Abstract
We present an approach that generates an oligomer-based library with minimal need for restriction site modification of sequences in the target vector. The technique has the advantage that it can be applied for generating peptide aptamer libraries at sites within proteins without the need for introducing flanking enzyme sites. As an example we present a phagemid retroviral shuttle vector that can be used to achieve stable expression of the library in mammalian cells for the purpose of screening for peptides with desired biological activity.
INTRODUCTION
A number of cloning strategies have evolved over the past three decades ranging from the conventional cassette cloning approach using restriction enzymes and ligase to more recent techniques involving cre recombinase (1) and integrase–exisionase systems (2). The latter techniques are adequate for basic cloning where the goal is to transfer a single insert into a vector. However, these techniques are less powerful when the number of unique sequences to be inserted increases. Such is the case with aptamer libraries that require the insertion of thousands or millions of different sequences into the same backbone vector.
At the DNA level an aptamer library can be defined by a constant region shared by all clones and a variable region that is unique for every member represented in the library. In order to achieve a high complexity library, it is necessary to insert a suitably large number of unique inserts into a specific site within the vector. This is applicable for peptide libraries and ribozyme libraries, among others (3–10). The number of unique clones within the library defines its complexity, and it is usually desirable to have a high complexity that represents as many different sequences as possible. The creation of such libraries can represent a significant share of the time invested in setting up a genetic screen using such libraries.
The construction of a random peptide expression library requires a random central region usually 27–45 nucleotides in length flanked by regions of defined sequence and the backbone vector chosen to carry the library. Examples of such libraries can be found in multiple publications (3,5,11–14). The process usually involves a modified version of the cassette cloning approach. In brief, a small oligonucleotide complementary to the non-random 3′ end of the library oligonucleotide is annealed to prime a polymerase reaction that makes the library insert double stranded (3). The now double-stranded insert is restricted with endonucleases, purified by gel electrophoresis and ligated into a vector previously digested with complementary restriction enzymes. Because the oligonucleotide is usually less than 100 bases long, it can be difficult to efficiently purify the double-stranded insert that was successfully cut with both restriction enzymes from incompletely digested material. Both the ligation of a small insert into a much larger vector and the inability to adequately purify the insert can result in loss of library complexity.
Here we have considered a different strategy: the creation of a single-stranded backbone vector that is compatible with a single-stranded insert containing the aptamer library. Although such an approach has previously been used primarily for the substitution or incorporation of one or a few nucleotides, we were encouraged that such site-directed mutagenesis has been used to successfully integrate sequences as large as 27 bases such as the HA1 epitope (15), a size equal to that of many libraries. However, conventional site-directed mutagenesis is an inefficient process that yields the desired product much less than 50% of the time (16), an efficiency too low for library generation of sufficient complexity. The more advanced QuikChange Mutagenesis method is still incapable of introducing sequences long enough to generate biologically active peptide libraries. When a 31 nucleotide sequence was introduced, more than 25% of the transformants failed to carry the insert even after substantial optimization (17). This procedure does not improve transformation efficiency, critical for complex library production.
The technique presented here uses a library oligonucleotide that hybridizes to the single-stranded vector, and primes a polymerase reaction that uses the vector strand as template. The newly synthesized library strand is covalently closed—creating a double-stranded DNA (dsDNA)—and purified from template materials. Modifications to the technique ensure that nearly 100% of the resulting vectors can contain inserts. We demonstrate that the procedure is sufficiently efficient to generate libraries of a complexity of at least 1 × 106. With optimization and increases in scale it should be possible to make libraries of 1 × 108. The approach should simplify the creation of high complexity oligomer-based libraries in a number of experimental settings.
MATERIALS AND METHODS
Strains
The Escherichia coli strain XL1-Blue was used in this study for transformation of plasmids and production of phage. This strain carries an F’ episome that confers tetracycline resistance and is required for pilus formation and phage infection. M13KO7 bacteriophage was used as the helper phage.
Purification of ssDNA template
Bacteria transformed with the phagemid were cultured in LB containing ampicillin (100 µg/ml) + tetracycline (50 µg/ml). This culture inoculated 2YT containing ampicillin (100 µg/ml) and helper phage. After 2 h of helper phage exposure, kanamycin (50 µg/ml) was added and the culture incubated at 32°C with agitation overnight. At stationary phase the bacteria were pelleted, the supernatant spun twice and filtered through a 0.45 µm Acrodisc filter. One hundred and fifty microliters of a 20% solution of PEG 8000 and 2.5 M NaCl was added per milliliter of supernatant and incubated at 4°C for 45 min to precipitate the phage. The mixture was centrifuged at 11 000 g to pellet the phage. The pellet was resuspended in EDTA and single-stranded DNA (ssDNA) extracted by phenol followed by precipitation with sodium acetate and ethanol. A 250 ml culture grown overnight at 32°C with agitation yielded ∼2 mg of ssDNA.
Library strand synthesis
The library oligonucleotide (50 pmol) and 2 µg of the ssDNA template was combined with 0.5 µl of ‘Platinum Taq High Fidelity’ (a mixture of heat-activated Taq polymerase and Pyrococcus species GB-D polymerase from Invitrogen) in 200 µM dNTP, 2 mM MgSO4, 60 mM Tris–SO4 (pH 8.9), 180 mM ammonium sulfate. This enzyme cocktail was chosen to minimize misincorporation and enzyme stalling. It was activated by incubation at 95°C for 50 s followed by ramping down to 60°C over 1 min to anneal the library oligonucleotide. The sample temperature was then increased to 68°C for 30 min. The reaction was quenched by addition of buffer-saturated phenol to minimize low temperature mispriming.
Library strand ligation
High efficiency T4 ligase (Promega) was employed and the ligation reaction was allowed to proceed in 30 mM Tris–HCl (pH 7.8), 10 mM MgCl2, 10 mM DTT and 1 mM ATP in a volume of 100 µl at 14°C overnight. The products of ligation were phenol extracted and precipitated with sodium acetate and ethanol.
Restriction digest of the kill site
SphI restriction digests were carried out prior to both transformation steps. Less than 1 µg of DNA was cut with 1 µl of the restriction enzyme SphI (NEB) carried out in 20 µl of NEB buffer 2 (10 mM Tris–HCl, 10 mM MgCl2, 50 mM NaCl, 1 mM DTT pH 7.9) at 37°C for 2 h. An additional 1 µl of SphI enzyme was added and the reaction incubated at 37°C for 2 h.
Transformation of the heteroduplex
The heteroduplex product was purified by agarose gel electrophoresis as the band that co-migrates with the covalently closed circular band of the double-stranded vector and was excised and phenol extracted. One microliter of the purified DNA was used to transform 20 µl of electrocompetent XL1-Blue E.coli per electroporation cuvette (0.1 cm). After rescuing for 2 h at 32°C in salt-optimized + carbon (SOC) broth with vigorous agitation the entire transformation was used to innoculate 20 ml of LB containing ampicillin (100 µg/ml). In addition, a small quantity of the transformation was plated on ampicillin plates to determine library complexity. The liquid culture was grown overnight and the plasmid DNA purified by alkaline lysis followed by binding to silica columns–Qiagen’s Miniprep kit.
Elimination of the template strand
One microgram of the plasmid prep was digested with SphI as above, visualized by agarose gel electrophoresis, and used to transform 20 µl of electrocompetent XL1-Blue E.coli. After rescuing for 2 h at 32°C in SOC with vigorous agitation the transformation was used to innoculate 20 ml of LB containing ampicillin (100 µg/ml). After incubation overnight at 32°C the culture was either prepped directly or used to innoculate 500 ml of ampicillin–LB.
RESULTS AND DISCUSSION
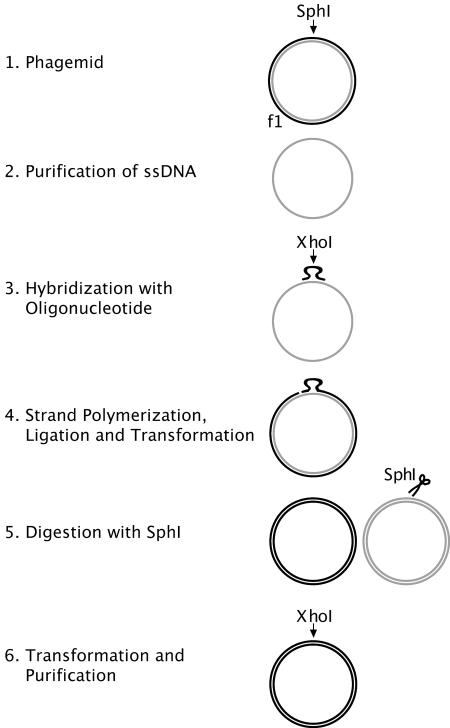
We outline the approach by which we generate oligonucleotide-directed site-specific libraries as follows: (i) purification of a single-stranded phagemid acceptor vector; (ii) design of a corresponding library oligonucleotide insert ssDNA; (iii) extension of the library ssDNA by priming; (iv) covalent closing of the extended library strand to create a dsDNA for transformation; and (v) purification of the double-stranded library (see Figure 1 for outline of protocol).
Figure 1.
Overview of the process. A vector containing the f1 region and a kill site such as SphI (step 1) is subjected to ssDNA purification (step 2). Oligonucleotides carrying the library or a XhoI site as tracker is hybridized to the ssDNA (step 3). Polymerization and ligation is followed by transformation (step 4). SphI restriction eliminates background (step 5) and transformation results in library dsDNA, marked by the XhoI site (step 6). Arrow depicts restriction enzyme site and scissors depicts actual restriction.
Phagemid and oligonucleotide design
This technique required the purification of one of two potential circular single strands of the vector used to express the library. We created a phagemid that contained both the E.coli origin of replication as well as the origin of replication and packaging site (f1) from a single-stranded phage (18) in addition to our required vector expression regions (in this case a retrovirus construct). The E.coli origin allows the phagemid to propagate in E.coli as a self-replicating, double-stranded, extrachromosomal element similar to most cloning and expression plasmid vectors. The viral origin of replication allows helper phage-directed, specific amplification of one strand of the phagemid when the bacterium carrying the vector is infected with M13KO7 bacteriophage. The packaging site allows the single-stranded, circular form of the vector to be encapsidated into phage particles that then bud off from the host bacterium and can be easily purified (19).
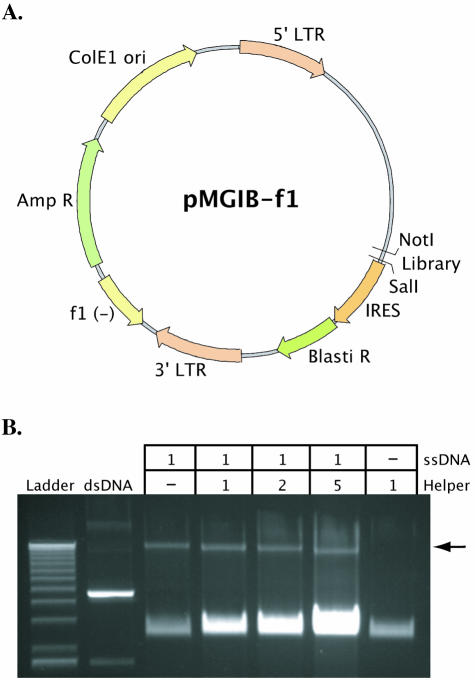
The murine leukemia virus-based vector, pMGIB, was chosen as the backbone for our phagemid. The use of this backbone enables the production of retroviral particles that will ultimately carry the library. This basic format is one that we and others have previously used in the generation of peptide libraries (20–22). To allow for conversion of the pMGIB retroviral vector into a phagemid format we subcloned in the bacteriophage f1 region from pBluescript II SK (–) (Fig. 2A). The orientation of the f1 determines which strand of a phagemid is specifically amplified by phage-directed replication and packaged into budding phage particles. The pMGIB-based phagemids used in these experiments allowed for the purification of the antisense or (–) strand. The vector also includes a unique restriction site in the position where the library is to be incorporated, the ‘kill site’. We chose SphI as it is not present in the backbone. This kill site was designed to enable destruction of any vector background resulting from transformation of the library-containing heteroduplex DNA (see Fig. 4) that does not incorporate an insert.
Figure 2.
Phagemid vector used for library insertion. (A) Map of phagemid vector. The filamentous phage intergenic region f1 was subcloned from pBluescript into the retroviral vector pMGIB. The plasmid constructed contains the functional components of both phagemid and retroviral shuttle vectors including: E.coli origin of replication, bacteriophage origin of replication (f1), ampicillin resistance, 5′ and 3′ retroviral LTRs, Psi packaging site, IRES and blasticidin resistance. (B) Purification of the antisense strand of the phagemid vector. Superinfection with helper phage is required for packaging of the antisense strand of the phagemid vector (arrow). The single-stranded helper phage genome is always present in the final ssDNA prep. The gel electrophoresis shows the separation of the library strand synthesis reactions where increasing amounts of single-stranded helper phage genome was added to a constant amount of ssDNA vector (lanes 3–6).
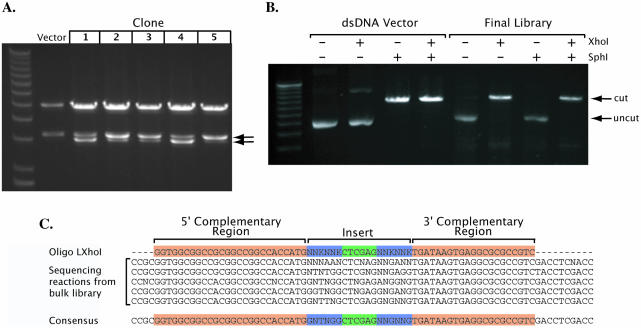
Figure 4.
Purification of dsDNA library. (A) Restriction analysis of clones from the first transformation. Gel electrophoresis shows a double digest with HindIII and XhoI. The XhoI site introduced by library insertion results in cleavage of the 3.2 kb HindIII fragment yielding a 3 kb band (arrows). The heteroduplex product of library strand synthesis is expected to yield two plasmid populations in each transformed bacterium. (B) Restriction analysis of the final library. The XhoI site replaces the SphI site. (C) Sequencing reaction using the final library as template. Several sequences obtained and the resulting consensus are shown.
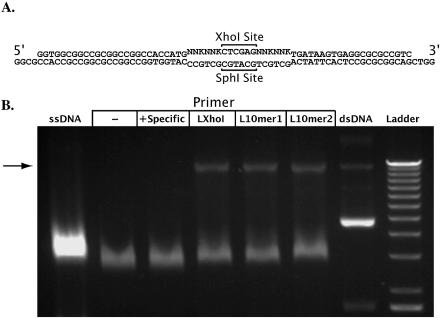
In order to efficiently anneal the random sequence to the purified ssDNA vector, the sequence is flanked by oligomers (26 and 21 bases, respectively) complementary to regions upstream and downstream of the SphI kill site (Fig. 3A). Two library oligonucleotides were used in these experiments, each with the same sequences complementary to the vector but flanking different random regions. The first library oligonucleotide is referred to as LXhoI and contains an XhoI site flanked by four codons of random sequence (Fig. 3A). Library insertion with LXhoI replaces the SphI site with the XhoI site—this can be demonstrated by restriction analysis of the final library. The second library oligonucleotide is referred to as L10mer and contains ten codons of random sequence. Fundamentally, a random oligonucleotide library allows the incorporation of any of the four DNA nucleotides at each position in the sequence. Here, a (NNK)n sequence (where N represents any nucleotide, K guanine or thymidine, and n the specified number of codons in the sequence) is used instead. The lack of an adenine in the third position of each codon rules out two of the three eukaryotic stop codons and results in a more balanced distribution of amino acids when translated (3). The ssDNA form of the phagemid used in these experiments co-migrates with the ssDNA genome of the helper phage and cannot be separated by agarose gel electrophoresis. It might also interfere with the generation of the library by reducing efficiency of transformation. To evaluate the effect of the helper ssDNA impurity, we added increasing quantities of single-stranded helper phage genome, holding the quantity of the ssDNA vector constant. The dilution did not significantly inhibit the library strand synthesis reaction (Fig. 2B).
Figure 3.
Synthesis of the library strand. (A) Oligonucleotide used for synthesis of the library strand. The bottom strand is the (–) strand of the phagemid vector where the library primer anneals. The non-complementary region contains an XhoI site flanked by four NNK codons. This region will replace the region of the vector that includes the SphI site. (B) Agarose gel electrophoresis of library strand synthesis reactions. Lane 1: ssDNA template prior to library strand synthesis reaction. Lane 2: no primer. Lanes 3–6: library strand synthesis reactions performed with primers listed. Lane 3: the primer is the reverse complement of LXhoI, shown to bind with high specificity to the (+) strand of the vector (data not shown). Lanes 4–6: library primers that bind with high efficiency and specificity to the ssDNA template. Lane 7: double-stranded vector. Arrow indicates nicked circular dsDNA (Form II).
Synthesis of the library strand
We used PCR to corroborate that the library oligonucleotides can bind specifically to the template target. PCR was performed using reverse primers that annealed at known distances from the kill site. We obtained large quantities of the expected size PCR products (data not shown). The reverse complement of the library oligonucleotides were also used with forward primers that annealed at known distances, again yielding products of expected size.
The library primers and their reverse complements were also effective as sequencing primers, further demonstrating the specificity and efficiency with which they annealed to the template. The primers were not only site-specific but also strand discriminatory as the library primer was effective at sequencing both double-stranded templates and the ssDNA prep, but the primer’s reverse complement only gave an efficient sequencing reaction with the double-stranded template. This further demonstrates the specificity of primer annealing to the correct ssDNA template, as no synthesis is obtained when the sense primer is used with (+) strand or antisense primer with (–) strand.
Following successful annealing of the library oligonucleotide with the ssDNA phagemid vector (illustrated in Fig. 3A), we proceeded with a polymerization reaction to create a DNA strand that contained the library insert. This reaction is referred to as ‘library strand synthesis’. The single-stranded vector serves as a template and is referred to as the ‘template strand’. Annealing of the library oligonucleotide to the template is facilitated by a ramped reduction in temperature from 95 to 60°C (see Materials and Methods). Primer extension and second-strand synthesis occurs during the 30 min incubation at 68°C, the optimal temperature for the enzyme cocktail. When visualized by agarose gel electrophoresis the product of this reaction co-migrates with a nicked form of the double-stranded vector (Fig. 3B).
In reactions without a primer (Fig. 3B, lane 2) the electrophoretic mobility of the ssDNA template was slightly greater than that of the template before the reaction. When the reverse complement of the library primer was used (Fig. 3B, lane 3) no change in electrophoretic mobility was observed. This is because the reverse complement of the library primer does not bind to the purified (–) strand of the vector. When the library strand synthesis reaction is carried out with the library primer the desired product of the reaction co-migrated with the nicked circular form (Form II) (23) of the uncut double-stranded vector (Fig. 3B, lanes 4–7). The above experiment suggested that the synthesis of dsDNA from ssDNA vector and library insert was specific and successful.
ssDNA background was eliminated via agarose gel electrophoresis followed by purification of the band that co-migrated with the covalently closed, relaxed form of the double-stranded vector after the second-strand synthesis reaction. The single-stranded vector and helper phage ssDNA had significantly higher electrophoretic mobilities than the double-stranded product so gel purification efficiently removed these contaminations (Fig. 3). This purification step was found to substantially increase the efficiency of the first transformation and thus increased the complexity of the final library (data not shown).
Ligation of the library strand and transformation
Library strand synthesis normally results in dsDNA molecules with a single-stranded nick 5′ to the library oligonucleotide. A subsequent ligation reaction is used to covalently close the remaining single-stranded nick in the library strand. Omitting this step or using a library oligonucleotide that is not 5′ phosphorylated results in substantially lower transformation efficiency (data not shown) (24).
Digestion of the kill site following the ligation reaction linearized the double-stranded phagemid molecules resulting from mispriming by the library oligonucleotide or DNA fragments. The remaining molecules are circular plasmids that are heteroduplex, containing both template and library strands. Resolution of the library strand into a pure, double-stranded form requires transformation of the mixture, culture of the bulk transformation, and plasmid purification of the culture. Restriction analysis of individual colonies from this transformation showed, as expected, that each clone contains a mixture of two populations of plasmids, one that carries the insert and the other that lacks it (Fig. 4A). The restriction enzyme HindIII cuts the vector twice yielding the fragment bearing the site for library insertion intact (a 3.2 kb band). XhoI, whose site is carried by the library primer LXhoI used here, will cut this band further to a 3 kb band if the library is introduced. The analysis did confirm heteroduplex plasmid formation with mismatches at the library integration site. Due to these mismatches the heteroduplex is not expected to be cut by SphI. Upon transformation with the heteroduplex plasmid rolling circle amplification would replicate each strand independently giving rise to two plasmid populations, one originating from the library strand and the other from the template strand (16).
The resulting template plasmid population was cut by SphI. Removal of linearized DNA and amplification of the library were achieved by transforming bacteria with the products of the digest and culturing the bulk transformation, considered here as transformation 2. Since linearized DNA will not efficiently transform nor propagate in bacteria, purification of the uncut DNA by agarose gel electrophoresis prior to transformation 2 was not critical but did increase transformation efficiency and thus the complexity of the final library (data not shown).
Plasmid purification following transformation 2 showed that there was no discernible vector without inserts in the final library (Fig. 4B). Double-stranded vector DNA used as negative control is not cut by the library XhoI site but it is cleaved by the killer enzyme SphI (Fig. 4B, lanes 2–5). In contrast, the final library is efficiently cleaved by XhoI but not by SphI (Fig. 4B, lanes 6–9). This library was sequenced to test the efficiency of the process. We successfully generated a random oligonucleotide library of 1 × 106 complexity that contained the XhoI site flanked by four codons of random sequence in accord with the designed oligomer (Fig. 4C).
As a proof that the final retroviral vector was functional the library was transfected into Phoenix retroviral packaging cells to produce infectious viral particles (25). Forty-eight hours post-transfection, the supernatant from the packaging cells was used to infect 293T target cells. Three days after infection blasticidin was added to the target cells to a final concentration of 50 µg/ml (the selectable marker for this phagemid was blasticidin resistance). Cell cultures infected with supernatant from packaging cells transfected with the library survived the blasticidin challenge. In contrast, blasticidin killed 100% of the cells in cultures treated with supernatant from packaging cells that had been mock-transfected. This demonstrated that the library was still capable of being packaged as infectious viral particles that conveyed blasticidin resistance to target cells. The successful retroviral delivery of the library, together with the sequencing results, provides proof of principle for the generation of aptamer libraries by second-strand synthesis.
CONCLUSION
Most existing library creation approaches share a common set of problems: (i) the multi-step modification of the library insert to make it compatible for cloning into the vector; (ii) requirement for considerable modification of the target insert site to allow acceptance of the insert, usually via restriction sites, and (iii) inherent issues with ‘unacceptable’ amino acids encoded by such modifications. Here we present a technique that uses the library oligonucleotide in its native, single-stranded form, and that instead minimally modifies the vector insertion site to achieve desired library motifs.
We introduced the aptamer library within a retroviral transfer vector that had been modified to contain the f1 phagemid region. The f1 region was required for generation of single-strand vector template DNA. The retroviral portion of the vector allows for stable expression of the peptide library members within mammalian cells (26–28). We introduced the f1 sequence, produced ssDNA in large amounts and subsequently produced dsDNA from the relatively large (8 kb) template with a library insert region as the primer. Using this approach we created a library of up to 106 independent sequences. Sequencing confirmed the inserts were as expected based on the aptamer design. The resulting library, used to transfect Phoenix packaging cells, was incorporated into infectious retroviral particles. These were able to efficiently infect target cells that transferred the selection element present within the retroviral backbone. This demonstrated that the technique did not mutate important vector sequences involved with retroviral transfer function or expression.
It is possible to use a ‘kill’ site that removed all of the unaltered vectors from the library. By requiring a kill site one must modify the vector insertion region, which is not always desirable. For instance, this might change the amino acid content of a protein that contains or presents the aptamer. However, if one is willing to accept the presence of background unaltered library members as up to 50% of the library contents the system we present here will accommodate such a possibility. Under circumstances where one has a potent and well-designed genetic screen such background library members will not be selected.
We anticipate this approach will be used to generate aptamer libraries in a variety of different settings, including those in which the presenting scaffold for library insertion has constraints regarding amino acid composition. The relative ease with which this library can be generated should increase the utility of the system and its availability to other researchers wishing to apply library selection schemes and dominant effector approaches (11,26,29–35).
Acknowledgments
ACKNOWLEDGEMENTS
We want to acknowledge Jeff Fortin for useful scientific discussions and Khoua Vang for administrative help. M.B.H. was supported as a National Science Foundation Graduate Research Fellow. G.P.N. was supported by NIH grants P01-AI39646, AR44565, AI35304, N01-AR-6-2227, A1/GF41520-01, NHLBI contract N01-HV-28183I, and the Juvenile Diabetes Foundation. R.W. was supported by the University of Michigan grant 2HPZ713 and NIH grant 2HPA412.
REFERENCES
- 1.Hatanaka K., Ohnami,S., Yoshida,K., Miura,Y., Aoyagi,K., Sasaki,H., Asaka,M., Terada,M., Yoshida,T. and Aoki,K. (2003) A simple and efficient method for constructing an adenoviral cDNA expression library. Mol. Ther., 8, 158–166. [DOI] [PubMed] [Google Scholar]
- 2.Ohara O. and Temple,G. (2001) Directional cDNA library construction assisted by the in vitro recombination reaction. Nucleic Acids Res., 29, e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wolkowicz R. and Nolan,G.P. (2003) Retroviral technology—applications for expressed peptide libraries. Front. Biosci., 8, d603–d619. [DOI] [PubMed] [Google Scholar]
- 4.Raffler N.A., Schneider-Mergener,J. and Famulok,M. (2003) A novel class of small functional peptides that bind and inhibit human alpha-thrombin isolated by mRNA display. Chem. Biol., 10, 69–79. [DOI] [PubMed] [Google Scholar]
- 5.Bupp K. and Roth,M.J. (2002) Altering retroviral tropism using a random-display envelope library. Mol. Ther., 5, 329–335. [DOI] [PubMed] [Google Scholar]
- 6.Felts K.A., Chen,K., Zaharee,K., Sundar,L., Limjoco,J., Miller,A. and Vaillancourt,P. (2002) Functional cloning using pFB retroviral cDNA expression libraries. Mol. Biotech., 22, 25–32. [DOI] [PubMed] [Google Scholar]
- 7.Wang W. and Saven,J.G. (2002) Designing gene libraries from protein profiles for combinatorial protein experiments. Nucleic Acids Res., 30, e120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tailor C.S., Nouri,A., Lee,C.G., Kozak,C. and Kabat,D. (1999) Cloning and characterization of a cell surface receptor for xenotropic and polytropic murine leukemia viruses. Proc. Natl Acad. Sci. USA, 96, 927–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cesareni G., Castagnoli,L. and Cestra,G. (1999) Phage displayed peptide libraries. Comb. Chem. High Throughput Screen., 2, 1–17. [PubMed] [Google Scholar]
- 10.Lieber A. and Strauss,M. (1995) Selection of efficient cleavage sites in target RNAs by using a ribozyme expression library. Mol. Cell. Biol., 15, 540–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buerger C., Nagel-Wolfrum,K., Kunz,C., Wittig,I., Butz,K., Hoppe-Seyler,F. and Groner,B. (2003) Sequence-specific peptide aptamers, interacting with the intracellular domain of the epidermal growth factor receptor, interfere with Stat3 activation and inhibit the growth of tumor cells. J. Biol. Chem., 278, 37610–37621. [DOI] [PubMed] [Google Scholar]
- 12.Tolstrup A.B., Duch,M., Dalum,I., Pedersen,F.S. and Mouritsen,S. (2001) Functional screening of a retroviral peptide library for MHC class I presentation. Gene, 263, 77–84. [DOI] [PubMed] [Google Scholar]
- 13.Xu X., Leo,C., Jang,Y., Chan,E., Padilla,D., Huang,B.C., Lin,T., Gururaja,T., Hitoshi,Y., Lorens,J.B., Anderson,D.C., Sikic,B., Luo,Y., Payan,D.G. and Nolan,G.P. (2001) Dominant effector genetics in mammalian cells. Nature Genet., 27, 23–29. [DOI] [PubMed] [Google Scholar]
- 14.Peelle B., Lorens,J., Li,W., Bogenberger,J., Payan,D.G. and Anderson,D.C. (2001) Intracellular protein scaffold-mediated display of random peptide libraries for phenotypic screens in mammalian cells. Chem. Biol., 8, 521–534. [DOI] [PubMed] [Google Scholar]
- 15.Swanson M.S., Carlson,M. and Winston,F. (1990) SPT6, an essential gene that affects transcription in Saccharomyces cerevisiae, encodes a nuclear protein with an extremely acidic amino terminus. Mol. Cell. Biol., 10, 4935–4941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Craik C.S. (1985) Use of oligonucleotides for site-specific mutagenesis. Biotechniques, 3, 12–19. [Google Scholar]
- 17.Wang W. and Malcolm,B.A. (1999) Two-stage PCR protocol allowing introduction of multiple mutations, deletions and insertions using QuikChange site-directed mutagenesis. Biotechniques, 26, 680–682. [DOI] [PubMed] [Google Scholar]
- 18.Zagursky R.J. and Berman,M.L. (1984) Cloning vectors that yield high levels of single-stranded DNA for rapid DNA sequencing. Gene, 27, 183–191. [DOI] [PubMed] [Google Scholar]
- 19.Vieira J. and Messing,J. (1987) Production of single-stranded plasmid DNA. Methods Enzymol., 153, 3–11. [DOI] [PubMed] [Google Scholar]
- 20.Kitamura T., Onishi,M., Kinoshita,S., Shibuya,A., Miyajima,A. and Nolan,G.P. (1995) Efficient screening of retroviral cDNA expression libraries. Proc. Natl Acad. Sci. USA, 92, 9146–9150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller A.D. and Rosman,G.J. (1989) Improved retroviral vectors for gene transfer and expression. Biotechniques, 7, 980–990. [PMC free article] [PubMed] [Google Scholar]
- 22.Weber-Benarous A., Cone,R.D., London,I.M. and Mulligan,R.C. (1988) Retroviral-mediated transfer and expression of human beta-globin genes in cultured murine and human erythroid cells. J. Biol. Chem., 263, 6142–6145. [PubMed] [Google Scholar]
- 23.Thorne H.V. (1967) Electrophoretic characterization and fractionation of polyoma virus DNA. J. Mol. Biol., 24, 203–211. [DOI] [PubMed] [Google Scholar]
- 24.Bebenek K. and Kunkel,T.A. (1989) The use of native T7 DNA polymerase for site-directed mutagenesis. Nucleic Acids Res., 17, 5408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Swift S., Lorens,J., Achacoso,P. and Nolan,G.P. (1999) Rapid production of retroviruses for efficient gene delivery to mammalian cells using 293T cell-based systems. Curr. Prot. Immun., unit 10.28, Suppl. 31. [DOI] [PubMed] [Google Scholar]
- 26.Kinsella T.M., Ohashi,C.T., Harder,A.G., Yam,G.C., Li,W., Peelle,B., Pali,E.S., Bennett,M.K., Molineaux,S.M., Anderson,D.A., Masuda,E.S. and Payan,D.G. (2002) Retrovirally delivered random cyclic peptide libraries yield inhibitors of interleukin-4 signaling in human B cells. J. Biol. Chem., 277, 37512–37518. [DOI] [PubMed] [Google Scholar]
- 27.Lorens J.B., Bennett,M.K., Pearsall,D.M., Throndset,W.R., Rossi,A.B., Armstrong,R.J., Fox,B.P., Chan,E.H., Luo,Y., Masuda,E., Ferrick,D.A., Anderson,D.C., Payan,D.G. and Nolan,G.P. (2000) Retroviral delivery of peptide modulators of cellular functions. Mol. Ther., 1, 438–447. [DOI] [PubMed] [Google Scholar]
- 28.Poritz M.A., Malmstrom,S., Schmitt,A., Kim,M.K., Zharkikh,L., Kamb,A. and Teng,D.H. (2003) Isolation of a peptide inhibitor of human rhinovirus. Virology, 313, 170–183. [DOI] [PubMed] [Google Scholar]
- 29.Chu P., Pardo,J., Zhao,H., Li,C.C., Pali,E., Shen,M.M., Qu,K., Yu,S.X., Huang,B.C., Yu,P., Masuda,E.S., Molineaux,S.M., Kolbinger,F., Aversa,G., De Vries,J., Payan,D.G. and Liao,X.C. (2003) Systematic identification of regulatory proteins critical for T-cell activation. J. Biol., Epub September 15. [DOI] [PMC free article] [PubMed]
- 30.Mèuller O.J., Kaul,F., Weitzman,M.D., Pasqualini,R., Arap,W., Kleinschmidt,J.A. and Trepel,M. (2003) Random peptide libraries displayed on adeno-associated virus to select for targeted gene therapy vectors. Nat. Biotechnol., 21, 1040–1046. [DOI] [PubMed] [Google Scholar]
- 31.Klevenz B., Butz,K. and Hoppe-Seyler,F. (2002) Peptide aptamers: exchange of the thioredoxin-A scaffold by alternative platform proteins and its influence on target protein binding. Cell. Mol. Life Sci., 59, 1993–1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wiesner C., Hoeth,M., Binder,B.R. and de Martin,R. (2002) A functional screening assay for the isolation of transcription factors. Nucleic Acids Res., 30, e80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dhiman A., Rodgers,M.E. and Schleif,R. (2001) Identification of oligomerizing peptides. J. Biol. Chem., 276, 20017–20021. [DOI] [PubMed] [Google Scholar]
- 34.Cho G., Keefe,A.D., Liu,R., Wilson,D.S. and Szostak,J.W. (2000) Constructing high complexity synthetic libraries of long ORFs using in vitro selection. J. Mol. Biol., 297, 309–319. [DOI] [PubMed] [Google Scholar]
- 35.Gallagher W.M., Cairney,M., Schott,B., Roninson,I.B. and Brown,R. (1997) Identification of p53 genetic suppressor elements which confer resistance to cisplatin. Oncogene, 14, 185–193. [DOI] [PubMed] [Google Scholar]