Abstract
Nonsense-mediated mRNA decay (NMD) directs rapid degradation of premature termination codon (PTC)-containing mRNAs, e.g. those containing frameshift mutations. Many viral mRNAs encode polycistronic messages where programmed –1 ribosomal frameshift (–1 PRF) signals direct ribosomes to synthesize polyproteins. A previous study, which identified consensus –1 PRF signals in the yeast genome, found that, in contrast to viruses, the majority of predicted –1 PRF events would direct translating ribosomes to PTCs. Here we tested the hypothesis that a –1 PRF signal can function as a cis-acting mRNA destabilizing element by inserting an L-A viral –1 PRF signal into a PGK1 reporter construct in the ‘genomic’ orientation. The results show that even low levels of –1 PRF are sufficient to target the reporter mRNA for degradation via the NMD pathway, with half-lives similar to messages containing in-frame PTCs. The demonstration of an inverse correlation between frameshift efficiency and mRNA half-lives suggests that modulation of –1 PRF frequencies can be used to post-transcriptionally regulate gene expression. Analysis of the mRNA decay profiles of the frameshift-signal- containing reporter mRNAs also supports the notion that NMD remains active on mRNAs beyond the ‘pioneer round’ of translation in yeast.
INTRODUCTION
Aberrant mRNAs containing premature termination codons (PTCs) are removed post-translationally from the mRNA pool by the nonsense-mediated decay (NMD) pathway (reviewed in 1–3). Many polycistronic viral messages contain termination codons well upstream of the 3′ untranslated region and presumably have evolved mechanisms to avoid NMD. However, there are an increasing number of eukaryote genes that have programmed ribosomal frameshift signals: programmed +1 frameshifts are known to be required for expression of the ABP140 gene of yeast (4), two genes encoding telomerase-associated proteins (5,6) and all of the known metazoan genes encoding the ornithine decarboxylase (ODC) antizyme (7). Additionally, it is becoming clear that there is a high frequency of programmed +1 frameshifting in ciliates of the Euplotes genus as a consequence of high frequencies of stop codon reassignment (8). Thus, although we typically think of programmed ribosomal frameshifting (PRF) as a minor, virus-specific contrivance, it also presents a potentially powerful molecular mechanism that could be used to control the expression of a significant subset of chromosomally encoded mRNAs. Although –1 PRF seems to be favored over +1 frameshifting in viruses, it has not yet been demonstrated that this mechanism is used as a general regulator of cellular gene expression. The only known example is in the mouse Edr gene (and perhaps the human KIAA1051), which utilizes a classical programmed –1 ribosomal frameshift to translate its C-terminus (9). However, there is a growing body of evidence that indirectly supports this hypothesis. For example, in yeast, cells harboring mutations that specifically promote increased –1 PRF efficiencies also have phenotypic defects normally associated with aberrant regulation of gene expression (10–12).
In an earlier computational screen we reported the presence of 260 consensus –1 PRF signals in the yeast genome, but the function of these motifs was open to speculation (13). In contrast to viral frameshifts, which generally result in the production of C-terminally extended fusion proteins, analyses of predicted genomic –1 PRF signals revealed that most of the predicted frameshift events would cause elongating ribosomes to encounter premature termination codons. In theory, such an event on an mRNA might serve to activate the NMD pathway, which would in turn promote the rapid degradation of that specific mRNA. Thus, PRF could be used to initiate mRNA suicide, i.e. programmed frameshifting might be used to control mRNA abundance. In this study we have rigorously tested this theory by combining the two most well characterized assay systems for programmed –1 ribosomal frameshifting and NMD. These proof-of-principle experiments convincingly demonstrate that a –1 PRF signal can act as an mRNA suicide element, and that there is an inverse correlation between programmed ribosomal frameshift efficiency and mRNA half-life.
MATERIALS AND METHODS
Strains, genetic manipulation and media
Escherichia coli DH5α was used to amplify plasmid DNA. Transformation of yeast and E.coli were performed as described previously (10). YPAD and synthetic complete medium (H–) were as reported previously (12). DNA- modifying enzymes were obtained from MBI Fermentas. Radioactive nucleotides were obtained from NEN. T7 Sequenase was obtained from USB and Sequagel-6 was obtained from National Diagnostics. DNA sequence analysis was performed by the UMDNJ–RWJMS DNA core facility. Oligonucleotide primers were purchased from IDT. Yeast strains used in this study were RY262+ (MATα his4-519 ura3-52 rpb1-1) (14) and RY262– (MATα his4-519 ura3-52 upf1::hisG rpb1-1) (15).
Mutagenesis and plasmid construction
pF′8 (16) served as the source of the wild-type L-A pro grammed –1 ribosomal frameshift signal. Oligonucleotide site-directed in vitro mutagenesis was carried out using the Bio-Rad Muta-Gene® kit. Oligonucleotide 5′-GCGTCG TACTCAGCAAGGGTTTAGGAGTGGTAGG-3′ was used to make pJD216 and oligonucleotide 5′-GCGTACTCAG CAGGGTCCAAGGAGTGGTAGGTCTTACG-3′ was used to construct pJD217. All oligonucleotides were phosphorylated with T4 DNA kinase, and annealing and synthesis of complementary strands were carried out according to the manufacturers’ instructions. To construct plasmids pJD255, pJD257 and pJD258, the L-A virus derived frameshift signals from pF′8, pJD216 and pJD217 were cloned by PCR using 5′ primer 5′-CCCCGGTACCGGTAGTGTGCGTCGTAC-3′ and 3′ primer 5′-CCCCGGTACCCTGCAGTATGTACCTTG-3′ which contain a KpnI restriction site. For pJD269, the 3′ primer 5′-CCCCGGTACCTATCTCTACTAAAACATTGTCC-3′ was used. The PCR fragments from pF′8, pJD216 and pJD217 were subcloned into the KpnI site of pPW9, which is pRS316 in which the KpnI site was deleted, and into which the full length of PGK1, including its promoter, was subsequently cloned. The resulting plasmids containing sense insertions of the different L-A derived frameshift signals were digested with HindIII and BamHI and the PGK1 fragments were subcloned into similarly digested p3131 [referred to as pRIPPGK(-AU) by Hagan et al. (17)]. The corresponding plasmids are pJD255, pJD257, pJD258 and pJD269. Plasmids p3131 and p3082 [also known as pRIPPGK(-AU) AspUGA] were generous gifts of Dr Stuart Peltz.
RNA preparation and decay analysis
mRNA decay rates were determined as described previously (17). Strains RY262+ (UPF+) or RY262– (Upf–) were transformed with the different synthetic PGK1 constructs and maintained on selective media. Mid-logarithmic yeast cultures (100 ml) were grown at 22°C, collected by centrifugation and resuspended in 15 ml of the same medium. After incubation at 22°C for 15 min, an equal volume of medium preheated to 60°C was mixed with the yeast cells, and 4 ml of culture was immediately withdrawn and frozen in either a dry ice–ethanol bath or liquid nitrogen and used as zero time controls. After the temperature shift, the cultures were maintained at 36°C with constant shaking. At 5 min intervals, 4 ml of the cultures were removed, cells were pelleted by centrifugation for 10 s and immediately frozen. Total RNAs were extracted by the hot phenol method (18). The isolated RNAs were denatured at 70°C for 20 min before determining the concentrations by OD260 readings. Equal amounts of total RNA (20 µg/sample) were analyzed by northern blot. For the polydAdT oligonucleotide probe (to detect synthetic PGK1 transcripts), pre-hybridization was performed in solution containing 200 µg of denatured salmon sperm DNA, 6× SSC, 1% SDS and 10× Denhardt’s solution for 1 h at 42°C. Hybridization was carried out overnight with fresh buffer containing 108 c.p.m. of 32P-labeled probe. Blots were washed once with 2× SSC and 0.1% SDS at room temperature for 20 min and once at 50°C for 15 min. Probes were washed off membranes by boiling in 0.1% SDS. Probes for CYH2 and U3 snRNA were generated from transcription of p3433 digested with HincII and from pJD161 digested with SspI, respectively (10). For CYH2 and U3 snRNA probed blots, membranes were pre-hybridized in 5× SSC, 50% formamide, 1% SDS and 10× Denhardt’s solution, 200 µg of denatured salmon sperm DNA and hybridized in a solution containing 5× SSC, 50% formamide, 1% SDS, 200 µg of denatured salmon sperm DNA and 2× Denhardt’s solution containing 108 c.p.m. of 32P-labeled probes at 55°C. Membranes were washed once at room temperature and twice at 65°C for 15 min. Radioactivity in sample lanes was determined using either Typhoon® or Storm® Phosphoimagers (Molecular Dynamics). All of the RNA half-life experiments were performed independently in two or more independent experiments and statistical analyses were performed using SPSS software.
NMD modeling
To separate the pioneer round of translation for each message from subsequent rounds, decay of cellular mRNAs was modeled according to each ‘round of translation’ as opposed to overall time of decay. One successful round of translation represents (in terms of our in silico model) the complete cycle of initiation, elongation and termination events. The following assumptions were devised for simplification and reduction of parameters: (i) rates of these individual translational events were considered uniform for each mRNA in the pool; (ii) premature termination of a ribosome directs an mRNA to the NMD pathway with an efficiency of 98% (19); (iii) mRNAs in the pool are exposed to a constitutive rate of decay of 1% (pCRD = 0.01) independent of translational accuracy or fidelity; (iv) the probability of frameshifting (pPRF) per translational round ranged from 1 to 4% for each message. Modeling of the pioneer round of translation was also done using the same parameters, except that pPRF was set to 0% after the first round of translation. The degradation of wild-type mRNAs assumed pPRF = 0% for those messages. A background rate of non-programmed frameshifting was introduced at 0.01% (pNPRF = 0.0001) (16). For each simulation, a starting pool of 10 000 mRNAs was used. The computational model was developed in PERL and the source code is available on request.
RESULTS
Programmed ribosomal frameshift signals can direct mRNAs to the NMD pathway
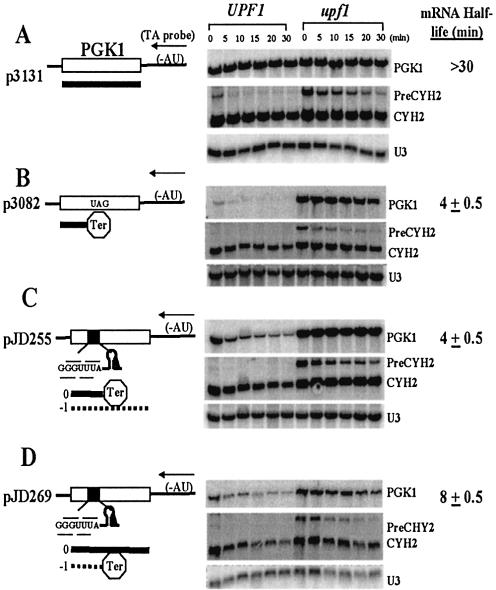
To test the hypothesis that a –1 PRF signal can function in cis to destabilize an otherwise normal transcript, we combined the standard yeast-based systems developed to study NMD and –1 PRF. The yeast PGK1 gene has been extensively used to delineate the cis-acting sequence elements and trans-acting factors of the NMD pathway. Recapitulation of previous experiments from other laboratories demonstrated that the PGK1 mRNA is very stable in yeast cells (p3131, t1/2 > 30 min, see Fig. 1A), and that introduction of a premature in-frame termination codon promotes its destabilization via the NMD pathway (p3082, t1/2 = 4.0 ± 0.5 min, see Fig. 1B). In pJD255, the L-A encoded programmed –1 ribosomal frameshift signal is oriented in the virus-typical context: continued translation through to the end of the PGK1 transcript requires a frameshift, whereas unshifted ribosomes encounter multiple 3′ proximal termination codons in the 0-frame. This construct encodes a nonsense-containing mRNA, as evidenced by its equally short half-life in wild-type cells (t1/2 = 4.0 ± 0.5 min, see Fig. 1C). The stability of this mRNA in an isogenic upf1Δ strain demonstrates that destabilization of this reporter mRNA is dependent on the NMD pathway in wild-type cells.
Figure 1.
Proof of principle: a –1 PRF signal can function as a cis-acting mRNA destabilizing element. (A) A PGK1 mRNA is stable in both wild-type and upf1Δ strains. Isogenic wild-type and upf1Δ cells harboring the rpb1-1 allele were transformed with a plasmid carrying the 3′ poly-AT tagged PGK1 reporter p3131 (17). At time 0, cells were shifted to 37°C to arrest RNA polymerase II-mediated transcription, total RNAs were harvested at the indicated time points, separated through a denaturing agarose gel, transferred to nitrocellulose, and hybridized with a 32P-labeled oligo-dTdA probe. mRNA half-life in wild-type cells is indicated on the right. The blot was stripped and hybridized with probes complementary to the endogenous CYH2 mRNA. The inefficiently spliced CYH2 precursor mRNA serves as an internal monitor of the status of the nonsense-mediated mRNA apparatus. The U3 snoRNA is used as a loading control. (B) Instability of a nonsense-containing PGK1 mRNA is dependent on the NMD pathway. The PGK1 reporter plasmid p3082 contains an in-frame opal codon inserted ∼39% into the PGK1 coding region (17). This was introduced into isogenic wild-type and upf1Δ strains and RNAs were harvested and processed as described above. The mRNA encoded by p3082 is unstable in UPF1, but is stable in upf1Δ cells. (C) A PGK1 mRNA containing an L-A viral –1 PRF signal in the viral context is unstable, and instability is NMD-dependent. In pJD255, the L-A –1 PRF signal was introduced into p3131 with the result that continued translation in the 0-frame results in a premature termination event. This plasmid was introduced into isogenic wild-type and upf1Δ strains and the RNAs were harvested and analyzed as described above. (D) A –1 PRF event that directs ribosomes to encounter a premature termination codon is sufficient to destabilize a PGK1 mRNA in an NMD-dependent manner. In pJD269, the L-A –1 PRF signal was introduced into p3131 with the result that a –1 PRF event results in a premature termination event. This plasmid was introduced into isogenic wild-type and upf1Δ strains and the RNAs were harvested and analyzed as described above.
As noted above, in contrast to the viral context, the majority of the putative –1 PRF signals that we previously found in chromosomally encoded genes are oriented so that continued translation in the 0-frame would result in production of the full-length protein, whereas a –1 PRF event would result in a ribosome encountering a premature termination codon (13). The construct pJD269 was designed to mimic this ‘genomic’ –1 PRF scenario. The results demonstrate that the mRNA encoded by this PGK1 allele was also unstable in wild-type cells, with a half-life of 8.0 ± 0.5 min (Fig. 1D). Notably, the instability of this mRNA was also NMD-dependent as evidenced by its stabilization in an isogenic upf1Δ strain. These findings provide proof-of-principle for the hypothesis that a transcript containing a –1 PRF signal is subject to the NMD pathway even though it does not contain any premature termination codons in the 0-frame.
Programmed frameshifting is required for mRNA destabilization
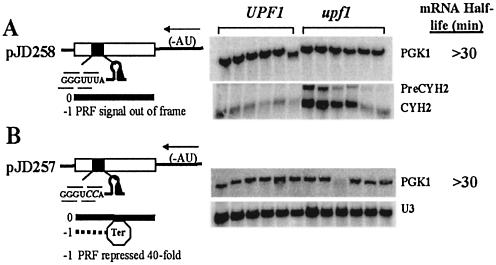
The mRNAs produced from two additional PGK1 alleles were tested to address the question of whether mRNA destabilization of pJD269 was a direct consequence of –1 PRF. Addition of an extra adenosine residue 5′ of the frameshift signal (pJD258) moved the slippery site out of phase with regard to incoming ribosomes, forestalling its ability to promote –1 frameshifting. This reporter mRNA was very stable in both wild-type and upf1Δ strains (t1/2 >30 min; Fig. 2A), demonstrating that destabilization of the reporter mRNA encoded by pJD269 was dependent on –1 PRF. This experiment also shows that neither the mRNA pseudoknot nor the L-A frameshift signal alone are capable of destabilizing the reporter mRNA.
Figure 2.
mRNA destabilization is dependent on efficient –1 PRF. (A) In pJD258, the slippery site was rendered non-functional by moving it out-of-frame. (B) In pJD257, the GGGUCCA heptamer decreases –1 PRF to ∼0.05% (40-fold less than wild-type). mRNAs were analyzed as described in Figure 1.
The GGGUCCA slippery site pJD257 was previously shown to reduce programmed –1 ribosomal frameshifting efficiencies by ∼40-fold (16). Substitution of this slippery site into the frameshift signal also yielded a stable PGK1 allele (half-life to >30 min, see Fig. 2B). A particularly interesting observation was that, though the –1 PRF efficiency promoted by the GGGUCCA slippery site is significantly less than that of the wild-type heptamer, it still promotes frameshifting at levels ∼5-fold greater than undirected frameshifting (16). Though this slippery site did not appear to affect the half-life of the reporter mRNA, it did affect mRNA steady state abundance (see below).
An inverse relationship between frameshifting efficiencies and mRNA half-lives
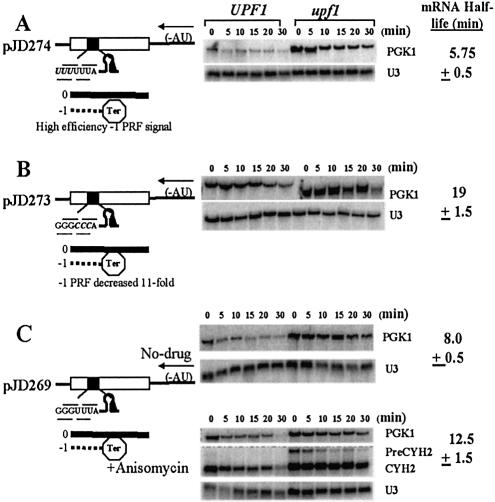
Corollary to the finding that PRF can destabilize mRNAs is the hypothesis that altering PRF efficiencies could be used to regulate mRNA half-lives. A series of experiments were designed to test this hypothesis. In the context of the L-A RNA pseudoknot, the slippery site UUUUUUA has been demonstrated to promote a 6-fold increase in programmed –1 ribosomal frameshifting [12% versus 2%, see Dinman et al. (16)]. Thus PGK1 alleles containing this frameshift signal should be less stable than those harboring the wild-type GGGUUUA slippery site. As predicted, in pJD274 the UUUUUUA slippery site reduced the half-life of the PGK1 reporter mRNA to 5.75 ± 0.5 min (Fig. 3A). It is interesting to note, however, that the magnitude of the effect on mRNA half-life was not as great as that on frameshifting efficiency, suggestive perhaps of a threshold effect of frameshifting efficiency on mRNA decay rates.
Figure 3.
An inverse relationship between –1 PRF efficiencies and mRNA half-lives. (A) The high efficiency UUUUUUA frameshift signal in pDJ274 promotes an ∼6-fold increase in –1 PRF efficiency and decreases the half-life of the PGK1 reporter from 8 min to 5.75 ± 0.5 min. (B) The GGGCCCA slippery site decreases –1 PRF by ∼11-fold as compared with wild-type. The half-life of the PGK1 reporter mRNA produced by pJD273 is increased to ∼20 min. (C) The half-life of the wild-type –1 PRF signal-containing reporter (pJD269) is 8 min in the absence of drug. Addition of anisomycin (4 µg/ml) to cells harboring pJD269 results in partial stabilization of the PKG1 reporter so that its half-life is 13.5 ± 1.5 min. mRNAs were analyzed as described in Figure 1.
In contrast, decreasing the efficiency of –1 PRF would be expected to stabilize an mRNA. Two experiments were conducted to test this hypothesis. First, substitution of the GGGCCCA slippery site into the L-A frameshift signal, which has been shown to promote an ∼11-fold decrease in –1 PRF efficiencies (16), resulted in an increase of the PGK1 reporter mRNA half-life to ∼19 min (pJD273; Fig. 3B). In a second experiment, half-lives of the pJD269-encoded PGK reporter mRNA were monitored in the presence or absence of anisomycin, an agent that significantly inhibits programmed –1 ribosomal frameshifting (20–22). Figure 3C shows that the half-life of the PGK1 reporter mRNA is increased to 12.5 ± 1.5 min at a drug concentration that promoted an ∼50% decrease in –1 PRF efficiency. Importantly, the CYH2 pre-mRNA remained unstable in wild-type cells in the presence of anisomycin as compared with the isogenic upf1Δ strain, demonstrating that the NMD pathway was not affected at the concentration of anisomycin used in this experiment. These experiments illuminate an inverse relationship between PRF efficiencies and mRNA half-lives, demonstrating the proof-of-principle that gene expression could be regulated by alterations in PRF efficiencies.
PRF has strong effects on mRNA steady-state abundance
In the end, the extent of protein expression is most strongly influenced by the overall abundance of its corresponding mRNA. To examine the effects of –1 PRF at this level, we monitored the steady state abundance of the PGK1 mRNAs expressed from the different constructs assayed in this study. The results show that directing ribosomes to encounter premature termination codons as a consequence of programmed frameshifting can have highly significant impacts on mRNA steady state abundance (Fig. 4). The mRNAs encoded by the wild-type and high efficiency –1 PRF signals (pJD269 and pJD274) are nearly undetectable: ∼30-fold less abundant than the 0-frame control (p3131). Interestingly, the inefficient frameshift signals also significantly reduced PGK1 mRNA steady state abundance (compare pJD257 and pJD273 with p3131), suggesting that even low levels of –1 PRF may be sufficient to control gene expression. That the observed effects of low levels of –1 PRF on mRNA abundance are dependent on –1 PRF is evidenced by the observation that an out-of-frame –1 PRF signal had no effect on PGK1 mRNA steady state abundance (see pJD258).
Figure 4.
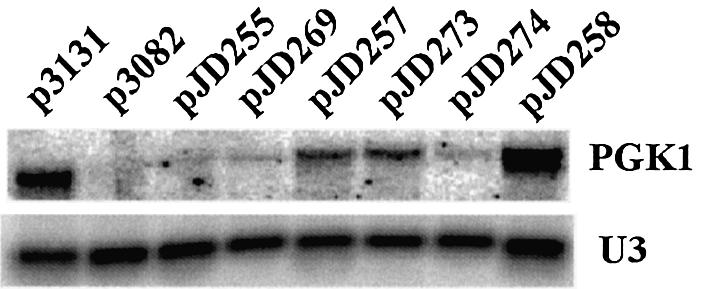
Programmed –1 ribosomal frameshifting affects mRNA steady-state abundance. Total RNAs were harvested from wild-type cells containing the indicated reporter plasmids and hybridized with probes to visualize PGK1 and U3snoRNA hybridizing bands.
Computational modeling of PRF-dependent NMD
Although the in vivo efficiency of the L-A frameshift is only ∼2–6% depending on the assay system used (16,23), this message was only 2-fold more stable than the 0-frame nonsense-containing mRNAs encoded by p3082 and pJD255. An in silico approach was devised to address how such low levels of frameshifting could have such strong effects on mRNA stability. Theoretical mRNA decay rates based on several different models were computationally generated to address this question. Beginning with several pools of 10 000 identical mRNAs of equal length, each mRNA in each pool was assumed to be subject to a constitutive rate of decay, independent of translation, that was arbitrarily set at 1% of the messages capable of entering a decay pathway after each successful round of translation. This model also assumes all messages in a given pool have a number of identical features, including maximal ribosome load, one –1 PRF signal (except wild-type, which has none), and that if any of the loaded ribosomes shifted frame there would be a 98% chance that the mRNA will be recognized as aberrant and degraded by NMD (19). Finally, the process of translational elongation was considered error-free for the purposes of this simulation.
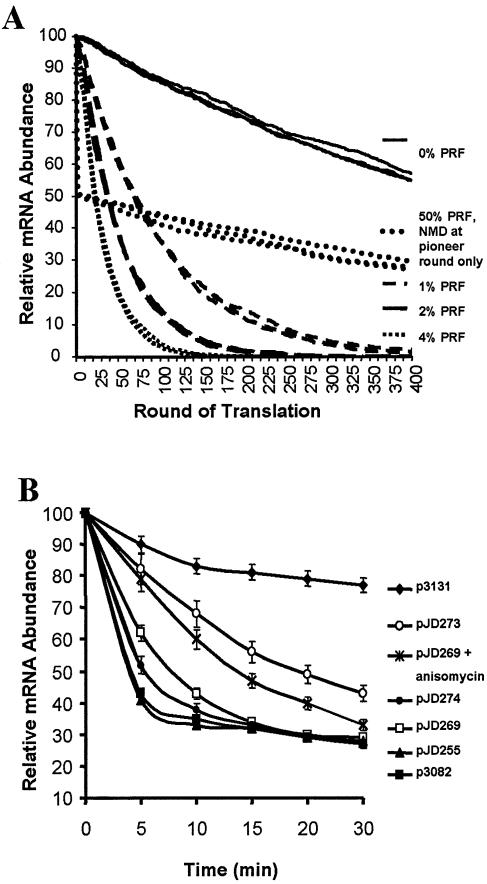
Several computationally generated solutions are depicted in Figure 5A, each depending on whether or not NMD remains active after the pioneer round of translation. The decay profile predicted for a wild-type message (no PRF) follows a shallow, approximately linear negative slope (solid lines). A previous in vitro study suggested that approximately half of ribosomes that pause at the wild-type L-A –1 PRF signal actually shifted (24). Assuming that 50% of ribosomes shift at the pioneer round, and that NMD was inactivated after this first round, the calculated decay profile shows that, although roughly half of the mRNAs are eliminated from the pool at the first round of translation, the trajectories and decay rates of the remaining mRNAs parallel that of the wild-type mRNA (heavy dotted lines). In contrast, if –1 PRF efficiency occurs at an efficiency of 2%, and if NMD remains active beyond the pioneer round, the calculated decay profile follows an exponential trajectory, and the theoretical decay rate of such an mRNA is significantly greater than that of the wild-type mRNA (heavy dashed lines). Further, this model predicts that rates of mRNA decay would follow an inverse proportionality relationship with –1 PRF efficiency (lightly dashed and dotted lines). This is based on the notion that efficient PRF signals would direct messages to the NMD pathway with greater effectiveness.
Figure 5.
Nonsense-mediated mRNA decay remains active after the pioneer round of translation. (A) Computationally predicted mRNA decay rates. Starting with several pools of 10 000 identical mRNAs of equal length, each mRNA in each pool was assumed to be subject to a constitutive rate of decay independent of translation; this rate was arbitrarily set to a 1% chance of any of the messages entering a decay pathway after each successful round of translation. The model also assumes that each message had the same number of ribosomes on it; that each mRNA had only one –1 PRF signal (except wild-type, which had none); that there would be a 98% chance that the mRNA will be degraded by NMD after a –1 PRF event; and a 0.01% frequency of non-programmed frameshifting. The predicted decay profile of an mRNA lacking a PRF signal (0% PRF) is shown as solid lines. The decay profile of an mRNA in which 50% of ribosomes shift at the pioneer round, and where NMD was inactivated after this first round, is shown as heavy dotted lines. The calculated decay profile of an mRNA where –1 PRF efficiency was set at 2% efficiency and NMD remained active after the pioneer round is depicted by heavy dashed lines. When NMD remained active after the pioneer round of translation and –1 PRF efficiencies were set at 4% or 1%, the calculated decay profiles followed the trajectories shown as lightly dotted and dashed lines as indicated. (B) Empirically derived mRNA decay profiles. Graphic representation of PGK1 reporter mRNA decay profiles corresponding to blots in Figures 1 and 3. Bands were quantitated using either Molecular Dynamics Typhoon or Storm Phosphoimagers. PGK1 signals were first normalized relative to U3 loading controls and the resulting ratios were normalized relative to the 0-min values. All of the mRNA half-life experiments were independently performed in triplicate and statistical analyses were performed using SPSS software.
Figure 5B plots the empirical decay profiles of the PGK1 reporter mRNAs shown in Figures 1 and 3. The data for the in-frame control (p3131) follows the predicted shallow linear negative slope. The decay profiles for the frameshift reporters (pJD269, pJD274 and pJD273), follow the typical ‘biphasic’ decay profiles of nonsense-containing mRNAs observed with the nonsense-containing controls (p3082 and pJD255) (25). The first phase of these decay profiles follow the logarithmic trajectories predicted by the model of continuous NMD, providing independent support for the model that the NMD apparatus remains active beyond the pioneer round of translation in yeast. It is notable, however, that whereas logarithmic mRNA decay proceeds to zero in the computational model, this decay function abates at ∼30% of time zero, after which the shallow negative linear function is observed. Given that NMD is only active on actively translating ribosomes, we suggest that only ∼70% of the PGK1 test mRNAs were initially present in the pool of actively translated mRNAs. The remaining 30% would not be actively translated, and thus only subject to degradation by non-NMD processes.
DISCUSSION
The relative simplicity of viral systems facilitates the discovery process. Thus, although programmed ribosomal frameshifting is generally considered to be virus-specific, there is an alluring possibility that this mechanism may also be used by chromosomally encoded mRNAs, and efforts are currently being devoted toward the creation of predictive software, identification and characterization of chromosomally encoded PRF signals (13,26–28). Combining the ‘gold standards’ of the yeast –1 PRF and NMD assay systems, we have demonstrated that a –1 PRF signal can function as a cis-acting mRNA destabilizing element. Further, the observed inverse correlation between –1 PRF efficiency and mRNA stability suggests that frameshifting could be used to post-transcriptionally regulate gene expression. Thus, in addition to its role in viral gene expression, PRF also signifies a potential general biological regulatory mechanism. In addition, a recent report showing that the EST3 mRNA, which contains a +1 PRF signal, is a substrate for NMD in yeast provides further support for this hypothesis (29). The broad aim of our future research will be to use a combination of computational, DNA microarray and molecular approaches to identify cellular mRNAs that utilize –1 PRF, and to characterize how modulation of –1 PRF efficiency may be used to regulate gene expression.
The extremely efficient nature of the NMD apparatus on nonsense-containing mRNAs has hampered our ability to determine whether the NMD can happen on mature mRNAs after the first round of translation (reviewed in 30). One approach to circumvent this problem was effected by repressing and subsequently inducing NMD in yeast (31). The results of that study showed that nonsense-containing mRNAs are available to the NMD pathway after the pioneer round of translation in yeast. Here, we have addressed the issue by inserting –1 PRF signals into a reporter mRNA so that ribosomes encounter nonsense codons at low frequencies. Comparison of the resulting PGK1 reporter mRNA decay profiles with computationally modeled ones supports the findings of Maderazo et al. (31) by providing an independent, less invasive way to address the question of whether or not NMD can remain active after the pioneer round of translation.
The observed mRNA decay profiles are also important because they address the question of whether an mRNA pseudoknot can reform on an mRNA after it has been denatured by an elongating ribosome. For example, a pioneer ribosome that does not shift and continues to translate the message in the 0-frame would denature the mRNA pseudoknot. If the pseudoknot were not able to reform, then the –1 PRF signal would be rendered non-functional and the mRNAs would be stable. Similar to the scenario described above for the case of NMD confined to the pioneer round of translation, if this were the case then the observed decay plots would follow linear rather than exponential trajectories. The observed mRNA decay profiles clearly show continuous frameshifting on –1 PRF-competent mRNAs. Thus, even if the first ribosome fails to shift and denatures the mRNA pseudoknot, the ability of subsequent ribosomes to shift demonstrates that the mRNA pseudoknot is able to reform. This hypothesis is also in accordance with recent findings that actively translated mRNAs are not maximally loaded with ribosomes (32) and that considerable secondary structure is present in the coding regions of actively translated mRNAs (33).
The results presented here also bring into question the primary function of the NMD apparatus. Has it evolved to clear the cell of rare nonsense-containing mRNAs, or does it exist to regulate cellular gene expression on a more global level? One could envision that the abundance of PRF signal-containing mRNAs could be regulated by changing PRF efficiencies and/or by altering the efficiency of the NMD apparatus to recognize premature termination codons. The ability of the Upf proteins to be phosphorylated (reviewed in 30,34) supports this latter notion, i.e. that regulation of NMD can be used to control gene expression.
Acknowledgments
ACKNOWLEDGEMENTS
We want to thank the members of the Peltz laboratory for p3131 and p3082. We also thank Jason W. Harger and Kristi L. Muldoon Jacobs for both technical and editorial assistance, and the Advanced Biomedical Computing Center at the National Cancer Institutes at Fredrick, MD. This work was supported by grants to J.D.D. from the NIH (R01 GM58859, R21 GM068123), the National Science Foundation (SGER MCB-0084559) and from the New Jersey State Commission on Cancer Research (96-62-CCR-00, 97-60-CCR).
REFERENCES
- 1.Maquat L.E. (1995) When cells stop making sense: effects of nonsense codons on RNA metabolism in vertebrate cells. RNA, 1, 453–465. [PMC free article] [PubMed] [Google Scholar]
- 2.Gonzalez C.I., Bhattacharya,A., Wang,W. and Peltz,S.W. (2001) Nonsense-mediated mRNA decay in Saccharomyces cerevisiae. Gene, 274, 15–25. [DOI] [PubMed] [Google Scholar]
- 3.Culbertson M.R. and Leeds,P.F. (2003) Looking at mRNA decay pathways through the window of molecular evolution. Curr. Opin. Genet. Dev., 13, 207–214. [DOI] [PubMed] [Google Scholar]
- 4.Asakura T., Sasaki,T., Nagano,F., Satoh,A., Obaishi,H., Nishioka,H., Imamura,H., Hotta,K., Tanaka,K., Nakanishi,H. et al. (1998) Isolation and characterization of a novel actin filament-binding protein from Saccharomyces cerevisiae. Oncogene, 16, 121–130. [DOI] [PubMed] [Google Scholar]
- 5.Lundblad V. and Morris,D.K. (1997) Programmed translational frameshifting in a gene required for yeast telomere replication. Curr. Biol., 7, 969–976. [DOI] [PubMed] [Google Scholar]
- 6.Aigner S., Lingner,J., Goodrich,K.J., Grosshans,C.A., Shevchenko,A., Mann,M. and Cech,T.R. (2000) Euplotes telomerase contains an La motif protein produced by apparent translational frameshifting. EMBO J., 19, 6230–6239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ivanov I.P., Matsufuji,S., Murakami,Y., Gesteland,R.F. and Atkins,J.F. (2000) Conservation of polyamine regulation by translational frameshifting from yeast to mammals. EMBO J., 19, 1907–1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Klobutcher L.A. and Farabaugh,P.J. (2002) Shifty ciliates: frequent programmed translational frameshifting in euplotids. Cell, 111, 763–766. [DOI] [PubMed] [Google Scholar]
- 9.Shigemoto K., Brennan,J., Walls,E., Watson,C.J., Stott,D., Rigby,P.W.J. and Reith,A.D. (2001) Identification and characterisation of a developmentally regulated mammalian gene that utilises –1 programmed ribosomal frameshifting. Nucleic Acids Res., 29, 4079–4088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cui Y., Dinman,J.D. and Peltz,S.W. (1996) mof4-1 is an allele of the UPF1/IFS2 gene which affects both mRNA turnover and –1 ribosomal frameshifting efficiency. EMBO J., 15, 5726–5736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dinman J.D. and Wickner,R.B. (1992) Ribosomal frameshifting efficiency and Gag/Gag-pol ratio are critical for yeast M1 double-stranded RNA virus propagation. J. Virol., 66, 3669–3676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dinman J.D. and Wickner,R.B. (1994) Translational maintenance of frame: mutants of Saccharomyces cerevisiae with altered –1 ribosomal frameshifting efficiencies. Genetics, 136, 75–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hammell A.B., Taylor,R.L., Peltz,S.W. and Dinman,J.D. (1999) Identification of putative programmed –1 ribosomal frameshift signals in large DNA databases. Genome Res., 9, 417–427. [PMC free article] [PubMed] [Google Scholar]
- 14.Nonet M., Scafe,C., Sexton,J. and Young,R. (1987) Eucaryotic RNA polymerase conditional mutant that rapidly ceases mRNA synthesis. Mol. Cell. Biol., 7, 1602–1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peltz S.W. and Jacobson,A.J. (1993) mRNA turnover in Saccharomyces cerevisiae. In Brawerman,G. and Belasco,J. (eds) Control of mRNA Stability. Academic Press, New York, NY, pp. 291–328. [Google Scholar]
- 16.Dinman J.D., Icho,T. and Wickner,R.B. (1991) A –1 ribosomal frameshift in a double-stranded RNA virus forms a Gag-pol fusion protein. Proc. Natl Acad. Sci. USA, 88, 174–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hagan K.W., Ruiz-Echevarria,M.J., Quan,Y. and Peltz,S.W. (1995) Characterization of cis-acting sequences and decay intermediates involved in nonsense-mediated mRNA turnover. Mol. Cell. Biol., 15, 809–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cui Y., Kinzy,T.G., Dinman,J.D. and Peltz,S.W. (1998) Mutations in the MOF2/SUI1 gene affect both translation and nonsense-mediated mRNA decay. RNA, 5, 794–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cao D. and Parker,R. (2003) Computational modeling and experimental analysis of nonsense-mediated decay in yeast. Cell, 113, 533–545. [DOI] [PubMed] [Google Scholar]
- 20.Dinman J.D., Ruiz-Echevarria,M.J., Czaplinski,K. and Peltz,S.W. (1997) Peptidyl transferase inhibitors have antiviral properties by altering programmed –1 ribosomal frameshifting efficiencies: development of model systems. Proc. Natl Acad. Sci. USA, 94, 6606–6611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dinman J.D., Ruiz-Echevarria,M.J. and Peltz,S.W. (1998) Translating old drugs into new treatments: Identifying compounds that modulate programmed –1 ribosomal frameshifting and function as potential antiviral agents. Trends Biotechnol., 16, 190–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pittman R.H., Andrews,M.T. and Setzer,D.R. (1999) A feedback loop coupling 5 S rRNA synthesis to accumulation of a ribosomal protein. J. Biol. Chem., 274, 33198–33201. [DOI] [PubMed] [Google Scholar]
- 23.Harger J.W. and Dinman,J.D. (2003) An in vivo dual-luciferase assay system for studying translational recoding in the yeast Saccharomyces cerevisiae. RNA, 9, 1019–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lopinski J.D., Dinman,J.D. and Bruenn,J.A. (2000) Kinetics of ribosomal pausing during programmed –1 translational frameshifting. Mol. Cell. Biol., 20, 1095–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leeds P., Peltz,S.W., Jacobson,A.J. and Culbertson,M.R. (1991) The product of the yeast UPF1 gene is required for rapid turnover on mRNAs containing a premature translational termination codon. Genes Dev., 5, 2303–2314. [DOI] [PubMed] [Google Scholar]
- 26.Shah A.A., Giddings,M.C., Parvaz,J.B., Gesteland,R.F., Atkins,J.F. and Ivanov,I.P. (2002) Computational identification of putative programmed translational frameshift sites. Bioinformatics, 18, 1046–1053. [DOI] [PubMed] [Google Scholar]
- 27.Gurvich O.L., Baranov,P.V., Zhou,J., Hammer,A.W., Gesteland,R.F. and Atkins,J.F. (2003) Sequences that direct significant levels of frameshifting are frequent in coding regions of Escherichia coli. EMBO J., 22, 5941–5950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gao X., Havecker,E.R., Baranov,P.V., Atkins,J.F. and Voytas,D.F. (2003) Translational recoding signals between gag and pol in diverse LTR retrotransposons. RNA, 9, 1422–1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.He F., Li,X., Spatrick,P., Castillo,R., Dong,S. and Jacobson,A. (2003) Genome-wide analysis of mRNAs regulated by the nonsense-mediated and 5′ to 3′ mRNA decay pathways in yeast. Mol. Cell, 12, 1439–1452. [DOI] [PubMed] [Google Scholar]
- 30.Maquat L.E. and Serin,G. (2001) Nonsense-mediated mRNA decay: insights into mechanism from the cellular abundance of human Upf1, Upf2, Upf3, and Upf3X proteins. Cold Spring Harb. Symp. Quant. Biol., 66, 313–320. [DOI] [PubMed] [Google Scholar]
- 31.Maderazo A.B., Belk,J.P., He,F. and Jacobson,A. (2003) Nonsense-containing mRNAs that accumulate in the absence of a functional nonsense-mediated mRNA decay pathway are destabilized rapidly upon its restitution. Mol. Cell. Biol., 23, 842–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arava Y., Wang,Y.L., Storey,J.D., Liu,C.L., Brown,P.O. and Herschlag,D. (2003) Genome-wide analysis of mRNA translation profiles in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA, 100, 3889–3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Katz L. and Burge,C.B. (2003) Widespread selection for local RNA secondary structure in coding regions of bacterial genes. Genome Res., 13, 2042–2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gonzalez C.I., Wang,W. and Peltz,S.W. (2001) Nonsense-mediated mRNA decay in Saccharomyces cerevisiae: a quality control mechanism that degrades transcripts harboring premature termination codons. Cold Spring Harb. Symp. Quant. Biol., 66, 321–328. [DOI] [PubMed] [Google Scholar]