Abstract
In the present study, the relationship between short interfering RNA (siRNA) sequence and RNA interference (RNAi) effect was extensively analyzed using 62 targets of four exogenous and two endogenous genes and three mammalian and Drosophila cells. We present the rules that may govern siRNA sequence preference and in accordance with which highly effective siRNAs essential for systematic mammalian functional genomics can be readily designed. These rules indicate that siRNAs which simultaneously satisfy all four of the following sequence conditions are capable of inducing highly effective gene silencing in mammalian cells: (i) A/U at the 5′ end of the antisense strand; (ii) G/C at the 5′ end of the sense strand; (iii) at least five A/U residues in the 5′ terminal one-third of the antisense strand; and (iv) the absence of any GC stretch of more than 9 nt in length. siRNAs opposite in features with respect to the first three conditions give rise to little or no gene silencing in mammalian cells. Essentially the same rules for siRNA sequence preference were found applicable to DNA-based RNAi in mammalian cells and in ovo RNAi using chick embryos. In contrast to mammalian and chick cells, little siRNA sequence preference could be detected in Drosophila in vivo RNAi.
INTRODUCTION
RNA interference (RNAi) is the process of double-stranded (ds) RNA-dependent, post-transcriptional gene silencing (1–4). dsRNA introduced into cells is digested by Dicer to yield short interfering RNA (siRNA) 21–23 nt in length (5,6). siRNA thus generated within cells or that synthesized in vitro and introduced into cells reacts directly or indirectly with PIWI protein and/or relevant proteins to give rise to the RNA-induced silencing complex (RISC), which is responsible for mRNA degradation (7–12). eIF2C1, a human counterpart of Drosophila PIWI protein, Argonaute 1 [Ago 1 (13,14)], was previously shown to be essential for siRNA-based RNAi in mammalian cells (15). Martinez et al. (16) observed eIF2C1 to be involved in active RISC prepared from HeLa cells. Active RISC includes siRNA antisense strands (AS) but not sense strands (SS) (16), thus indicating that double-stranded siRNA undergoes denaturation via helicase (8) either prior to or at an early stage in RISC formation. Dicer, an enzyme which possesses both RNase III and helicase domains, may be essential for siRNA-based mammalian RNAi (15). In Drosophila and Caenorhabditis elegans, spindle E (17) and mut-14 (18), both encoding helicase, have also been shown to be involved in RNAi. Target mRNA is cleaved at a specific site corresponding to the center of the siRNA AS in mammalian and Drosophila cells (9,16,19,20).
The introduction of long dsRNA into mammalian cells frequently induces a fatal interferon response (21), and thus siRNA should be a more promising reagent for mammalian RNAi (19) than long dsRNA (22–26). siRNA-based RNAi, however, may not be readily usable for the large-scale gene silencing essential for mammalian functional genomics, since only a limited fraction of siRNAs appear capable of producing highly effective RNAi in mammalian cells [(27,28) see also Fig. 2A].
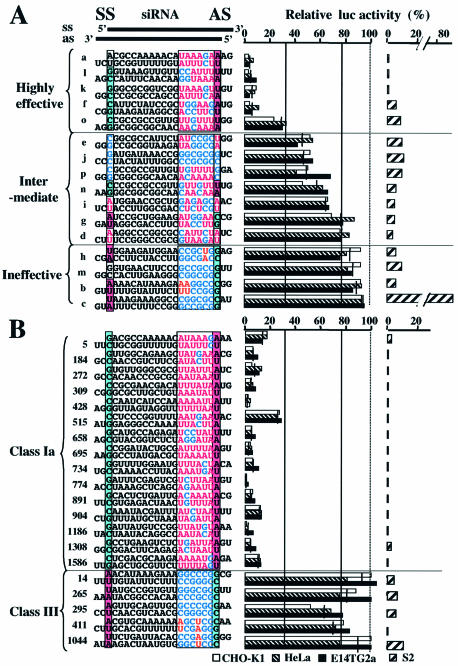
Figure 2.
Relationship between luc siRNA sequence and induced luc gene silencing (RNAi) activity. AS and SS, respectively, in the upper margin, indicate siRNA ends with the 5′ antisense strand and 5′ sense strand ends. (A) Classification of 16 luc siRNAs. siRNA-dependent reduction in firefly luciferase activity in three mammalian (CHO-K1, HeLa and E14TG2a) and Drosophila (S2) cells was examined using 50 nM of 16 siRNAs, a–p, shown in Figure 1. Sixteen siRNAs were aligned according to their RNAi activity in mammalian cells from top to bottom. siRNAs were classified into three groups depending on RNAi or reduction in relative luciferase activity. (B) RNAi activity caused by siRNAs designed using our sequence preference rules. Using the rules, 15 class Ia and five class III siRNAs were designed and their capability to bring about RNAi in CHO-K1, HeLa, E14TG2a and S2 cells was examined. The siRNA number indicates the nucleotide position within the luc coding region, corresponding to the 3′ end of the siRNA AS. The concentration of siRNA was 50 nM and RNAi effects were observed 24 h after transfection. Data obtained from 2–4 experiments were averaged and are shown. Thin vertical lines indicate the average of three mammalian cells. The 7 bp terminal region with the 5′ AS end is boxed. While A/U and G/C in the boxed region are colored in red and blue, respectively, highly conserved, 5′-terminal bases are shown on red (A/U) or blue (G/C) backgrounds. Note that, in class Ia and highly effective siRNAs, the 5′ AS and SS ends, respectively, are almost exclusively A/U and G/C.
The relationship between the siRNA sequence and its capability to bring about RNAi in human, Chinese hamster and mouse embryonic stem (ES) cells as well as Drosophila cells was examined in detail in the present study. Highly effective RNAi was found to occur in mammalian cells if siRNA satisfying the four following sequence conditions at the same time is used: (i) A/U at the 5′ end of the AS; (ii) G/C at the 5′ end of the SS; (iii) AU-richness in the 5′ terminal, 7 bp long region of the AS; and (iv) the absence of any long GC stretch of more than 9 bp in length. All siRNAs opposite in sequence features except for the fourth condition brought about the least levels of RNAi in mammalian cells. Essentially the same rules for siRNA sequence preference appeared to apply for siRNA-based RNAi in chick embryos and DNA-based RNAi in mammalian cells.
During the course of the preparation of this manuscript, Schwarz et al. (29) showed sequence conditions resembling the above to some extent to be necessary for RISC formation and subsequent target RNA cleavage in Drosophila embryonic extracts. Khvorova et al. (30) indicated that both the enhanced flexibility at the siRNA end including the 5′ AS end and low internal energy across the duplex are strongly correlated with siRNA and microRNA (miRNA) functions. The similarity in siRNA sequence requirements for in vivo RNAi in mammalian cells [this work, (30)] and in vitro Drosophila RNAi (29) could reflect underlying similarities between the RNAi mechanisms in insects and animals. The findings in the present study are thus evaluated and discussed from the standpoints of RISC formation and siRNA unwinding.
MATERIALS AND METHODS
Cell culture
Drosophila S2 cells were cultured in Schneider’s Drosophila medium (Gibco-BRL) at 25°C. Chinese hamster CHO-K1 (RIKEN Cell Bank) and human HeLa cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Gibco-BRL) at 37°C. Both media were supplemented with 10% heat-inactivated fetal bovine serum (FBS; Mitsubishi Kagaku) and antibiotics [10 U/ml of penicillin (Meiji) and 50 µg/ml of streptomycin (Meiji)]. E14TG2a (mouse ES) cells were cultured in DMEM supplemented with 20% heat-inactivated FBS (Hyclone), 0.1 mM 2-mercaptoethanol (Wako), 8 µg/ml of adenosine, 8.5 µg/ml of guanosine, 7.3 µg/ml of cytidine, 7.3 µg/ml of uridine, 2.4 µg/ml of thymidine, 0.1 mM each non-essential amino acid and 1000 U/ml of leukemia inhibitory factor (CHEMICON International).
Preparation of siRNA
RNA oligonucleotides were synthesized by Proligo. Double-stranded siRNA was prepared as described previously (22). The concentration of siRNA is shown based on that of the AS. When necessary, siRNAs were numbered based on the nucleotide position within the coding region of the target mRNA, corresponding to the 3′ siRNA AS end.
luc RNAi assay
A 1 ml aliquot of S2 (1 × 106 cells/ml), CHO-K1 (3 × 105 cells/ml), HeLa (1 × 105 cells/ml) or E14TG2a (2 × 105 cells/ml) cell suspension was inoculated into a 1.5 cm well 24 h prior to transfection. Cells were transfected with pGL3-Control DNA (1 µg, Promega) encoding the firefly luciferase gene, and pRL-TK DNA (0.1–1 µg, Promega) or pRL-SV40 DNA (0.1–1 µg, Promega), both encoding the Renilla luciferase gene, with or without siRNA. The calcium phosphate precipitation method (22) was used for transfection for S2, HeLa or CHO-K1 cells, while DMRIE C reagent (Invitrogen) was used for E14TG2a transfection. Cells were harvested 24 h after transfection and luciferase activity was measured using the Dual-Luciferase Reporter Assay System (Promega).
Vimentin RNAi and immunostaining
A 1 ml aliquot of HeLa cell suspension (1 × 105 cells/ml) was inoculated into a 1.5 cm well 24 h prior to the first transfection. Cells were treated with three cycles of transfection carried out in 24 h intervals with vimentin siRNA at 50 nM. Lipofectamine 2000 (Invitrogen) was used for transfection. The estimated transfection efficiency was >95%. Cells were fixed with 3.7% formaldehyde in phosphate-buffered saline (PBS) and permeabilized 24 h after the last transfection. After washing with PBS, cells were doubly stained with anti-porcine vimentin antibody (Oncogene Research Products), Cy3- conjugated second antibody (Jackson Immuno Research) and anti-human Yes antibody (Upstate Biotechnology), with Cy5-conjugated second antibody (Jackson Immuno Research).
Oct 4 RNAi assay
Using Lipofectamine 2000 (Invitrogen), E14TG2a cells (2 × 105 cells/ml) were co-transfected with 50 nM Oct 4 siRNA shown in Figure 3B and pCAGIPuro-EGFP (0.5 µg/ml), encoding enhanced green fluorescent protein (EGFP) and puromycin-resistant genes. Puromycin (2 µg/ml; Clontech) was added to the medium 24 h after transfection, and morphological change was observed under a phase contrast microscope 3 days after transfection. RNA was also extracted 3 days after transfection using RNeasy (QIAGEN) and was subjected to RT–PCR using the RNA LA-PCR kit (Takara). Almost all cells were found to express EGFP 3 days after transfection. The following primers were used for RT–PCR to measure the concentration of glyceraldehyde-3-phosphate dehydrogenase (Gapd) and Oct 4 mRNA. Gapd, 5′-GCCTCATCCGGTAGACAAAA and 5′-ACCGTGGTCATGAGTCCTTC; Oct-4, 5′-AGCTGCTGAAGCAGAAGAGG and 5′-TGTCTACCTCCCTTGCCTTG.
Figure 3.
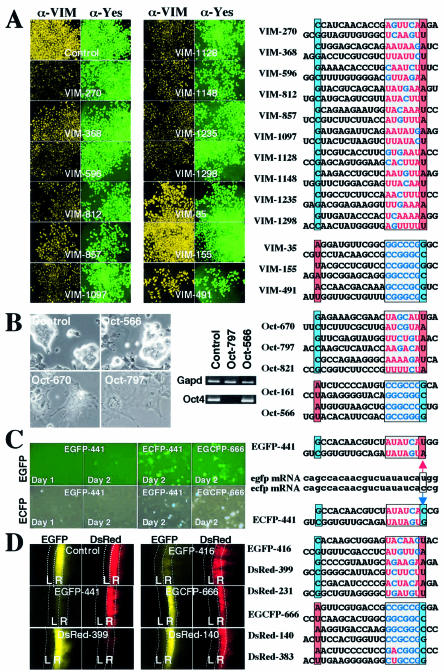
Highly effective silencing of endogenous genes by class Ia siRNAs (A and B), RNAi caused by an non-cognate siRNA (C) and class Ia siRNA-dependent RNAi in chick embryos (D). The sequences of siRNAs examined here are depicted in the right margin (see Fig. 2 for coloration and box). (A) Silencing of vimentin, a human endogenous gene, by class Ia and class III siRNAs. Ten class Ia (VIM-270, -368, -596, -812, -857, -1097, -1128, -1148, -1235 and -1298) and three class III siRNAs (VIM-35, -155 and -491) were designed and their RNAi activity was examined in HeLa cells subjected to three cycles of 50 nM siRNA transfection. On day 3, cells were stained for vimentin (target) and Yes (control). Vimentin and Yes protein signals are colored in yellow and green, respectively. Note that all class Ia but not class III siRNAs significantly reduced vimentin signals. (B) Effects of siRNA transfection on the expression of Oct 4, a mouse endogenous gene. E14TG2a (mouse ES) cells were transfected with class Ia (Oct-670, -797 and -821) or class III (Oct-161 and -566). The four pictures on the left are phase contrast photographs. Note that class Ia siRNAs (Oct-670 and -797) induced a flat morphology typical of trophectoderm cells. Class Ia siRNA-specific degradation of Oct 4 mRNA visualized with RT–PCR is also shown. Gapd was used as a control. (C) ECFP RNAi caused by an non-cognate EGFP siRNA. EGFP-441 is a class Ia EGFP siRNA but not identical in sequence to ECFP-441, a class II ECFP siRNA possessing G at the 5′ AS end. While ECFP-441 did not abolish ECFP signals 2 days after transfection (on day 2 of transfection), non-cognate EGFP-441 virtually completely eliminated ECFP signals. Note that RNAi effects 1 day after transfection (on day 1) indicate that EGFP is a better target for EGFP-441 than ECFP. Blue and red arrows indicate that EGFP and ECFP mRNA possess a base substitution at the position corresponding to the 5′ AS end of EGFP-441 or ECFP-441. (D) In ovo RNAi in chick embryo. EGFP and DsRed expression plasmids were co-electroporated into chick spinal cord with class Ia siRNAs (EGFP-416, -441 and DsRed-399) or class III siRNAs (EGCFP-666 and DsRed-383). Class Ia siRNAs significantly abolished the targeted fluorescence, while class III siRNAs did not. Expression of EGFP and DsRed is colored in yellow and red, respectively. Dotted lines show left (L) and right (R) portions of the spinal cords. The 5′ ends of SS and AS, and the 5′ region of the AS (7 bp) are boxed and colored as described in Figure 2.
RNAi assay for EGFP, ECFP and DsRed
HeLa cells (1 × 105 cells/ml) were transfected with pCAGGS-EGFP (0.25 µg/well), pCAGGS-DsRed [0.25 µg/well (15)] and siRNA (50 nM) for EGFP RNAi. For enhanced cyan fluorescent protein (ECFP) RNAi, HeLa cell transfection was carried out with pECFP-N1 (0.25 µg/well; Clontech), pCAGGS-DsRed (0.25 µg/well) and siRNA (50 nM). Transfection was carried out using Lipofectamine 2000 (Invitrogen). RNAi activity was estimated by counting EGFP- or ECFP-positive cells among DsRed-positive cells under a fluorescence microscope (Zeiss). pCAGGS-EGFP was constructed by inserting an EGFP fragment of pEGFP-N1 (Clontech) into the EcoRI site of pCAGGS (31).
In ovo electroporation
Fertile chick eggs obtained from a local farm were incubated at 37°C for 2 days. The eggs were windowed, and 0.1–0.5 µl of PBS containing pCAGGS-EGFP (0.1 µg/µl) and pCAGGS-DsRed (0.1 µg/µl) and siRNA (5 µg/µl) along with 0.01% of luxol fast blue was injected into the central canal of the spinal cord at the wing level using a glass capillary with a tip diameter of 50–100 µm. A pair of platinum electrodes 4 mm apart (Nepagene) was used for electroporation. Transfection occurred exclusively on the right hemilateral side of the neural tube. Five timed pulses of 50 ms duration at 20 mV were used. Embryos were incubated at 37°C for 2 days and killed. EGFP and DsRed expression was observed under a fluorescence microscope on embryonic day 4.
Construction of siRNA expression plasmids for DNA-based RNAi
Single-stranded DNA oligonucleotides, ∼80 nt in length and encoding, in order: (i) a 21 nt siRNA SS; (ii) a human miRNA loop; and (iii) the 19 nt AS of the identical siRNA, minus 3′ overhangs, were annealed with corresponding complementary single-stranded DNA oligonucleotides. The resultant dsDNA was inserted into the BamHI–HindIII site of pSilencer 3.0-H1 (Ambion) to generate FLx-m23L or FLx-m212L plasmids, where x indicates the position of the corresponding target sequence in the firefly luc gene. In FL826-m212L, the order of SS and AS was reversed. As human miRNA loops, m23L and m212L, derived from miR-23 (32,33) and miR-212 (34), respectively, were used. Escherichia coli XL1-Blue competent cells (Gibco-BRL) were transformed with the resultant plasmids. Plasmid DNA was purified using a commercial DNA purification kit (QIAGEN). HeLa cells (1 × 105 cells/ml) were transfected with 150 ng of the plasmid DNA along with pGL3-Control (1 µg) and pRL-SV40 (0.1 µg, Promega). pSilencer with no insert was used as a control. Luciferase activity was measured using the Dual-Luciferase Reporter Assay System (Promega) 3 days following transfection.
Free energy calculation
Standard Gibbs free energies, which reflect the stability of pentamer subsequences, were calculated from the siRNA duplex end containing the 5′ AS end (position 1) according to the nearest-neighbor method described by Freier et al. (35). The values from positions 16 to 19 were not calculated because of the absence of available pentamer subsequences.
RESULTS
Strong siRNA sequence preference in mammalian RNAi
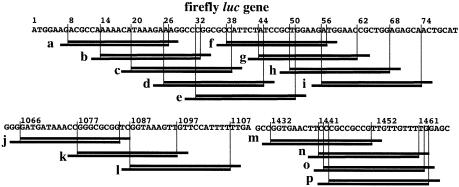
RNAi in mammalian cells was previously noted to vary considerably depending on the siRNA sequence (27,28). To examine this point in greater detail, 16 siRNAs targeting for the firefly luciferase gene (luc) were prepared (Fig. 1) and assessed for their ability to produce RNAi in human (HeLa), Chinese hamster (CHO-K1), mouse ES (E14TG2a) and Drosophila (S2) cells by dual luciferase assay (22). Figure 1 shows the nine luc target sequences, corresponding to siRNA a–i, to be spaced 6 nt apart, while three of the remaining (corresponding to siRNA n–p) are spaced only 1 nt apart. Cells were simultaneously transfected with plasmid DNA encoding the firefly luc gene (target), plasmid DNA with the Renilla luciferase gene (reference) and 50 nM cognate siRNA, and luciferase activity was measured 24 h later (Fig. 2A). In Figure 2A, siRNA sequences are listed in rank, in order of average RNAi activity in three mammalian cells, so as to obtain some clarification of the relationship between siRNA sequence and the resultant reduction in firefly luc gene activity.
Figure 1.
Location of 16 cognate siRNAs for silencing of the firefly luc gene.
In mammalian cells, RNAi activity varied significantly depending on the siRNA employed. Use of five highly effective siRNAs (a, l, k, f and o) resulted in a 70–95% reduction in relative firefly luciferase activity, while use of four highly ineffective siRNAs (h, m, b and c) resulted in <20% reduction. Even a 1 nt variation in the target sequence had a considerable effect on RNAi activity in mammalian cells (compare RNAi effects of siRNA n and o). In contrast, firefly luciferase activity was always abolished at >85% upon transfecting Drosophila cells with any siRNA other than siRNA c. Thus, most, if not all, siRNAs should be capable of producing highly effective RNAi in Drosophila cells, at least under given conditions. Three of the four siRNAs (a, l and k) giving rise to the highest levels of RNAi in mammalian cells were also noted to bring about the highest levels of RNAi in Drosophila cells.
siRNA sequence requirement for highly effective and ineffective RNAi in mammalian cells
The values in Figure 2A for reduction in relative firefly luciferase activity in CHO-K1, HeLa and E14TG2a cells can be seen to be virtually the same, suggesting that siRNA-based RNAi in mammalian cells is in accordance with the same rules for siRNA sequence preference.
Three immediately apparent features of the siRNA sequence may possibly serve to discriminate highly effective siRNAs from those that are ineffective. First, the 5′ AS end of highly effective siRNAs may always be A or U, with the counterpart of ineffective siRNAs being G or C. A/U and G/C residues were respectively found to be present at the 5′ AS ends of all five highly effective and all four ineffective siRNAs. Secondly, the 5′ SS ends of highly effective siRNAs are preferably G or C, with the counterpart of ineffective siRNAs being A or U. Thirdly, in the case of highly effective siRNAs, at least four out of seven nucleotides in the 5′-terminal AS are A or U while the corresponding region of ineffective siRNAs are GC rich. Most, if not all, siRNAs associated with mixed features appear to belong to an siRNA class with intermediate RNAi activity. A possible molecular basis for the effectiveness of siRNA a is discussed below.
siRNAs may be grouped into three classes, I–III, based on combinations of terminal base sequences. Class I consists of siRNAs possessing A/U at the 5′ AS end, G/C at the 5′ SS end and at least four A/U nucleotides in a 7 nt 5′-terminal end of the AS, whereas those with opposite features are class III siRNAs. All other siRNAs are considered to belong to class II. Class I siRNAs may be subdivided into two classes, Ia and Ib. Class I siRNAs with 5–7 A/U residues in a 7 nt 5′-terminal end of the AS are presumed to belong to class Ia siRNAs; the remainder belong to class Ib.
It is possible to generate 1631 different siRNAs based on the firefly luc coding sequence. The number of class I siRNAs was calculated as 275 (17% of the total) and that of class Ia siRNAs as 154 (9%). To test the validity of the above rules for siRNA sequence preference, assessment was made of the ability of 15 different class Ia and five class III siRNAs to give rise to RNAi using three mammalian and Drosophila S2 cells (Fig. 2B). All class Ia siRNAs brought about highly effective RNAi in all three mammalian cells as well as Drosophila cells, while little or no effective RNAi resulted via transfection of class III siRNAs in the mammalian cells. We thus conclude that the rules stipulated here for siRNA sequence preference predict sequences for highly effective and ineffective siRNAs for mammalian RNAi at least in the case of the exogenous firefly luc gene.
Silencing of mammalian endogenous genes by siRNA transfection
Examination was carried out to determine whether the rules for siRNA sequence preference would be applicable for designing highly effective and ineffective siRNAs for RNAi of mammalian endogenous genes. The right margin of Figure 3A and B shows class Ia and class III siRNAs, designed for highly effective and ineffective RNAi, respectively, of vimentin and Oct 4 in mammalian cells (HeLa and E14TG2a). Candidate siRNAs designed by the present rules were further selected by Blast search so that the activity of any gene other than the target would not be affected by siRNA introduced into cells. Class Ia siRNAs unique to vimentin and Oct 4, respectively, were found to represent 5% (n = 64) and 3% (n = 37) of all possible siRNAs estimated based on vimentin and Oct 4 gene sequences.
The vimentin gene codes for an intermediate filament protein. It has been reported that reduction in vimentin gene activity by cognate siRNA transfection is difficult (19). Three cycles of siRNA transfection (one transfection/day) were thus carried out on HeLa cells prior to staining for vimentin and Yes (control). All 10 vimentin class Ia siRNAs were found to significantly reduce vimentin protein, but not Yes signals (Fig. 3A). Little or no reduction in vimentin or Yes signals could be detected when using class III vimentin siRNAs for RNAi. RT–PCR results (K. Ui-Tei and K. Saigo, unpublished data) indicated 70–95% of vimentin mRNA to be degraded by class Ia vimentin siRNA, while virtually no vimentin mRNA cleavage occurred by class III siRNA.
Oct 4 is a POU transcription factor encoded by the Pou5f1 (Oct 4) gene and is considered to be a regulator of ES cell pluripotency (36). A 50–100% increment in Oct 4 expression may cause the differentiation of pluripotent ES cells into primitive endoderm and mesoderm, while reduction in Oct 4 expression induces loss of pluripotency to differentiate ES cells into trophectoderm, which is characterized by flat morphology and induced expression of Hand 1 and Psx (36,37). Three class Ia siRNAs (Oct-670, -797 and -821) and two class III siRNAs (Oct-161 and -566) for Oct 4 RNAi were prepared and examined for change in cell morphology and gene expression 3 days following transfection of 50 nM cognate siRNA. As partly shown in Figure 3B, the pluripotent ES cells treated with cognate class Ia siRNAs, Oct-670, -797 and -821, had flattened out over the culture surface, with enlarged nuclei acquired in many cases. Oct 4 expression was virtually eliminated (Fig. 3B) while the expression of trophectoderm markers, Hand 1 and Psx, was induced (K.Ui-Tei, unpublished data). In contrast, no apparent change in morphology or gene expression could be found to result from class III Oct 4 siRNAs, Oct-566 and -161 (Fig. 3B). Our rules for siRNA sequence preference are thus shown to serve quite well for identifying highly effective and ineffective siRNAs for RNAi of endogenous genes in mammals.
Thirty-two class Ia siRNAs for firefly luc, vimentin and Oct 4 were examined, 31 (97%) of which were found to be capable of giving rise to highly efficient RNAi in human, Chinese hamster and mouse cells. Virtually all of the investigated class Ia siRNAs were thus shown to be highly efficient RNAi reagents for mammalian cells. Thus, it is concluded that our rules for siRNA sequence preference may be highly useful for the design of effective siRNAs for RNAi of both exogenous and endogenous genes in mammalian cells.
siRNAs with long stretches of G/C residues are incapable of bringing about high levels of RNAi in both mammalian and Drosophila cells
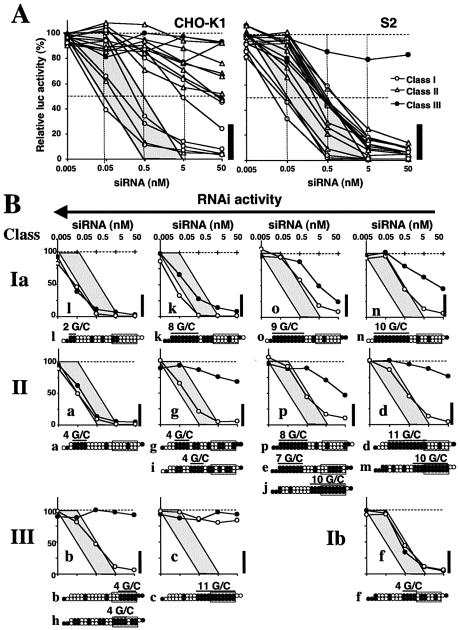
siRNA n may be an exceptional member of class Ia siRNAs in that, unlike any others which we evaluated, it was incapable of giving rise to high levels of RNAi in mammalian cells when transfected at 50 nM (see Fig. 2A). An investigation was thus undertaken to clarify in greater detail relationships among the siRNA sequence, siRNA concentration and RNAi activity in CHO-K1 or S2 cells using the 16 siRNAs shown in Figure 2A (Fig. 4A). With siRNA at 0.005–5 nM, most graph points for siRNAs which gave rise to effective RNAi in CHO-K1 or S2 cells after transfection at 50 nM overlapped or were situated near the shaded area bounded by two lines, intersecting, respectively, with the horizontal axis at 0.5 and 5 and the 50% line of relative luciferase activity at 0.05 and 0.5. The vertical bars in Figure 4A show the relative luciferase or RNAi activity range for siRNAs which give rise to effective RNAi in CHO-K1 or S2 cells subsequent to transfection at 50 nM. siRNAs that bring about highly effective RNAi on transfection at 50 nM would thus appear comprised of heterogenous members with over 10 times the capacity to bring about RNAi.
Figure 4.
Dose dependency of RNAi effects in CHO-K1 and S2 cells. The shaded area is the region bounded by two lines, intersecting, respectively, with the horizontal axis at 0.5 and 5 and the 50% line of luc activity at 0.05 and 0.5. The thick vertical bar at the right of each panel indicates the region with >77% reduction in luc activity. (A) Change in luc gene silencing activity with siRNAs ranging from 0.005 to 50 nM in CHO-K1 (left) and S2 (right) cells. siRNAs a–p in Figure 1 are grouped into three classes, I (open circles), II (open triangles) and III (closed circles). (B) Direct comparisons of RNAi activity curves in S2 (open circles) and CHO-K1 cells (filled circles). The sequences of corresponding or similar siRNAs are schematically shown in the lower margin. siRNAs i, e, j, m and h behave the same way as their corresponding examples above. Filled circles, G/C; open circles, A/U. The 7 bp duplex region containing the 5′ end is boxed. Eleven pictures are positioned based on siRNA classification and induced RNAi activity levels.
A comparison of RNAi effects due to individual siRNA in CHO-K1 and S2 cells is presented in each of the 11 pictures in Figure 4B. The pictures are arranged according to siRNA classification and order of RNAi activity. Maximum levels of RNAi resulted from the transfection of siRNA l, a class Ia siRNA, in both CHO-K1 and S2 cells. Note that suppression due to siRNA l in S2 cells was virtually the same as in CHO-K1 cells. We interpret this finding as suggesting that virtually all siRNA l molecules incorporated into cells become fully functional in both Drosophila and mammals. Hardly any RNAi occurred with transfection in siRNA c, a class III siRNA, to S2 and CHO-K1 cells. Mammalian and Drosophila cells would thus appear to possess virtually the same capacity of siRNA-mediated RNAi induction, the maximum and the minimum limits of which are determined by the transfection of siRNA l and c, respectively. Although within each class, siRNA-dependent RNAi activity in S2 cells increases with increasing RNAi activity in CHO-K1 cells, our rules for siRNA sequence preference may not be applicable for predicting highly effective and ineffective siRNAs for RNAi in S2 cells. RNAi-inducing capability in S2 cells was much the same for two class Ia siRNAs (o and n) and two class III siRNAs (b and h). Three class II siRNAs (a, i and g) were found to be much more effective in S2 cells compared with two class Ia siRNAs (o and n).
We noted that siRNA n, the most ineffective class Ia siRNA, possesses a long GC stretch extending from the 5′ end of the SS and that class Ia siRNA-dependent RNAi activity in S2 and CHO-K1 cells is negatively correlated with the length of the GC stretch extending from the 5′ end of the SS. Similar negative effects of a long GC stretch on RNAi were also evident in class II- or class III-dependent RNAi in CHO-K1 and S2 cells. In contrast, the average GC content in the 11 bp region adjacent to the 5′ SS end was ∼50% in the case of the 31 highly effective class Ia siRNAs (Fig. 5). It may thus follow that a long GC stretch in the siRNA sequence serves as a suppressor of RNAi, the extent depending on the length of the stretch.
Figure 5.
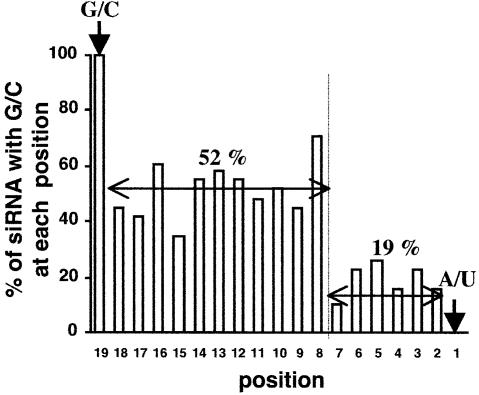
GC content distribution of highly effective class Ia siRNAs. The distribution of the GC content of 31 highly effective class Ia siRNAs shown in Figures 2 and 3 is presented. Position 1 corresponds to the siRNA duplex end including the 5′ AS end. The 5′ ends of AS and SS of all class Ia siRNAs causing highly effective RNAi in mammalian cells were A/U and G/C, respectively. The average GC content in the region 2–7 was 19%, while that of the region 8–18 was 52%.
Possible dual functions of the 5′ end of the siRNA antisense strand
During RNAi of EGFP and ECFP (a derivative of EGFP), EGFP-441, an siRNA homologous in sequence to the EGFP but not completely to the ECFP gene, was noted to be capable of effectively inactivating ECFP. HeLa cells were transfected simultaneously with DsRed plasmid DNA (control), EGFP or ECFP plasmid DNA (target) and siRNA, and the relative number of target gene-expressing cells was counted at various times. As shown in Figure 3C, nearly all EGFP signals from EGFP-expressing cells were abolished 24 h after transfection, when EGFP-441, a cognate class Ia siRNA, was transfected, while EGCFP-666, a class III siRNA completely homologous in sequence to EGFP and ECFP genes, could reduce only a few EGFP signals 2 days following transfection. EGFP-441 is homologous in sequence to ECFP mRNA except for the position corresponding to the 5′ AS end (see the right margin of Fig. 3C). Figure 3C shows that EGFP-441 is capable of more effectively bringing about ECFP RNAi than ECFP-441, a class II siRNA completely identical in sequence to the target (ECFP mRNA). EGFP-441 abolished nearly 70% of ECFP signals at 24 h following transfection and the rest was almost entirely eliminated at 2 days after transfection. On challenging ECFP with the cognate siRNA, ECFP-441 (class II), most of the ECFP signals could still be detected 2 days following transfection. The presence of A/U at the 5′ end of the siRNA AS would thus appear essential for some RNAi process other than mRNA recognition. That EGFP mRNA is a better target for EGFP-441 than ECFP would indicate that the 5′ end of the siRNA AS is also involved in hydrogen bonding between the target mRNA and the siRNA AS. Accordingly, the 5′ end of the AS would probably be involved in two separate RNAi processes, RISC formation, which includes siRNA unwinding, and mRNA recognition.
The time course of RNAi, as followed using several highly effective EGFP or ECFP siRNAs, showed target gene activity abolishment to remain at >70% for 7 days, at least starting from day 2. In contrast, little or no RNAi effects were evident on using ineffective class III siRNAs (data not shown).
siRNA sequence requirement for DNA vector-based RNAi
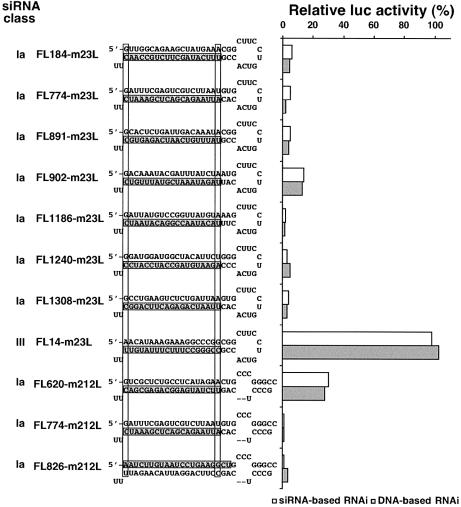
To determine whether target sequence preference in mammalian siRNA-based RNAi is intrinsic to the RNAi mechanism, a study was carried out to clarify whether similar rules for target sequence preference would hold for DNA-based mammalian RNAi, in which siRNA is produced via cleavage of hairpin-type RNA first transcribed and then transported from nuclei (38–40). pSilencer and firefly luc were used as vector and target genes, respectively. The profiles of RNAi activity change in DNA-induced RNAi can be seen from Figure 6 to be basically the same as siRNA-based RNAi. That is, all the pSilencer with the DNA insert encoding hairpin-type class Ia siRNA (shRNA) induced highly efficient RNAi in mammalian cells 3 days following transfection. In contrast, little or no RNAi was induced by transfection of pSilencer with the DNA insert encoding the hairpin of class III siRNA (FL14-m23L). siRNA sequence preference in mammalian siRNA-based RNAi may thus be concluded to hold for DNA-based RNAi in mammalian cells and, accordingly, should be a reflection of the intrinsic features of RNAi.
Figure 6.
Comparison of siRNA-based RNAi and DNA-based RNAi in HeLa cells. The predicted sequences of hairpin-type transcripts are shown on the left, while induced RNAi activity (reduction in relative luciferase activity) is shown by open boxes on the right. Stippled boxes indicate relative luciferase activity reduction due to cognate siRNA in HeLa cells. On the left, predicted AS are shaded. Note that the sequence preference of DNA-based RNAi is essentially identical to that of siRNA-based RNAi. Data obtained from 2–4 experiments were averaged and are shown.
siRNA sequence requirement for RNAi in chick embryos
The siRNA sequence preference rules presented here may be applicable to RNAi in vertebrates other than mammals and may prove useful in the design of siRNAs for gene silencing in individuals. To confirm these possibilities, siRNAs designed by the present rules were introduced into the right half of the spinal cord of day 2 chick embryos by in ovo electroporation, and the change in target gene activity on embryonic day 4 was examined (Fig. 3D). EGFP and DsRed expression served as criteria for assessing RNAi effects brought on by transfected siRNAs. EGFP-441, EGFP-416, DsRed-399 (Fig. 3D) and DsRed-231 (data not shown), all being class Ia siRNAs, were clearly shown to be capable of bringing about highly effective RNAi in the spinal cord of chick embryos. EGCFP-666, DsRed-140 (Fig. 3D) and DsRed-383 (data not shown), all belonging to class III, were found to be ineffective in this regard. Thus, our rules for siRNA sequence preference would certainly appear quite useful for the design of effective siRNAs in chick embryos.
Free energy calculation of siRNAs
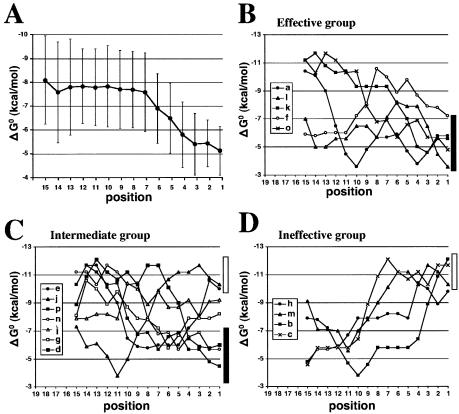
The enhanced flexibility at the siRNA end containing the 5′ AS end and low internal energy across the duplex (especially at the region 9–14) have recently been shown to be strongly correlated with siRNA function (30). Thus, internal stability reflecting the stability of pentamer subsequences was estimated in each of the 16 luc siRNAs shown in Figure 2A, using the nearest-neighbor method (35). ΔG° at position 1 of five highly effective siRNAs varied from –3.6 to –7.2 kcal/mol (Fig. 7B), whereas for seven siRNAs causing intermediate levels of RNAi, from –4.5 to –10.3 kcal/mol (Fig. 7C) and for highly ineffective siRNAs, the values exceeded –9.8 kcal/mol (Fig. 7D). These values would support the notion that the duplex end containing the 5′ AS end of highly effective siRNAs is considerably less thermostable. However, our data disclosed no clear reduction in the absolute values of ΔG° in the region 9–14. To further examine this point, value distribution across the duplex was studied using 32 highly effective siRNAs shown in Figures 2 and 3, but again there was no apparent low internal energy across the duplex (Fig. 7A). Thus, the notion proposed by Khvorova et al. (30) was partly supported by our study.
Figure 7.
Internal stability profiles of siRNAs. Thermodynamic profiles of highly effective 32 siRNAs (A), and 16 luc siRNAs shown in Figure 1 are composed of three groups giving rise to highly effective, intermediate and ineffective RNAi in mammalian cells (B–D). The vertical bars indicated in (A) show the standard deviation of 32 highly effective siRNAs. Thick and open vertical bars, respectively, indicate free energy change ranges at position 1 of highly effective and ineffective siRNAs.
The experimental results in Figure 7B and C indicate ΔG° at position 1 of three siRNAs that give rise to intermediate levels of RNAi in mammalian cells (p, n and d) to be within the range of those of five highly effective siRNAs (a, f, k, l and o). Thus, based on thermodynamic stability calculation, the selection of highly effective siRNAs from a random siRNA set may be quite possible, but only at a probability of 60%.
DISCUSSION
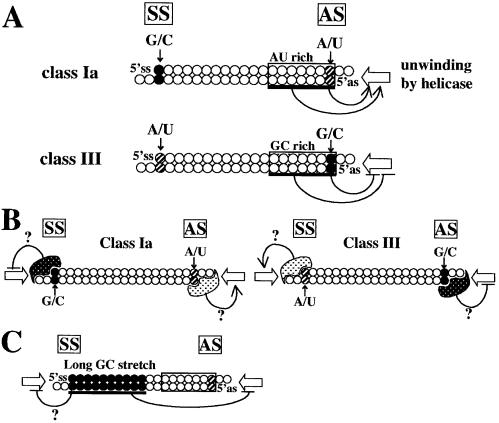
The relationship between siRNA sequence and its ability to give rise to RNAi in mammalian cells was extensively examined here and, on the basis of the results, rules were established for siRNA sequence preference and are schematically presented in Figure 8A. The rules predict that siRNAs satisfying all the four following sequence conditions at the same time give rise to highly effective RNAi in mammalian cells and possibly also in chick embryos: A/U at the 5′ AS end; G/C at the 5′ SS end; at least five A/U residues in the 5′ terminal one-third of the AS, and the absence of any GC stretch of >9 nt in length. siRNAs opposite in features with respect to the first three conditions bring about little or no gene silencing. A total of 57 highly effective and 16 ineffective siRNA candidates have been designed for four exogenous and 23 endogenous genes to date based on these rules (this work and our unpublished data), and all have been found to produce the anticipated RNAi activity in mammalian cells and chick embryos.
Figure 8.
Possible models of siRNA-based RNAi in mammalian cells. The rules for siRNA sequence preference are schematically shown in (A). siRNA unwinding might be effectively initiated from the AU-rich AS end in the case of class Ia siRNA, lacking a long GC stretch. On the other hand, siRNA duplex unwinding might be suppressed from the GC-rich class III AS end. G/C at the 5′ SS end of class Ia and the 5′ AS end of class III siRNAs might provide a site for binding of an unidentified protein possibly suppressing siRNA unwinding (B). Alternatively, A/U at the 5′ SS end of class III and the 5′ AS end of class Ia siRNAs might serve as a binding site for putative unwinding stimulation factors other than helicase. A long GC stretch such as that found in siRNA n (see Fig. 4B) might prevent the elongation of siRNA duplex denaturation from the AS end (C). A/U at the 5′ AS and SS ends and their counterparts in the SS and AS, respectively, are shown as hatched circles; G or C, closed circles. Terminal AU-rich or GC-rich regions are boxed. Open arrows indicate the direction of siRNA unwinding due to a hypothetical siRNA helicase.
Recently, Holen et al. pointed out that siRNA-based RNAi in mammalian cells varied considerably depending on target sequences (27). Their experimental results are clearly explained based on our rules. They showed that only four of 11 siRNAs examined could give rise to effective RNAi in HeLa, 293, Cos-1 and HaCaT cells. Our rules show that only these four effective siRNAs belong to class Ia or Ib, highly effective siRNA classes. Thus, the rules here may be concluded to be very useful for designing highly effective and ineffective siRNAs for silencing of mammalian and chick genes. However, it should be pointed out that, while the four conditions above are almost entirely sufficient for highly effective gene silencing, some may possibly be replaced by other functionally redundant conditions (see below for example).
The secondary structure of target RNA has been shown to be important for target mRNA recognition by siRNAs (41,42). However, at variance with these considerations, our results would indicate that target sequences are much more essential for target recognition by siRNAs than the secondary structure. No special secondary structure of the target can be deduced from our rules. Possibly, the frequency of serious secondary structure occurrence may be quite low in protein-coding regions of mRNA used here as targets.
EGFP/ECFP RNAi experiments (see Fig. 3C) indicated the presence of A/U at the 5′ AS end to possibly be required not only for target recognition but RISC formation as well, which includes siRNA unwinding. The step size of unwinding for UvrD DNA helicase is 5 bp (43) and thus a one-step motor function of putative siRNA helicase may unwind several base pairs from one of the two siRNA ends at the earliest stage in RISC formation. The 7 bp AS terminal duplex regions of highly effective and ineffective siRNAs are AU rich and GC rich, respectively, and 5′ AS ends of highly effective and ineffective siRNAs are A/U and G/C, respectively. It would thus follow that the putative siRNA helicase preferably initiates unwinding of the RNA duplex in an AU-rich terminal region with A/U at its 5′ free end, while RNA duplex unwinding from the GC-rich terminal region with G/C at its 5′ free end is blocked. Our unpublished experiments (Y.Naito, K.Ui-Tei and K.Saigo) have indicated that while virtually no degradation of the sense target RNA (vimentin mRNA) is brought about by VIM-35, a class III vimentin siRNA, ∼80% of antisense target RNA is cleaved by the same siRNA, which serves as class Ib siRNA for antisense target silencing. These considerations would appear consistent with the asymmetric RISC formation model recently proposed by Schwarz et al. (29) for in vitro RNAi in Drosophila embryonic extracts. This model predicts that siRNA unwinding preferably occurs at an ‘easier’ duplex end, possessing A:U, G:U or unpaired bases at its 5′ end position and being thermodynamically less stable, and that the strand with the 5′ end serves as a single-stranded guide RNA assembled into RISC. The importance of thermodynamically unstable or flexible base pairs at or near the AS end for siRNA unwinding in HEK 293 cells has also been pointed out by Khvorova et al. (30). A RISC formation mechanism similar to that proposed for the Drosophila in vitro system should thus also be applicable to mammalian and chick in vivo RNAi (see Fig. 8A).
According to the rules established here, 5′ AS and SS ends of highly effective siRNAs should be A/U and G/C, respectively, with the counterparts of ineffective siRNAs being G/C and A/U (see Fig. 8B). This terminal base compositional asymmetry may be important for determining the direction of siRNA unwinding. Recently, two Drosophila PIWI proteins have been shown to be capable of binding to a 5 bp single-stranded RNA or siRNA duplex (44–46). We found that the PAZ domain of eIF2C1, a human PIWI protein, binds to dsRNA with a 2 nt 3′ overhang but not to those with blunt or 5′ overhang ends (N. Doi, K. Ui-Tei and K. Saigo, unpublished data). In plant cells infected with tombusvirus, p19 may bind to siRNA ends and inhibit post-transcriptional gene silencing (47). Thus, a protein or protein complex, possibly not relevant to helicase but capable of binding preferentially to G/C or A/U at siRNA ends, might be involved in early strand separation of siRNA so as to either suppress or stimulate siRNA duplex unwinding.
Helicase functions might be doubly suppressed by G/C at the 5′ AS end position and an adjacent GC-rich sequence in highly ineffective siRNAs, while helicase functions appear blocked only by a single G/C pair at the 5′ SS end position (Fig. 8), suggesting that a single G/C pair at the 5′ SS end position and a GC-rich sequence near the 5′ SS end might be functionally redundant with each other and, accordingly, the latter might serve as a substitute for the former. We consider that this might be the reason why siRNA a (a class II siRNA) is capable of acting as a highly effective siRNA (see Figs 2A and 4B).
The results in Figure 2A indicate that siRNA n, possessing a 10 bp G/C stretch extending from the SS end, is incapable of giving rise to highly effective RNAi in mammalian cells, although it belongs to class Ia. Complete strand separation of siRNA appears to be required for active RISC formation (16) and, consequently, a long G/C stretch extending from the SS end may prevent helicase from unwinding not only from the SS end but from the AS end as well in a G/C stretch length-dependent manner (Fig. 8C).
In contrast to in vitro RNAi in Drosophila (29), in vivo Drosophila RNAi was far less sensitive to the siRNA sequence (see Fig. 2); virtually all siRNAs gave rise to effective RNAi in S2 cells when used at 50 nM. Our siRNA sequence preference rules established based on mammalian RNAi data were found to be not directly applicable to in vivo Drosophila (Fig. 2). Unlike mammalian cells, Drosophila cells might produce more protein components required for RISC formation and, hence, be capable of accumulating a considerable amount of RISC with a less efficient siRNA strand, i.e. asymmetric RISC formation may possibly not be a rate-limiting step in RNAi in Drosophila cells.
Figure 4 also indicates highly effective class Ia siRNAs to be comprised of heterogeneous members with over 10 times the capacity to bring about RNAi and maximum gene silencing activity to be induced by siRNA l transfection to CHO-K1 and S2 cells. Schwarz et al. (29) indicated gene silencing activity of siRNAs in the Drosophila in vitro system to be improved by the introduction of a U:G pair or unpaired bases at the 5′ AS end position. There may thus be the possibility of converting almost all class Ia siRNAs to siRNAs capable of inducing maximum levels of RNAi or RNAi levels brought about by siRNA l in mammalian cells via a change in terminal base pairing.
In a separate study, 19 986 human and 16 256 murine sequences registered in the NCBI Reference Sequence (RefSeq) database were examined using the siRNA sequence preference rules established here, and 92 and 99% of human and mouse sequences, respectively, were noted to possess at least one unique potential target for class Ia siRNA without a long G/C stretch (Y. Naito, K. Ui-Tei and K. Saigo, unpublished data). Our rules should thus find a wide scope of application to the design of siRNAs that are highly effective for mammalian RNAi including systematic mammalian functional genomics.
Acknowledgments
ACKNOWLEDGEMENTS
We thank H. Sasaki and T. Hamada for supplying HeLa cells, K. Nakamura for ES cells, J. Miyazaki for pCAGGS vector, S. Zenno, T. Noce, H. Niwa and N. Doi for helpful discussions and comments, H.Kaji for experiments at the initial stages, and R. Eda and A. Tanaka for technical assistance. We also thank John Rose for critical reading of the manuscript and for discussion. This work was partially supported by a Special Coordination Fund for promoting Science and Technology to K.S., and grants from the Ministry of Education, Culture, Sports, Science and Technology of Japan to K.S. and K.U.-T.
REFERENCES
- 1.Fire A., Xu,S., Montgomery,M.K., Kostas,S.A., Driver,S.E. and Mello,C.C. (1998) Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature, 391, 806–811. [DOI] [PubMed] [Google Scholar]
- 2.McManus M.T. and Sharp,P.A. (2002) Gene silencing in mammals by small interfering RNAs. Nature Rev. Genet., 3, 737–747. [DOI] [PubMed] [Google Scholar]
- 3.Hammond S.M., Caudy,A.A. and Hannon,G.J. (2002) Post-transcriptional gene silencing by double-stranded RNA. Nature Rev. Genet., 2, 110–119. [DOI] [PubMed] [Google Scholar]
- 4.Hannon G.J. (2002) RNA interference. Nature, 418, 244–251. [DOI] [PubMed] [Google Scholar]
- 5.Bernstein E., Caudy,A.A., Hammond,S.M. and Hannon,G.J. (2001) Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature, 409, 363–366. [DOI] [PubMed] [Google Scholar]
- 6.Ketting R.F., Fischer,S.E.J., Bernstein,E., Sijen,T., Hannon,G.J. and Plasterk,R.H.A. (2001) Dicer functions in RNA interference and in synthesis of small developmental timing in C.elegans. Genes Dev., 15, 2654–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hammond S.M., Bernstein,E., Beach,D. and Hannon,G.J. (2000) An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature, 404, 293–296. [DOI] [PubMed] [Google Scholar]
- 8.Nykänen A., Haley,B. and Zamore,P.D. (2001) ATP requirements and small interfering RNA structure in the RNA interference pathway. Cell, 107, 309–321. [DOI] [PubMed] [Google Scholar]
- 9.Elbashir S.M., Lendeckel,W. and Tuschl,T. (2001) RNA interference is mediated by 21 and 22 nt RNAs. Genes Dev., 15, 188–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hammond S.M., Boettcher,S., Caudy,A.A., Kobayashi,R. and Hannon,G.J. (2001) Argonaute2, a link between genetic and biochemical analyses of RNAi. Science, 293, 1146–1150. [DOI] [PubMed] [Google Scholar]
- 11.Caudy A.A., Myers,M., Hannon,G.J. and Hammond,S.M. (2002) Fragile X-related protein and VIG associated with the RNA interference machinery. Genes Dev., 16, 2491–2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zamore P.D., Tuschl,T., Sharp,P.A. and Bartel,D.P. (2000) RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell, 101, 25–33. [DOI] [PubMed] [Google Scholar]
- 13.Kataoka Y., Takeichi,M. and Uemura,T. (2001) Developmental roles and molecular characterization of a Drosophila homologue of Arabidopsis Argonaute1, the founder of a novel gene superfamily. Genes Cells, 6, 313–325. [DOI] [PubMed] [Google Scholar]
- 14.Williams R.W. and Rubin,G.M. (2002) ARGONAUTE1 is required for efficient RNA interference in Drosophila embryos. Proc. Natl Acad. Sci. USA, 14, 6889–6894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doi N., Zenno,S., Ueda,R., Ohki-Hamazaki,H., Ui-Tei,K. and Saigo,K. (2003) Requirement of Dicer and eIF2C translation initiation factors for short-interfering-RNA-mediated gene silencing in mammalian cells. Curr. Biol., 13, 41–46. [DOI] [PubMed] [Google Scholar]
- 16.Martinez J., Patkaniowska,A., Urlaub,H., Lührmann,R. and Tuschl,T. (2002) Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell, 110, 563–574. [DOI] [PubMed] [Google Scholar]
- 17.Kennerdell J.R., Yamaguchi,S. and Carthew,R.W. (2002) RNAi is activated during Drosophila oocyte maturation in a manner dependent on aubergine and spindle-E. Genes Dev., 16, 1884–1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tijsterman M., Ketting,R.F., Okihara,K.L., Sijen,T. and Plasterk,R.H.A. (2002) RNA helicase MUT-14-dependent gene silencing triggered in C.elegans by short antisense RNAs. Science, 295, 694–697. [DOI] [PubMed] [Google Scholar]
- 19.Elbashir S.M., Harborth,J., Lendeckel,W., Yalcin,A., Weber,K. and Tuschl,T. (2001) Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature, 411, 494–498. [DOI] [PubMed] [Google Scholar]
- 20.Elbashir S.M., Martinez,J., Patkaniowska,A., Lendeckel,W. and Tuschl,T. (2001) Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J., 20, 6877–6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stark G.R., Kerr,I.M., Williams,B.R., Silverman,R.H. and Schreiber,R.D. (1998) How cells respond to interferons. Annu. Rev. Biochem., 67, 277–264. [DOI] [PubMed] [Google Scholar]
- 22.Ui-Tei K., Zenno,S., Miyata,Y. and Saigo,K. (2000) Sensitive assay of RNA interference in Drosophila and Chinese hamster cultured cells using firefly luciferase gene as target. FEBS Lett., 479, 79–82. [DOI] [PubMed] [Google Scholar]
- 23.Billy E., Brondani,V., Zhang,H., Müller,U. and Filipowicz,W. (2001) Specific interference with gene expression induced by long, double-stranded RNA in mouse embryonal teratocarcinoma cell lines. Proc. Natl Acad. Sci. USA, 98, 14428–14433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paddison P.J., Caudy,A.A. and Hannon,G.J. (2002) Stable suppression of gene expression by RNAi in mammalian cells. Proc. Natl Acad. Sci. USA, 99, 1443–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang S., Tutton,S., Pierce,E. and Yoon,K. (2001) Specific double-stranded RNA interference in undifferentiated mouse embryonic stem cells. Mol. Cell. Biol., 21, 7807–7816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wianny F. and Zemicka-Goetz,M. (1999) Specific interference with gene function by double-stranded RNA in early mouse development. Nature Cell Biol., 2, 70–75. [DOI] [PubMed] [Google Scholar]
- 27.Holen T., Amrzguioui,M., Wiiger,M.T., Babaie,E. and Prydz,H. (2002) Positional effects of short interfering RNAs targeting the human coagulation trigger tissue factor. Nucleic Acids Res., 30, 1757–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harborth J., Elbashir,S.M., Vandenburgh,K., Manninga,H., Scaringe,S.A., Weber,K. and Tuschl,T. (2003) Sequence, chemical and structural variation of small interfering RNAs and short hairpin RNAs and the effect on mammalian gene silencing. Antisense Nucleic Acid Drug Dev., 13, 83–105. [DOI] [PubMed] [Google Scholar]
- 29.Schwarz D.S., Hutvágner,G., Du,T., Xu,Z., Aronin,N. and Zamore,P.D. (2003) Asymmetry in the assembly of the RNAi enzyme complex. Cell, 115, 199–208. [DOI] [PubMed] [Google Scholar]
- 30.Khvorova A., Reynolds,A. and Jayasena,S.D. (2003) Functional siRNAs and miRNAs exhibit strand bias. Cell, 115, 209–216. [DOI] [PubMed] [Google Scholar]
- 31.Niwa H., Yamamura,K. and Miyazaki,J. (1991) Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene, 108, 193–200. [DOI] [PubMed] [Google Scholar]
- 32.Lagos-Quintana M., Rauhut,R., Lendeckel,W. and Tuschl,T. (2001) Identification of novel genes coding for small expressed RNA. Science, 294, 852–858. [DOI] [PubMed] [Google Scholar]
- 33.Mourelatos Z., Dostie,J., Paushkin,S., Sharma,A., Charroux,B., Abel,L., Rappsilber,J., Mann,M. and Dreyfuss,G. (2002) miRNPs: a novel class of ribonucleoproteins containing numerous microRNAs. Genes Dev., 16, 720–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lim L.P., Glasner,M.E., Yekta,S., Gurge,C.B. and Bartel,D.P. (2003) Vertebrate microRNA genes. Science, 299, 1540. [DOI] [PubMed] [Google Scholar]
- 35.Freier S.M., Kierzek,R., Jaeger,J.A., Sugimoto,N., Caruthers,N.M., Neilson,T. and Turner,D.H. (1986) Improved free-energy parameters for predictions of RNA duplex stability. Proc. Natl Acad. Sci. USA, 83, 9373–9377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Niwa H., Miyazaki,J. and Smith,A.G. (2000) Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nature, 24, 372–376. [DOI] [PubMed] [Google Scholar]
- 37.Chun J.-Y., Han,Y.-J. and Ahn,K.-Y. (1999) Psx homeobox gene is X-linked and specifically expressed in trophoblast cells of mouse placenta. Dev. Dyn., 216, 257–266. [DOI] [PubMed] [Google Scholar]
- 38.Shi Y. (2003) Mammalian RNAi for the masses. Trends Genet., 19, 9–12. [DOI] [PubMed] [Google Scholar]
- 39.Brummelkamp T.R., Bernards,R. and Agami,R. (2002) A system for stable expression of short interfering RNAs in mammalian cells. Science, 296, 550–552. [DOI] [PubMed] [Google Scholar]
- 40.Kawasaki H. and Taira,K. (2003) Short hairpin type of dsRNAs that are controlled by tRNAVal promoter significantly induce RNAi-mediated gene silencing in the cytoplasm of human cells. Nucleic Acids Res., 31, 700–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vickers T.A., Koo,S., Bennett,C.F., Crooke,S.T., Dean,N.M. and Baker,B.F. (2003) Efficient reduction of target RNAs by small interfering RNA and RNase H-dependent antisense agents. J. Biol. Chem., 278, 7108–7118. [DOI] [PubMed] [Google Scholar]
- 42.Kretschmer-KazemiFar R. and Schzakiel,G. (2003) The activity of siRNA in mammalian cells is related to structural target accessibility: a comparison with antisense oligonucleotides. Nucleic Acids Res., 31, 4417–4424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ali J.A. and Lohman,T.M. (1997) Kinetic measurement of the step size of DNA unwinding by Escherichia coli UvrD helicase. Science, 275, 377–380. [DOI] [PubMed] [Google Scholar]
- 44.Lingel A., Simon,B., Izaurralde,E. and Sattler,M. (2003) Structure and nucleic-acid binding of the Drosophila Argonaute 2 PAZ domain. Nature, 426, 465–469. [DOI] [PubMed] [Google Scholar]
- 45.Yan K.S., Yan,S., Farooq,A., Han,A., Zeng,L. and Zhou,M.-M. (2003) Structure and conserved RNA binding of the PAZ domain. Nature, 426, 469–474. [DOI] [PubMed] [Google Scholar]
- 46.Song J.-J., Liu,J., Tolia,N.H., Schneiderman,J., Smith,S.K., Martienssen,R.A., Hannon,G.J. and Joshua-Tor,L. (2003) The crystal structure of the Argonaute2 PAZ domain reveals an RNA binding motif in RNAi effector complexes. Nature Struct. Biol., 10, 1026–1032. [DOI] [PubMed] [Google Scholar]
- 47.Silhavy D., Molnár,A., Lucioli,A., Szittya,G., Hornyik,C., Tavazza,M and Burgyán. (2002) A viral protein suppresses RNA silencing and binds silencing-generates, 21- to 25-nucleotide double-stranded RNAs. EMBO J., 21, 3070–3080. [DOI] [PMC free article] [PubMed] [Google Scholar]