Abstract
Coffin–Lowry syndrome (CLS) is caused by mutations in the RSK2 gene encoding a protein kinase of the Ras signalling pathway. We have studied two point mutations which cause aberrant splicing but do not concern the invariant GT or AG nucleotides of splice sites. The first, an A→G transition at position +3 of the 5′ splice site of exon 6, results in vivo and in vitro in exon skipping and premature translation termination. The natural 5′ splice site, although intrinsically weak, is not transactivated under normal conditions. Consequently, replacement of an A/U by a G/U base pairing with U1 snRNA reduces its strength below a critical threshold. The second mutation, an A→G transition 11 nt upstream of exon 5, creates a new AG near the natural 3′ splice site. In vitro this synthetic 3′ AG is used exclusively by the splicing machinery. In vivo this splicing event is also observed, but is underestimated because the resulting RSK2 mRNA contains premature stop codons which trigger the nonsense-mediated decay process. We show that a particular mechanism is involved in the aberrant splicing of exon 5, implying involvement of the natural 3′ AG during the first catalytic step and the new 3′ AG during the second step. Thus, our results explain how these mutations cause severe forms of CLS.
INTRODUCTION
Coffin–Lowry syndrome (CLS) is a syndromic form of X-linked mental retardation. Affected males are of short stature and present moderate to severe psychomotor retardation, associated with typical facial and hand aspects and severe skeletal alterations which appear progressively through the course of the disease (1–3). In most female carriers, only minimal findings are observed (4). CLS is caused by mutations in the RSK2 (ribosomal S6 kinase 2) gene, encoding a serine/threonine kinase acting in the Ras/MAP-ERK signalling pathway (5).
To date >90 distinct mutations have been described, and splicing defects account for 20% of them (6–8). Interestingly, some splicing mutations are unusual since they do not affect the consensus GT or AG nucleotides found in the 5′ and 3′ splice sites. In one patient, an A→G transition was observed at position +3 of the 5′ splice site of RSK2 exon 6. This mutation resulted in complete skipping of exon 6 (8). This was surprising, since A and G nucleotides are found with similar frequencies at position +3 in 5′ splice sites (60 and 40%, respectively). A second patient carried an A→G transition 11 bp upstream from RSK2 exon 5. In a cell line derived from this patient, two aberrant transcripts were observed: in the first one, 10 intronic nucleotides were included between exons 4 and 5, whereas the second one resulted from skipping of exon 5 (8).
Pre-mRNA splicing is performed within a macromolecular complex, the spliceosome (9,10). Correct splicing requires recognition of consensus sequences at the intron/exon boundaries, including 5′ and 3′ splice sites, the branch site and auxiliary elements such as intronic or exonic splicing silencers and enhancers. Factors acting in trans, including SR proteins which are implicated in constitutive as well as alternative splicing processes, bind to such auxiliary elements (11,12).
In the present study we have investigated the mechanisms underlying the two aberrant splicing events described above. The 5′ splice site of exon 6 was interesting to consider because under its natural form it represents an intrinsically weak splice site, which could be activated by cis-acting enhancers. The splicing events occurring in vivo on the transcripts containing the mutation upstream of exon 5 could not be assessed precisely due to involvement of the nonsense-mediated decay (NMD) process and required further analysis. Importantly, CLS clinical manifestations appear progressively during the life of patients. Thus, CLS could be a good candidate disease for gene therapy. Therefore, it is important to understand the molecular basis of the splicing alterations occurring in the two patients analysed. Our results give an insight into how two unusual nucleotide substitutions, which would probably not be considered as pathogenetic mutations in a screening by single-strand conformation polymorphism analysis and without further analysis, dramatically affect pre-mRNA splicing of the RSK2 gene and result in severe forms of CLS.
MATERIALS AND METHODS
Cell culture and RT–PCR analysis
Patient and control lymphoblastoid cell lines were grown in RPMI medium supplemented with 10% fetal calf serum. When required, they were treated with 7.5 µg/ml cycloheximide for 6 h or incubated at 20 or 24°C before harvesting. Total RNA was extracted from cells using the RNA-Solv reagent (Omega Biotek). The first-strand cDNA was synthesized using random hexamers and oligo(dT) as primers. Primers used for PCR amplification of the RSK2 cDNA sequence between exons 1 and 7 were: forward primer 5′-TGGCGCAGCTGGCGGA-3′; reverse primer 5′-ATTATTCCCAGGCTATGTAG-3′. The human HPRT (hypoxanthine guanine phosphoribosyl transferase) cDNA used as a gene control was amplified using: forward primer 5′-CGTGGGGTCCTTTTCACCAGCAAG-3′ and reverse primer 5′-AATTATGGACAGGACTGAACGTC-3′. PCRs for the RSK2 and HPRT cDNAs were run for 32 and 22 cycles, respectively, and PCR products were resolved on 3% ethidium bromide-stained agarose gels.
Constructs
A wild-type minigene to study RSK2 exon 6 expression in transfected cells, pRSK2(6)wt, was obtained by insertion of a PCR-amplified fragment containing human genomic sequences from RSK2 exons 5–7 between the XhoI and BamHI sites of the pXJ42 plasmid (13). The corresponding mutated minigenes, pRSK2(6)A→G, corresponding to patient 1, pRSK2(6)mt+T→A, pRSK2(6)mt+A→G, pRSK2(6)mt+ TA→AG, pRSK2(6)mt+insTCTC, pRSK2(6)Δ1–4 and pRSK2(6)4mm (see the resulting sequences in Figs 2A and 3A and C), were obtained by site-directed mutagenesis (QuickChange; Stratagene) using primers containing the relevant mutations. The constructs for in vitro splicing assays were obtained by insertion of PCR-amplified exon 6 (wild-type, patient 1, Δ1–Δ4 and 4mm), flanked by its upstream and downstream intronic sequences (40 and 42 bp, respectively), between the XbaI and SmaI sites of the BSSK HG2 vector containing genomic sequences from exons 2–3 of the rabbit β-globin gene (14). PCR-amplified exon 5 (wild-type and patient 2), flanked by its surrounding intronic nucleotides (205 and 236 bp, respectively), was cloned in the BSSK HG2 plasmid as described above. Corresponding constructs in which the natural AG of patient 2 exon 5 was altered (constructs ΔAG and mtpolyPy/AG; see resulting sequences in Fig. 5A) were obtained by site-directed mutagenesis by overlap extension, using appropriate primers. Minigenes for exon 5 used in in vivo splicing assays [pRSK2(5)wt and pRSK2(5)A→G] were obtained by insertion of the PCR-amplified insert for in vitro splicing assays between the XhoI and EcoRI sites of the pXJ42 plasmid. All constructs were verified by sequencing.
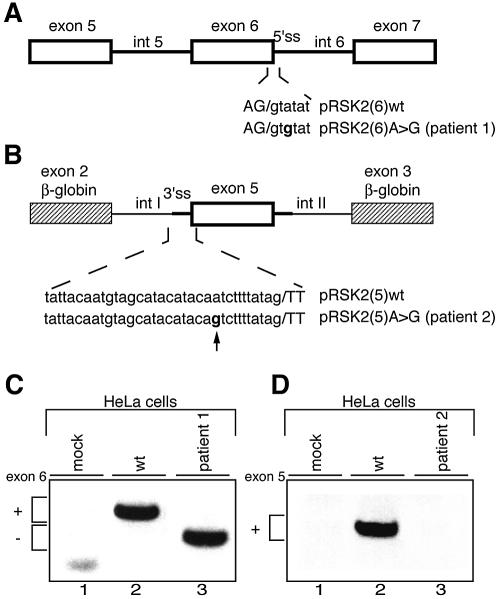
Figure 2.
Effect of the IVS6 +3 A→G and the IVS4 –11 A→G mutations in in vivo splicing assays. (A and B) Schematic representation of minigenes for RSK2 exons 6 and 5, respectively. Annotations are as in Figure 1. In (B), the genomic region of exon 5 was inserted between β-globin exons 2 and 3 represented by striped boxes. Chimeric introns upstream and downstream of exon 5, arbitrarily designated introns I and II, are 384 and 653 nt long, respectively. (C and D) RT–PCR analysis on RNA from HeLa cells transfected with exon 6 and exon 5 minigenes. In mock assays, cells were transfected with an empty vector. In (C), pRSK2(6)A→G recapitulated the splicing pattern of endogenous similarly mutated RSK2, producing complete skipping of exon 6 (lane 3). In (D), no splicing product was detectable when pRSK2(5)A→G was transfected into HeLa cells (lane 3).
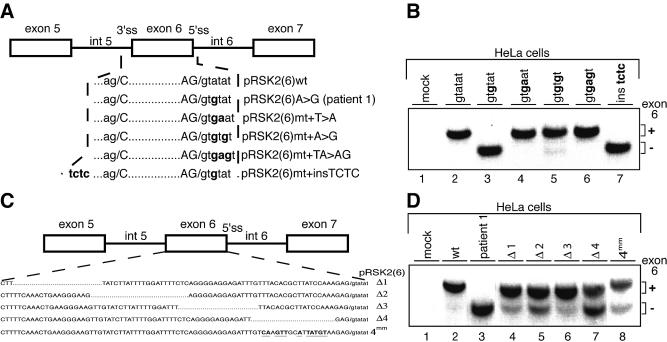
Figure 3.
Splicing pattern in HeLa cells transfected with minigene constructs harbouring mutations in the exon 6 region. (A) Schematic representation of mutations generated at positions +4 and +5 (or both) of the 5′ splice site and the insertion upstream of the 3′ splice site of exon 6. Modified nucleotides are indicated in bold. (B) RT–PCR analysis on RNA from HeLa cells transfected with the minigenes described in (A). The minigenes used for transfection are indicated above the lanes. (C) Representation of the minigenes carrying deletions Δ1–Δ4 (dotted lines) or multiple nucleotide changes (in bold and underlined) in RSK2 exon 6. (D) RT–PCR analysis on RNA from HeLa cells transfected with the minigenes (indicated above the lanes) described in (C). In (C) and (D), mock assays correspond to cells transfected with an empty vector.
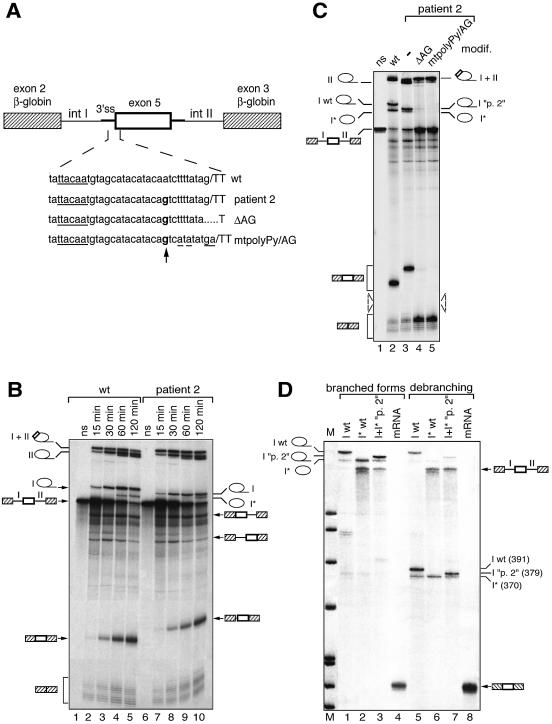
Figure 5.
In vitro splicing assays of wild-type and patient 2 (IVS4 –11 A→G) transcripts. (A) Chimeric constructs used in in vitro splicing assays are as in Figure 2B. Chimeric introns upstream and downstream of exon 5, arbitrarily designated introns I and II, are 384 and 653 nt long, respectively. The site of the mutation is indicated in bold and the consensus defining the branch site is underlined. The mutations altering the natural 3′ AG of patient 2 are also indicated. (B) Kinetic analysis of splicing. Splicing reactions were run for 15, 30, 60 and 120 min and RNA products were analysed by PAGE. Selected splicing products are indicated along the gel panel. (C) Comparison of splicing products generated after 120 min of splicing from the wild-type transcript (wt) and patient 2 transcripts carrying (lanes 4–5) or not (lane 3) mutations in the natural 3′ AG. (D) Comparative analysis of lariat intron I molecules produced from wild-type (wt) and mutant (“p. 2”) transcripts. Isolated putative intron I wt (I wt), shortened intron I wt (I* wt) as well as intact and shortened forms of intron I “p. 2” together (I+I* “p. 2”) were analysed directly (lanes 1–3) or after debranching (lanes 5–7). A linear three-exon mRNA was also analysed as a control before (lane 4) or after (lane 8) debranching. The size of the debranched RNAs was determined using DNA size markers (pBR322 cut by MspI), run in parallel.
In vivo splicing assays
HeLa cells were grown in DMEM supplemented with 2.5% calf serum and 2.5% fetal calf serum. Cells were transfected with 10 µg of the indicated RSK2 minigenes using the CaCl2 procedure. Total RNA was isolated 48 h after transfection using the RNA-Solv Reagent (Omega Biotek) and treated with DNase. Reverse transcription was performed in a total volume of 50 µl using 5 µg of total RNA, 1× first strand buffer (Invitrogen Life Technologies), 0.25 mM each dNTP, 5 mM DTT, 2.5 mM MgCl2, 100 U RNase inhibitor (Promega) and 200 U SuperScript II reverse transcriptase (Invitrogen Life technologies), for 40 min at 37°C. To ensure that only plasmid-derived RSK2 transcript was detected, the reverse transcription was performed with a vector-specific primer (5′-AACCATTATAAGCTGCAAT-3′) and the PCR with a RSK2 forward primer (5′-TCGAGACCGAGTTCGGAC-3′) and a vector-specific reverse primer (5′-AACCATTATAAGCTGCAATAAAC-3′). The cycle number was kept to 30 and the products were resolved on 3% ethidium bromide-stained agarose gels.
In vitro splicing assays
HeLa nuclear extracts and cytoplasmic S100 extracts were prepared as previously described and RNA substrates were synthesized by T3 polymerase transcription of the cognate DNA plasmids linearized with XhoI (15). In vitro splicing was performed in 25 µl assays containing 33 fmol of 32P-labelled pre-mRNA, 7 µl of nuclear extract, 5 µl of S100 and the basic components for the splicing reaction in the presence of 48 mM KCl and 2.6 mM MgCl2, for 2 h at 30°C unless otherwise stated (16). Splicing products were analysed on 5.2% denaturing polyacrylamide gels and visualized by autoradiography.
Quantification of splicing reactions in Table 1 was performed as follows. Radioactive bands corresponding to the lariat intron II and the two-exon mRNA were quantified with a Typhoon 8600 Imager (Amersham). Data were corrected with respect to the number of cytosine nucleotides in each splicing product, relative to the initial pre-mRNA. Estimation of the recognition of exon 6 was expressed as indicated in Table 1.
Table 1. Quantification of exon 6 inclusion from various mutant constructs in in vitro splicing assays.
| Construct |
Experiment | Mean |
SD |
||
|---|---|---|---|---|---|
| 1 | 2 | 3 | |||
| Wild-type |
1 |
1 |
1 |
1 |
– |
| Δ1 |
0.89 |
1.55 |
1.35 |
1.26 |
0.34 |
| Δ2 |
0.89 |
1.30 |
1.41 |
1.20 |
0.27 |
| Δ3 |
0.87 |
1.30 |
1.35 |
1.17 |
0.26 |
| Δ4 |
0.55 |
0.6 |
– |
0.58 |
0.04 |
| 4mm | – | 0.80 | 0.71 | 0.76 | 0.06 |
Columns 1–3 are the ratios between values of normalized radioactivity obtained for lariat intron II (exon 6 inclusion) and the sum of lariat intron II and the two-exon mRNA (exon 6 skipping), from three independent experiments. To allow comparison between individual experiments, the value of the ratio obtained for the wild-type construct in each experiment was set to 1. Mean is the mean value for all experiments; SD is the standard deviation from the mean value. –, data not available.
Debranching experiments were performed after in vitro splicing. RNA products were separated on 6% denaturing polyacrylamide gels and isolated by electroelution. The debranching reaction was carried out in the presence of HeLa nuclear extracts as described previously (17). Products were resolved on 5.2% denaturing polyacrylamide gels and visualized by autoradiography.
RESULTS
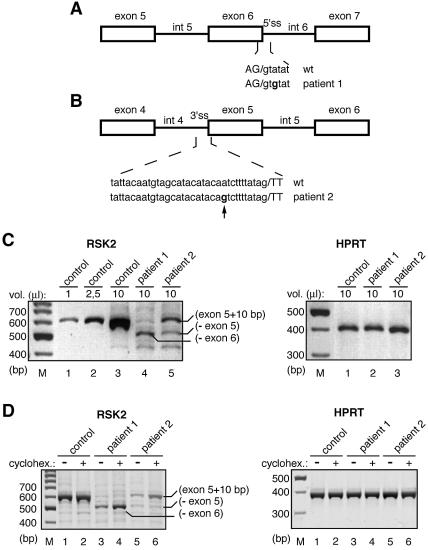
Mutations causing aberrant splicing of RSK2 exons 6 and 5 were described in CLS patients 1 and 2, respectively (Fig. 1A and B). RT–PCR analysis of RNA from a cell line derived from patient 1 revealed complete skipping of exon 6, whereas for patient 2, two abnormal transcripts were detected, one with an insertion of 10 intronic nucleotides and one resulting from skipping of exon 5 (Fig. 1C) (8). Both skipping of exon 6 and insertion of 10 nt in front of exon 5 result in a frameshift and premature translation termination. For both patients, the resulting abnormal transcripts were present at lower amounts compared with wild-type transcripts (Fig. 1C, compare lanes 1–3 with lanes 4 and 5). In patient 2, a small amount of a shorter mRNA resulting from in-phase skipping of exon 5 was also observed (Fig. 1C, lane 5). To determine whether the low amounts of abnormal transcripts in patients 1 and 2 are due to NMD (18,19), we exposed both patient and control cell lines to a translation inhibitor which is known to inhibit this process (20). RT–PCR analysis of RNA from cycloheximide-treated or untreated cells is presented in Figure 1D. Cycloheximide treatment had no significant effect on the wild-type RSK2 mRNA (lanes 1 and 2 of the RSK2 panel) and on the HPRT mRNA of all cell lines (lanes 1–6 of the HPRT panel). In contrast, we observed a significant increase (roughly 3-fold) in the amount of the skipped exon 6 transcripts of patient 1 and of the abnormal exon 5-containing transcripts of patient 2 (Fig. 1D, lanes 3–4 and 5–6). As expected, the transcript lacking exon 5 of patient 2, in which the natural coding phase is preserved, was not affected by cycloheximide treatment (Fig. 1D, lanes 5–6). These results strongly suggest that the major splicing event in patient 1 is efficient skipping of exon 6, whereas strong splicing of the abnormal exon 5 occurs in patient 2.
Figure 1.
Effect of the IVS6 +3 A→G and the IVS4 –11 A→G mutations in the patient cell lines. (A and B) Schematic diagrams of the localization of the mutations in patients 1 and 2, respectively. Exons are indicated by boxes and introns by lines. Lower case letters are intron sequences and capital letters are exon sequences. The arrow indicates the cryptic 3′ AG generated by the mutation found in patient 2. (C) RT–PCR experiments on RNA from lymphoblastoid cell lines derived from a control individual and patients 1 and 2. The primers used for PCR analysis are found in RSK2 exons 1 and 7 and they generate a 562 bp cDNA fragment from the control RNA. The size and relative abundance of RT–PCR products for RSK2 and HPRT transcripts in patients were compared with those of a control individual. Between 1 and 10 µl of PCR assays were analysed on agarose gels, as indicated above the lanes. The PCR fragment of ∼450 bp detected in lanes 4–6 could result from skipping of both exons 5 and 6 from the original mRNA molecule. (D) RT–PCR experiments on RNA from lymphoblastoid cell lines derived from patient and control individuals and treated (+) or not (–) with cycloheximide. The analysis was carried out as in (C).
To check for the presence of truncated forms of RSK2 in the cell lines of patients 1 and 2, we used antibodies directed against the N- or C-terminus of the protein. In both cases, no protein products were detected, indicating that if such truncated RSK2 forms exist, they are either produced at low levels and/or are unstable (8; data not shown).
To further understand the mechanisms underlying the aberrant splicing events occuring in patients 1 and 2, we first analysed splicing in vivo in HeLa cells transfected with constructs containing exon 6 or exon 5. Splicing of the pRSK2(6)A→G minigene (Fig. 2A) recapitulated the splicing of the endogenous RSK2 pre-mRNA observed in the cell line from patient 1, since complete skipping of exon 6 was observed (Fig. 2C). Note that since the two transcripts do not contain long open reading frames, they should not be subject to NMD. The wild-type sequence of the 5′ splice site of RSK2 exon 6 is AG/GTAtaT, where the residues at positions +4 and +5 (lower case letters) represent discordant nucleotides against the 5′ end of U1 snRNA, keeping the number of successive base pairs with U1 snRNA to only five. We hypothesized that the change of the A/U base pairing at position +3 to a weaker G/U base pairing in the mutant decreases the stability of the 5′ splice site/U1 snRNA hybrid below a critical threshold. To test this hypothesis, we replaced the nucleotides at positions +4 and +5 by nucleotides which are expected to reinforce the stability of the hybrid, in the context of the mutant 5′ splice site, and expressed the resulting minigenes in HeLa cells (Fig. 3A and B). Introduction of the concordant A nucleotide at position +4 or the concordant G nucleotide at position +5 or both fully prevented aberrant splicing (Fig. 3B, lanes 4–6). Conversely, increasing the length of the polypyrimidine tract (insertion of a TCTC sequence) upstream of the 3′ splice site of exon 6 had no effect (Fig. 3B, lane 7). These results strongly indicate that skipping of exon 6 in the mutated transcript results exclusively from a decrease in the strength of the 5′ splice site below a threshold which precludes its efficient recognition by the splicing machinery.
There is now emerging evidence that alternative as well as constitutive splicing processes may require, in addition to 5′ and 3′ splice sites, the presence of sequences that act as enhancers. Particular attention has been given to exonic splicing enhancers (ESEs) and mutations in such enhancers have been associated with several human diseases (21–24). Since, apparently, the strength of the 5′ splice site of exon 6 lies just above a critical threshold, we asked whether splicing of wild-type exon 6 requires the presence of a splicing enhancer which might be present in exon 6 itself. To test this, we performed in vivo splicing assays with four distinct constructs, derived from pRSK2(6)wt. They include deletions of four regions within exon 6, as indicated in Figure 3C. Only a marginal increase in exon 6 skipping was observed when regions 1, 2 and 3 were absent (Fig. 3D, compare lanes 4–6 with lane 2). This result was rather unexpected, especially in the case of region 3, since it contains a GGAGGAGA motif, which resembles exonic purine-rich sequences acting as ESEs (25,26). In contrast, deletion of region 4 led to a significant decrease in exon 6 inclusion (Fig. 3D, lane 7). To further investigate the existence of a putative splicing enhancer in the 3′ part of exon 6, we mutated numerous residues in region 4 (Fig. 3C). However, this mutagenesis altered correct splicing of exon 6 to a lesser extent (Fig. 3D, compare lanes 7 and 8), a result which does not favour the existence of a strong splicing enhancer in the 3′ part of exon 6. In agreement with this, co-transfection of the mutated minigene with expression vectors encoding various SR proteins failed to restore correct splicing of exon 6 (data not shown).
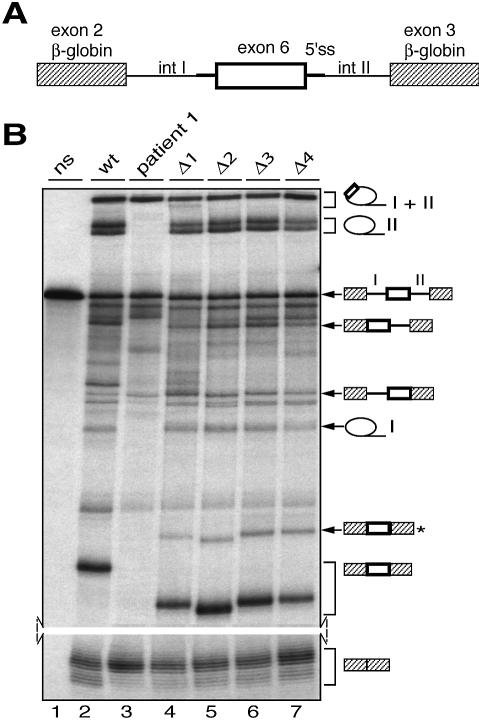
As splicing of exon 6 may be strongly dependent on the flanking genomic sequences, it was important to analyse splicing of wild-type and mutant exon 6 outside the RSK2 gene context. To this end, chimeric substrates containing exon 6 flanked by short intronic sequences (described in Fig. 4A) were processed in in vitro splicing assays. In vitro splicing of the wild-type chimeric substrate (Fig. 4B, lane 2) showed that exon 6 can be included (in a three-exon mRNA) or skipped (in a two-exon mRNA) during splicing and that these events are in competition. However, the behaviour of the mutated or deleted chimeric constructs, relative to the wild-type construct, was very similar to that observed in vivo (Fig. 4B, lanes 2–7). Whereas the mutation present in patient 1 resulted in complete skipping of exon 6, little or no effect was observed with deletions of regions 1–3 of exon 6. In contrast, deletion of region 4 reduced the efficiency of exon 6 splicing by 2-fold. Quantitative analysis of the in vitro splicing of all constructs, shown in Table 1, is given as the ratio between splicing products resulting from the recognition of exon 6 and the sum of the splicing products resulting from recognition or skipping of exon 6. Interestingly, results indicate that mutagenesis of region 4 affects exon 6 recognition to a lesser extend compared with deletion of the same region (Table 1), in agreement with the results obtained in vivo (Fig. 3D). Therefore, region 4 does not appear to function as a classical splicing enhancer whose action is mediated by a trans-acting factor. In agreement with this, in vitro splicing of the wild-type construct was not altered by the presence of a high excess (250-fold relative to the pre-mRNA) of a competitor RNA corresponding to exon 6 region 4 (data not shown). Finally, we have also verified by mutational analysis that the intronic sequences immediately flanking the 5′ splice site of exon 6 do not exhibit enhancer activity (data not shown).
Figure 4.
In vitro splicing assays of wild-type, patient 1 and deleted transcripts specific for RSK2 exon 6. (A) RSK2 exon 6 and surrounding intronic sequences were introduced between exons 2 and 3 of the β-globin gene (striped boxes). The resulting introns upstream and downstream of exon 6 were arbitrarily called introns I and II and are 218 and 457 nt long, respectively. (B) In vitro splicing assays with transcripts including wild-type exon 6 (wt), exon 6 mutated at the 5′ splice site (patient 1) or with deletions Δ1–Δ4 as described in Figure 3C, followed by PAGE analysis of the RNA products. Lane ns corresponds to non-spliced RNA. Selected splicing products are indicated along the gel panel. The three-exon mRNA product marked with an asterisk results from the use of a cryptic 3′ splice site located 42 nt upstream of exon 6.
For detailed analysis of altered splicing of RSK2 exon 5 in patient 2, the large size of intron 4 (>10 kb) excluded the insertion of the whole exon 4–exon 6 region in an expression vector. Therefore, we have constructed a chimeric minigene in which exon 5, flanked by at least 200 bp of intronic sequences, was inserted between exons 2 and 3 of the β-globin gene (Fig. 2B). Intriguingly, whereas the control chimeric pre-mRNA was efficiently spliced in transfected HeLa cells (Fig. 2D, lane 2), no spliced mRNA was detected in the presence of the patient 2 mutation (Fig. 2D, lane 3). The absence of mRNA was repeatedly obtained with two DNA clones selected among clones from two independent construct preparations. This result indicated that, for reasons that are not well understood, the chimeric transcripts containing the patient 2 mutation are either poorly synthesized and/or unstable in HeLa cells.
To analyse further the splicing events occurring in patient 2, we used an in vitro approach. The same kind of wild-type and mutant substrates as for transfection experiments (Fig. 5A) were spliced in vitro for between 0 and 120 min, the splicing kinetics allowing a better identification of intermediates and final splicing products (Fig. 5B). The splicing products designed along the gel have been unambiguously identified according to: (i) the size for linear or debranched RNA and the apparent mobility for lariat species; (ii) the kinetics of accumulation; (iii) the sequencing of final mRNA products after RT–PCR analysis. Note that introns I and II are submitted to 3′→5′ exonuclease digestion, resulting in shortening of their 3′ tail and generation of an additional band for each after 60 min of incubation (Fig. 5B, lanes 4, 5, 9 and 10).
Whereas the majority of splicing products generated from both substrates have the same electrophoretic mobility, the three-exon mRNA (including RSK2 exon 5) resulting from the mutated substrate is 10–15 nt longer than that resulting from the wild-type substrate, whereas the mobility of intron I from the mutated transcript is increased (Fig. 5B, compare lanes 5 and 10). Sequencing of the three-exon mRNA resulting from the mutated substrate revealed that the splicing machinery selected the AG generated by the mutation 10 nt upstream of the wild-type 3′ splice site, in agreement with one of the splicing events detected in the cell line derived from the patient (Fig. 1C).
Further analysis of the splicing kinetics of both wild-type and mutated substrates showed additional interesting features (Fig. 5B). First, although a cryptic AG is used in the mutated transcript for splicing of RSK2 exon 5, the efficiency of inclusion of this exon is very similar for both transcripts. Indeed, for each transcript accumulation of the three-exon mRNA or intron II is roughly equivalent (Fig. 5B, lanes 5 and 10; Fig. 5C, lanes 2 and 3). Second, in the mutated substrate use of the normal AG is abolished, whereas accumulation of the two-exon mRNA resulting from skipping of exon 5 does not change under our splicing conditions (Fig. 5B and C). Both these observations indicate that the major splicing event occurring in patient 2 is splicing of the abnormal exon 5 rather than skipping of this exon. Importantly, the shift from the natural to the cryptic AG occurs although the latter is not preceded by a pyrimidine-rich sequence (only two uridines and three cytosines present in the 10 nt upstream of it). This strongly suggests that occurrence of the cryptic AG does not create a classical 3′ splice site but rather, along with the natural 3′ splice site, a novel functional 3′ region in which the natural 3′ splice site serves during the branching reaction whereas the cryptic AG is selected only for the second step of splicing. This mechanism, evident only in a few natural examples (27,28), has two direct implications: (i) the natural 3′ AG, although not physically used, should be absolutely required for splicing of the mutated exon 5; (ii) the same branch site should be used for splicing of the wild-type and the mutated exon 5.
To address the first statement, we studied the consequences of two types of mutations altering the natural 3′ AG of exon 5 in patient 2 (see Fig. 5A). Introduction of these two mutations fully inhibited recognition of the mutated exon 5 (Fig. 5C, compare lanes 4–5 with lane 3), demonstrating that presence of the natural 3′ AG is absolutely required for efficient use of the cryptic AG found in patient 2. Concerning the second statement, we recognized a putative TACAAT branch site 27 nt upstream of the intron/exon 5 junction (Fig. 5A). The expected close proximity of the cryptic AG to the putative branch site (16 nt) precluded the use of reverse transcription to identify the branch site. Instead, we used a different approach based on the observation that the aberrant migration of lariat splicing products is very sensitive to the actual size of the RNA loop and of the 3′ tail. Indeed, we and others had previously observed that lariats of the same intron for which several branch sites spaced by only 2–4 nt are used could be easily separated by PAGE (29,30). As shown in Figure 5C (lanes 2 and 3), the putative intact intron I generated from the mutant substrate migrated faster than the lariat intron I obtained from the wild-type substrate, most likely due to its smaller size. However, their putative shortened forms showed exactly the same migration, suggesting that the same branch site is selected for splicing of intron I of both transcripts and that the 3′→5′ exonuclease digestion transforms them into the same shortened lariat form.
To validate these results, we isolated the RNA bands corresponding to the putative intact and shortened forms of wild-type intron I separately, as well as the corresponding species of mutant intron I together, submitted them to debranching and analysed them by PAGE (Fig. 5D). We observed that the linear forms of intact intron I, generated from wild-type or mutant transcripts, have apparent sizes of 391 and 379 nt (difference 12 nt), which are very close to the theoretical sizes of 384 and 374 nt, respectively (Fig. 5D, lanes 5 and 7). In contrast, after debranching the shortened intron I was the same size (370 nt) for both transcripts, indicating that the same branch site is used for splicing of both wild-type and mutated exon 5 and that the selected branch site lies at least 22 nt upstream of the natural AG, a distance compatible with the 27 nt proposed above (Fig. 5D, lanes 6 and 7). Taken together, these results reinforce the notion that a switch in recognition of the natural and cryptic 3′ AG pairs occurs during splicing of exon 5 in the presence of the mutation of patient 2.
DISCUSSION
This report provides insights into the mechanisms underlying splicing defects caused by two deleterious mutations in the RSK2 gene. The first mutation (in patient 1) is an A→G transition at position +3 of the 5′ splice site downstream of RSK2 exon 6. The resulting skipping of exon 6 was surprising, since A and G nucleotides are found at similar frequencies at this position of 5′ splice sites (31,32). Our results indicate that the strength of the wild-type 5′ splice site of RSK2 exon 6, which matches only five consecutive residues of the consensus sequence, lies just above a critical threshold. Indeed, replacement of an A/U pairing by a less stable G/U pairing during hybridization with U1 snRNA is sufficient to shift the splicing pattern towards weaker recognition and skipping of exon 6. Several other intrinsically weak 5′ splice sites with similar mutations have been described, but detailed analysis of the splicing events, which suggested that a 5′ site with a G residue at position +3 is efficiently used only if two of the residues at positions +4 to +6 may base pair with U1 snRNA, was only performed for one of them (32). Our results are in agreement with this conclusion, since in the mutant 5′ splice site of RSK2 exon 6 the residues at positions +4 and +5 differ from the consensus. Recently, Roca et al. (33) have compared, by several scoring methods, the relative strengths of authentic and cryptic 5′ splice sites. By using these methods, we have observed that the scores obtained for the wild-type 5′ splice site of exon 6 are between the average values obtained for authentic and cryptic 5′ splice sites. Furthermore, the scores for the exon 6 5′ splice site of patient 1 are even lower than those obtained for cryptic 5′ splice sites.
Because the 5′ splice site of exon 6 is intrinsically weak, it was possible that RSK2 exon 6 splicing is activated by splicing enhancers present in exon 6 itself or in surrounding sequences. Indeed, such elements may be present in pre-mRNA regions apparently submitted to constitutive splicing, such as the β-globin or SMN pre-mRNAs (23,34,35). Analysis of exon 6 mutation/deletion constructs was not in favour of the involvement of a classical splicing enhancer (Fig. 3). Taken together, our results showed that splicing of exon 6 occurs essentially according to a constitutive mode and that base pairing with U1 snRNA by at least five consecutive residues (or a base pairing with equivalent stability) is required to promote efficient inclusion of internal exons in the absence of additional cis-activation. In some cases of mutations which affect the strength of 5′ splice sites or which activate cryptic splice sites, it has been shown that incubation of cells derived from patients carrying such mutations at 31 or 20–24°C instead of 37°C partially reversed abnormal splicing (36,37). However, this was not observed with RSK2 transcripts when lymphoblastoid cells derived from patient 1 were incubated at 20–24°C (data not shown).
The mutation of patient 2 creates a new AG preceded by a C residue upstream of exon 5 which is efficiently recognized by the splicing machinery as a 3′ splice site. We observed quantitative differences in splicing between the lymphoblastoid cell line of patient 2 and in vitro assays. Indeed, the splicing machinery in vitro highly favoured the inclusion of a longer exon 5 containing 10 intronic nucleotides relative to skipping of exon 5, whereas both events are detected in vivo (Figs 5 and 1, respectively). However, we show that in vivo inclusion of 10 additional nucleotides upstream of exon 5, which introduces a premature stop codon, triggers the NMD process on the resulting mRNA, whereas the skipped exon 5 mRNA is unaffected by this process since the initial frame is preserved. As a consequence, accumulation of the first mRNA species in vivo is strongly decreased and underestimated relative to the second species. This is in agreement with the fact that total RSK2 transcripts are less abundant in the cell line derived from patient 2 compared with that of a normal individual, as shown in Figure 1. Such a phenomenon was also observed recently for a mutation in the fibrillin-1 gene and the acetylcholine receptor ε subunit gene (38,39).
The fact that the cryptic AG is not preceded by a pyrimidine-rich tract, that use of the cryptic 3′ AG is fully dependent on the natural 3′ AG and that the same branch site is used for splicing of wild-type and mutated transcripts strongly indicates that splicing in the context of the patient 2 mutation occurs through a particular mechanism. According to this mechanism, the natural AG with its polypyrimidine tract is involved in the first catalytic step of splicing, leading to use of the natural branch site, whereas the AG introduced by the mutation is selected only for the second catalytic step. The occurrence of such a mechanism has been evident in only a few natural splicing models. In the human β-globin gene, the mutation β110 creates a new AG 18 nt upstream of the 3′ end of intron 1 which is selected by the splicing machinery according to this mechanism (27). The regulation of Drosophila Sxl exon 3 alternative splicing depends on the immediately upstream intronic region, which contains two 3′ splice site AGs. Each of these AGs has a specific role during each step of splicing (28). Other mutations which create selected cryptic AG pairs upstream of a natural AG have been identified in human genes, but their splicing was not further analysed (31). The mechanisms of selection of an AG site (generally the one which is closest to the branch site), when several AG pairs are in competition, have mainly been defined on chimeric transcripts (40,41). However, how the AG is selected during the second step of splicing is not well known. Available data suggest that various splicing factors, including SLU7 and SPF45, are involved in the selection of constitutive or alternative 3′ splice site AG pairs (42,43).
Analysis of splicing alterations caused by mutations allows consideration of strategies for restoring correct splicing. The pioneer work of Kole and colleagues, who used antisense oligonucleotides to reduce the selection of cryptic splice sites generated by mutations in the β-globin gene, was the first therapeutic approach to genetic diseases caused by aberrant splicing (44,45). For instance, in the case of the β110 mutation in the β-globin gene, efficient reversion of selection of the cryptic AG was obtained in vitro by reducing the accessibility of the natural branch site rather than that of the cryptic AG (44). This strategy could be tested in the context of the mutation of patient 2, although in this case the relative positions of the branch site and the natural and cryptic AG pairs are different.
Another completely different approach, based on the property of SR proteins to activate weak splice sites, has recently been developed to restore in vitro the wild-type splicing pattern of BRCA1 exon 18 and SMN2 exon 7, both of which are skipped due to natural mutations in ESEs controlling their splicing (46). The method involves the use of small chimeric effectors containing a minimal RS domain covalently linked to an antisense nucleic acid moiety that targets the exonic region flanking a defective splice site. Interestingly, this strategy could be used to correct splicing of exon 6 in patient 1. However, this strategy would not be appropriate to correct splicing of exon 5 in patient 2 since SR proteins are apparently not involved in selection of the 3′ AG during the second step of splicing.
Detailed analysis of the consequences of mutations implicated in human disorders on the molecular mechanisms of splicing is important for confirming a clinical diagnosis, proper management of patients and genetic counselling. In some cases, the nucleotide changes detected are not obvious deleterious mutations and without further investigation they could be considered as polymorphisms. Such appreciation errors can have dramatic outcomes. We here report two such mutations that result in severe forms of CLS. More importantly, we suggest a mechanism by which these aberrant splicing events occur. In view of our results, more attention should be given to unusual nucleotide changes where molecular diagnosis of genetic diseases is concerned.
Acknowledgments
ACKNOWLEDGEMENTS
We thank L.Kister for excellent technical assistance and the Institut de Génétique et de Biologie Moléculaire et Cellulaire (IGBMC) core facilities for providing materials and technical assistance. This work was supported by grants from the Centre National de la Recherche Scientifique (CNRS), the Institut National de la Santé et de la Recherche Médicale (INSERM), the Hôpital Universitaire de Strasbourg and the Association Française pour la Recherche contre le Cancer (ARC).
REFERENCES
- 1.Coffin G.S., Siris,E. and Wegienka,L.C. (1966) Mental retardation with osteocartilaginous anomalies. Am. J. Dis. Child., 112, 205–213. [Google Scholar]
- 2.Lowry B., Miller,J.R. and Fraser,F.C. (1971) A new dominant gene mental retardation syndrome. Association with small stature, tapering fingers, characteristic facies and possible hydrocephalus. Am. J. Dis. Child., 121, 496–500. [PubMed] [Google Scholar]
- 3.Temtamy S.A., Miller,J.D. and Hussels-Maumenee,I. (1975) The Coffin–Lowry syndrome: an inherited faciodigital mental retardation syndrome. J. Pediatr., 86, 724–731. [DOI] [PubMed] [Google Scholar]
- 4.Plomp A.S., De Die-Smulders,C.E., Meinecke,P., Ypma-Verhulst,J.M., Lissone,D.A. and Fryns,J.P. (1995) Coffin-Lowry syndrome: clinical aspects at different ages and symptoms in female carriers. Genet. Couns., 6, 259–268. [PubMed] [Google Scholar]
- 5.Trivier E., De Cesare,D., Jacquot,S., Pannetier,S., Zackai,E., Young,I., Mandel,J.L., Sassone-Corsi,P. and Hanauer,A. (1996) Mutations in the kinase Rsk-2 associated with Coffin-Lowry syndrome. Nature, 384, 567–570. [DOI] [PubMed] [Google Scholar]
- 6.Delaunoy J., Abidi,F., Zeniou,M., Jacquot,S., Merienne,K., Pannetier,S., Schmitt,M., Schwartz,C. and Hanauer,A. (2001) Mutations in the X-linked RSK2 gene (RPS6KA3) in patients with Coffin-Lowry syndrome. Hum. Mutat., 17, 103–116. [DOI] [PubMed] [Google Scholar]
- 7.Jacquot S., Zeniou,M., Touraine,R. and Hanauer,A. (2002) X-linked Coffin-Lowry syndrome (CLS, MIM 303600, RPS6KA3 gene, protein product known under various names: pp90(rsk2), RSK2, ISPK, MAPKAP1). Eur. J. Hum. Genet., 10, 2–5. [DOI] [PubMed] [Google Scholar]
- 8.Zeniou M., Pannetier,S., Fryns,J.P. and Hanauer,A. (2002) Unusual splice-site mutations in the RSK2 gene and suggestion of genetic heterogeneity in Coffin-Lowry syndrome. Am. J. Hum. Genet., 70, 1421–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kramer A. (1996) The structure and function of proteins involved in mammalian pre-mRNA splicing. Annu. Rev. Biochem., 65, 367–409. [DOI] [PubMed] [Google Scholar]
- 10.Burge C.B., Tuschl,T., Sharp,P. A (1999) Splicing of precursors to messenger RNAs by the spliceosome. In Gesteland,R.F., Cech,T.R. and Atkins,J.F. (eds), The RNA World, 2nd edn. Cold Sping Harbor Laboratory Press, Cold Sping Harbor, NY, pp. 525–560. [Google Scholar]
- 11.Black D.L. (2003) Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem., 72, 291–336. [DOI] [PubMed] [Google Scholar]
- 12.Graveley B.R. (2000) Sorting out the complexity of SR protein functions. RNA, 6, 1197–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xiao J.H., Davidson,I., Matthes,H., Garnier,J.M. and Chambon,P. (1991) Cloning, expression and transcriptional properties of the human enhancer factor TEF-1. Cell, 65, 551–568. [DOI] [PubMed] [Google Scholar]
- 14.Sureau A., Gattoni,R., Dooghe,Y., Stevenin,J. and Soret,J. (2001) SC35 autoregulates its expression by promoting splicing events that destabilize its mRNAs. EMBO J., 20, 1785–1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gattoni R., Chebli,K., Himmelspach,M. and Stevenin,J. (1991) Modulation of alternative splicing of adenoviral E1A transcripts: factors involved in the early-to-late transition. Genes Dev., 5, 1847–1858. [DOI] [PubMed] [Google Scholar]
- 16.Bourgeois C.F., Popielarz,M., Hildwein,G. and Stevenin,J. (1999) Identification of a bidirectional splicing enhancer: differential involvement of SR proteins in 5′ or 3′ splice site activation. Mol. Cell. Biol., 19, 7347–7356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ruskin B. and Green,M.R. (1985) An RNA processing activity that debranches RNA lariats. Science, 229, 135–140. [DOI] [PubMed] [Google Scholar]
- 18.Hilleren P. and Parker,R. (1999) Mechanisms of mRNA surveillance in eukaryotes. Annu. Rev. Genet., 33, 229–260. [DOI] [PubMed] [Google Scholar]
- 19.Maquat L.E. and Carmichael,G.G. (2001) Quality control of mRNA function. Cell, 104, 173–176. [DOI] [PubMed] [Google Scholar]
- 20.Carter M.S., Doskow,J., Morris,P., Li,S., Nhim,R.P., Sandstedt,S. and Wilkinson,M.F. (1995) A regulatory mechanism that detects premature nonsense codons in T-cell receptor transcripts in vivo is reversed by protein synthesis inhibitors in vitro. J. Biol. Chem., 270, 28995–29003. [DOI] [PubMed] [Google Scholar]
- 21.Hofmann Y., Lorson,C.L., Stamm,S., Androphy,E.J. and Wirth,B. (2000) Htra2-beta 1 stimulates an exonic splicing enhancer and can restore full-length SMN expression to survival motor neuron 2 (SMN2). Proc. Natl Acad. Sci. USA, 97, 9618–9623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Faustino N.A. and Cooper,T.A. (2003) Pre-mRNA splicing and human disease. Genes Dev., 17, 419–437. [DOI] [PubMed] [Google Scholar]
- 23.Cartegni L. and Krainer,A.R. (2002) Disruption of an SF2/ASF-dependent exonic splicing enhancer in SMN2 causes spinal muscular atrophy in the absence of SMN1. Nature Genet., 30, 377–384. [DOI] [PubMed] [Google Scholar]
- 24.Liu H.X., Cartegni,L., Zhang,M.Q. and Krainer,A.R. (2001) A mechanism for exon skipping caused by nonsense or missense mutations in BRCA1 and other genes. Nature Genet., 27, 55–58. [DOI] [PubMed] [Google Scholar]
- 25.Manley J.L. and Tacke,R. (1996) SR proteins and splicing control. Genes Dev., 10, 1569–1579. [DOI] [PubMed] [Google Scholar]
- 26.Lejeune F., Cavaloc,Y. and Stevenin,J. (2001) Alternative splicing of intron 3 of the serine/arginine-rich protein 9G8 gene. Identification of flanking exonic splicing enhancers and involvement of 9G8 as a trans-acting factor. J. Biol. Chem., 276, 7850–7858. [DOI] [PubMed] [Google Scholar]
- 27.Zhuang Y. and Weiner,A.M. (1990) The conserved dinucleotide AG of the 3′ splice site may be recognized twice during in vitro splicing of mammalian mRNA precursors. Gene, 90, 263–269. [DOI] [PubMed] [Google Scholar]
- 28.Penalva L.O., Lallena,M.J. and Valcarcel,J. (2001) Switch in 3′ splice site recognition between exon definition and splicing catalysis is important for sex-lethal autoregulation. Mol. Cell. Biol., 21, 1986–1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chebli K., Gattoni,R., Schmitt,P., Hildwein,G. and Stevenin,J. (1989) The 216-nucleotide intron of the E1A pre-mRNA contains a hairpin structure that permits utilization of unusually distant branch acceptors. Mol. Cell. Biol., 9, 4852–4861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cote J. and Chabot,B. (1997) Natural base-pairing interactions between 5′ splice site and branch site sequences affect mammalian 5′ splice site selection. RNA, 3, 1248–1261. [PMC free article] [PubMed] [Google Scholar]
- 31.Krawczak M., Reiss,J. and Cooper,D.N. (1992) The mutational spectrum of single base-pair substitutions in mRNA splice junctions of human genes: causes and consequences. Hum. Genet., 90, 41–54. [DOI] [PubMed] [Google Scholar]
- 32.Ohno K., Brengman,J.M., Felice,K.J., Cornblath,D.R. and Engel,A.G. (1999) Congenital end-plate acetylcholinesterase deficiency caused by a nonsense mutation and an A→G splice-donor-site mutation at position +3 of the collagenlike-tail-subunit gene (COLQ): how does G at position +3 result in aberrant splicing? Am. J. Hum. Genet., 65, 635–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roca X., Sachidanandam,R. and Krainer,A.R. (2003) Intrinsic differences between authentic and cryptic 5′ splice sites. Nucleic Acids Res., 31, 6321–6333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mayeda A., Screaton,G.R., Chandler,S.D., Fu,X.D. and Krainer,A.R. (1999) Substrate specificities of SR proteins in constitutive splicing are determined by their RNA recognition motifs and composite pre-mRNA exonic elements. Mol. Cell. Biol., 19, 1853–1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schaal T.D. and Maniatis,T. (1999) Multiple distinct splicing enhancers in the protein-coding sequences of a constitutively spliced pre-mRNA. Mol. Cell. Biol., 19, 261–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee B., Vitale,E., Superti-Furga,A., Steinmann,B. and Ramirez,F. (1991) G to T transversion at position +5 of a splice donor site causes skipping of the preceding exon in the type III procollagen transcripts of a patient with Ehlers-Danlos syndrome type IV. J. Biol. Chem., 266, 5256–5259. [PubMed] [Google Scholar]
- 37.Gemignani F., Sazani,P., Morcos,P. and Kole,R. (2002) Temperature-dependent splicing of β-globin pre-mRNA. Nucleic Acids Res., 30, 4592–4598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Caputi M., Kendzior,R.J.,Jr and Beemon,K.L. (2002) A nonsense mutation in the fibrillin-1 gene of a Marfan syndrome patient induces NMD and disrupts an exonic splicing enhancer. Genes Dev., 16, 1754–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ohno K., Milone,M., Shen,X.M. and Engel,A.G. (2003) A frameshifting mutation in CHRNE unmasks skipping of the preceding exon. Hum. Mol. Genet., 12, 3055–3066. [DOI] [PubMed] [Google Scholar]
- 40.Smith C.W., Chu,T.T. and Nadal-Ginard,B. (1993) Scanning and competition between AGs are involved in 3′ splice site selection in mammalian introns. Mol. Cell. Biol., 13, 4939–4952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chua K. and Reed,R. (2001) An upstream AG determines whether a downstream AG is selected during catalytic step II of splicing. Mol. Cell. Biol., 21, 1509–1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chua K. and Reed,R. (1999) The RNA splicing factor hSlu7 is required for correct 3′ splice-site choice. Nature, 402, 207–210. [DOI] [PubMed] [Google Scholar]
- 43.Lallena M.J., Chalmers,K.J., Llamazares,S., Lamond,A.I. and Valcarcel,J. (2002) Splicing regulation at the second catalytic step by Sex-lethal involves 3′ splice site recognition by SPF45. Cell, 109, 285–296. [DOI] [PubMed] [Google Scholar]
- 44.Dominski Z. and Kole,R. (1993) Restoration of correct splicing in thalassemic pre-mRNA by antisense oligonucleotides. Proc. Natl Acad. Sci. USA, 90, 8673–8677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sierakowska H., Sambade,M.J., Agrawal,S. and Kole,R. (1996) Repair of thalassemic human beta-globin mRNA in mammalian cells by antisense oligonucleotides. Proc. Natl Acad. Sci. USA, 93, 12840–12844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cartegni L. and Krainer,A.R. (2003) Correction of disease-associated exon skipping by synthetic exon-specific activators. Nature Struct. Biol., 10, 120–125. [DOI] [PubMed] [Google Scholar]