(dT) as substrate.
Site-directed mutagenesis of Rnh2Ap
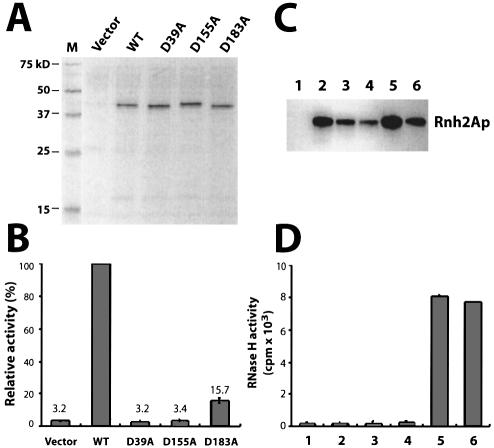
To demonstrate that Rnh2Ap is necessary for the RNase H activity seen in preparations such as those shown in Figure 1, we generated three Rnh2A proteins with substitutions of D39A, D155A or D183A. These mutations were chosen because they are known to affect the catalytic activity of archaeal RNases HII (3,4,20). Expression and purification of the tagged mutant Rnh2Ap were the same as for the wild-type polypeptide.
Both wild-type and mutant proteins were analyzed by SDS–PAGE and western blot analysis, with all proteins present in the partially purified fractions at similar levels (Fig. 2A). RNase H assays measured using poly(rA)–poly(dT) as substrate revealed that two of the three mutant proteins (D39A and D155A) were almost completely inactive in degradation of the labeled hybrid, but the D183A mutant protein retained ∼15% of the activity of the wild-type enzyme (Fig. 2B). These findings are consistent with the results from the archaeal RNases HII (3) and indicate that Rnh2Ap is necessary for the RNase H activity seen in the glycerol gradient fractions.
Figure 2.
Requirement for Rnh2Ap and two additional polypeptides for RNase H2 activity. (A) Silver-stained SDS–PAGE of samples. FLAG- immunopurified Asp→Ala mutant proteins of Rnh2Ap from S.cerevisiae expressed from a multicopy plasmid were analyzed by SDS–PAGE. (B) RNase H assay of Asp→Ala substitution mutants. Similar amounts of Rnh2Ap were used for assays based on quantification of total protein. Activity is from the mean of three replicate assays and is expressed as a percentage of the values for the wild-type enzyme. Error bars represent standard deviation. (C) Western blot analysis of purified recombinant Rnh2Ap in deletion strains: ydr279wΔ (lane 2), ylr154cΔ (lane 3) ydr279wΔ ylr154CΔ (lane 4), rnh2Δ (lane 5) and wild type (lane 6). Lane 1 is a control assay with an equivalent volume of an extract from the ydr279wΔ strain containing the vector. Anti-HA antibody was used as probe for western blotting. (D) RNase H assays of partially purified RNase H2 from various deletion strains. The amounts of Rnh2Ap used for assays were adjusted based on western blotting intensity. Lane numbers correspond to those of (C), and the values represent the mean of three assays.
Saccharomyces cerevisiae RNase H2 requires three polypeptides for activity
The tagged Rnh2Ap enabled us to rapidly and significantly purify an active RNase H2 enzyme with the most highly enriched fractions consistently containing additional polypeptides. Preparations for identification of proteins co-purifying with Rnh2Ap were not subjected to glycerol gradient fractionation and often contained additional protein bands. Two proteins with molecular masses of ∼37 and 15 kDa were always present. These two polypeptides were identified from three preparations as Ydr279p and Ylr154p by N-terminal microsequencing and mass spectrometry. Several other polypeptides that were found in some but not all of our preparations were also identified. Most of these were proteins that are highly abundant in S.cerevisiae and represent residual contaminants (data not shown).
The epitope-tagged Rnh2Ap was partially purified from rnh2AΔ, ydr279wΔ, ylr154cΔ and ydr279wΔylr154cΔ strains, with each strain bearing a plasmid expressing the tagged Rnh2Ap. Western blot analysis indicates that the Rnh2Ap was present in slightly different quantities in each preparation (Fig. 2C). We assayed these fractions for RNase H activity using poly(rA)–poly(dT) as substrate (Fig. 2D), adjusting the amount of protein added based upon the relative intensities of the bands seen (Fig. 2C) except for the vector control. RNase H2 activity was detected in protein preparations from strains containing all three genes but not from the ydr279wΔ, ylr154cΔ and ydr279wΔylr154cΔ strains (Fig. 2D). In these experiments, tagged Rnh2Ap was supplied from a plasmid, even when the chromosome contained the wild-type RNH2A gene. These results demonstrate that the activity we see in these partially purified extracts is due to RNase H2 and requires at least three polypeptides.
Rnh2Ap, Ydr279p and Ylr154p are sufficient to form an active RNase H2
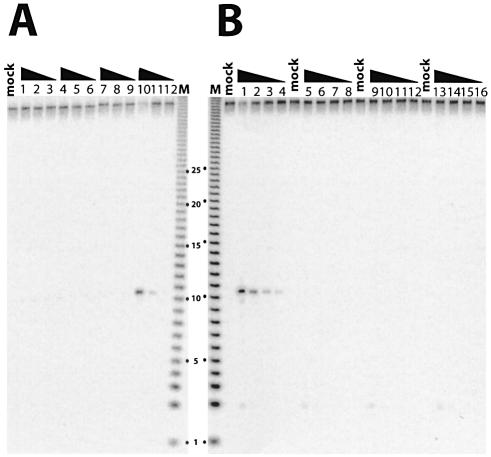
In order to determine whether the three polypeptides, Rnh2Ap, Ydr279p and Ylr154p, are sufficient to form an enzymatically active RNase H2, we expressed all three proteins in various combinations in E.coli. We employed a single ribonucleotide embedded in a duplex DNA as substrate since the endogenous E.coli RNase HI would not hydrolyze it. To use this substrate, it was necessary to partially purify the proteins by affinity chromatography to avoid degradation of the substrate in crude extracts (data not shown). Only when Rnh2Ap, Ydr279p and Ylr154p were simultaneously expressed in E.coli did we observe significant amounts of all proteins in the soluble fraction (data not shown). When all three polypeptides were co-expressed in a soluble form, RNase H2 activity was detected in the affinity-purified preparations (Fig. 3A, lanes 10–12). In all other instances, when one or more of the polypeptides were missing, no degradation was observed (Fig. 3A, lanes 1–9). Unlike as found with E.coli crude extracts, the single ribonucleotide-embedded DNA was stable in crude extracts of S.cerevisiae (14). Using crude extracts of wild-type and deletion mutants assayed for degradation of the DNA12–RNA1–DNA27/DNA40 substrate, we could confirm the requirement for each of the genes encoding the three polypeptides for formation of an active RNase H2 (Fig. 3B).
Figure 3.
RNase H assays using monoribonucleotide-embedded double-stranded DNA as substrate. DNA12–RNA1–DNA27/DNA40 was used as substrate and similar amounts of protein (determined by protein assays) were assayed. The largest amount of protein assayed was 10 times that used in the adjacent lane, and so forth. The triangle indicates decreasing concentrations. Mock is with no enzyme added. (A) His tag affinity purification of E.coli expressed Rnh2Ap (lanes 1–3), Ydr279p (lanes 4–6), Ylr154p (lanes 7–9) and all three polypeptides expressed together (lanes 10–12). (B) Crude extracts of S.cerevisiae from wild-type (lanes 1–4), rnh2AΔ (lanes 5–8), ydr279wΔ (lanes 9–12) and ylr154cΔ (lanes 13–16).
RNase H2 and RNase H1 produce different cleavage products
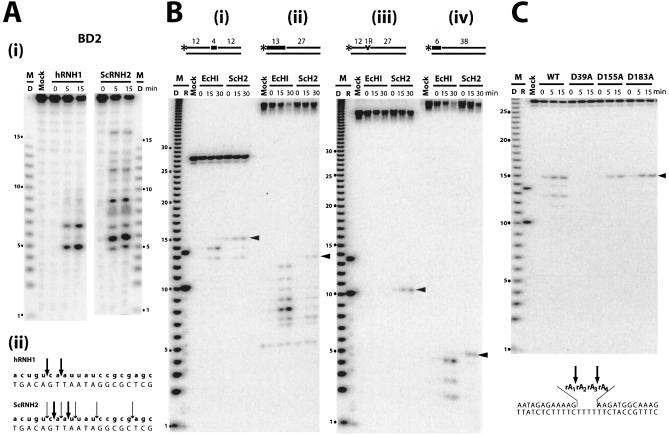
RNases H1 and H2 are reported to cleave the same RNA–DNA hybrid substrates but almost always at different positions in the RNA strand. Pileur et al. employed a 20 bp RNA–DNA substrate (BD2), which readily distinguishes between human RNases H1 and H2 (23). To test if purified S.cerevisiae RNase H2 produces cleavage patterns resembling that reported for human RNase H2, we digested the 20 bp substrate with human RNase H1 and S.cerevisiae RNase H2. Human RNase H1 cleaves the 20 bp 5′-RNA-labeled RNA–DNA, producing oligoribonucleotides of five and seven residues (Fig. 4A i). Purified tagged S.cerevisiae RNase H2 gave products distinct from those of human RNase H1 but also different from that reported for highly purified human RNase H2 [Fig. 4A i, and figs 3 and 4 of Pileur et al. (23)]. Human RNase H2 yields the 17 oligoribonucleotide also observed [figs 3 and 4 in Pileur et al. (23)] with S.cerevisiae RNase H2 (Fig. 4A i), but, unlike human RNase H2, the yeast enzyme generates numerous shorter products.
Figure 4.
Comparison of cleavage patterns of RNase H2 and RNases HI/H1. Digests were for the times (min) indicated above each lane. Molecular size markers are indicated as D [products of digestion of 32P-labeled poly(rA)–poly(dT) by E.coli RNase HI] and R synthetic oligoribonucleotide markers of 10 and 13 nucleotides. Arrowheads mark the cleavages leaving a single ribonucleotide attached to the downstream DNA. (A) (i) Digestion of BD2 substrate by human RNase H1 and S.cerevisiae RNase H2 analyzed by 12% TBE–urea PAGE. (ii) Sites of cleavage are marked by arrows, with the relative frequency indicated by the size of the arrow. (B) Comparison of digestion patterns generated by E.coli RNase HI and S.cerevisiae RNase H2 on (i) DNA12–RNA4–DNA12/DNA28; (ii) RNA13–DNA27/DNA40; (iii) DNA12–RNA1–DNA27/DNA40; and (iv) RNA6–DNA38/DNA40 as substrates. (C) Digestion of DNA12–RNA4–DNA12/DNA28 as substrate by S.cerevisiae RNase H2 and the three D→A mutant proteins (D39A, D155A and D183A). The four internal rA residues are shown as rA1rA2rA3rA4.
Sequences used to mimic RNA primed DNA fragments can vary widely, and we chose to employ the sequences used by others to examine cleavage products of RNase H2 (11,17,20). We examined the cleavage products of both classes of RNases H. Escherichia coli RNases HI cleaved the RNA–DNA/DNA substrates at multiple sites (Fig. 4B i–iv), with human RNase H1 producing products very similar to the E.coli enzyme (data not shown). There was little or no degradation at the junction between the ribonucleotide and deoxyribonucleotide for the type I RNases H (Fig. 4B). In contrast, S.cerevisiae RNase H2 preferentially cleaved the hybrid leaving a single ribonucleotide attached to the downstream DNA (Fig. 4B sites marked with arrowhead) regardless of the length of the RNA or the distance of the RNA–DNA junction from either end of the substrate (Fig. 4B). As mentioned in the previous section, a DNA duplex with a single embedded ribonucleotide is cleaved by RNase H2 but not E.coli RNase HI (Fig. 4B iii).
A DNA12–RNA4–DNA12/DNA28 substrate is cleaved by both E.coli RNase HI and S.cerevisiae RNase H2, but at different sites (Fig. 4B i). Escherichia coli RNase HI cleaves at rA1–rA2 and rA2–rA3, while S.cerevisiae RNase H2 cleaves at rA3–rA4 as well as rA1–rA2. Interestingly, the catalytically compromised mutant RNases H2 either do not cleave the DNA12–RNA4–DNA12/DNA28 substrate (Fig. 4C, D39A) or cleave at a single site (Fig. 4C, D155A and D183A).
Limited distribution of Rnh2Bp and Rnh2Cp in other species
Rnh2Ap has homologs in species from all kingdoms and, within the Saccharomyces sensu stricto (24,25), 54% identity among all amino acids is found (Supplementary Fig. 1S). The identity between S.cerevisiae Rnh2Ap and each individual homolog of the other species is extremely high (Table 1). In contrast, Rnh2Bp and Rnh2C have very limited distribution. Rnh2Bp has similar proteins in other Saccharomyces sensu stricto species and a protein with marginal similarity in Candida albicans (Supplementary Fig. 1S), but no related protein in any other organism is found using the BLAST program. Rnh2Cp of S.cerevisiae has highly related proteins in S.paradoxous, S.mikatae and S.kudriavzev, while in S.castellii and S.kluyveri, the conservation of proteins related to Rnh2Cp is significantly lower (Table 1) with identities confined mainly to the N- and C-termini (Supplementary Fig. 1S). The most highly purified fractions of human RNase H2 contain two polypeptides of 32 and 21 kDa, the former being the counterpart of the S.cerevisiae Rnh2Ap (16,26). The small subunit (21 kDa) of human RNase H2 (accession No. AF312034) has no obvious sequence similarity to either Rnh2Bp or Rnh2Cp of S.cerevisiae RNase H2. It seems likely that the composition of proteins constituting eukaryotic RNases H2 evolved independently and may allow the enzymes to associate with other different proteins.
Table 1. Identities between RNase H2 subunits of Saccharomyces sensu stricto species.
| Species | Rnh2Ap | Rnh2Bp | Rnh2Cp |
|---|---|---|---|
|
S.cerevisiae |
307 |
350 |
110 |
|
S.paradoxus |
288 (93%) |
286 (81%) |
92 (83%) |
|
S.bayanus |
281 (91%) |
247 (70%) |
80 (72%) |
|
S.mikatae |
276 (89%) |
276 (78%) |
86 (78%) |
|
S.kudriavzevii |
271 (88%) |
256 (73%) |
81 (73%) |
|
S.castellii |
201 (67%) |
152 (43%) |
35 (31%) |
| S.kluyveri | 188 (61%) | 144 (41%) | 32 (29%) |
Amino acid lengths of S.cerevisiae RNase H2 subunits are given in the S.cerevisiae row. The number of identical amino acids is shown, with the percentage identity in parentheses. See Supplementary Figure 1S for sequences and alignments.
DISCUSSION
Saccharomyces cerevisiae RNase H2 comprises three polypeptides
Bacterial and archaeal RNases HII are active as single polypeptides (2,4,6,20). However, the composition of eukaryotic RNase H2 has been a long-standing issue, with reports describing RNase H2 comprising either a single or two distinct polypeptides (15). By expressing an epitope-tagged Rnh2Ap, the S.cerevisiae homolog of bacterial and archaeal RNase HII, we were able to obtain an enzymatically active RNase H2 from yeast that contains two additional polypeptides, Ydr279p and Ylr154p. Deletion studies and the ability to form a catalytically active complex when all three subunits are co-expressed in E.coli led us to conclude that Rnh2Ap, Ydr279p and Ylr154p are necessary and sufficient to make functional S.cerevisiae RNase H2. Therefore, we suggest the protein subunits of RNase H2 be named Rnh2Ap (Ynl072p), Rnh2Bp (Ydr279p) and Rnh2Cp (Ylr154p).
Rnh2Ap is important for catalytic activity of S.cerevisiae RNase H2
The catalytic residues in S.cerevisiae reside at least in part in Rnh2Ap, as demonstrated by the loss or decrease in RNase H2 activity when mutant Rnh2Ap proteins are examined (Figs 2A and B, and 4C). Expression of S.cerevisiae Rnh2Ap or human RNASEH2A (data not shown) in E.coli does not yield an active enzyme in our hands, a result consistent with that reported by Lima et al. (27) for the human protein. However, Qiu et al. describe recovering RNase H activity of Rnh2Ap after expression in E.coli in an insoluble form followed by renaturation (11). We employed one of the substrates used in their experiments (Fig. 4B) and observed cleavage at the same sites as did Qiu et al. (11,20). Moreover, Frank et al. report activity associated with the large subunit of RNase H2 purified from calf thymus using the gel renaturation assay for RNase H (26). The simplest explanation to reconcile these differences is to assume that the catalytic activity resides in the Rnh2Ap subunit and can inefficiently be made active upon renaturation when expressed in E.coli or possibly exhibit activity when there is additional in vivo modification of the bovine enzyme. Interestingly, the compromised D155A and D183A mutant proteins cleave the DNA12–RNA4–DNA12/DNA28 substrate almost exclusively to form an RNA1–DNA12 fragment, suggesting a strong preference for cleavage adjacent to an RNA–DNA junction (Fig. 4C), a hallmark of RNase HII/2 proteins (12,13,17). RNases H1 do not cleave at this position (Fig. 4B).
Phenotypes associated with RNase H2 mutations
Removal of RNA primers during lagging strand DNA synthesis seems to involve Rad27p (S.cerevisiae Flap endonuclease) and Dna2p, but some evidence suggests RNase H2 might aid in this process (28,29). Two studies examined the effect of deleting rnh2A on expansion of triplet repeats and chromosome breakage at the triplet repeats and found either no or only modest effects. Qiu et al. observed a small increase in frequency of canavanine-resistant mutants as well as an increase in a frameshift mutation (CAN1 forward mutation assay) (11). The combination of rad27Δ and rnh2AΔ increased the frequencies in a manner suggesting that Rad27p and RNase H2 may both recognize and resolve RNA-primed Okazaki fragments (11). In a genome-wide screen of yeast mutants whose deletion results in an increase in mutagenesis using the CAN1 forward mutation assay, RNH2C was one of 33 genes of S.cerevisiae identified as necessary to keep mutation frequencies at normal levels (30). Mutation frequencies for rnh2AΔ and rnh2CΔ in the two different reports are very similar. Of the 33 strains identified in the genome-wide study, the rnh2CΔ strain exhibited mutation frequencies about two to three times that of the wild-type strain. Failure to identify RNH2A or RNH2B in the genome-wide screen may reflect the difficulty in including or excluding candidates with such small increases in resistance to canavanine.
Deletions of any of the genes encoding the three subunits of RNase H2 exhibit a synthetic fitness phenotype with an sgs1Δ (31). Sgs1p is a helicase of the RecQ family, with several of the members of this family known to be involved in human diseases (32). Each of the sgs1Δrnh2 (A, B or C)Δ double deletion strains has a doubling time of ∼140 min compared with 90–100 min for each single deletion strain. The slow growth rates of the double mutants were partially reversed in all cases by introduction of a rad51Δ. The similarity in the synthetic fitness properties conferred on sgs1Δ strains by rnh2Δ mutations is readily explained by the results we report here that the loss of any of the three subunits of RNase H2 results in the absence of RNase H2 activity. The synthetic fitness results have been interpreted as an indication that the Sgs1p helicase can aid in resolving problems created by the absence of RNase H2, particularly resolution of recombination intermediates and damaged replication forks (31).
Potential for additional subunits to interact with other cellular components
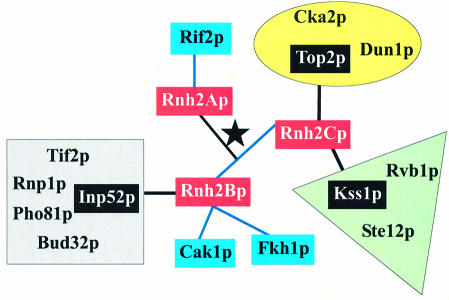
Additional proteins in eukaryotic RNases H2 may serve two functions. First, eukaryotic Rnh2A-like proteins are longer at both the N- and C-termini than the bacterial and archaeal proteins, and Rnh2Bp and Rnh2Cp might be required to fold Rnh2Ap into an active conformation by proper positioning of the N- and C-terminal amino acids. Secondly, the accessory proteins may provide a means by which RNase H2 interacts with other cellular components. In some of our analyses of the components of the RNase H2 complex, we have detected other proteins, most of which are abundant in the cell, and none seems likely to be related to RNase H2 function. However, large-scale studies detecting interactions amongst proteins in S.cerevisiae have indicated several interactions of Rnh2Bp and Rnh2Cp (Fig. 5), including with one another (33–36). There is no previous description of Rnh2Ap interacting with either Rnh2Bp or Rnh2Cp as we report here. Saccharomyces cerevisiae RNase H2 is cell cycle regulated, having its highest levels during S and G2/M phases where it could be important in DNA replication and repair. Interestingly, Rnh2Bp and Rnh2Cp are suggested to interact with a variety of proteins some of which are involved in transcription (Fkh1p), replication/repair (Top2p), and cell cycle progression during S and G2/M phase (Cak1p). Many of the proteins are kinases and phosphatases that have the potential to modify any or all of the RNase H2 subunits.
Figure 5.
Interactions of RNase H2 subunits. The three subunits of RNase H2 are indicated in red and connected by a star. Interactions obtained using the yeast two-hybrid system are indicated by blue connecting lines; note that Rnh2B and Rnh2C are connected by blue lines. Complexes in which Rnh2B and Rnh2C were found by mass spectrometric analysis are shown in groups by color. The proteins used to isolate the complexes are indicated in black and connected by lines to the RNase H2 subunits. The Top2p complex is in the yellow oval, the Kss1p complex is in the green triangle, and the Inp52p complex is in the gray square.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Susana M. Cerritelli for many helpful comments and critical reading of the manuscript as well as supplying many useful reagents. RNase H1 was kindly supplied by Sergei Gaidamakov, and Arulvathani Arudchandran provided the FLAG-tagged RNH2A plasmid. Also, we thank Siew Ooi and Jef Boeke for communicating their data prior to publication.
REFERENCES
- 1.Crouch R.J. and Cerritelli,S.M. (1998) RNases H of S.cerevisiae, S.pombe, C.fasciculata, and N.crassa. In Crouch,R.J. and Toulmé,J.J. (eds), Ribonucleases H. INSERM, Paris, pp. 79–100. [Google Scholar]
- 2.Ohtani N., Haruki,M., Morikawa,M., Crouch,R.J., Itaya,M. and Kanaya,S. (1999) Identification of the genes encoding Mn2+-dependent RNase I–III and Mg2+-dependent RNase HIII from Bacillus subtilis: classification of RNases H into three families. Biochemistry, 38, 605–618. [DOI] [PubMed] [Google Scholar]
- 3.Muroya A., Tsuchiya,D., Ishikawa,M., Haruki,M., Morikawa,M., Kanaya,S. and Morikawa,K. (2001) Catalytic center of an archaeal type 2 ribonuclease H as revealed by X-ray crystallographic and mutational analyses. Protein Sci., 10, 707–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chapados B.R., Chai,Q., Hosfield,D.J., Qiu,J.Z., Shen,B.H. and Tainer,J.A. (2001) Structural biochemistry of a type 2 RNase H: RNA primer recognition and removal during DNA replication. J. Mol. Biol., 307, 541–556. [DOI] [PubMed] [Google Scholar]
- 5.Ohtani N., Haruki,M., Morikawa,M. and Kanaya,S. (1999) Molecular diversities of RNases H. J. Biosci. Bioeng., 88, 12–19. [DOI] [PubMed] [Google Scholar]
- 6.Itaya M. (1990) Isolation and characterization of a second RNase H (RNase HII) of Escherichia coli K12 encoded by the rnhB gene. Proc. Natl Acad. Sci. USA, 87, 8587–8591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arudchandran A., Cerritelli,S.M., Narimatsu,S.K., Itaya,M., Shin,D.Y., Shimada,Y. and Crouch,R.J. (2000) The absence of ribonuclease H1 or H2 alters the sensitivity of Saccharomyces cerevisiae to hydroxyurea, caffeine and ethyl methanesulphonate: implications for roles of RNases H in DNA replication and repair. Genes Cells, 5, 789–802. [DOI] [PubMed] [Google Scholar]
- 8.Ray D.S. and Hines,J.C. (1995) Disruption of the Crithidia fasciculata RNH1 gene results in the loss of 2 active forms of ribonuclease H. Nucleic Acids Res., 23, 2526–2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cerritelli S.M., Frolova,E.G., Feng,C.G., Grinberg,A., Love,P.E. and Crouch,R.J. (2003) Failure to produce mitochondrial DNA results in embryonic lethality in Rnaseh1 null mice. Mol. Cell, 11, 807–815. [DOI] [PubMed] [Google Scholar]
- 10.Filippov V., Filippova,M. and Gill,S.S. (2001) Drosophila RNase H1 is essential for development but not for proliferation. Mol. Gen. Genet., 265, 771–777. [DOI] [PubMed] [Google Scholar]
- 11.Qiu J.Z., Qian,Y., Frank,P., Wintersberger,U. and Shen,B.H. (1999) Saccharomyces cerevisiae RNase H(35) functions in RNA primer removal during lagging-strand DNA synthesis, most efficiently in cooperation with Rad27 nuclease. Mol. Cell. Biol., 19, 8361–8371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eder P.S., Walder,R.Y. and Walder,J.A. (1993) Substrate specificity of human RNase H1 and its role in excision repair of ribose residues misincorporated in DNA. Biochimie, 75, 123–126. [DOI] [PubMed] [Google Scholar]
- 13.Eder P.S. and Walder,J.A. (1991) Ribonuclease H from K562 human erythroleukemia cells—purification, characterization, and substrate specificity. J. Biol. Chem., 266, 6472–6479. [PubMed] [Google Scholar]
- 14.Rydberg B. and Game,J. (2002) Excision of misincorporated ribonucleotides in DNA by RNase H (type 2) and FEN-1 in cell-free extracts. Proc. Natl Acad. Sci. USA, 99, 16654–16659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Büsen W. and Frank,P. (1998) Bovine RNases H. In Crouch,R.J. and Toulmé,J.J. (eds), Ribonucleases H. INSERM, Paris, pp. 113–146. [Google Scholar]
- 16.Frank P., Albert,S., Cazenave,C. and Toulmé,J.J. (1994) Purification and characterization of human ribonuclease HII. Nucleic Acids Res., 22, 5247–5254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murante R.S., Henricksen,L.A. and Bambara,R.A. (1998) Junction ribonuclease: an activity in Okazaki fragment processing. Proc. Natl Acad. Sci. USA, 95, 2244–2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frank P., Braunshofer-Reiter,C. and Wintersberger,U. (1998) Yeast RNase H(35) is the counterpart of the mammalian RNase HI, and is evolutionarily related to prokaryotic RNase HII. FEBS Lett., 421, 23–26. [DOI] [PubMed] [Google Scholar]
- 19.Crouch R.J., Arudchandran,A. and Cerritelli,S.M. (2003) RNase H1 of Saccharomyces cerevisiae: methods and nomenclature. Methods Enzymol., 341, 395–413. [DOI] [PubMed] [Google Scholar]
- 20.Chai Q., Qiu,J., Chapados,B.R. and Shen,B.H. (2001) Archaeoglobus fulgidus RNase HII in DNA replication: enzymological functions and activity regulation via metal cofactors. Biochem. Biophys. Res. Commun., 286, 1073–1081. [DOI] [PubMed] [Google Scholar]
- 21.Muroya A., Nakano,R., Ohtani,N., Haruki,M., Morikawa,M. and Kanaya,S. (2002) Importance of an N-terminal extension in ribonuclease HII from Bacillus stearothermophilus for substrate binding. J. Biosci. Bioeng., 93, 170–175. [DOI] [PubMed] [Google Scholar]
- 22.Carl P.L., Bloom,L. and Crouch,R.J. (1980) Isolation and mapping of a mutation in Escherichia coli with altered levels of ribonuclease H. J. Bacteriol., 144, 28–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pileur F., Toulmé,J.J. and Cazenave,C. (2000) Eukaryotic ribonucleases HI and HII generate characteristic hydrolytic patterns on DNA–RNA hybrids: further evidence that mitochondrial RNase H is an RNase HII. Nucleic Acids Res., 28, 3674–3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cliften P., Sudarsanam,P., Desikan,A., Fulton,L., Fulton,B., Majors,J., Waterston,R., Cohen,B.A. and Johnston,M. (2003) Finding functional features in Saccharomyces genomes by phylogenetic footprinting. Science, 301, 71–76. [DOI] [PubMed] [Google Scholar]
- 25.Kellis M., Patterson,N., Endrizzi,M., Birren,B. and Lander,E.S. (2003) Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature, 423, 241–254. [DOI] [PubMed] [Google Scholar]
- 26.Frank P., Braunshofer-Reiter,C., Wintersberger,U., Grimm,R. and Büsen,W. (1998) Cloning of the cDNA encoding the large subunit of human RNase HI, a homologue of the prokaryotic RNase HII. Proc. Natl Acad. Sci. USA, 95, 12872–12877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lima W.F., Wu,H.J. and Crooke,S.T. (2001) Human RNases H. Methods Enzymol., 341, 430–440. [DOI] [PubMed] [Google Scholar]
- 28.Bae S.H., Kim,D.W., Kim,J., Kim,J.H., Kim,D.H., Kim,H.D., Kang,H.Y. and Seo,Y.S. (2002) Coupling of DNA helicase and endonuclease activities of yeast Dna2 facilitates Okazaki fragment processing. J. Biol. Chem., 277, 26632–26641. [DOI] [PubMed] [Google Scholar]
- 29.Ayyagari R., Gomes,X.V., Gordenin,D.A. and Burgers,P.M.J. (2003) Okazaki fragment maturation in yeast—I. Distribution of functions between FEN1 AND DNA2. J. Biol. Chem., 278, 1618–1625. [DOI] [PubMed] [Google Scholar]
- 30.Huang M.E., Rio,A.G., Nicolas,A. and Kolodner,R.D. (2003) A genomewide screen in Saccharomyces cerevisiae for genes that suppress the accumulation of mutations. Proc. Natl Acad. Sci. USA, 100, 11529–11534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ooi S.L., Shoemaker,D.D. and Boeke,J.D. (2003) DNA helicase gene interaction network defined using synthetic lethality analyzed by microarray. Nature Genet., 35, 277–286. [DOI] [PubMed] [Google Scholar]
- 32.Hickson I.D. (2003) RecQ helicases: caretakers of the genome. Nature Rev. Cancer, 3, 169–178. [DOI] [PubMed] [Google Scholar]
- 33.Ho Y., Gruhler,A., Heilbut,A., Bader,G.D., Moore,L., Adams,S.L., Millar,A., Taylor,P., Bennett,K., Boutilier,K. et al. (2002) Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature, 415, 180–183. [DOI] [PubMed] [Google Scholar]
- 34.Gavin A.C., Bosche,M., Krause,R., Grandi,P., Marzioch,M., Bauer,A., Schultz,J., Rick,J.M., Michon,A.M., Cruciat,C.M. et al. (2002) Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature, 415, 141–147. [DOI] [PubMed] [Google Scholar]
- 35.Ito T., Chiba,T., Ozawa,R., Yoshida,M., Hattori,M. and Sakaki,Y. (2001) A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl Acad. Sci. USA, 98, 4569–4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ito T., Tashiro,K., Muta,S., Ozawa,R., Chiba,T., Nishizawa,M., Yamamoto,K., Kuhara,S. and Sakaki,Y. (2000) Toward a protein–protein interaction map of the budding yeast: a comprehensive system to examine two-hybrid interactions in all possible combinations between the yeast proteins. Proc. Natl Acad. Sci. USA, 97, 1143–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.