Abstract
Osteogenesis imperfecta (OI) is a heritable bone dysplasia characterized by bone fragility and deformity and growth deficiency. Most cases of OI (classical types) have autosomal dominant inheritance and are caused by mutations in the type I collagen genes. During the past several years, a number of noncollagenous genes whose protein products interact with collagen have been identified as the cause(s) of rare forms of OI. This has led to a paradigm shift for OI as a collagen-related condition. The majority of the non-classical OI types have autosomal recessive inheritance and null mutations in their respective genes. The exception is a unique dominant defect in IFITM5, which encodes Bril and leads to hypertrophic callus and interosseous membrane ossification. Three recessive OI types arise from defects in any of the components of the collagen prolyl 3-hydroxylation complex (CRTAP, P3H1, CyPB), which modifies the collagen α1(I)Pro986 residue. Complex dysfunction leads to delayed folding of the procollagen triple helix and increased helical modification. Next, defects in collagen chaperones, HSP47 and FKBP65, lead to improper procollagen folding and deficient collagen cross-linking in matrix, respectively. A form of OI with a mineralization defect is caused by mutations in SERPINF1, whose protein product, PEDF, is a well-known antiangiogenesis factor. Defects in the C-propeptide cleavage enzyme, BMP1, also cause recessive OI. Additional genes, including SP7 and TMEM38B, have been implicated in recessive OI but are as yet unclassified. Elucidating the mechanistic pathways common to dominant and recessive OI may lead to novel therapeutic approaches to improve clinical manifestations.
Osteogenesis imperfecta (OI), commonly known as “brittle bone disease,” is a clinically and genetically heterogeneous connective tissue disorder associated with skeletal fragility, deformity, and growth deficiency. It has an etiology related directly or indirectly to type I collagen, the most abundant protein of bone extracellular matrix (ECM). Type I collagen is a triple helical molecule synthesized as procollagen from 2 genes, COL1A1 and COL1A2. It is assembled as a heterotrimer, composed of two proα1(I) and one proα2(I) chains, which undergo extensive post-translational modifications (ie, hydroxylation, glycosylation) in the endoplasmic reticulum during chain synthesis and helix formation. The central helical domain is composed of uninterrupted repeats of the tripeptide Gly-X-Y, where X and Y are often proline and hydroxyproline, respectively. The procollagen triple helical domain is flanked by globular N- and C-propeptide domains that undergo cleavage before secretion into the extracellular space. Defects in either type I collagen structure or synthesis can give rise to OI.
Currently, a molecular genetic classification of OI contains 12 types that display either autosomal-dominant or autosomal-recessive patterns of inheritance and exhibit broad variations in clinical severity (Table 1). The majority of OI cases (types I–IV) are dominantly inherited. OI phenotypes were originally classified by Sillence in 1979 based on clinical and radiological criteria (1). The classical OI types I–IV are caused by mutations in COL1A1 or COL1A2, leading to defects in type I collagen synthesis or structure. In 2000, type V OI was defined based on histological and clinical findings (2), and in 2002, type VI OI was delineated based on histological findings (3). These additions to the nosology were made before molecular and genetic discoveries showing that the mutations responsible for these OI types are contained in noncollagenous genes. More recently, additional OI cases lacking COL1 gene mutations have been discovered to arise from mutations in noncollagenous genes whose protein products interact with type I collagen for modification and/or folding. These cases have been subsequently classified into consecutive types VII–XII OI (Table 1) based on distinct clinical and bone histological features, inheritance pattern, and genetic findings.
Table 1.
Classification of OI Types
| OI Type | Gene Defect | Phenotype |
|---|---|---|
| Dominant inheritance | ||
| Classical Sillence types | ||
| I | COL1A1 null allele | Mild, nondeforming |
| II | COL1A1 or COL1A2 | Lethal perinatal |
| III | COL1A1 or COL1A2 | Progressively deforming |
| IV | COL1A1 or COL1A2 | Moderately deforming |
| COL1-mutation negative | ||
| V | IFITM5 | Distinct histology |
| Recessive inheritance | ||
| Mineralization defect | ||
| VI | SERPINF1 | Distinct histology |
| 3-Hydroxylation defects | ||
| VII | CRTAP | Severe to lethal |
| VIII | LEPRE1 | Severe to lethal |
| IX | PPIB | Moderate to lethal |
| Chaperone defects | ||
| X | SERPINH1 | Severe |
| XI | FKBP10 | Progressive deforming, Bruck syndrome 1 |
| C-Propeptide cleavage defect | ||
| XII | BMP1 | Severe, high bone mass case |
| Unclassified | ||
| Zinc-finger transcription factor defect | SP7 | Moderate |
| Cation channel defect | TMEM38B | Moderate to severe |
| WNT signaling pathway defect | WNT1 | Moderate, progressively deforming |
Autosomal Dominant OI
Classical Sillence types: types I–IV OI
The phenotypes of types I–IV OI (Table 1) are classified as mild, nondeforming type I; perinatal lethal type II; progressively deforming type III; and moderately deforming type IV (1). Type I OI, the mildest form, exhibits the triad of features first described by Van der Hoeve and de Kleyn (4)—fractures, blue sclerae, and hearing loss. These individuals typically have near normal stature, minimal bone deformation, and fractures that typically lessen in frequency after puberty. Type II OI, the most severe form, is generally perinatal lethal. Fractures are typically detected in utero. Infants who survive the perinatal period often succumb in the first year of life, most commonly due to cardiopulmonary causes. Type III OI is progressively deforming and the most severe nonlethal form. Patients generally exhibit gray or blue sclerae and extreme short stature and sustain frequent fractures; they often have dentinogenesis imperfecta. About half of the individuals with type III OI exhibit radiographic “popcorn” calcifications at the distal femoral growth plates (5). Patients with type IV, moderately severe OI display a broad range of phenotypes and may or may not exhibit dentinogenesis imperfecta. Individuals typically achieve ambulation but incur frequent long bone fractures; final stature is typically comparable to the prepubertal height of unaffected children (6).
Dominant types I–IV OI are caused by mutations in either of the 2 type I collagen genes, COL1A1 or COL1A2. Mutations can be divided into 2 categories: quantitative defects, in which type I collagen is structurally normal but synthesized in about half the normal amount, and structural defects. The matrix insufficiency of type I OI is due to mutations causing premature termination codons (PTCs) in COL1A1, which activates nonsense-mediated mRNA decay (NMD) of defective transcripts. Because most transcripts from the mutant COL1A1 allele are degraded, only about half the normal amount of matrix is deposited, and it contains almost entirely structurally normal collagen with α1(I) chains from the normal COL1A1 allele. The resulting matrix insufficiency is responsible for the mild phenotype of type I OI. Interestingly, homozygosity for null mutations in COL1A2 transcripts does not cause OI; these mutations lead to the formation of α1(I) homotrimers resulting in mild Ehlers-Danlos syndrome (EDS) with hypermobility and cardiac valve disease (6).
Types II–IV OI are caused by mutations that alter type I collagen structure. Over 80% of these mutations are single base pair changes resulting in substitutions of glycine residues (7). Glycine substitutions in either α1(I) or α2(I) lead to a delay in helix folding, causing post-translational overmodification (7). Phenotypic severity can range from mild to lethal. Only one-fifth of glycine substitutions in α2(I) are lethal, whereas nearly one-third of all glycine substitutions in α1(I) are lethal. Glycine substitutions by branched nonpolar or charged amino acids, specifically glutamate, aspartic acid, arginine, and valine, are most detrimental (7). In the α1(I) chain, lethal substitutions were identified in the major ligand-binding regions (MLBR2 and MLBR3), indicating the importance of interactions between the collagen monomer and noncollagenous proteins, such as integrins, matrix metalloproteinases, fibronectin, and decorin (6, 7). Clusters of lethal glycine substitutions along the α2(I) chain largely align with known regions for proteoglycan binding sites on the collagen fibril (7). Non-glycine substitutions occurring at X- and Y-positions along the collagen triple helical domain have also been described to cause OI/EDS conditions. Arginine to cysteine substitutions at the Y-position can induce a significant register shift along the length of the helix, which impedes N-propeptide processing and causes a range of phenotypes including mild OI, hyperextensibility, and Caffey disease (6).
The second most frequent type of mutation altering type I collagen structure is splice site mutations, which can lead to exon skipping, intronic retention, or activation of cryptic splice sites from intronic or exonic sequences (7). Often, splice site mutations introduce frameshifts that lead to PTCs and result in a mild phenotype; most mutations are not lethal (7).
Mutations at the propeptide domain cleavage sites can disrupt the processing of procollagen to collagen. Exon 6 of procollagen α-chains contains the N-proteinase cleavage site. Deletion of exon 6 and consequent elimination of N-propeptide processing leads to EDS VII with hyperextensibility of large and small joints and inexorable scoliosis (8). Interference with N-propeptide processing may lead to combined OI/EDS. Glycine substitutions within the first 85 helical residues of α1(I) collagen (a region with high local melting temperature that serves as an anchor for stabilizing collagen folding at the amino end of the triple helix) are associated with the combined OI/EDS phenotype (8). The helical location of these glycine substitutions leads to OI symptoms, including bone fragility, growth deficiency, and intensely blue sclerae, whereas the unfolding of the N-propeptide cleavage site causes EDS symptoms similar to EDS VII. Incorporation of pN-collagen into fibrils results in decreased fibril diameter and ECM with compromised mechanical strength (9). Substitutions at the procollagen C-terminal cleavage sites do not significantly alter collagen post-translational modification but do cause incorporation of uncleaved pC-collagen into fibrils. Incorporation of pC-collagen into fibrils leads to increased mineral deposition, resulting in a high bone mass phenotype, with increased mineralization detected by Fourier transform infrared spectroscopy and bone mineral density distribution analyses (10).
Autosomal dominant type V OI (IFITM5)
In 2000, a group of COL1 mutation-negative OI patients, originally characterized as type IV OI, were reclassified as a novel OI group, type V. Type V OI, much like the classical OI types, exhibits a dominant pattern of inheritance, yet the affected individuals exhibit distinguishing clinical features not observed in other OI types. Individuals with type V OI have a moderate to severe skeletal phenotype with variable scleral hue and a typical triad of findings: calcification of the forearm interosseous membrane, a radiodense metaphyseal band at growth plates of long bones (2), and a tendency to form hyperplastic callus after surgical intervention or fracture (11). Over 85% of patients also experience radial-head dislocation (12). In the original report, the definitive feature of type V OI was based on histological observations. Specifically, there is an irregular “mesh-like” pattern of lamellar bone unique to type V OI (2).
The etiology of type V OI was recently demonstrated to be a unique defect in the gene encoding interferon-induced transmembrane protein 5 (IFITM5), alternatively called bone-restricted ifitm-like protein (Bril) (Table 1 and Figure 1). In 2012, two independent research groups utilized whole exome sequencing to identify a heterozygous de novo mutation in the 5′-untranslated region of IFITM5 located 14 base pair (bp) upstream from the translation initiation codon (c.-14C>T) (13, 14). In several reports and unpublished data, all type V OI patients exhibit the same defect in the causative gene, a feature that is unique among OI types. This mutation causes an in-frame alternative initiation codon upstream of the annotated translation initiation codon and is predicted to add 5 residues to the amino terminus of the protein.
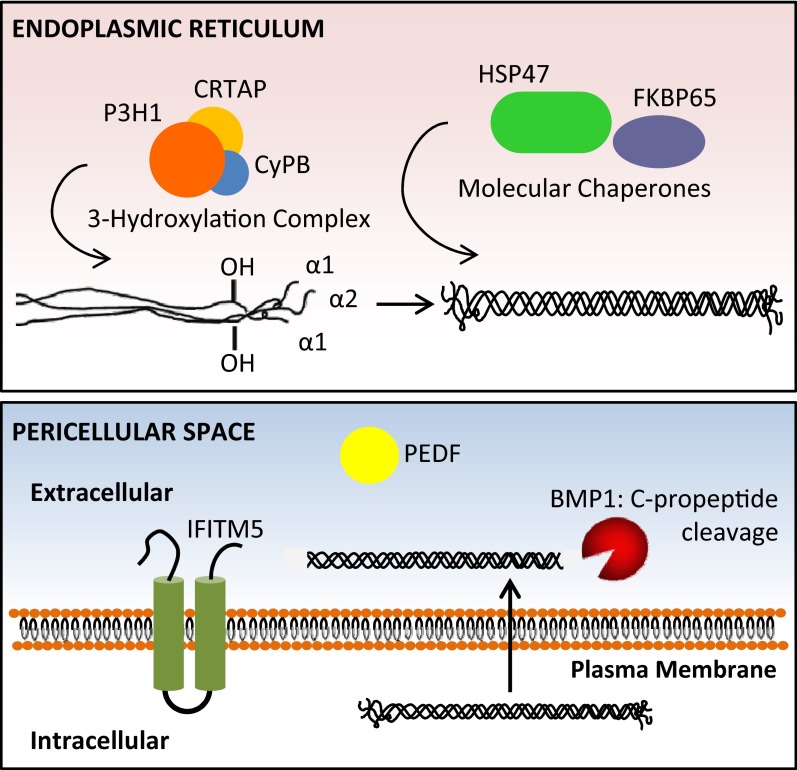
Figure 1.
Defects in noncollagenous proteins causing OI have been identified. Top, Recessive OI is caused by defects in CRTAP, P3H1, and CyPB [forming the ER-resident, collagen prolyl 3-hydroxylation complex responsible for the modification of α1(I) proline 986], as well as HSP47 and FKBP65 (ER-resident collagen chaperones primarily involved in proline isomerization required for collagen helix folding). Bottom, Defects in IFITM5 (implicated in regulating mineralization), PEDF (a well-known inhibitor of angiogenesis), and BMP1 (responsible for procollagen C-propeptide cleavage) have recently been found to cause OI. HSP47, heat shock protein 47.
IFITM5 is a 14.8-kDa transmembrane protein. Similar to all members of the IFITM family, it contains 2 transmembrane domains, an intracellular domain and amino- and carboxyl-terminal sequences that are positioned in the extracellular space (Figure 1). IFITM5 is a known osteoblast marker; it is most highly expressed during early mineralization and is thought to play a critical role in bone formation. Skeletons of Ifitm5 knockout mice displayed stunted growth, particularly smaller long bones as compared to heterozygous and wild-type littermates, but no significant difference in bone morphometric parameters (15). This suggests that loss of Ifitm5 expression does not alter osteoblastogenesis or osteoclastogenesis (15). In mouse embryos, Ifitm5 transcripts were reported to be restricted to skeletal tissue, particularly in long bones (16), and were first observed at embryonic day 14.5 when undifferentiated cells differentiate into osteoblasts and begin to form mineralized structures (15, 16). Immunohistochemical reactivity for IFITM5 localized it to regions of endochondral and intramembranous ossification (15). In addition, Ifitm5 expression declined in rodents after 8 months of age (16). These studies implicate a role for IFITM5 in early mineralization. However, the mechanism by which IFITM5 regulates collagen mineralization is not known.
IFITM5 coprecipitates with FK506-binding protein 11 (FKBP11) (15). As a member of the FKBP family, FKBP11 has peptidyl-prolyl cis-trans isomerase activity, with an implication of a functional role in protein folding. IFITM5 truncation mutants were used to localize the binding domain of IFITM5 to FKBP11 to the first transmembrane domain and intracellular region. Interestingly, mice with defects in Fkbp11 show increased bone density (15). It is not yet known whether IFITM5 and FKBP11 have cooperative or independent roles with respect to bone formation.
Autosomal Recessive OI
Mineralization defects—type VI OI (SERPINF1)
Type VI OI was originally delineated as a mineralization defect in a group of COL1 mutation-negative individuals, based on unique histological features (3). To date, 16 patients have been reported with SERPINF1-deficient OI (17, 18). Previously, diagnosis required assessment of iliac crest biopsy samples, which display undermineralization and a loss of lamellar orientation, as well as a “fish-scale” pattern distinct from other OI types when assessed under polarized light (3). In addition, quantitative histomorphometric analysis of type VI OI trabecular bone shows significantly elevated osteoid volume with decreased bone formation parameters including mineralizing surface, adjusted apposition rate, and osteoblast surface (3). Individuals with type VI OI have a moderate to severe skeletal phenotype (17).
In 2011, whole exome sequencing was used to identify SERPINF1 as the causative gene for type VI OI (Table 1 and Figure 1) (17). SERPINF1 encodes pigment epithelium-derived factor (PEDF), a member of the serine protease inhibitor serpin superfamily. In reported patients, SERPINF1 mutations led to PTCs resulting in NMD of mRNA transcripts and subsequent loss of SERPINF1 expression (17). Type I collagen secretion and post-translational modification was reported to be normal in OI patients with SERPINF1 mutations (17). Recently, the determination of serum PEDF levels was proposed as a diagnostic tool for type VI OI. Patients diagnosed with type VI OI have undetectable levels of circulating PEDF, while all other tested groups, including healthy controls, patients with type I, III, and IV OI, and patients with hypophosphatemic rickets, have measurable PEDF levels (18).
The mechanism by which loss of PEDF results in undermineralized bone ECM is as yet unknown. PEDF is a 50-kDa secreted glycoprotein and one of the strongest known inhibitors of angiogenesis (17, 18). It is expressed in a variety of tissues and binds to ECM components including collagens and glycosaminoglycans. In vitro studies elucidated the ECM-binding sites on PEDF and demonstrate that binding to type I collagen was due mainly to ionic interactions (19). Importantly, loss of collagen binding in PEDF leads to loss of antiangiogenic properties (20). In bone, PEDF binds to type I collagen with high affinity and is highly expressed in osteoblasts in regions of active bone formation, indicating an important functional role of PEDF in bone angiogenesis and matrix remodeling (21). Interestingly, a skeletal phenotype was not reported in Serpinf1 knockout mice (22), but it remains to be determined whether the mice have a subtle phenotype.
Collagen 3-hydroxylation defects—types VII, VIII, and IX OI (CRTAP, LEPRE1, PPIB)
Recessive types VII, VIII, and IX OI are caused by defects in cartilage-associated protein (CRTAP), prolyl 3-hydroxylase 1 (P3H1), and cyclophilin B (CyPB), respectively (Table 1 and Figure 1). These 3 proteins are components of an endoplasmic reticulum (ER)-resident complex, the collagen prolyl 3-hydroxlyation complex, in a 1:1:1 ratio. The 3-hydroxylation complex modifies a limited number of helical proline residues, most significantly proline 986 of collagen α1(I) and α1(II) and proline 707 in α2(I) (6). The biological significance of these hydroxylation events remains to be elucidated.
Beyond prolyl 3-hydroxylase activity, the 3-hydroxylation complex functions as a peptidyl prolyl cis-trans isomerase (PPIase) and molecular chaperone for collagen. In addition, CRTAP, P3H1, and CyPB may also serve important functional roles independent of complex activities. Within the 3-hydroxylation complex, CRTAP is considered to be a helper protein. CRTAP is expressed in a variety of tissues. In mouse, immunohistochemical analyses showed CRTAP to be highly expressed at growth plates in proliferating chondrocytes and to be largely restricted to intracellular regions, often colocalizing with the ER; low expression levels were observed in the matrix (23). In a cell-based assay, up to 12% of total CRTAP protein was detected in conditioned media after 24 hours of culture, indicating a potential role in the ECM (24). CRTAP shares a high degree of homology with the P3H family of proteins (P3H1, P3H2, P3H3), predominantly at the N-terminal region.
The enzymatic 3-hydroxylase activity for the complex is harbored in P3H1. P3H1, encoded by LEPRE1 (leucine- and proline-enriched proteoglycan 1) was originally identified as the matrix proteoglycan leprecan. P3H1 contains both an RGD cell attachment motif and a KDEL ER-retrieval signal, implying its dual role in the extracellular matrix and the ER/Golgi circuit, respectively (25). Studies on fibroblasts from patients with type VII and type VIII OI show that protein levels of CRTAP and P3H1 are reduced or negligible in cells containing null mutations for either gene. However, transcript levels of CRTAP and LEPRE1 were normal in LEPRE1- or CRTAP-null cells, respectively, indicating mutual stabilization of CRTAP and P3H1 (24). The third component of the 3-hydroxylation complex, CyPB, is transcribed from the PPIB gene (peptidyl-prolyl cis-trans isomerase B). CyPB is ubiquitously expressed and independent of CRTAP or P3H1 levels. Conversely, CRTAP and P3H1 protein levels are moderately decreased in PPIB-null cells, indicating a role for CyPB in 3-hydroxylation complex stabilization, although not absolutely required for complex stability (24, 26). In addition to its well-studied function as a PPIase, CyPB is thought to serve as a chaperone for proteins destined for the plasma membrane (26).
The association between OI and the members of the 3-hydroxylation complex was discovered due to a convergence of findings. In 2002, a form of recessive OI with rhizomelia in a First Nations people in Northern Quebec was mapped to chromosome 3p22 (27). Incidentally, this chromosomal region includes CRTAP, but its importance was not appreciated at that time. Four years later, in parallel investigations, Crtap-null mice were discovered to exhibit recessive chondro-osseous dysplasia with rhizomelia and severe osteopenia (23). Also, CRTAP was a candidate gene for screening individuals with severe OI without collagen mutations, but with overmodified collagen because it had been identified in the 3-hydroxylation complex (23, 28, 29).
In humans, the phenotype of type VII OI (CRTAP deficiency) overlaps Sillence types II and III but has distinctive features. CRTAP deficiency causes severe to lethal osteochondrodysplasia with rhizomelia, neonatal fractures, broad undertubulated long bones, frail ribs, and relatively normal head circumference. Sclerae are white or, rarely, light gray. Individuals who survive into childhood have severe growth deficiency and exhibit “popcorn” calcifications of epiphyses, which are also observed in about half of type III OI patients (25). Most mutations causing type VII OI lead to a loss of CRTAP transcripts by NMD. Interestingly, the mutations causing almost all reported cases occur in the first or fourth exon (or adjacent intronic sequences) (25). Absence of CRTAP abolishes 3-hydroxylation complex function and results in the absence of α1(I) Pro986 hydroxylation. An additional consequence of CRTAP deficiency is overmodification of the type I collagen helix by lysyl hydroxylase and prolyl 4-hydroxylase. The extent of overmodification is comparable to that caused by C-terminal collagen structural defects and implies that folding of the collagen helix is delayed (28).
Individuals with types VII and VIII OI exhibit a similar phenotype due to the mutual stabilization between CRTAP and P3H1 (24). Similar to type VII OI, type VIII OI (LEPRE1 deficiency) is caused by homozygous or compound heterozygous mutations that lead to null alleles in most cases. In addition, type I collagen synthesized by LEPRE1-null cells is overmodified. An unexpected consequence of LEPRE1 deficiency is increased collagen production. LEPRE1-null fibroblasts from type VIII OI individuals displayed a slight delay in collagen secretion rate, yet total collagen secretion was increased up to 50% as compared to control cells (30). Mutations in LEPRE1 causing type VIII OI have been identified in 13 of 15 exons or surrounding intronic regions. However, the most common LEPRE1 mutation is a splice site defect in intron 5 (c.1080+1G>T) resulting in 5 alternatively spliced transcripts, each of which contains a PTC. All individuals reported to carry this mutation are African American or of West African descent. Haplotype analysis shows this is a founder mutation that originated in West Africa about 600–900 years ago and was brought to the Americas via the Atlantic slave trade (31).
The phenotype of the small number of individuals reported with type IX OI (PPIB deficiency) is variable. Several reported cases describe phenotypes that largely overlap types VII and VIII, except without rhizomelia (32). Two moderately severe cases have also been reported (26). The more severe phenotypes had mutations predicted to lead to truncated proteins, which might lead to negative interactions within the complex (32). The patients with the most severe phenotypes of type IX OI have decreased, but not absent, 3-hydroxylation of α1(I)P986 (about 30% of normal levels) (32). In addition, collagen secreted from cells of individuals with a severe phenotype exhibit delayed gel migration indicating overmodification of helical lysine and proline residues similar to types VII and VIII OI, consistent with 3-hydroxylation complex dysfunction (32). However, in 1 case with a moderate phenotype, the PPIB mutation affected the start codon and led to undetectable CyPB levels and normally modified collagen (26).
Isomerase activity is known to be the rate-limiting step for collagen helix formation due to the necessity of cis-proline conversion to trans-proline for proper helical folding. The collagen overmodification observed in some CyPB-deficient cells may be caused by the absence of the functional role of CyPB as a PPIase. However, not all cases of type IX OI exhibit collagen overmodification (26), indicating that CyPB may not be the sole PPIase contributing to collagen folding. Mutation location on the PPIB gene may determine whether CyPB is expressed as a dysfunctional protein or is absent altogether, potentially allowing a secondary PPIase to provide isomerase activity.
Collagen chaperone defects—types X and XI OI (SERPINH1, FKBP10)
Defects in genes SERPINH1 and FKBP10 cause types X and XI OI, respectively (Table 1 and Figure 1). These genes were excellent candidate genes for recessive OI because their protein products, HSP47 and FKBP65, respectively, exhibit collagen chaperone activity essential in the proper folding of triple helical procollagen molecules. It has been postulated that HSP47 and FKBP65 form an ER resident complex, although no evidence has been presented. In addition to chaperone function, FKBP65 also exhibits PPIase activity (6).
An HSP47 defect was first characterized in dachshunds using homozygosity mapping. Affected dogs were homozygous for a missense mutation (c.977T>C) that resulted in expression of mutant mRNA and predicted to alter 3-dimensional protein structure (33). A year later, in 2010, the only report of a type X OI patient, a child with severe progressive OI who died at 3 years of age, delineated a recessive missense mutation (c.233T>C, p.Leu78Pro) in SERPINH1 (34). The mutation resulted in stable mutant transcripts, but proteosomal degradation of HSP47 protein. Type I procollagen modification was normal in patient cells, including Pro986 3-hydroxylation (34). In addition, the amount of collagen secreted by patient cells was similar to controls, but secretion rate was slightly delayed. Type I procollagen localized predominantly in the Golgi in SERPINH1-mutant cells, whereas control cells distribute type I procollagen in both ER and Golgi. Procollagen secreted from SERPINH1-mutant cells is susceptible to protease digestion at discrete sites within its helical region (34). Also, studies on Serpinh1 knockout mice show abnormal processing of collagen. Taken together, these data suggest improper helical folding in the absence of normal HSP47.
In 2010, the first FKBP10 mutations were identified as the cause of moderately severe type XI OI in a group of consanguineous Turkish families and a Mexican-American family (35). All affected Turkish individuals were homozygous for the same mutation in FKBP10, indicating a founder mutation within the population. The mutation consists of a 33-bp deletion (c.321_353del) that is predicted to result in the deletion of 11 amino acids in the first PPIase domain of FKBP65, as supported by truncated mRNA transcripts in patient cells. In the Mexican-American family, affected individuals were homozygous for an insertion mutation (c.831_832insC) in FKBP10 leading to a translational frameshift and a null allele (35). The range of type XI OI phenotypes broadened when several individuals with Bruck syndrome I, a recessive condition with severe OI and congenital contractures, were demonstrated to have FKBP10 mutations (36). However, both unrelated individuals and siblings with the same mutation may or may not have contractures, indicating that contractures are a variable manifestation of the same allele (36). Interestingly, an FKBP10 mutation that does not cause a null allele (deletion of a conserved tyrosine in the third PPIase domain) was found to cause Kuskokwim syndrome among the Yup'ik people of Alaska. This syndrome is characterized predominantly by congenital contractures and osteopenia, but not OI (37). Therefore, the phenotypic range for FKBP10 defects includes OI, OI plus contractures, and contractures without OI.
In cell-based assays, collagen secreted from FKBP10-defective cells displayed normal Pro986 3-hydroxylation and helical post-translational modifications, indicated by normal electrophoretic migration of α-chains (35). Collagen deposition in matrix of FKBP10-mutant cells is drastically reduced (38). In normal dermal fibroblasts, collagen C-telopeptide lysine residues are typically approximately 60% hydroxylated, while collagen from mutant cells showed less than 1% hydroxylation. Furthermore, the lack of telopeptide hydroxylation in collagen was verified in bone tissue (39). Telopeptide lysyl hydroxylation is known to be catalyzed by lysyl hydroxylase 2 (LH2), not the isomerase FKBP65. Two possible explanations have been proposed regarding impaired telopeptide hydroxylation and the role of FKBP65. FKBP65 may be required to allow LH2 access to the collagen substrate by isomerizing nearby proline residues, or alternatively, FKBP65 may be required for activity or stabilization of LH2 (38, 39). Further investigation is needed to understand the interaction between LH2 and FKBP65.
C-Propeptide cleavage enzyme defect—type XII OI (BMP1)
Recently, a mutation in BMP1 (bone morphogenetic protein 1) was identified as a cause of recessive OI; in the genetic nosology, this is classified as type XII OI (Table 1 and Figure 1). Homozygosity mapping and sequencing analysis revealed a homozygous missense mutation (c.747C>G) in BMP1 in 2 affected children of a consanguineous Egyptian couple (40). The skeletal phenotype included severe generalized deformities in all long bones, with hyperextensibility of elbows, wrists, and interphalangeal joints (40). BMP1 is a known protease, which cleaves the C-propeptide of type I procollagen. The mutation identified in type XII OI leads to an amino acid substitution within the catalytic peptidase domain of BMP1. Cell culture studies indicate a reduction in procollagen I C-propeptide peptidase activity in homozygous mutant fibroblasts as compared to control cells (40). An additional mutant BMP1 allele, a homozygous missense mutation (c.34G>C), has been reported. The causative mutation is located within the BMP1 signal peptide and leads to increased bone mineral density and recurrent fractures (41). Interestingly, this recessive condition complements the high bone mass phenotype observed in dominant cases of OI with mutations in the type I procollagen C-propeptide cleavage site (10). However, BMP1 has other substrates in addition to type I collagen, and it is reasonable that a more severe phenotype would result from the recessive conditions (BMP1 absence or dysfunction) than from the corresponding dominant substrate mutations in the type I procollagen C-propeptide.
Unclassified OI types (SP7, TMEM38B, and WNT1)
A homozygous frameshift mutation (c.1052delA) in SP7 was reported as the cause of recessive OI in 1 Egyptian child born to consanguineous parents (42). The mutant SP7 allele is predicted to synthesize a truncated protein product lacking the final 81 normal residues and, instead adding 18 novel residues including a premature stop codon within the final exon. The mutant transcript is predicted to escape NMD. SP7 encodes the protein osterix, a zinc-finger transcription factor that is important in the regulation of osteoblast differentiation (42). The mechanism by which this mutation causes OI symptoms is yet unknown.
Two recent reports identified a single mutant allele of TMEM38B as a cause of recessive OI. The identical mutation occurs in all affected individuals in 3 consanguineous families from Saudi Arabia (43) and 3 consanguineous Bedouin families in southern Israel (44). Because the affected individuals have a shared haplotype in the 2 MB region containing the TMEM38B mutation, it is likely a founder mutation originating from the Arabian Peninsula (44). The mutation (c.455_542del) results in complete loss of TMEM38B exon 4 and would presumably lead to a truncated protein (p.Gly152Alafs*5) (43). In the Israeli study, TMEM38B mRNA transcript levels were significantly reduced (44). TMEM38B encodes a ubiquitously expressed monovalent cation channel protein, TRICB (trimeric intracellular cation channel type B), important for regulating the release of calcium from the ER/sarcoplasmic reticulum, and could represent a novel OI mechanism (43, 44).
Mutations causing OI have recently been identified in WNT1 (45–47). The genetic variability of reported mutations is broad, including a variety of homozygous (missense, nonsense, deletion, splice-site, frameshift) and heterozygous (missense and compound) mutations. The phenotype associated with these mutations is primarily moderately severe and progressively deforming, although early-onset osteoporosis is also described. The presumption of 2 inheritance patterns has left open the question of mechanism. Functional studies in transfected cells have shown a failure to activate the canonical LRP5-mediated, WNT-regulated, β-catenin signaling pathway, but still lack direct demonstration of Wnt1 expression in normal osteoblasts (45).
Conclusions
Since 2006, a series of exciting discoveries has revealed multiple new genes responsible for rare forms of OI. The protein products of these genes interact with collagen post-translationally for folding, modification, or cross-linking, resulting in a collagen-related paradigm for OI. Comparison of the mechanistic pathways in recessive and dominant types of OI will guide investigators to critical common pathways that can be targeted with novel therapeutic approaches.
Acknowledgments
This work was supported by National Institute of Child Health and Human Development intramural funding (to J.C.M.).
Disclosure Summary: The authors have nothing to declare.
Footnotes
- BMP1
- bone morphogenetic protein 1
- CRTAP
- cartilage-associated protein
- CyPB
- cyclophilin B
- ECM
- extracellular matrix
- EDS
- Ehlers-Danlos syndrome
- ER
- endoplasmic reticulum
- FKBP
- FK506-binding protein
- IFITM5
- interferon-induced transmembrane protein 5
- LH2
- lysyl hydroxylase 2
- NMD
- nonsense-mediated mRNA decay
- OI
- osteogenesis imperfecta
- PEDF
- pigment epithelium-derived factor
- P3H1
- prolyl 3-hydroxylase 1
- PPIase
- peptidyl prolyl cis-trans isomerase
- PTC
- premature termination codon.
References
- 1. Sillence DO, Senn A, Danks DM. Genetic heterogeneity in osteogenesis imperfecta. J Med Genet. 1979;16(2):101–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Glorieux FH, Rauch F, Plotkin H, et al. Type V osteogenesis imperfecta: a new form of brittle bone disease. J Bone Miner Res. 2000;15(9):1650–1658 [DOI] [PubMed] [Google Scholar]
- 3. Glorieux FH, Ward LM, Rauch F, Lalic L, Roughley PJ, Travers R. Osteogenesis imperfecta type VI: a form of brittle bone disease with a mineralization defect. J Bone Miner Res. 2002;17(1):30–38 [DOI] [PubMed] [Google Scholar]
- 4. Van der Hoeve J, de Kleyn A. Blaue Scleren, Knochenbrüchigkeit und Schwerhörigkeit. Arch Ophthalmol. 1918;95:81–93 [Google Scholar]
- 5. Obafemi AA, Bulas DI, Troendle J, Marini JC. Popcorn calcification in osteogenesis imperfecta: incidence, progression, and molecular correlation. Am J Med Genet A. 2008;146A(21):2725–2732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Forlino A, Cabral WA, Barnes AM, Marini JC. New perspectives on osteogenesis imperfecta. Nat Rev Endocrinol. 2011;7(9):540–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Marini JC, Forlino A, Cabral WA, et al. Consortium for osteogenesis imperfecta mutations in the helical domain of type I collagen: regions rich in lethal mutations align with collagen binding sites for integrins and proteoglycans. Hum Mutat. 2007;28(3):209–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cabral WA, Makareeva E, Colige A, et al. Mutations near amino end of alpha1(I) collagen cause combined osteogenesis imperfecta/Ehlers-Danlos syndrome by interference with N-propeptide processing. J Biol Chem. 2005;280(19):19259–19269 [DOI] [PubMed] [Google Scholar]
- 9. Makareeva E, Cabral WA, Marini JC, Leikin S. Molecular mechanism of alpha 1(I)-osteogenesis imperfecta/Ehlers-Danlos syndrome: unfolding of an N-anchor domain at the N-terminal end of the type I collagen triple helix. J Biol Chem. 2006;281(10):6463–6470 [DOI] [PubMed] [Google Scholar]
- 10. Lindahl K, Barnes AM, Fratzl-Zelman N, et al. COL1 C-propeptide cleavage site mutations cause high bone mass osteogenesis imperfecta. Hum Mutat. 2011;32(6):598–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cheung MS, Glorieux FH, Rauch F. Natural history of hyperplastic callus formation in osteogenesis imperfecta type V. J Bone Miner Res. 2007;22(8):1181–1186 [DOI] [PubMed] [Google Scholar]
- 12. Fassier AM, Rauch F, Aarabi M, Janelle C, Fassier F. Radial head dislocation and subluxation in osteogenesis imperfecta. J Bone Joint Surg Am. 2007;89(12):2694–2704 [DOI] [PubMed] [Google Scholar]
- 13. Cho TJ, Lee KE, Lee SK, et al. A single recurrent mutation in the 5′-UTR of IFITM5 causes osteogenesis imperfecta type V. Am J Hum Genet. 2012;91(2):343–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Semler O, Garbes L, Keupp K, et al. A mutation in the 5′-UTR of IFITM5 creates an in-frame start codon and causes autosomal-dominant osteogenesis imperfecta type V with hyperplastic callus. Am J Hum Genet. 2012;91(2):349–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hanagata N, Li X, Morita H, Takemura T, Li J, Minowa T. Characterization of the osteoblast-specific transmembrane protein IFITM5 and analysis of IFITM5-deficient mice. J Bone Miner Metab. 2011;29(3):279–290 [DOI] [PubMed] [Google Scholar]
- 16. Moffatt P, Gaumond MH, Salois P, et al. Bril: a novel bone-specific modulator of mineralization. J Bone Miner Res. 2008;23(9):1497–1508 [DOI] [PubMed] [Google Scholar]
- 17. Becker J, Semler O, Gilissen C, et al. Exome sequencing identifies truncating mutations in human SERPINF1 in autosomal-recessive osteogenesis imperfecta. Am J Hum Genet. 2011;88(3):362–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rauch F, Husseini A, Roughley P, Glorieux FH, Moffatt P. Lack of circulating pigment epithelium-derived factor is a marker of osteogenesis imperfecta type VI. J Clin Endocrinol Metab. 2012;97(8):E1550–E1556 [DOI] [PubMed] [Google Scholar]
- 19. Yasui N, Mori T, Morito D, et al. Dual-site recognition of different extracellular matrix components by anti-angiogenic/neurotrophic serpin, PEDF. Biochemistry. 2003;42(11):3160–3167 [DOI] [PubMed] [Google Scholar]
- 20. Hosomichi J, Yasui N, Koide T, Soma K, Morita I. Involvement of the collagen I-binding motif in the anti-angiogenic activity of pigment epithelium-derived factor. Biochem Biophys Res Commun. 2005;335(3):756–761 [DOI] [PubMed] [Google Scholar]
- 21. Tombran-Tink J, Barnstable CJ. Osteoblasts and osteoclasts express PEDF, VEGF-A isoforms, and VEGF receptors: possible mediators of angiogenesis and matrix remodeling in the bone. Biochem Biophys Res Commun. 2004;316(2):573–579 [DOI] [PubMed] [Google Scholar]
- 22. Doll JA, Stellmach VM, Bouck NP, et al. Pigment epithelium-derived factor regulates the vasculature and mass of the prostate and pancreas. Nat Med. 2003;9(6):774–780 [DOI] [PubMed] [Google Scholar]
- 23. Morello R, Bertin TK, Chen Y, et al. CRTAP is required for prolyl 3-hydroxylation and mutations cause recessive osteogenesis imperfecta. Cell. 2006;127(2):291–304 [DOI] [PubMed] [Google Scholar]
- 24. Chang W, Barnes AM, Cabral WA, Bodurtha JN, Marini JC. Prolyl 3-hydroxylase 1 and CRTAP are mutually stabilizing in the endoplasmic reticulum collagen prolyl 3-hydroxylation complex. Hum Mol Genet. 2010;19(2):223–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Marini JC, Cabral WA, Barnes AM. Null mutations in LEPRE1 and CRTAP cause severe recessive osteogenesis imperfecta. Cell Tissue Res. 2010;339(1):59–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Barnes AM, Carter EM, Cabral WA, et al. Lack of cyclophilin B in osteogenesis imperfecta with normal collagen folding. N Engl J Med. 2010;362(6):521–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ward LM, Rauch F, Travers R, et al. Osteogenesis imperfecta type VII: an autosomal recessive form of brittle bone disease. Bone. 2002;31(1):12–18 [DOI] [PubMed] [Google Scholar]
- 28. Barnes AM, Chang W, Morello R, et al. Deficiency of cartilage-associated protein in recessive lethal osteogenesis imperfecta. N Engl J Med. 2006;355(26):2757–2764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Marini JC, Cabral WA, Barnes AM, Chang W. Components of the collagen prolyl 3-hydroxylation complex are crucial for normal bone development. Cell Cycle. 2007;6(14):1675–1681 [DOI] [PubMed] [Google Scholar]
- 30. Cabral WA, Chang W, Barnes AM, et al. Prolyl 3-hydroxylase 1 deficiency causes a recessive metabolic bone disorder resembling lethal/severe osteogenesis imperfecta. Nat Genet. 2007;39(3):359–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cabral WA, Barnes AM, Adeyemo A, et al. A founder mutation in LEPRE1 carried by 1.5% of West Africans and 0.4% of African Americans causes lethal recessive osteogenesis imperfecta. Genet Med. 2012;14(5):543–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. van Dijk FS, Nesbitt IM, Zwikstra EH, et al. PPIB mutations cause severe osteogenesis imperfecta. Am J Hum Genet. 2009;85(4):521–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Drögemüller C, Becker D, Brunner A, et al. A missense mutation in the SERPINH1 gene in dachshunds with osteogenesis imperfecta. PLoS Genet. 2009;5(7):e1000579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Christiansen HE, Schwarze U, Pyott SM, et al. Homozygosity for a missense mutation in SERPINH1, which encodes the collagen chaperone protein HSP47, results in severe recessive osteogenesis imperfecta. Am J Hum Genet. 2010;86(3):389–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Alanay Y, Avaygan H, Camacho N, et al. Mutations in the gene encoding the RER protein FKBP65 cause autosomal-recessive osteogenesis imperfecta. Am J Hum Genet. 2010;86(4):551–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kelley BP, Malfait F, Bonafe L, et al. Mutations in FKBP10 cause recessive osteogenesis imperfecta and Bruck syndrome. J Bone Miner Res. 2011;26(3):666–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Barnes AM, Duncan G, Weis M, et al. Kuskokwim syndrome, a recessive congenital contracture disorder, extends the phenotype of FKBP10 mutations [published online July 8, 2013]. Hum Mutat. doi:10.1002/humu.22362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Barnes AM, Cabral WA, Weis M, et al. Absence of FKBP10 in recessive type XI osteogenesis imperfecta leads to diminished collagen cross-linking and reduced collagen deposition in extracellular matrix. Hum Mutat. 2012;33(11):1589–1598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Schwarze U, Cundy T, Pyott SM, et al. Mutations in FKBP10, which result in Bruck syndrome and recessive forms of osteogenesis imperfecta, inhibit the hydroxylation of telopeptide lysines in bone collagen. Hum Mol Genet. 2013;22(1):1–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Martínez-Glez V, Valencia M, Caparrós-Martín JA, et al. Identification of a mutation causing deficient BMP1/mTLD proteolytic activity in autosomal recessive osteogenesis imperfecta. Hum Mutat. 2012;33(2):343–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Asharani PV, Keupp K, Semler O, et al. Attenuated BMP1 function compromises osteogenesis, leading to bone fragility in humans and zebrafish. Am J Hum Genet. 2012;90(4):661–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lapunzina P, Aglan M, Temtamy S, et al. Identification of a frameshift mutation in osterix in a patient with recessive osteogenesis imperfecta. Am J Hum Genet. 2010;87(1):110–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Shaheen R, Alazami AM, Alshammari MJ, et al. Study of autosomal recessive osteogenesis imperfecta in Arabia reveals a novel locus defined by TMEM38B mutation. J Med Genet. 2012;49(10):630–635 [DOI] [PubMed] [Google Scholar]
- 44. Volodarsky M, Markus B, Cohen I, et al. A deletion mutation in TMEM38B associated with autosomal recessive osteogenesis imperfecta. Hum Mutat. 2013;34(4):582–586 [DOI] [PubMed] [Google Scholar]
- 45. Keupp K, Beleggia F, Kayserili H, et al. Mutations in WNT1 cause different forms of bone fragility. Am J Hum Genet. 2013;92(4):565–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Fahiminiya S, Majewski J, Mort J, Moffatt P, Glorieux FH, Rauch F. Mutations in WNT1 are a cause of osteogenesis imperfecta. J Med Genet. 2013;50(5):345–348 [DOI] [PubMed] [Google Scholar]
- 47. Pyott SM, Tran TT, Leistritz DF, et al. WNT1 mutations in families affected by moderately severe and progressive recessive osteogenesis imperfecta. Am J Hum Genet. 2013;92(4):590–597 [DOI] [PMC free article] [PubMed] [Google Scholar]