Lissencephaly-1 promotes the interaction of dynein with dynactin and facilitates motor complex association with mRNA cargos.
Abstract
Microtubule-based transport mediates the sorting and dispersal of many cellular components and pathogens. However, the mechanisms by which motor complexes are recruited to and regulated on different cargos remain poorly understood. Here we describe a large-scale biochemical screen for novel factors associated with RNA localization signals mediating minus end–directed mRNA transport during Drosophila development. We identified the protein Lissencephaly-1 (Lis1) and found that minus-end travel distances of localizing transcripts are dramatically reduced in lis1 mutant embryos. Surprisingly, given its well-documented role in regulating dynein mechanochemistry, we uncovered an important requirement for Lis1 in promoting the recruitment of dynein and its accessory complex dynactin to RNA localization complexes. Furthermore, we provide evidence that Lis1 levels regulate the overall association of dynein with dynactin. Our data therefore reveal a critical role for Lis1 within the mRNA localization machinery and suggest a model in which Lis1 facilitates motor complex association with cargos by promoting the interaction of dynein with dynactin.
Introduction
Transport by cytoskeletal motors is critical for the subcellular localization of many cellular constituents and pathogens. Structural and single-molecule studies have provided remarkable insights into the mechanochemical properties of different motor proteins. However, much less is known about the mechanisms underlying motor recruitment to specific cargos and how the transport of cargo–motor complexes is regulated in vivo.
mRNAs are one important cargo for cytoskeletal motors, with transport of these macromolecules dictating the site of synthesis and action of proteins in many cell types (Holt and Bullock, 2009; Martin and Ephrussi, 2009). The microtubule (MT)-based transport of mRNAs to the apical cytoplasm of the Drosophila syncytial blastoderm embryo is a valuable system for elucidating mechanisms of mRNA transport. This is largely because gene perturbation can be combined with imaging of microinjected, fluorescent mRNAs moving on a MT cytoskeleton that is highly polarized, with minus-ends nucleated apically and plus-ends extending basally (Wilkie and Davis, 2001; Bullock et al., 2006). Injection experiments have revealed that all mRNA species move bidirectionally, with individual ribonucleoprotein particles (RNPs) exhibiting frequent reversals in their direction of movement (Bullock et al., 2006; Vendra et al., 2007). Uniformly distributed mRNAs undergo bidirectional transport with little net bias, presumably facilitating their dispersal in the crowded cytoplasm (Bullock et al., 2006). In contrast, mRNAs that accumulate in the apical cytoplasm contain RNA elements, called “localization signals,” which strongly increase the probability of bidirectional complexes initiating and maintaining minus end–directed runs (Bullock et al., 2006). These runs are driven by the cytoplasmic dynein motor (Wilkie and Davis, 2001; Bullock et al., 2006), a large complex containing a heavy chain with force-generating ATPase activity and intermediate, light intermediate, and light chains that have roles in cargo recruitment (Vallee et al., 2012).
RNA localization signals are specialized stem–loop structures of ∼40–60 nucleotides (nts) that are bound by the noncanonical RNA-binding protein Egalitarian (Egl; Dienstbier et al., 2009). Egl also binds Bicaudal-D (BicD; Dienstbier et al., 2009), a protein with roles in the transport of a subset of dynein’s cargos (Dienstbier and Li, 2009). Both Egl and BicD physically interact with components of the dynein complex and/or its accessory complex dynactin (Hoogenraad et al., 2001; Navarro et al., 2004; Splinter et al., 2012). Dynactin participates in most cellular transport events mediated by dynein and has been implicated in increasing the processivity of the motor and mediating its recruitment to cargos (King and Schroer, 2000; Ross et al., 2006; Kardon et al., 2009; Schroer and Cheong, 2012). The functional role of dynactin in promoting apical mRNA transport in the Drosophila embryo has been confirmed through analysis of a dominant mutation in the p150Glued subunit of the complex (Vendra et al., 2007).
Recent in vitro experiments have provided evidence that nonlocalizing RNAs also associate with dynein–dynactin and that localization signals, through Egl and BicD, increase the probability of persistent minus end–directed motion by recruiting additional copies of dynein–dynactin complexes to individual RNPs (Amrute-Nayak and Bullock, 2012). However, it remains unclear whether localization signals recruit other proteins that further bias transport in the minus-end direction by regulating the mechanochemical properties of individual motors. The mechanistic details of many other steps in the RNA localization process also remain poorly defined, including how the assembly of dynein and dynactin on RNPs is controlled.
In this study we sought to better understand the mechanisms of mRNA transport by screening for additional factors recruited to RNA localization signals and characterizing their functions. This led to the identification of the highly conserved Lissencephaly-1 (Lis1) protein as a novel component of RNA–motor complexes. Haploinsufficiency for loss-of-function mutations in human LIS1 causes type 1 lissencephaly (Reiner et al., 1993), a brain disorder associated with aberrant neuronal migration during development (Hirotsune et al., 1998; Cahana et al., 2001; Tsai et al., 2005).
Lis1 functions in many dynein-dependent processes, including nucleokinesis (Xiang et al., 1995; Lei and Warrior, 2000; Lee et al., 2003; Tsai et al., 2005), centrosome separation (Cockell et al., 2004; Siller et al., 2005), nucleus–centrosome coupling (Shu et al., 2004; Tanaka et al., 2004; Rehberg et al., 2005), mitotic chromosome congression (Faulkner et al., 2000), mitotic spindle orientation (Faulkner et al., 2000; Siller and Doe, 2008; Yingling et al., 2008), and the transport of checkpoint proteins from the mitotic kinetochore (Siller et al., 2005). However, there are conflicting reports on the importance of Lis1 in the transport of endosomes and lysosomes, as well as in Golgi positioning (Faulkner et al., 2000; Smith et al., 2000; Rehberg et al., 2005; Lenz et al., 2006; Lam et al., 2010; Zhang et al., 2010; Pandey and Smith, 2011; Yi et al., 2011; Egan et al., 2012). Indeed, whether Lis1 regulates most dynein-dependent transport processes or participates in a specialized set of motor machinery functions, such as high-load transport events (McKenney et al., 2010; Yi et al., 2011), is still under debate.
Lis1 binds to the motor domain of Dynein heavy chain (Dhc; Sasaki et al., 2000; Tai et al., 2002; McKenney et al., 2010), with a recent low-resolution structure indicating an interaction site between the MT-binding domain and the AAA3/4 domains (Huang et al., 2012). In vitro experiments using purified proteins have highlighted the ability of Lis1 to regulate mechanochemical aspects of dynein function, including the capacity to enhance the interaction of the motor domain with MTs (McKenney et al., 2010; Huang et al., 2012; Vallee et al., 2012). However, it is not known to what extent this function accounts for Lis1’s regulation of dynein-mediated cargo transport in vivo.
In this study we reveal an important functional requirement for Lis1 in augmenting minus end–directed travel distances of localizing RNAs and find that this protein promotes the recruitment of dynein and dynactin complexes to RNPs. Furthermore, we provide evidence that Lis1 enhances the overall association of the dynein complex with dynactin. These data lead us to propose a model in which Lis1 promotes the association of dynein with cargos by regulating the binding of the motor complex to dynactin.
Results
Lis1 and CLIP-190 are specifically recruited to RNA localization signals
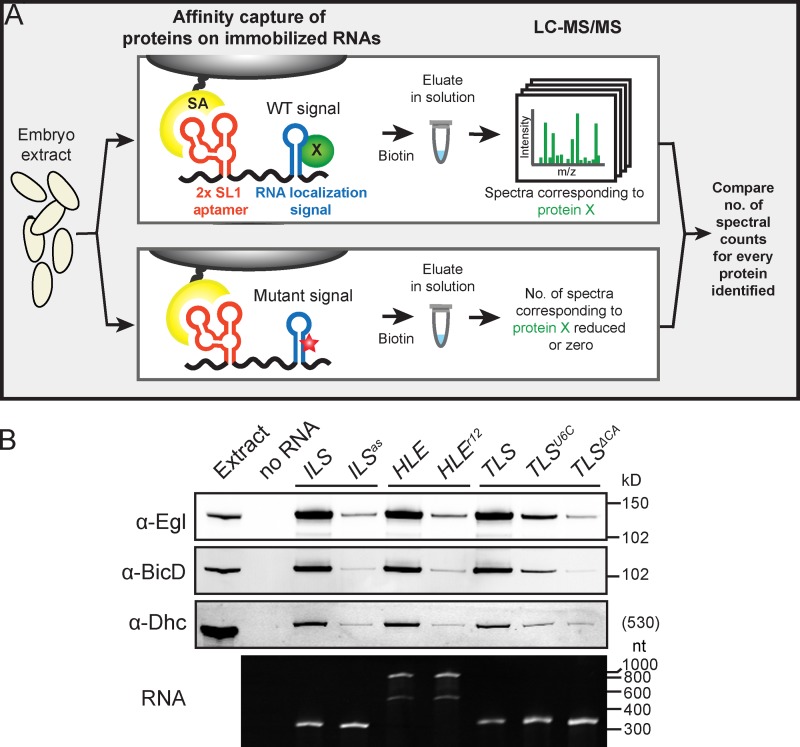
To identify novel components of mRNA localization machinery we developed methodology for assembling transport complexes on RNA localization signals and determining their compositions by mass spectrometry (MS; Fig. 1 A and Materials and methods). Drosophila embryo extract was incubated with in vitro–transcribed RNAs containing localization signals fused to streptavidin-binding RNA aptamers, which had been immobilized on streptavidin-coupled beads. Assembled RNPs were then washed briefly and eluted using biotin, which competes for the interaction of streptavidin with the aptamer. This methodology was adapted from a study analyzing the interactions of Egl and BicD with RNA signals using more stringent conditions that precluded the stable association of other components of the localization machinery, including dynein (Dienstbier et al., 2009).
Figure 1.
A biochemical screen for novel components of mRNA transport complexes. (A) Cartoon depicting methodology used to identify novel components of mRNA transport complexes. For simplicity only one of the two streptavidin-binding aptamers in the in vitro transcribed RNA is shown. SA, streptavidin; red star, mutation in localization signal; green circle, protein enriched on WT signal. (B) Top panels, immunoblots showing that Egl, BicD, and Dhc are strongly enriched on the ILS, HLE, and TLS signals compared with mutated nonlocalizing equivalents (ILSas, HLEr12, TLSU6C, and TLSΔCA). Bottom panel, ethidium bromide–stained TBE-urea gel showing equivalent amounts of WT and mutant RNAs after coupling to beads, washing, and elution under the same conditions as the pull-downs from extract. Note the two bands of aptamer-HLE RNA and its mutant equivalent, which have sizes consistent with monomeric and dimeric forms (see Materials and methods).
We used three RNA localization signals necessary and sufficient for Egl–BicD–dynein-dependent transport of associated transcripts in the Drosophila blastoderm embryo or during oogenesis (Fig. S1, A–C). These were the single stem–loop signals ILS (Van De Bor et al., 2005) and TLS (Serano and Cohen, 1995; Bullock and Ish-Horowicz, 2001) from the retrotransposon RNA I factor and fs(1)K10 (K10) mRNA, respectively, and the double stem–loop signal HLE from the pair-rule transcript hairy (h) (Bullock et al., 2003). As specificity controls we used subtly mutated versions of each signal that abolish apical localization activity but are predicted to retain stem–loop structures (Fig. S1, A–C) (Dienstbier et al., 2009).
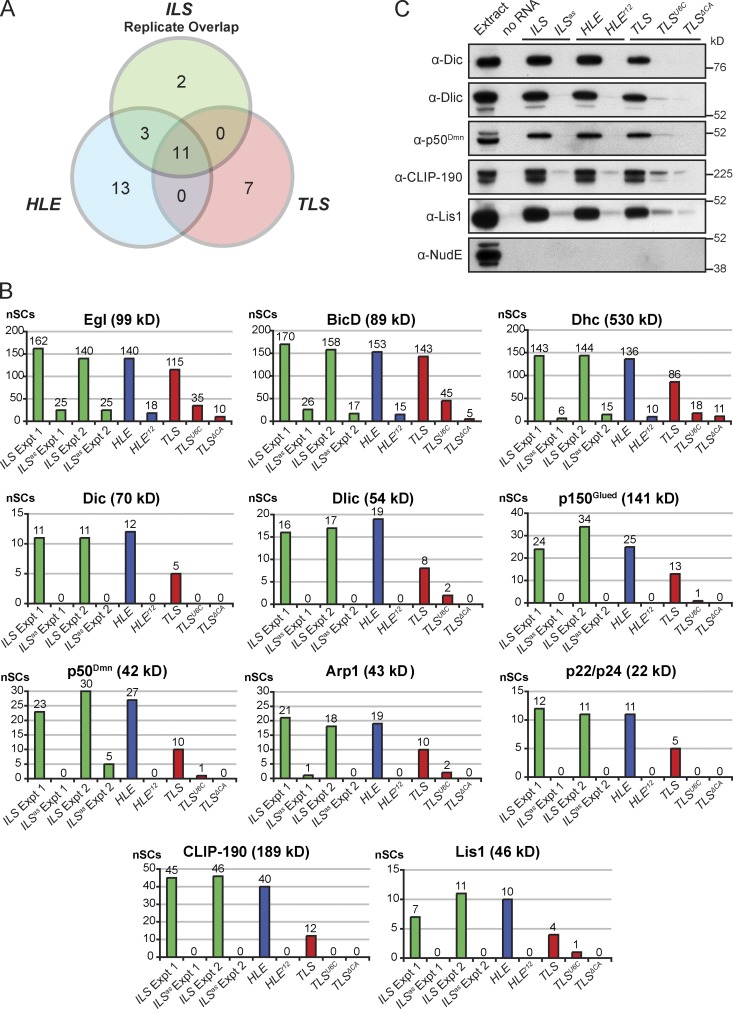
Using the modified assay conditions we detected enrichment of not only Egl and BicD, but also Dhc, on all WT localization signals tested, as assessed by both immunoblotting (Fig. 1 B) and silver staining (Fig. S1 D). The presence of Dhc demonstrated that RNA–motor complexes were assembled on at least a subset of localization signals. Due to the low stringency conditions used there was a large number of proteins in each experiment that bound to a similar degree to wild-type (WT) and mutant signals (Fig. S1 D). To enable identification of other proteins enriched on localization signals we performed liquid chromatography–mass spectrometry/mass spectrometry (LC-MS/MS) directly on eluates from the pull-downs and used spectral counting (Liu et al., 2004; Zhu et al., 2010) to compare the relative abundance of proteins between paired WT and mutant samples generated in parallel (Table S3 and Fig. 2 A).
Figure 2.
Identification of known and novel components of mRNA transport complexes by mass spectrometry. (A) Venn diagram showing the number of proteins classed as enriched on the ILS, HLE, and TLS localization signals by MS/MS using our selection criteria (>4 normalized spectral counts [nSCs] present on WT signal and >80% of the total normalized spectral counts for a WT and paired mutant signal present on the WT signal; nSCs are spectral counts normalized to correct for differences in the total number of spectra between individual MS runs; see Materials and methods and Table S3 legend for details). Proteins were only classed as enriched on the ILS if they fulfilled the selection criteria in the two independent experiments. Proteins classed as enriched on the TLS in A are from the comparison of the WT element to the mutant ΔCA, which has a stronger inhibitory effect than the U6C mutant on recruitment of known components of the localization machinery (B). (B) nSCs observed for the 11 proteins enriched on all three localization signals tested. The nSCs for the two biological replicates of ILS vs. ILSas experiments are shown separately for comparison, as are the data for the TLSU6C mutant. Note that differences in nSCs between different proteins are not a good reflection of abundance, as these values are heavily influenced by the protein’s molecular mass and how well individual peptides are detected by MS. (C) Immunoblot showing the enrichment of Dic, Dlic, p50Dmn, CLIP-190, and Lis1 on active localization signals. The Lis1-interacting protein NudE is not detectable on any signal.
Consistent with the immunoblotting results, spectral counting revealed clear enrichment of Egl, BicD, and Dhc on all three elements tested in all biological and technical replicates (Fig. 2 B). This was also the case for other dynein–dynactin subunits, namely Dynein intermediate chain (Dic), Dynein light intermediate chain (Dlic), and the dynactin components p150Glued, p50Dmn (Dynamitin), Actin-related protein 1 (Arp1), and p22/24 (Fig. 2 B). The availability of specific antibodies allowed us to confirm the enrichment of Dic, Dlic, and p50Dmn on all three localization signals compared with their mutant equivalents (Fig. 2 C). We also detected two other dynein or dynactin subunits (Dynein light chain LC7/Roadblock and the p62 subunit of dynactin) enriched on a subset of localization signals by MS (Table S1). These proteins may be ubiquitous components of transport complexes not detected in every MS experiment for technical reasons (Table S1, footnotes). Collectively, these results reveal that our experimental pipeline is able to detect components of RNA–motor complexes, and shed light on at least some of the dynein–dynactin subunits recruited to RNPs.
We detected a number of other proteins enriched on a subset of localization signals in the MS experiments (Fig. 2 A; Table S1) that are candidates for future investigations. In the current study we focused on the two factors that were detectably enriched on all WT localization signals in all biological and technical replicates and were thus strong candidates to be novel components of RNA–motor complexes. These proteins were cytoplasmic linker protein 190 (CLIP-190; called CLIP-170 in vertebrates) and Lis1 (Fig. 2 B). The specific enrichment of these factors on all three WT localization signals was confirmed by immunoblotting (Fig. 2 C). Intriguingly, we did not detect the single Drosophila orthologue of the Lis1-binding protein NudE (Vallee et al., 2012) on localization signals by either MS (Table S3) or immunoblotting (Fig. 2 C). NudE is therefore unlikely to be a component of apical RNA transport complexes.
CLIP-190 is dispensable for apical mRNA transport in the Drosophila embryo
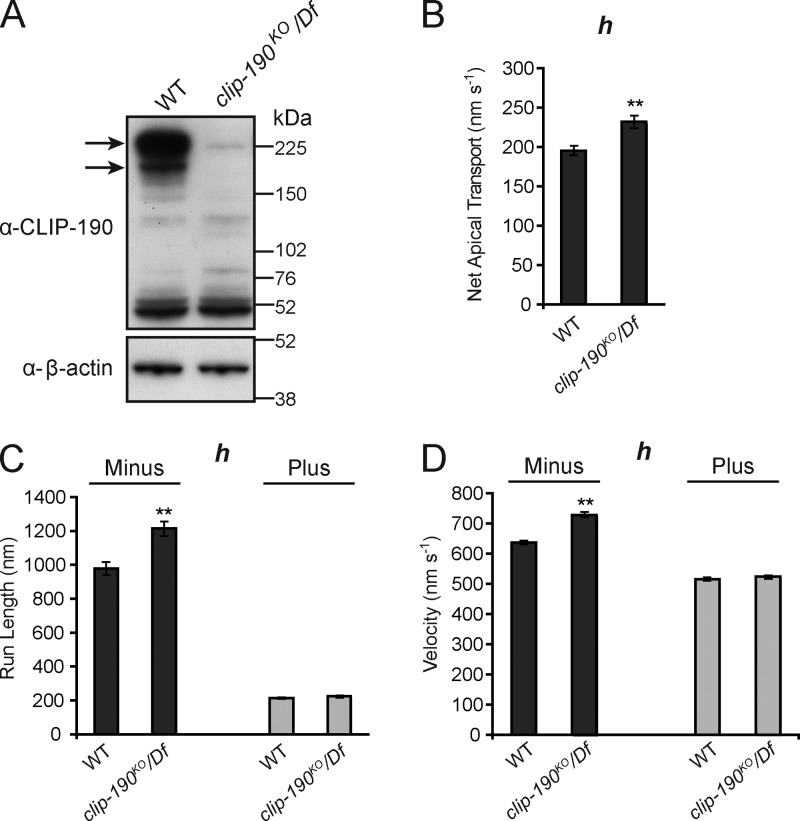
CLIP-190/170 is a component of MT plus-end tracking complexes (Akhmanova and Steinmetz, 2008) and has close links with the dynein–dynactin pathway at both MT plus ends and at kinetochores (Dujardin et al., 1998; Coquelle et al., 2002; Tai et al., 2002; Lansbergen et al., 2004; Dzhindzhev et al., 2005). The strong and reproducible enrichment of CLIP-190 on RNA localization signals suggested that it might participate in minus end–directed cargo transport in vivo. To test this hypothesis we used homologous recombination in Drosophila to generate a null allele of the clip-190 gene (clip-190KO; Fig. 3 A). Surprisingly, clip-190–null flies were viable. Microinjection of fluorescently labeled h mRNA, followed by quantification of motile RNP properties using automatic tracking software (Bullock et al., 2006), revealed no gross change in RNA transport in clip-190 mutant embryos (Video 1; Fig. 3, B–D; and Table S2). We did observe a modest but statistically significant increase in the rate of net apical transport of h in clip-190 mutant embryos compared with WT (Fig. 3 B), which was associated with a small increase in the velocity and length of minus end–directed runs (Fig. 3, C and D). Overall, however, we can conclude that CLIP-190 function is not strongly required for apical mRNA transport in embryos. We also did not observe defects in the apical anchorage of RNPs (a process that is also dependent on dynein; Delanoue and Davis, 2005) in clip-190 mutants (Videos 1 and 2). Long-term studies will be required to determine whether CLIP-190 acts redundantly with other components of mRNA transport or anchorage complexes.
Figure 3.
CLIP-190 is not required for apical mRNA transport in embryos. (A) Immunoblot confirming loss of CLIP-190 protein (arrows) in clip-190 mutant ovary extracts. The clip190KO allele was generated by homologous recombination; Df is the genomic deficiency Df(2L)BSC294, which removes the clip-190 locus. (B–D) Motile properties of particles of the localizing h RNA after injection into embryos of the indicated maternal genotypes. See Table S2 for full details of motile properties and number of particle tracks analyzed. **, P < 0.01 (ANOVA test). Error bars represent SEM.
Lis1 is strongly required for net minus end–directed transport of apically localized transcripts
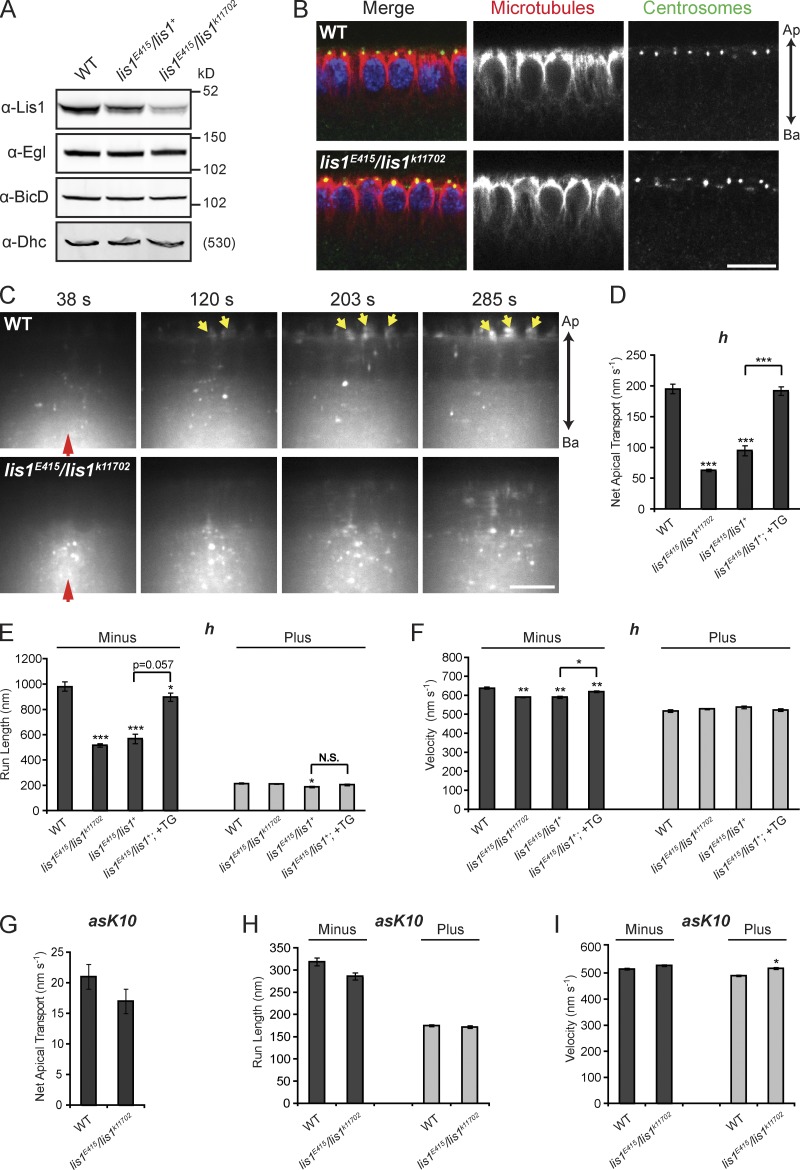
It is currently unclear whether Lis1 participates in the transport of most, or only a subset, of dynein’s cargos (see Introduction). Defects in the positioning of axis-determining mRNAs during Drosophila mid-oogenesis were previously observed in strong hypomorphic lis1 mutants (Swan et al., 1999; Lei and Warrior, 2000). However, abnormalities in MT organization and nuclear positioning in oocytes of these genotypes have precluded assessment of a direct role of Lis1 in RNA transport. Our finding that Lis1 is recruited to RNA signals suggests that it could dictate the motile properties of RNPs directly. To investigate this possibility we identified a weaker maternal lis1 mutant genotype (trans-heterozygous for the strong hypomorphic P-element allele lis1E415 [Swan et al., 1999] and a weaker P-element allele lis1k11702 [Lei and Warrior, 2000]) that has ∼25% of WT Lis1 protein levels (Fig. 4 A and Fig. S2 A). The vast majority of blastoderm embryos of this genotype had no overt morphological abnormalities. Furthermore, immunostaining did not reveal detectable defects in centrosome positioning or the integrity of the MT cytoskeleton in the mutant embryos (Fig. 4 B). The orientation of MTs in WT and lis1 mutant embryos was also indistinguishable (i.e., with the vast majority of plus ends extending basally), as assessed by live imaging of a marker of growing plus ends, EB1-GFP (Video 3). Consistent with the normal MT orientation, basal enrichment of lipid droplets—a process mediated by net plus end–directed transport by the kinesin-1 motor (Shubeita et al., 2008)—was observed in lis1 mutant embryos (Fig. S2 B).
Figure 4.
Net minus end–directed transport of apically localizing transcripts requires Lis1. (A) Fluorescent immunoblot showing equivalent levels of Egl, BicD, and Dhc between WT and lis1 mutant ovaries. Comparison of the fluorescence intensity in Lis1 blots reveals that Lis1 levels in the heterozygous and trans-heterozygous genotype are ∼65% and ∼25% of WT, respectively (for quantification see Fig. S2 A). (B) Immunostaining of MTs (α-tubulin, red) and centrosomes (α-centrosomin, green) in cycle 14 blastoderm embryos from WT and lis1E415/lis1k11702 mutant mothers, revealing that centrosome position and MT organization and length are not perturbed by reduced Lis1 levels. Nuclei, blue (DAPI). Ap, apical; Ba, basal. (C) Still images from time-lapse movies of injected Alexa Fluor 488–labeled h RNA, revealing a strong defect in apical transport in embryos from lis1E415/lis1k11702 mothers compared with WT (see corresponding Video 4). Red and yellow arrows mark injection site and apically localized RNA, respectively. Time after injection is shown. (D–I) Motile properties of h (D–F) and asK10 (G–I) RNA particles after injection into the stated maternal genotypes. In D–F, +TG indicates the presence of two copies of a weakly expressed lis1 transgene. See Table S2 for full details of motile properties and number of particle tracks analyzed. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (ANOVA test). Error bars represent SEM. Bars: (B and C) 10 µm.
We observed a striking reduction in the rate of apical accumulation of injected h RNA in lis1E415/lis1k11702 embryos compared with WT (Fig. 4 C; Video 4). Levels of Egl, BicD, and dynein–dynactin subunits were not reduced in lis1 mutants (Fig. 4 A; see also Figs. 6 and 7), indicating that defective mRNA motility is not due to diminished levels of these transport components. Automatic tracking revealed a greater than threefold decrease in the rate of net apical transport of h RNA in lis1E415/lis1k11702 embryos (Fig. 4 D). The mutants exhibited a strong decrease in minus-end run lengths (Fig. 4 E), a substantial increase in the frequency of both pauses and plus-end transport (Table S2), and a modest but statistically significant decrease in the velocity of minus end–directed runs (Fig. 4 F). Very similar defects in RNP motility were seen for K10 RNA injected into lis1E415/lis1k11702 embryos (Fig. S2, C–E), revealing that the role of Lis1 in apical mRNA transport is not transcript specific.
Figure 6.
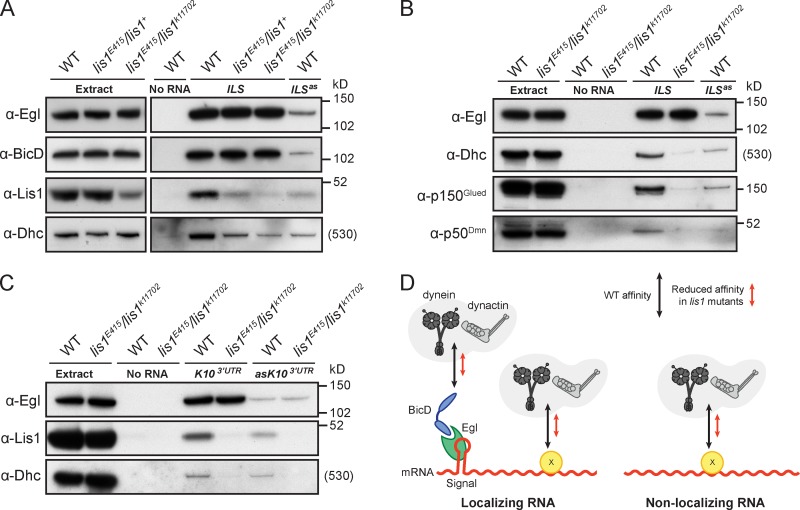
Lis1 promotes the recruitment of dynein and dynactin to RNAs. (A) Immunoblots showing that the recruitment of Dhc, but not Egl and BicD, to the ILS is dramatically reduced when complexes are assembled from lis1 mutant ovary extract using aptamer-based RNA pull-downs. Recruitment of proteins to the ILSas mutant from WT extract is shown as a specificity control. Note that reduced Lis1 levels in the starting extract are not apparent for the heterozygous lis1E415/lis1+ combination due to nonlinear enzyme-coupled chemiluminescent detection (reduction of Lis1 levels in heterozygotes is revealed by nonenzymatic detection with fluorescent antibodies [Figs. 4 A, 7 C, and Fig. S4 D]). (B) Immunoblots showing a reduced association of p150Glued and p50Dmn with the aptamer-associated ILS when complexes are assembled from lis1 mutant ovary extract compared with WT. Blots for Egl and Dhc recruitment to signals are shown as controls. (C) Immunoblots assessing protein recruitment from WT and lis1 mutant ovary extracts to a localizing 800-nt piece of the K10 3′UTR (K103′UTR) and an antisense nonlocalizing mutant version (asK103′UTR). Reduced Lis1 levels have no effect on Egl binding, but clearly decrease the association of Dhc with both RNA species. We confirmed that the association of p150Glued with asK103′UTR was also reduced using the lis1 mutant versus WT extract (Fig. S3 D). (D) Cartoon summarizing the results from A–C and Fig. S3 D. Localizing RNAs recruit dynein–dynactin complexes to sites on localization signals bound by Egl–BicD complexes, and to additional sites on the RNA bound via an unknown protein(s) (factor X), also present on nonlocalizing RNAs (see also Bullock et al., 2006). Reduced Lis1 levels decrease the affinity of dynein and dynactin complexes for both signals and nonsignal sites.
Figure 7.
Lis1 regulates the association of dynein and dynactin components. (A) Immunoblots showing that reduced Lis1 levels do not perturb the composition of dynein complexes immunoprecipitated from ovary extracts using anti-Dic antibodies. Anti-GFP antibodies were used as a control. Note that Lis1 is not detectably associated with dynein complexes immunoprecipitated from lis1E415/lis1k11702 extract, suggesting that the Lis1 concentration in these extracts is limiting for efficient binding to dynein. Although Dlic levels appeared different in WT and lis1 mutant extracts in the experiment shown, this difference did not affect Dlic incorporation into dynein complexes. (B) Immunoblots showing reduced levels of p150Glued precipitated with GFP-Dlic (using GBP pull-downs) in lis1E415/lis1k11702 versus WT ovary extract (see Fig. S4 B for quantification of the reduced signal in multiple trials). (C) Immunoblots showing reduced levels of Dhc and Dic precipitated with GFP-p50Dmn (using GBP pull-downs) in lis1E415/lis1+ versus WT embryo extract (see Fig. S4 C for quantification of the reduced Dhc signal in multiple trials). Fluorescent detection was used for the Lis1 blot in this panel. The lower molecular weight species of GFP-p50Dmn precipitated in this experiment is presumably a degradation product. (D) Approximately fourfold overexpression (OE) of Lis1 (quantification not depicted) increases the amount of Dhc, Dic, and BicD coprecipitated with GFP-p50Dmn from ovary extract. Lis1 was overexpressed by driving UAS-lis1 in the germ line with MTD-GAL4. Note that, unlike in embryo extract (C), association of Lis1 with GFP-p50Dmn is not readily detectable in WT ovary extract (D). This may be due to tissue-specific differences in the abundance, affinities, or stoichiometries of the proteins participating in the interaction.
Similar but less severe defects in h transport were observed in embryos heterozygous for the stronger lis1E415 allele (lis1E415/lis1+; Fig. 4, D–F; Table S2), which have ∼65% of WT Lis1 levels (Fig. 4 A and Fig. S2 A). The RNA transport defects in lis1E415/lis1+ embryos were suppressed by ubiquitous expression of a Lis1 transgene (Fig. 4, D–F; and Fig. S2, F and G; Table S2), confirming that they are due to altered Lis1 levels. Taken together, these data highlight the critical importance of Lis1 function during apical mRNA transport.
We did not observe substantial changes in the run length or velocity of plus end–directed movements of h or K10 RNAs in lis1 mutant embryos compared with WT (Fig. 4, E and F; Fig. S2, D and E; Table S2). This is interesting because inhibition of dynein affects minus end– and plus end–directed transport (Bullock et al., 2006), presumably due to functional interplay between dynein and an unidentified plus end–directed motor. We also found that RNPs that reached the apical cytoplasm were retained there in lis1 mutants (Video 4), indicating that dynein-dependent RNP anchorage is not overtly affected. Thus, either a subset of dynein functions during mRNA transport involves Lis1, or there is differential sensitivity of Lis1/dynein-mediated processes to a partial reduction in Lis1 concentration.
There were also no significant differences in the mean minus- or plus-end run lengths of an injected nonlocalizing mRNA (the antisense version of the K10 transcript lacking the TLS [asK10]) between lis1E415/lis1k11702 and WT embryos (Fig. 4, G–I; and Table S2), even though travel distances of these cargos are strongly reduced in both directions when dynein is inhibited (Bullock et al., 2006). However, there was a modest but statistically significant increase in the frequency of pauses of asK10 in lis1E415/lis1k11702 embryos compared with WT, accompanied by decreased frequencies of minus end– and plus end–directed transport events (Table S2). A subtle but statistically significant increase in the plus-end velocity of asK10 particles was also observed in lis1 mutants (Fig. 4 I), consistent with the modest elevation in plus-end velocities of late endosomes/lysosomes in mammalian COS-7 cells after inhibition of Lis1 (Yi et al., 2011). Nonetheless, we can conclude that the motile properties of nonlocalizing RNAs show a significantly lower sensitivity to reduced Lis1 levels than those of localizing RNAs.
RNP size is not a key determinant of sensitivity of transport to reduced Lis1 levels
Biophysical experiments with purified proteins have shown that Lis1 allows dynein to engage in a persistent force-producing state on MTs (McKenney et al., 2010). This finding has led to the hypothesis that Lis1 functions in a specific subset of dynein transport events in which a high load is imparted on the motor (McKenney et al., 2010). In support of this notion, reducing Lis1 levels/activity in mammalian cells was found to strongly affect the transport of large but not small membranous cargos (Pandey and Smith, 2011; Yi et al., 2011).
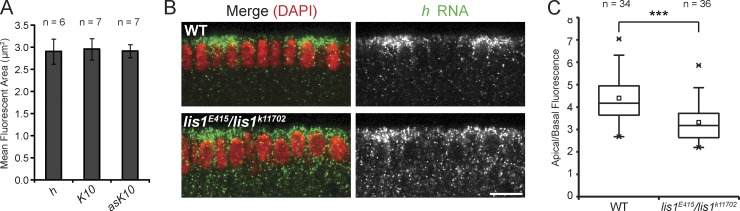
However, cargo size does not appear to dictate sensitivity of RNP transport to reduced Lis1 levels in our assays. Nonlocalizing and localizing RNAs—which are moderately and strongly affected by reduced Lis1 levels, respectively—form RNPs with indistinguishable mean sizes (Fig. 5 A). Furthermore, Lis1’s function in the transport of localizing mRNAs is not restricted to the large RNPs formed after injection, which are likely to contain many RNA molecules (Bullock et al., 2003). Fluorescent in situ hybridization revealed a highly statistically significant reduction in the apical enrichment of endogenous h mRNA in lis1E415/lis1k11702 embryos compared with WT (Fig. 5, B and C), even though this transcript appears to be transported in RNPs containing a single RNA molecule (Amrute-Nayak and Bullock, 2012).
Figure 5.
Size is not the sole factor dictating the sensitivity of RNP transport to reduced Lis1 levels. (A) Cross-sectional fluorescent area of RNPs containing different RNAs. Values shown are means of the mean area of RNPs per WT embryo. In A and C, n is number of embryos analyzed. Note that the values obtained are unlikely to represent the true size of RNPs due to light scattering in the embryo, but allow the relative size of RNPs formed by different species to be evaluated (no overt differences in the size of fluorescent RNPs were observed between WT and lis1 mutant genotypes, e.g., Video 4). (B) Fluorescent, nonenzymatic in situ hybridization demonstrating a reduction in the apical accumulation of endogenous h transcripts (green) in lis1E415/lis1k11702 cycle 14 blastoderm embryos compared with WT. Nuclei, red (DAPI). Note that h is transcribed in stripes. Bar, 10 µm. (C) Box-and-whisker plot revealing a statistically significant decrease in the apical/basal fluorescence of endogenous h signal in lis1 mutant embryos compared with WT. Whiskers mark the first and 95th percentile, box marks 50th percentile, crosses mark the maximum and minimum values, and the hollow square and horizontal line represents the mean and median, respectively. ***, P < 0.001 (unpaired t test).
Lis1 is required for the recruitment of dynein and dynactin, but not Egl and BicD, to RNA localization signals
Next, we further explored the mechanistic basis of Lis1’s function in mRNA transport. Interestingly, inhibition of Lis1 within cells has been reported to reduce the accumulation of dynein at kinetochores (Dzhindzhev et al., 2005; Siller et al., 2005), the nuclear envelope (Cockell et al., 2004; Sitaram et al., 2012; Splinter et al., 2012), spindle poles (Sitaram et al., 2012), and its association with membrane-bound organelles (Lam et al., 2010). Although regulation of dynein mechanochemistry by Lis1 could influence trafficking of the motor to these subcellular sites, it is also possible that Lis1 has a direct role in promoting the binding of dynein to cargos including RNPs. To test this notion we took advantage of our aptamer-based RNA pull-down assay, which reports on the de novo association of motor complexes with RNAs of interest.
We performed a series of experiments comparing the ability of dynein to be recruited to RNA localization signals from WT and lis1 mutant ovary extracts. Although the recruitment of Egl and BicD to ILS and TLS signals was not affected when lis1E415/lis1k11702 extracts were used, Dhc association with both RNAs was dramatically reduced in the lis1 mutant condition compared with the WT (Fig. 6, A and B; and Fig. S3 A). A substantial but less severe reduction in Dhc association with the ILS was also observed when complexes were assembled using extract from the heterozygous genotype lis1E415/lis1+ (Fig. 6 A), consistent with the intermediate mRNA transport phenotype (Fig. 4, D–F). The reduced association of Dhc with RNA from lis1E415/lis1+ extract could be suppressed by a Lis1 transgene, confirming the role of Lis1 in dynein recruitment (Fig. S3 B).
We also found that the association of the dynactin components p150Glued and p50Dmn with the ILS was strongly reduced when complexes were assembled from lis1 mutant extract compared with WT (Fig. 6 B). This finding indicates that reported dynactin–BicD interactions (Hoogenraad et al., 2001; Splinter et al., 2012) are not sufficient for dynactin recruitment to RNA localization complexes in lis1 mutants, consistent with recent evidence from mammalian cells that dynein and dynactin complexes mutually depend on each other for cargo association (Splinter et al., 2012).
Collectively, these data reveal that Lis1 promotes the assembly of dynein–dynactin complexes with RNA localization signals bound by Egl and BicD. Given evidence for the importance of dynein–dynactin copy number on individual RNPs for net minus end–directed motion (Bullock et al., 2006; Amrute-Nayak and Bullock, 2012), such a role may make a substantial contribution to the defective transport of localizing RNAs in lis1 mutant embryos (see Discussion).
Lis1 promotes dynein and dynactin recruitment to nonlocalizing RNAs
The results from our RNP assembly experiments raised the possibility that Lis1 functions as a direct physical bridge between Egl–BicD and dynein–dynactin complexes. However, we found no evidence that this was the case from yeast two-hybrid assays; we did not detect binding of Lis1 to Egl or BicD, whereas the Lis1–NudE interaction was observed (Fig. S3 C).
We next assessed if Lis1’s role in controlling dynein recruitment to cargo is restricted to facilitating interactions with Egl–BicD complexes. We have previously shown that dynein-dependent bidirectional transport of long, nonlocalizing transcripts is largely independent of Egl and BicD activity (Bullock et al., 2006), indicating that other factors mediate motor complex recruitment to these transcripts through “nonsignal” RNA sites. To test if Lis1 plays a role in recruitment of dynein to nonsignal sites, RNA pull-downs were performed from ovary extracts using an aptamer-linked nonlocalizing RNA.
The antisense version of an 800-nt piece of the K10 3′UTR with a mutated TLS (asK103′UTR) was used as a nonlocalizing RNA, with the equivalent sense K10 RNA containing an intact TLS (K103′UTR) selected as a localizing control. As expected, Egl was very strongly enriched on K103′UTR compared with asK103′UTR when WT extracts were used (Fig. 6 C). In contrast, Dhc was only modestly enriched on the localizing RNA relative to the nonlocalizing RNA (Fig. 6 C), consistent with photobleaching analysis of the relative copy number of the motor complex on localizing and nonlocalizing RNPs in vitro (Amrute-Nayak and Bullock, 2012). The nonproportional nature of Egl and Dhc association with localizing and nonlocalizing RNAs in our RNP assembly assay provides further evidence that Egl–BicD complexes are not responsible for binding of the motor to nonsignal sites.
Consistent with previous results using isolated localization signals, the recruitment of Egl to the longer RNAs was not affected by reduced Lis1 levels (Fig. 6 C). In contrast, Dhc association with both localizing and nonlocalizing transcripts was clearly decreased in the lis1 mutant condition compared with WT (Fig. 6 C). We confirmed that the association of the nonlocalizing RNA with the p150Glued subunit of dynactin was also reduced when lis1 mutant extract was used (Fig. S3 D). We therefore conclude that Lis1 increases the affinity of dynein and dynactin for both localization signals bound by Egl and BicD and for nonsignal sites on RNA (Fig. 6 D).
Because visualizing dynein on RNPs in embryos is currently not possible (presumably due to a large excess of unbound motor), we cannot determine to what degree the reduced affinity of the motor for nonsignal sites in lis1 mutants affects binding of the motor to nonlocalizing RNPs in vivo. However, the altered transport properties of nonlocalizing mRNAs in lis1 mutant embryos are compatible with a partial reduction in motor association. Increased pausing of asK10 in lis1 mutants (Table S2) may reflect a reduced likelihood of initiating runs when the average dynein number per RNP is decreased. By lowering the amount of drag on a tightly coupled plus-end motor, reduced dynein number may also be responsible for the increased plus end–directed velocity of this RNA in lis1 mutants (Fig. 4 I and Table S2). Potential reasons for the distinct sensitivities of the transport of localizing and nonlocalizing RNAs to reduced dynein affinity are explored in the discussion.
Lis1 promotes complex formation between dynein and dynactin components
Our observation that Lis1 increases the association of dynein with both localizing and nonlocalizing RNAs suggests that it facilitates the interaction of the motor with distinct molecular adaptors. This, together with Lis1’s implication in the dynein-dependent transport of a wide range of cargos, led us to hypothesize that reduced Lis1 levels have a general effect on the dynein complex or its interactions that attenuates its ability to bind cargos.
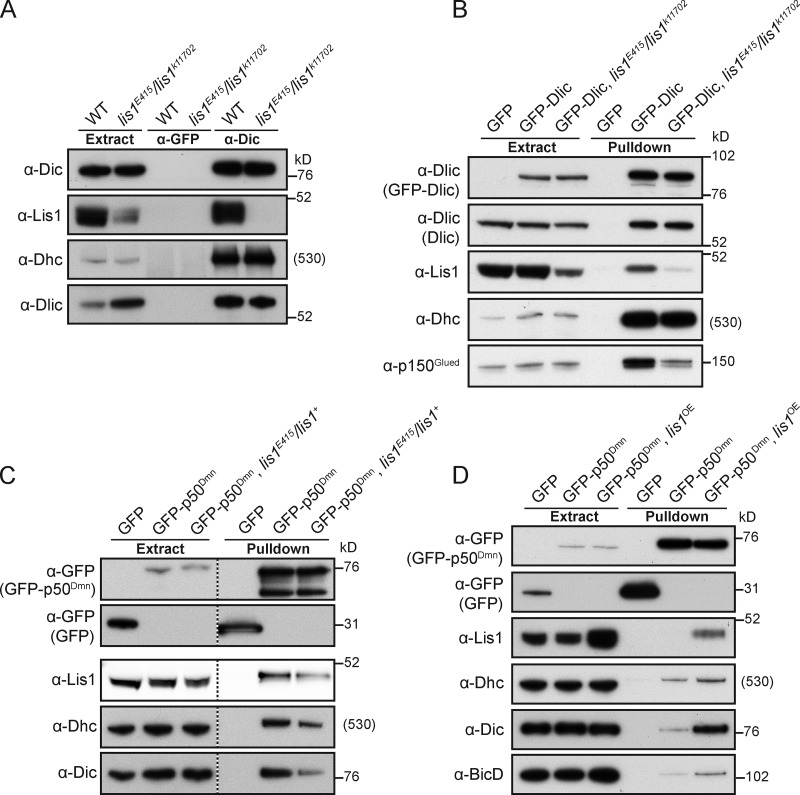
Gross differences in the cellular distribution of Dhc were not observed in lis1E415/lis1k11702 embryos (Fig. S4 A). This suggests that defective dynein recruitment to localization signals from mutant extract is not a result of widespread motor misfolding and/or aggregation. We also did not detect changes in dynein complex composition in WT and lis1E415/lis1k11702 mutants by immunoprecipitating Dic from ovary extracts and blotting with available antibodies to other dynein subunits (Fig. 7 A). Thus, reduced recruitment of dynein to RNPs in lis1 mutants is not associated with a gross defect in dynein complex assembly.
Association with the dynactin complex is reportedly crucial for association of dynein with several cargos, in some cases due to direct interactions between dynactin components and cargo-associated proteins (Echeverri et al., 1996; Burkhardt et al., 1997; Steffen et al., 1997; Roghi and Allan, 1999; Schroer and Cheong, 2012; Splinter et al., 2012). We therefore asked whether Lis1 plays a role in facilitating the interaction of dynein with dynactin by performing pull-downs from Drosophila extracts expressing GFP-tagged versions of Dlic or p50Dmn. Both fusion proteins are expressed at modest levels, are readily incorporated into dynein–dynactin complexes (see Materials and methods), and can be efficiently captured from extracts using a high affinity GFP-binding protein (GBP).
We found that the amount of the dynactin component p150Glued that coprecipitated with GFP-Dlic was consistently and strongly reduced in lis1E415/lis1k11702 ovary extract compared with ovary extract with WT Lis1 levels (Fig. 7 B; see Fig. S4 B for quantification). As expected from our previous observations (Fig. 7 A), the association of GFP-Dlic with Dhc was not affected in the lis1 mutant extract (Fig. 7 B; see Fig. S4 B for quantification). The impaired association of dynein and dynactin components when Lis1 levels are lowered was corroborated by the reduced coprecipitation of Dhc and Dic with GFP-p50Dmn in lis1E415/lis1+ versus WT embryo extract (Fig. 7 C; see Fig. S4 C for quantification). Anti-Dic immunoprecipitations confirmed that expression of GFP-p50Dmn did not have deleterious effects on dynein complex composition in the lis1 mutant embryo extract (Fig. S4 D). Taken together, these data indicate that normal Lis1 levels are required for efficient association of dynein with dynactin components, but not for overall dynein complex assembly.
Further supporting a role for Lis1 in promoting the interaction of dynein with dynactin, overexpression of Lis1 increased the association of Dhc and Dic with GFP-p50Dmn compared with that observed with WT Lis1 levels (Fig. 7 D). Interestingly, overexpression of Lis1 also enhanced the coprecipitation of BicD with GFP-p50Dmn (Fig. 7 D). This finding is compatible with the notion that the increased association of dynein with dynactin induced by Lis1 promotes the association of dynein with its cargos (in this case via the adaptor BicD).
Discussion
Lis1 promotes apical mRNA transport and the association of dynein and dynactin with RNPs
We have identified Lis1 as a novel factor recruited to RNA localization signals and shown that it is critical for net minus end-directed transport of localizing RNAs. Since the integrity and orientation of the MT cytoskeleton appears normal in the hypomorphic lis1 mutants, these observations provide the first direct evidence for Lis1’s role as a component of RNA transport machinery, as well as the first insights into which aspects of RNP motility are regulated by this protein. Even a relatively subtle reduction in cellular Lis1 concentration (in lis1E415 heterozygotes) reduces the efficiency of minus end-directed mRNA transport. This finding raises the possibility that mislocalization of functionally important RNAs contributes to lissencephaly in humans, which is associated with loss of a single LIS1 allele (Reiner et al., 1993). Consistent with this notion, there is mounting evidence that mRNA localization plays important roles in neuronal morphogenesis (Doyle and Kiebler, 2011; Jung et al., 2012).
We identified CLIP-190 as another novel component of protein complexes assembled on localization signals. Interestingly, experiments in other organisms have shown that CLIP-190 and Lis1 can associate directly (Coquelle et al., 2002; Sheeman et al., 2003). However, we show that—unlike Lis1—CLIP-190 is dispensable for apical mRNA localization. This finding is compatible with the finding that the CLIP190 homologue in the fungus Ustilago maydis is not required for Lis1-dependent early endosome transport (Lenz et al., 2006).
Reducing Lis1 levels suppresses apical transport of both large RNA particles formed after injection, likely containing many RNA molecules, and endogenous RNPs, which appear to contain a single RNA copy. These data are consistent with complete loss of Lis1 in Aspergillus nidulans inhibiting the transport of both small and large endosomal cargos away from hyphal tips (Egan et al., 2012). It is conceivable that small endosomes depend on Lis1 for transport in Aspergillus because there is still a substantial load on dynein (Yi et al., 2011), for instance due to cytoplasmic flows in the opposing direction (Lew, 2011). However, our finding that the motility of equivalently sized localizing and nonlocalizing RNA particles is differentially sensitive to reduced Lis1 levels in the same cell type provides compelling evidence that the amount of load on dynein is not the sole determinant of Lis1-dependence of cargo transport.
We have used an assay for de novo assembly of localization complexes on RNAs to demonstrate that Lis1 promotes the association of dynein and dynactin with Egl-BicD complexes bound to RNA localization signals. Although we cannot rule out additional contributions of Lis1 to mechanochemical regulation of dynein during RNA transport, the defects in motility of localizing mRNAs in lis1 mutant embryos are entirely compatible with reduced recruitment of the motor. Minus end runs of localizing RNAs in the mutants have strongly reduced travel distances while their velocities are slightly attenuated, consistent with the effects of reducing the copy number of dynein on bead transport in vitro (Mallik et al., 2005).
The RNP assembly assay also identified a role for Lis1 in promoting the association of dynein and dynactin with nonsignal sites in RNAs. Thus, Lis1 appears to facilitate the recruitment of dynein–dynactin to distinct molecular adaptors. This function could contribute to the reduced targeting of dynein to diverse subcellular sites observed in several studies when Lis1 is inhibited (Cockell et al., 2004; Dzhindzhev et al., 2005; Siller et al., 2005; Lam et al., 2010; Sitaram et al., 2012; Splinter et al., 2012). Interestingly, in some studies, perturbations of Lis1 function did not abolish (Tai et al., 2002) or only partially inhibited (Cockell et al., 2004) the recruitment of dynein to cargos. This apparent discrepancy could be explained by differences in the methods used to perturb Lis1 function, including the degree to which Lis1 function is inhibited, or redundant mechanisms for motor recruitment on different cargos or in different cell types.
Sensitivity of transport of different cargos to reduced Lis1 levels
We found that a partial reduction in Lis1 levels affects dynein-dependent motility of localizing RNPs more strongly than that of nonlocalizing RNPs. This is surprising given our evidence that Lis1 augments the affinity of dynein–dynactin for both mRNA species. It is conceivable that transport of localizing and nonlocalizing RNPs are differentially sensitive to reduced dynein–dynactin affinity due to the way in which the opposing motor systems operate on each cargo. For example, because nonlocalizing RNPs predominantly undergo brief bidirectional excursions, dynein copy number may be less limiting because runs are frequently cut short by engagement of a functionally coupled plus end motor (i.e., before the processivity of the minus end motors becomes limiting). An analogous mechanism has been proposed to explain why a decrease in the number of engaged kinesin-1 motors does not reduce travel distances of bidirectional lipid droplets in the Drosophila embryo (Shubeita et al., 2008).
RNA localization signals might overcome tight coupling between opposite polarity motors by recruiting additional dynein–dynactin complexes without plus end-directed motors, a scenario consistent with our failure to detect enrichment of a kinesin protein on RNA localization signals in our proteomic screen. These additional dynein–dynactins could give rise to long minus end-directed runs, by engaging multiple dyneins either simultaneously or consecutively, which are less likely to be cut short by engagement of plus end motors. Such runs would therefore be more sensitive to a reduction in dynein copy number in lis1 mutants than those of nonlocalizing RNPs.
More generally, our results indicate that partially reduced Lis1 function could predominantly affect the transport of those Lis1-associated cargos that rely on particularly efficient recruitment of dynein–dynactin. Scenarios where this may be the case include when many dynein–dynactin complexes are needed to translocate a cargo—such as when the consignment is large or transport occurs in a viscous cytoplasm or constricted area—or when the activity of dynein–dynactin is uncoupled from plus end motors, as proposed here for localizing RNPs.
Lis1’s role in dynein–dynactin association and the potential relationship to cargo binding
We found that cellular levels of Lis1 regulate the extent of coprecipitation of dynein and dynactin components from Drosophila extracts. These data suggest that Lis1 facilitates the overall association of the dynein complex with dynactin. It is conceivable that this reflects an indirect consequence of Lis1’s function in promoting dynein binding to cargo, for example if dynactin interacts preferentially with dynein that is in the cargo-bound state. However, several studies have provided evidence that the dynactin complex promotes the binding of dynein to its cargos (Echeverri et al., 1996; Burkhardt et al., 1997; Steffen et al., 1997; Roghi and Allan, 1999; Schroer and Cheong, 2012), including the finding that dynactin is required for association of dynein with a purified mammalian BicD protein (Splinter et al., 2012). Hence, we currently favor the model that reduced binding of dynein to dynactin in lis1 mutants makes a direct contribution to the defect in association of the motor with RNPs.
How might Lis1 influence dynein’s interaction with dynactin? In addition to the well-established ability of Lis1 to bind the motor domain of Dhc (Sasaki et al., 2000; Tai et al., 2002; McKenney et al., 2010; Huang et al., 2012) there is evidence that Lis1 can bind to Dic and the p50Dmn subunit of dynactin (Smith et al., 2000; Tai et al., 2002). It is therefore conceivable that Lis1 could act as a direct molecular link facilitating the interaction of dynein complexes with dynactin. Alternatively, the binding of Lis1 to the Dhc motor domain could stimulate a conformation change within dynein that promotes its stable association with dynactin. For example, it is possible that Lis1 binding to the dynein motor domain overcomes the proposed autoinhibitory interaction with the tail (Markus and Lee, 2011), freeing the latter element to interact via associated dynein subunits with dynactin (Schroer and Cheong, 2012). Long-term experiments will be required to discriminate between these and other possibilities.
Finally, we wish to emphasize that our data do not contradict the proposed role of Lis1 in regulating the mechanochemical properties of dynein once it is bound to cargos, including the ability of the motor to withstand load. Indeed, it is tempting to speculate that a single Lis1-induced conformational change in dynein could be the basis of both the regulation of motor mechanochemistry and dynactin association. This could couple the ability of dynein to associate with cargo with its competence to move productively on MTs. Nevertheless, our results suggest that defective association of dynein with dynactin could make an important contribution to cellular defects caused by experimental inhibition of Lis1 in different model organisms, as well as by haploinsufficiency for the human gene in lissencephaly.
Materials and methods
RNA affinity purifications
Inserts encoding RNA localization signals or UTR sequences were cloned into the pTRAPv5 vector (Cytostore), allowing a fusion RNA to be transcribed from the T7 promoter that contains two copies of the S1 streptavidin-binding aptamer 5′ to the RNA of interest (Srisawat and Engelke, 2001). pTRAPv5 vectors containing ILS, ILSas, TLS, TLSU6C, or TLSΔCA (previously known as TLSΔbub) sequences inserted into the polylinker were produced in an earlier study (Dienstbier et al., 2009). The WT ILS and TLS sequences inserted are 58 and 44 bp, respectively. pTRAP-HLE contains sequences encoding the 125-nt localization element (Bullock et al., 2003) plus 50 nt of flanking sequence. The HLEr12 mutation (Bullock et al., 2003) was introduced into this vector by QuikChange mutagenesis (Agilent Technologies). Sequences of WT and mutant localization elements used in this study are reproduced in Fig. S1. pTRAP-K103′UTR is described in Amrute-Nayak and Bullock (2012). asK103′UTR corresponds to the antisense version of the same region as in K103′UTR containing a randomized sequence of the TLS (the mutation in the full-length K10scram construct in Bullock et al. [2010]). Vectors were linearized with unique restriction enzymes in the pTRAPv5 polylinker. Uncapped aptamer-tagged RNAs were transcribed from these templates using the T7 Megascript or Megashortscript kits (Ambion) and unincorporated nucleotides removed using mini Quick Spin RNA columns (Roche). This resulted in WT aptamer-ILS, aptamer-HLE, and aptamer-TLS RNAs that were 325, 472, and 311 nt, respectively.
RNA affinity experiments were performed using a protocol modified from Dienstbier et al. (2009). All steps were performed using Drosophila extraction buffer (DXB; 25 mM Hepes, pH 6.8, 50 mM KCl, 1 mM MgCl2, 2 mM DTT, 250 mM sucrose, and 2× complete EDTA-free protease inhibitor tablets [Roche]) supplemented with 10 µM MgATP (DXB + MgATP), unless otherwise stated. For experiments assessing RNP assembly from embryo extract by immunoblotting, 180 µl of streptavidin-conjugated magnetic beads (Dynabeads M-280; Invitrogen) were blocked with 0.5 mg ml−1 BSA in PBS for 30 min at 4°C. Beads were then incubated with 15 µg aptamer-linked RNA and 150 U RNase inhibitor (RNasin; Promega) in 300 µl DXB + MgATP for 2 h at 4°C. Extracts were prepared from frozen 0–8- or 0–16-h collections of dechorionated Drosophila embryos. 150 mg embryos were homogenized in 300 µl prechilled DXB + MgATP per sample, using a motor-driven pestle in a prechilled glass dounce at 4°C. Extracts were cleared twice by centrifugation for 10 min at 16,000 g in a pre-chilled tabletop centrifuge. 40 U RNasin was added per 100 µl extract. Embryo extract (total protein ranging from 4 to 7.5 mg per sample) was incubated with RNA-coupled streptavidin beads for 15 min at room temperature (RT), followed by 30 min at 4°C. Beads were rinsed three times, and washed five times for 5 min in 1 ml DXB + MgATP. Complexes were eluted in 60 µl 10 mM D-biotin (Invitrogen) in DXB for 20 min at 20°C. LDS loading buffer (Invitrogen) was added to 1× and DTT to 50 mM. Typically, the extract loaded on each immunoblot corresponded to ∼0.3% of the extract loaded onto the beads, and ∼20% of the total eluate from the beads was loaded per lane. In cases where lower or higher volumes of eluate were loaded (i.e., 15–25%), extract loading was scaled proportionally. Note that this modified protocol uses a less stringent washing buffer and a reduced number of washes compared with Dienstbier et al. (2009). We found that although our washing conditions increased the level of background binding (i.e., the indiscriminate binding of proteins to WT and mutant signals), they enabled us to detect strong and reproducible enrichment of dynein and dynactin components on localization signals not observed previously.
Large-scale experiments for MS analysis were performed in 15-ml falcon tubes using a large magnetic rack; bead volume was 1.5 ml; 120 µg RNA was coupled to beads in 2.4 ml DXB + MgATP containing 1,100 U RNAsin; 1.6 g embryos were homogenized in 3 ml DXB + MgATP per sample; washes consisted of three rinses and seven washes of 5 min with 12 ml buffer; elution was in 600 µl 10 mM D-biotin in DXB for 20 min at RT with rotation. A 40-µl sample was taken for analysis by immunoblotting and silver staining, and the remainder was flash frozen in liquid nitrogen and stored at −80°C for subsequent MS analysis.
The procedure was scaled down for loss-of-function experiments using ovary extracts (ovary rather than embryo extracts were used for these experiments to avoid the small proportion of lis1E415/lis1k11702 embryos that are unfertilized or develop abnormally and to facilitate acquisition of sufficient material). The bead volume was 60 µl and 5 µg RNA was coupled in 100 µl DXB containing 50 U RNAsin. Ovaries were dissected from female flies fed on fresh yeast (overnight to three days), and flash frozen and stored at −80°C. For each sample, 100 ovaries were homogenized in 300 µl DXB + MgATP using a Kontes pestle. Triton X-100 was added to 0.2% and the extracts incubated on ice for 5 min. Extracts were cleared by centrifugation at 5,000 g for 5 min. The ovary extract added to beads per experiment contains ∼1–2 mg of total protein. Washes consisted of three rinses in 1 ml DXB + MgATP; elution was in 20 µl 10 mM D-biotin in DXB for 20 min at 20°C. LDS loading buffer (Invitrogen) was added to 1× and DTT to 50 mM. Typically, the extract loaded for each immunoblot corresponded to ∼1–1.5% of the extract loaded onto the beads, with 60–80% of the eluate from the beads loaded per lane. This applies to all blots except for those for BicD in Fig. 6 A and Egl in Fig. 6 B, where ∼20% of the eluate was loaded, as well as all the blots in Fig. 6 C and Fig. S3 D, where the entire eluate was loaded.
Mass spectrometry and data analysis
Protein samples were prepared for analysis using a filter-aided sample preparation method, essentially as described previously (Wiśniewski et al., 2009). In brief, the protein sample was reduced in a buffer containing 8 M urea, 0.1 M Tris-HCl, pH 8.5, 10 mM DTT, and 0.05% (wt/vol) Rapigest (Waters). The protein mixture was placed in an Amicon Ultra 0.5-ml centrifugal filter unit (EMD Millipore) and then alkylated with 55 mM iodoacetamide. After alkylation the proteins were buffer exchanged into 8 M urea, 0.1 M Tris-HCl, pH 8. Proteins were digested on the membrane for 18 h at 37°C with 800 ng endoproteinase Lys C (Roche). The sample was subsequently digested with 800 ng trypsin (Promega) for 4 h at RT. The filter unit was then centrifuged at 14,000 g for 20 min and the peptide filtrate lyophilized. The peptides were reconstituted in 200 µl of Milli-Q water and 10 µl of this mixture was desalted on stage tips prepared in-house (3M Empore HP C18-SD). The eluate was diluted to a volume of 21 µl in 3% (vol/vol) acetonitrile/0.1% (vol/vol) formic acid before LC-MS/MS. The peptide fraction (7 µl) was analyzed by nano-scale capillary LC-MS/MS using an UltiMate U3000 HPLC (Thermo Fisher Scientific) to deliver a flow of ∼300 nL/min. A µ-precolumn cartridge, C18 Acclaim PepMap 100 (5 µm, 300 µm × 5 mm; Thermo Fisher Scientific), trapped the peptides before separation on a C18 Acclaim PepMap100 (3 µm, 75 µm × 250 mm; Thermo Fisher Scientific). Peptides were eluted with a 4-h gradient of acetonitrile (5–50% [vol/vol]). The analytical column outlet was directly interfaced via a modified nano-flow electrospray ionization source, with a hybrid linear quadrupole ion trap mass spectrometer (Orbitrap LTQ XL; Thermo Fisher Scientific).
Data-dependent analysis was performed using a resolution of 60,000 for the full MS spectrum, followed by 10 MS/MS spectra in the linear ion trap. MS spectra were collected over a m/z range of 200 to 1,800. MS/MS scans were collected using threshold energy of 35 for collision-induced dissociation. LC-MS/MS data were then searched against the National Centre for Biotechnology Information database of Drosophila melanogaster protein sequences using the Mascot program (Matrix Science, UK; Perkins et al., 1999). A precursor tolerance of 5 ppm and a fragment ion mass tolerance of 0.8 D were applied during searching. Two missed enzyme cleavages were allowed and variable modifications for oxidized methionine, carbamidomethyl cysteine, and methyl and dimethyl lysine were included. MS/MS-based peptide and protein identifications were validated with Scaffold version 3 (Proteome Software, Inc.). Peptide and protein identifications were generated by the Peptide Prophet (Keller et al., 2002) and Protein Prophet (Nesvizhskii et al., 2003) algorithms, respectively. Peptide identifications were accepted if established at greater than 80.0% probability. Protein identifications were accepted if established at greater than 90.0% probability and provided they contained more than one identified peptide.
Spectral counting data were exported directly to Microsoft Excel and values normalized across all samples to correct for differences in the total number of spectra between individual MS runs. This allowed accurate comparisons between samples in pairwise experiments, and the application of global selection criteria for enriched factors across all experiments. The spectral counts in each experiment were adjusted with a normalization factor, which was determined by comparing the total spectral counts for each individual sample (MS run) with the average of the total spectral count from all experiments (see also Table S3 legend). Note that the total raw spectral count values were very similar between the different samples from individual comparative experiments (e.g., ILS vs. ILSas), and between experiments of the same type (e.g., comparing ILS experiments 1 and 2).
Drosophila strains
Oregon-R was used as the WT strain. The lis1E415 (Swan et al., 1999) and lis1k11702 (Lei and Warrior, 2000) alleles contain P-element insertions in the lis1 locus. The strains lis1E415/CyO and P(mat-tub-α4-GFP::Dmn) (Januschke et al., 2002) were gifts from A. Guichet (Institut Jacques Monod, Paris, France). The latter line expresses p50Dmn with an N-terminal GFP tag from a maternal α-tubulin promoter. P(Ubi-GFP::Dlic), driving Dlic with an N-terminal GFP tag from the ubiquitin promoter (Pandey et al., 2007), was a gift of J. Raff (University of Oxford, Oxford, England, UK). GFP-Dlic or GFP-p50Dmn transgenes were recombined onto the lis1E415 chromosome to enable assessment of the association of dynein and dynactin components when Lis1 levels are lowered. lis1k11702/CyO (BL#10179), Sco/CyO P(Actin5C-GFP) (BL#4533; used for expression of GFP alone in pull-down experiments), and Df(2L)BSC294 (BL#23679; which uncovers the clip-190 locus) were acquired from the Bloomington Drosophila Stock Center at Indiana University. P(Ubi-EB1::GFP), driving EB1 with a C-terminal GFP tag from the ubiquitin promoter, is described in Parton et al. (2011) and was generated by the laboratory of H. Ohkura. The P(α-tubulin-Lis1) and P(UASp-lis1) constructs, which allow ubiquitous and tissue-specific expression of Lis1, respectively, were generated by cloning the Lis1-coding sequence amplified from the Lis1 cDNA (LD11219; obtained from the Drosophila Genome Resource Center) into the appropriate vectors. Inserts were sequenced to confirm their integrity. Transgenic flies harboring these constructs were generated by P-element–mediated transposition, with injections performed by Bestgene, Inc. Assessing the effect of lis1 overexpression on the association of dynein and dynactin components involved recombining the UASp-lis1 and GFP-p50Dmn transgenes onto the same chromosome, followed by crossing to the maternal triple driver (MTD)-GAL4 line (P(otu-GAL4-VP16);P(GAL4-nosNGT);P(GAL4-VP16-nos)) (BL#31777; which drives strong expression of GAL4 throughout oogenesis).
Lis1 antibody generation
The first 213 codons of the Drosophila Lis1 open reading frame were cloned into the pQE31 vector (QIAGEN) to produce an N-terminally His-tagged protein in which the tag is followed by a spacer encoded by the pSL1180/90 polylinker sequence (Sac1–Nde1; GE Healthcare). The Escherichia coli–overexpressed protein was purified from inclusion bodies, enriched on Ni-NTA beads, and further purified through an SDS gel. The electro-eluted protein, mixed with Freund’s adjuvant (1:1), was injected into rabbits at 240 µg ml−1. Serum was affinity purified against the N-terminal 213 amino acids of Drosophila Lis1 expressed from a pMal vector (New England Biolabs, Inc.).
Generation of a clip-190 mutant allele
The clip-190KO allele was generated by homologous recombination with ends-out targeting (Gong and Golic, 2004). In brief, sequences corresponding to the region between 17,406,580 bp and 17,409,639 bp from the Drosophila melanogaster genome assembly were amplified from genomic DNA purified from Oregon-R flies using the following primers: 5′-AGTTCTGTGGGTACCAAGTCGTACAAGTACAACTAGAGCAACTCC-3′ and 5′-AGTTCTGTGGCGGCCGCATTGTGTGTTTGCCACGAATCGCATTCAAT-3′. These were cloned into the NotI–Acc65I restriction sites of the downstream polylinker of the pW25 vector (Gong and Golic, 2004). In a second step, sequences corresponding to the region between 17,385,290 bp and 17,389,389 bp from the genome assembly were amplified from the same source using the following primers: 5′-AGTTGTGTGCGTACGGAAGAAAAGAAATCTGCTCTCAGTCAGCTG-3′ and 5′-AGTTCTGTGGGCGCGCCATTCCGCGTCTGAAACACGTCCGCCTCGCG-3′. These were cloned into the BsiWI–AscI restriction sites of the upstream polylinker of the same vector. This results in the creation of P-donor-clip-190 construct. After the generation of transgenic P-donor-clip-190 fly lines, the donor construct was remobilized to target the endogenous clip-190 gene as described previously (Gong and Golic, 2004). The clip-190KO allele was verified by immunoblotting. The allele comprises a deletion of bp 17,389,389–17,406,580 on chromosome 2L, which removes the majority of the clip-190 gene, including the CAP-Gly domain.
RNA injections
Capped Alexa Fluor 488–labeled RNAs were transcribed from linearized plasmid templates using 0.4 mM ATP, 0.4 mM CTP, 0.3 mM UTP, 0.1 mM Alexa Fluor 488-UTP (Invitrogen), 0.12 mM GTP, and 0.3 mM 7mG(5′)pppG cap analogue (Agilent Technologies) using T7 or T3 RNA polymerases (Roche) according to the manufacturer’s instructions. Unincorporated nucleotides were removed with mini Quick Spin RNA columns (Roche). Injected hairy RNA corresponds to a 661-nt piece representing the majority of the transcript’s 3′UTR; K10 corresponds to the full 1,432-bp 3′UTR and an 860-bp portion of the adjacent genomic sequence, as described previously (Bullock et al., 2010); asK10 corresponds to an antisense version of the same portion of the K10 transcript harboring a randomized TLS (see RNA affinity purifications, above). Embryos were injected with a 1-µg µl−1 solution of fluorescent RNA. Embryos of the stated maternal genotypes were fertilized with heterozygous males. We did not observe two classes of embryos in our subsequent mRNA motility analysis, indicating that there is no significant paternal influence of these proteins on transport at this stage of development. Injection of cycle 14 blastoderm embryos with fluorescent RNA followed by time-lapse imaging (3.3 frames s−1) and centroid-based automatic particle tracking was as described previously (Bullock et al., 2006). RNA was not injected into morphologically abnormal lis1E415/lis1k11702 embryos, which occurred at a low frequency. In these and other live-imaging experiments, embryos were mounted in 10S halocarbon oil and data captured with a spinning disk imaging system (Ultraview ERS; PerkinElmer), using a CCD camera (Orca ER; Hamamatsu Photonics) and an inverted microscope body (IX71; Olympus) equipped with a 60×/1.2 NA UPlanApo water objective. Embryos were maintained at ∼22°C during data acquisition.
Assessment of relative sizes of fluorescent RNPs containing different RNAs
mRNAs were synthesized in parallel using the same master mix of reaction components. We confirmed with a spectrophotometer that the Alexa Fluor 488/base ratio was very similar for all fluorescent RNAs (∼1:40). After RNA injection and time-lapse imaging, the sizes of RNPs containing different injected RNAs were determined using the Analyze Particles tool in ImageJ (National Institutes of Health). Movies were cropped to remove the apical region where individual RNPs congregate. A threshold was applied that allowed the fluorescent signal from RNPs to be detected above background fluorescence, and automatic quantification of the fluorescence of RNPs in each frame of the movie performed. 6 or 7 injected embryos were analyzed per mRNA species, with a mean fluorescence area determined per movie. The data in Fig. 5 A represent the mean of these means.
Pull-down and immunoprecipitation experiments
Dynein complexes were immunoprecipitated from ovary or embryo extracts using mouse anti-Dic 74.1 antibodies (EMD Millipore), with mouse anti-c-myc (9E10; Santa Cruz Biotechnology, Inc.) or mouse anti-GFP (clones 7.1 and 13.1; Roche) as controls. 4 µg of antibodies were coupled to 30 µl Protein G Dynabeads (Invitrogen) in 400 µl PBS + 0.1% Tween 20 for 2 h at 4°C. Antibodies were chemically cross-linked to the beads using 20 mM dimethyl pimelimidate in 0.2 M triethanolamine, pH 8.2, and quenched with 50 mM Tris, pH 7.5. Embryo and ovary extracts were prepared as described for RNA affinity experiments (except in DXB without MgATP), with the amount of total protein added to the beads in the same range. Extracts were incubated on the beads overnight at 4°C, beads washed four times with 1 ml ice-cold DXB, and complexes eluted by boiling in 50 µl 2x LDS loading buffer (Invitrogen) with 50 mM DTT. Typically, the extract loaded corresponds to ∼1% of the extract loaded onto the beads. The amount of eluate from the beads loaded was 6% for blots of dynein subunits and 30% for Lis1 blots.
GFP-binding protein (GBP) pull-downs were performed with transgenic fly lines expressing GFP-Dlic (Pandey et al., 2007) or GFP-p50Dmn (Januschke et al., 2002). Both fusion proteins are expressed at similar levels to their endogenous counterparts (Amrute-Nayak and Bullock, 2012) and show comparable localization to them (Januschke et al., 2002; Pandey et al., 2007). Furthermore, we found that the fusion proteins do not cause detectable morphological defects, consistent with previous observations (Januschke et al., 2002; Pandey et al., 2007). Both proteins are incorporated into microtubule-associated dynein–dynactin complexes in accordance with their abundance in total extract (Amrute-Nayak and Bullock, 2012), providing evidence that the GFP-tagged proteins can be used to reliably assess association of dynein and dynactin components. We also determined that neither fusion protein inhibits apical accumulation of injected localizing mRNAs in the Drosophila embryo.
Ovary extracts expressing GFP-Dlic were prepared as described for RNA affinity purifications, except that 150 dissected ovaries were homogenized in 450 µl DXB per genotype per experiment. Extract containing ∼4 mg of total protein was incubated with 30 µl of GBP-Agarose beads (ChromoTek), and incubated overnight at 4°C. Beads were then washed 5 times with 1 ml ice-cold DXB + 100 mM NaCl with rotation at 4°C, and complexes eluted by boiling in 50 µl 2× LDS-loading buffer (Invitrogen) containing 50 mM DTT. The extract loaded on the immunoblots corresponds to ∼1% of the extract loaded onto the beads, with 10% of the eluate loaded for α-Dlic blots and 30% loaded for all other blots (which were part of a single membrane).
Extracts from embryos expressing GFP-p50Dmn were prepared from 0–4-h collections in DXB. GBP pull-downs were performed as described above for GFP-Dlic, except that 4–7.5 mg of total protein was loaded onto 30 µl GBP-Agarose beads. The extract loaded on the immunoblots corresponds to ∼0.3% of the extract loaded onto the beads, with 10% of the eluate loaded for α-GFP blots and 30% loaded for all other blots (which were part of a single membrane).
For experiments involving overexpression of Lis1, ovary extracts were made from 200 ovaries per genotype per condition, homogenized in 350 µl DXB. GFP-p50Dmn precipitations were performed essentially as described above, with ∼4.5 mg of total protein loaded on the beads. The extract loaded corresponds to 0.6% of the extract added to the beads, with 20% of the eluate loaded for α-GFP blots and 30% loaded for all other blots (which were part of a single membrane).
Quantification of band densities on immunoblots was performed for GFP-Dlic and GFP-p50Dmn pull-downs using the Gel Analysis tool in ImageJ. Raw images of scanned immunoblots were exported into ImageJ software, and rectangular selections made around gels bands using the Gel Lanes selection tool. Background noise was removed from the profile plots using the Straight-line tool, and the peak areas (corresponding to signal densities) exported to Excel for analysis. Within individual experiments, the densities for bands of each protein of interest (i.e., Dhc and/or p150Glued) were normalized to correct for any subtle difference in the densities of the primary target of the pull-down (either GFP-Dlic or GFP-p50Dmn) in that sample. For each protein of interest, the corrected values for WT and lis1 samples were then expressed as a percentage of the WT corrected values.
Immunofluorescence, lipid droplet staining, and in situ hybridization
2–4-h embryos were collected, dechorionated, and formaldehyde fixed according to standard procedures. For immunofluorescence, embryos were stained with mouse anti-Dhc 2C11-2 (Sharp et al., 2000); raised against bulk preparations of Drosophila MT-associated proteins, followed by identification of specificity of this clone for Dhc; provided by the Developmental Studies Hybridoma Bank; 1:500 dilution), rabbit anti-Centrosomin (Cnn; Dobbelaere et al., 2008; raised against Drosophila Cnn; provided by J. Raff, University of Oxford, Oxford, UK; 1:500 dilution) or mouse anti-α-tubulin (DM1α, 1:1,000; Sigma-Aldrich). Lipid droplets were stained using Nile Red, as described previously (Yu et al., 2011). In brief, embryos were fixed in 4% formaldehyde in PBS for 15–20 min and stained with a 20 µg ml−1 solution of Nile Red. Secondary antibodies, purchased from Invitrogen, were conjugated to Alexa Fluor 488 or 555. Non-enzymatic RNA in situ hybridizations were performed as described previously (Amrute-Nayak and Bullock, 2012), except that probes were labeled with dinitrophenol (DNP)-UTP (PerkinElmer) and detected with rabbit anti-DNP (1:1,000) and Alexa Fluor 555–conjugated anti–rabbit (1:500) antibodies (both from Invitrogen). In all cases, embryos were mounted in Vectashield (Vector Laboratories) and imaged with a 780 confocal microscope (Carl Zeiss) using a Plan Apochromat 63×/1.4 NA oil-immersion objective. Quantification of apical h RNA enrichment in embryos subjected to in situ hybridization was performed as follows. Mean values for apical/basal fluorescence per embryo were calculated from measurements of apical/basal fluorescence for three individual stripes of h expression from a single confocal image of an embryo. Apical and basal fluorescence measurements were made with Solis software (Andor Technology). A rectangular region of interest (ROI) was drawn around a stripe of h RNA in the apical cytoplasm and mean apical fluorescence value determined after background subtraction. The same size ROI was then positioned directly basal to the apical ROI, just beneath the peripheral nuclei (stained with DAPI), followed by determination of the mean basal fluorescence value after background subtraction. These values were used to determine the apical/basal fluorescence per stripe.
Yeast two-hybrid interaction assays
All methods were based on the Matchmaker system (Takara Bio Inc.) and were performed as described previously (Dienstbier et al., 2009). The Drosophila Lis1 and NudE genes were cloned from cDNAs into the pGBKT7 and pGAD424 vectors, respectively. Inserts were sequenced to confirm their integrity. Interactions with full-length and truncated Drosophila Egl and BicD proteins were tested using previously described constructs (Dienstbier et al., 2009). Plasmids were transformed into the yeast two-hybrid strains PJ69-4a or PJ69-4α (James, 2001), which were subsequently mated. Interactions were tested on −HIS/−LEU/−TRP plates, and mating controls on −LEU/−TRP plates. Images were taken 72 h after mating.
Immunoblotting
Immunoblotting was performed using mouse anti-BicD 1B11 (Suter and Steward, 1991; raised against full-length Drosophila BicD; used at 1:1,000 dilution); rabbit anti–CLIP-190 ((Dzhindzhev et al., 2005; raised against residues 851–1468 of Drosophila CLIP-190; 1:500 dilution); rabbit anti-Cnn (see above; 1:1,000 dilution); mouse anti-Dhc 2C11-2 (see above; 1:1,000); mouse anti-Dic 74.1 (see above; 1:1,000 dilution); mouse anti-Dlic P5F5 (Mische et al., 2008; raised against a synthetic peptide encoding Drosophila Dlic residues 378–405; provided by T. Hays, University of Minnesota, Minneapolis, MN; 1:1,500 dilution); rat anti-p50Dmn (Duncan and Warrior, 2002; raised against full-length Drosophila p50Dmn; provided by R. Warrior, University of California, Irvine, Irvine, CA; 1:2,000 dilution); rabbit anti-Egl (Mach and Lehmann, 1997; raised against residues 1–415 of Drosophila Egl; provided by C. Navarro [Boston University, Boston, MA] and R. Lehmann [Skirball Institute, New York, NY]; 1:3,000 dilution); rabbit anti-Lis1 (see above; 1:1,000 dilution); rabbit anti-p150Glued (Kim et al., 2007; raised against residues 1073–1280 of Drosophila p150Glued; provided by V. Gelfand, Northwestern University, Chicago, IL; 1:5,000 dilution) and rabbit anti-NudE (Wainman et al., 2009; raised against full-length Drosophila NudE; provided by M. Goldberg, Cornell University, Ithaca, NY; 1:1,000 dilution). Secondary antibodies were HRP conjugated except for those used in fluorescent blotting, which were coupled to fluorophores excited at 680 or 800 nm (Invitrogen and Rockland ImmunoChemicals, respectively). Fluorescent Westerns were scanned using the Odyssey infrared imaging system (LI-COR Biosciences), and protein abundance quantified using the Odyssey software version 3.0. The integrated intensity of a defined area (expressed as counts/mm2) was determined and background measurements subtracted manually. Lis1 protein abundance in specific genotypes was compared with a series of dilutions of WT extract. Sizes of proteins were estimated by comparison to Full Range Rainbow markers (GE Healthcare). In all immunoblotting experiments in the study, the data for each vertically aligned composite in the panels derive from the same protein samples. Each experiment typically includes blotting for several proteins on the same membrane. We only probed for proteins on a second membrane when it was necessary, i.e., when two proteins migrated similarly on the gel or when equivalent loading would produce a saturated signal for one of the components. Composites of blot images were assembled using Adobe Illustrator, with a dashed line used to demarcate where lanes not relevant to this study had been cropped out.
Statistics
Information on statistical tests is provided in the figure legends.
Online supplemental material
Fig. S1 shows predicted secondary structures of WT and mutant localization elements and a representative total protein stain for RNA affinity purification experiments. Fig. S2 documents additional characterization of lis1 mutants. Fig. S3 documents additional characterization of Lis1’s role in promoting dynein–dynactin recruitment to RNAs. Fig. S4 provides additional data on Lis1’s role in promoting association of dynein and dynactin components. Videos 1 and 2 show time-lapse movies of h RNA motility in WT and clip190 mutant embryos. Video 3 shows time-lapse movies of EB1-GFP behavior in WT and lis1 mutant embryos. Video 4 shows time-lapse movies of h RNA motility in WT and lis1 mutant embryos. Table S1 documents proteins enriched on WT localization signals in a subset of MS experiments. Table S2 provides overall quantification of motile properties of injected RNAs in different genetic backgrounds. Table S3 is an Excel spreadsheet of the MS data. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201211052/DC1.
Supplementary Material
Acknowledgments
We are very grateful to B. Fievet and M. Skehel for help with MS data analysis; H. Doerflinger, I. Torres, and D. St Johnston for providing a cleaned up clip-190KO chromosome; V. Gelfand, M. Goldberg, A. Guichet, T. Hays, R. Lehmann, C. Navarro, J. Raff, and R. Warrior for providing reagents; and A. Carter and F. Port for comments on the manuscript.
This work was supported by the UK Medical Research Council (Project U105178790 to S.L. Bullock), a Lister Prize Fellowship (to S.L. Bullock), and the Wellcome Trust (grants 081849 and 092076 to H. Ohkura).
Footnotes
Abbreviations used in this paper:
- HLE
- hairy localization element
- ILS
- I factor localization element
- LC-MS/MS
- liquid chromatography-mass spectrometry/mass spectrometry
- MS
- mass spectrometry
- MT
- microtubule
- nSC
- normalized spectral counts
- nt
- nucleotide
- RNP
- ribonucleoprotein particle
- TLS
- transport and localization signal (K10)
- UTR
- untranslated region
- WT
- wild-type
References
- Akhmanova A., Steinmetz M.O. 2008. Tracking the ends: a dynamic protein network controls the fate of microtubule tips. Nat. Rev. Mol. Cell Biol. 9:309–322 10.1038/nrm2369 [DOI] [PubMed] [Google Scholar]
- Amrute-Nayak M., Bullock S.L. 2012. Single-molecule assays reveal that RNA localization signals regulate dynein–dynactin copy number on individual transcript cargoes. Nat. Cell Biol. 14:416–423 10.1038/ncb2446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullock S.L., Ish-Horowicz D. 2001. Conserved signals and machinery for RNA transport in Drosophila oogenesis and embryogenesis. Nature. 414:611–616 10.1038/414611a [DOI] [PubMed] [Google Scholar]
- Bullock S.L., Zicha D., Ish-Horowicz D. 2003. The Drosophila hairy RNA localization signal modulates the kinetics of cytoplasmic mRNA transport. EMBO J. 22:2484–2494 10.1093/emboj/cdg230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullock S.L., Nicol A., Gross S.P., Zicha D. 2006. Guidance of bidirectional motor complexes by mRNA cargoes through control of dynein number and activity. Curr. Biol. 16:1447–1452 10.1016/j.cub.2006.05.055 [DOI] [PubMed] [Google Scholar]
- Bullock S.L., Ringel I., Ish-Horowicz D., Lukavsky P.J. 2010. A’-form RNA helices are required for cytoplasmic mRNA transport in Drosophila. Nat. Struct. Mol. Biol. 17:703–709 10.1038/nsmb.1813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhardt J.K., Echeverri C.J., Nilsson T., Vallee R.B. 1997. Overexpression of the dynamitin (p50) subunit of the dynactin complex disrupts dynein-dependent maintenance of membrane organelle distribution. J. Cell Biol. 139:469–484 10.1083/jcb.139.2.469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahana A., Escamez T., Nowakowski R.S., Hayes N.L., Giacobini M., von Holst A., Shmueli O., Sapir T., McConnell S.K., Wurst W., et al. 2001. Targeted mutagenesis of Lis1 disrupts cortical development and LIS1 homodimerization. Proc. Natl. Acad. Sci. USA. 98:6429–6434 10.1073/pnas.101122598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockell M.M., Baumer K., Gönczy P. 2004. lis-1 is required for dynein-dependent cell division processes in C. elegans embryos. J. Cell Sci. 117:4571–4582 10.1242/jcs.01344 [DOI] [PubMed] [Google Scholar]
- Coquelle F.M., Caspi M., Cordelières F.P., Dompierre J.P., Dujardin D.L., Koifman C., Martin P., Hoogenraad C.C., Akhmanova A., Galjart N., et al. 2002. LIS1, CLIP-170’s key to the dynein/dynactin pathway. Mol. Cell. Biol. 22:3089–3102 10.1128/MCB.22.9.3089-3102.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delanoue R., Davis I. 2005. Dynein anchors its mRNA cargo after apical transport in the Drosophila blastoderm embryo. Cell. 122:97–106 10.1016/j.cell.2005.04.033 [DOI] [PubMed] [Google Scholar]
- Dienstbier M., Li X. 2009. Bicaudal-D and its role in cargo sorting by microtubule-based motors. Biochem. Soc. Trans. 37:1066–1071 10.1042/BST0371066 [DOI] [PubMed] [Google Scholar]
- Dienstbier M., Boehl F., Li X., Bullock S.L. 2009. Egalitarian is a selective RNA-binding protein linking mRNA localization signals to the dynein motor. Genes Dev. 23:1546–1558 10.1101/gad.531009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobbelaere J., Josué F., Suijkerbuijk S., Baum B., Tapon N., Raff J. 2008. A genome-wide RNAi screen to dissect centriole duplication and centrosome maturation in Drosophila. PLoS Biol. 6:e224 10.1371/journal.pbio.0060224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle M., Kiebler M.A. 2011. Mechanisms of dendritic mRNA transport and its role in synaptic tagging. EMBO J. 30:3540–3552 10.1038/emboj.2011.278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dujardin D., Wacker U.I., Moreau A., Schroer T.A., Rickard J.E., De Mey J.R. 1998. Evidence for a role of CLIP-170 in the establishment of metaphase chromosome alignment. J. Cell Biol. 141:849–862 10.1083/jcb.141.4.849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan J.E., Warrior R. 2002. The cytoplasmic dynein and kinesin motors have interdependent roles in patterning the Drosophila oocyte. Curr. Biol. 12:1982–1991 10.1016/S0960-9822(02)01303-9 [DOI] [PubMed] [Google Scholar]
- Dzhindzhev N.S., Rogers S.L., Vale R.D., Ohkura H. 2005. Distinct mechanisms govern the localisation of Drosophila CLIP-190 to unattached kinetochores and microtubule plus-ends. J. Cell Sci. 118:3781–3790 10.1242/jcs.02504 [DOI] [PubMed] [Google Scholar]
- Echeverri C.J., Paschal B.M., Vaughan K.T., Vallee R.B. 1996. Molecular characterization of the 50-kD subunit of dynactin reveals function for the complex in chromosome alignment and spindle organization during mitosis. J. Cell Biol. 132:617–633 10.1083/jcb.132.4.617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan M.J., Tan K., Reck-Peterson S.L. 2012. Lis1 is an initiation factor for dynein-driven organelle transport. J. Cell Biol. 197:971–982 10.1083/jcb.201112101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulkner N.E., Dujardin D.L., Tai C.Y., Vaughan K.T., O’Connell C.B., Wang Y., Vallee R.B. 2000. A role for the lissencephaly gene LIS1 in mitosis and cytoplasmic dynein function. Nat. Cell Biol. 2:784–791 10.1038/35041020 [DOI] [PubMed] [Google Scholar]
- Gong W.J., Golic K.G. 2004. Genomic deletions of the Drosophila melanogaster Hsp70 genes. Genetics. 168:1467–1476 10.1534/genetics.104.030874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirotsune S., Fleck M.W., Gambello M.J., Bix G.J., Chen A., Clark G.D., Ledbetter D.H., McBain C.J., Wynshaw-Boris A. 1998. Graded reduction of Pafah1b1 (Lis1) activity results in neuronal migration defects and early embryonic lethality. Nat. Genet. 19:333–339 10.1038/1221 [DOI] [PubMed] [Google Scholar]
- Holt C.E., Bullock S.L. 2009. Subcellular mRNA localization in animal cells and why it matters. Science. 326:1212–1216 10.1126/science.1176488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoogenraad C.C., Akhmanova A., Howell S.A., Dortland B.R., De Zeeuw C.I., Willemsen R., Visser P., Grosveld F., Galjart N. 2001. Mammalian Golgi-associated Bicaudal-D2 functions in the dynein–dynactin pathway by interacting with these complexes. EMBO J. 20:4041–4054 10.1093/emboj/20.15.4041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Roberts A.J., Leschziner A.E., Reck-Peterson S.L. 2012. Lis1 acts as a “clutch” between the ATPase and microtubule-binding domains of the dynein motor. Cell. 150:975–986 10.1016/j.cell.2012.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- James P. 2001. Yeast two-hybrid vectors and strains. Methods Mol. Biol. 177:41–84 [DOI] [PubMed] [Google Scholar]
- Januschke J., Gervais L., Dass S., Kaltschmidt J.A., Lopez-Schier H., St Johnston D., Brand A.H., Roth S., Guichet A. 2002. Polar transport in the Drosophila oocyte requires Dynein and Kinesin I cooperation. Curr. Biol. 12:1971–1981 10.1016/S0960-9822(02)01302-7 [DOI] [PubMed] [Google Scholar]
- Jung H., Yoon B.C., Holt C.E. 2012. Axonal mRNA localization and local protein synthesis in nervous system assembly, maintenance and repair. Nat. Rev. Neurosci. 13:308–324 10.1038/nrn3274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kardon J.R., Reck-Peterson S.L., Vale R.D. 2009. Regulation of the processivity and intracellular localization of Saccharomyces cerevisiae dynein by dynactin. Proc. Natl. Acad. Sci. USA. 106:5669–5674 10.1073/pnas.0900976106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller A., Nesvizhskii A.I., Kolker E., Aebersold R. 2002. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 74:5383–5392 10.1021/ac025747h [DOI] [PubMed] [Google Scholar]
- Kim H., Ling S.C., Rogers G.C., Kural C., Selvin P.R., Rogers S.L., Gelfand V.I. 2007. Microtubule binding by dynactin is required for microtubule organization but not cargo transport. J. Cell Biol. 176:641–651 10.1083/jcb.200608128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King S.J., Schroer T.A. 2000. Dynactin increases the processivity of the cytoplasmic dynein motor. Nat. Cell Biol. 2:20–24 10.1038/71338 [DOI] [PubMed] [Google Scholar]
- Lam C., Vergnolle M.A., Thorpe L., Woodman P.G., Allan V.J. 2010. Functional interplay between LIS1, NDE1 and NDEL1 in dynein-dependent organelle positioning. J. Cell Sci. 123:202–212 10.1242/jcs.059337 [DOI] [PubMed] [Google Scholar]
- Lansbergen G., Komarova Y., Modesti M., Wyman C., Hoogenraad C.C., Goodson H.V., Lemaitre R.P., Drechsel D.N., van Munster E., Gadella T.W., Jr, et al. 2004. Conformational changes in CLIP-170 regulate its binding to microtubules and dynactin localization. J. Cell Biol. 166:1003–1014 10.1083/jcb.200402082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W.L., Oberle J.R., Cooper J.A. 2003. The role of the lissencephaly protein Pac1 during nuclear migration in budding yeast. J. Cell Biol. 160:355–364 10.1083/jcb.200209022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei Y., Warrior R. 2000. The Drosophila Lissencephaly1 (DLis1) gene is required for nuclear migration. Dev. Biol. 226:57–72 10.1006/dbio.2000.9848 [DOI] [PubMed] [Google Scholar]
- Lenz J.H., Schuchardt I., Straube A., Steinberg G. 2006. A dynein loading zone for retrograde endosome motility at microtubule plus-ends. EMBO J. 25:2275–2286 10.1038/sj.emboj.7601119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lew R.R. 2011. How does a hypha grow? The biophysics of pressurized growth in fungi. Nat. Rev. Microbiol. 9:509–518 10.1038/nrmicro2591 [DOI] [PubMed] [Google Scholar]
- Liu H., Sadygov R.G., Yates J.R., III 2004. A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal. Chem. 76:4193–4201 10.1021/ac0498563 [DOI] [PubMed] [Google Scholar]
- Mach J.M., Lehmann R. 1997. An Egalitarian-BicaudalD complex is essential for oocyte specification and axis determination in Drosophila. Genes Dev. 11:423–435 10.1101/gad.11.4.423 [DOI] [PubMed] [Google Scholar]
- Mallik R., Petrov D., Lex S.A., King S.J., Gross S.P. 2005. Building complexity: an in vitro study of cytoplasmic dynein with in vivo implications. Curr. Biol. 15:2075–2085 10.1016/j.cub.2005.10.039 [DOI] [PubMed] [Google Scholar]
- Markus S.M., Lee W.L. 2011. Regulated offloading of cytoplasmic dynein from microtubule plus ends to the cortex. Dev. Cell. 20:639–651 10.1016/j.devcel.2011.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin K.C., Ephrussi A. 2009. mRNA localization: gene expression in the spatial dimension. Cell. 136:719–730 10.1016/j.cell.2009.01.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenney R.J., Vershinin M., Kunwar A., Vallee R.B., Gross S.P. 2010. LIS1 and NudE induce a persistent dynein force-producing state. Cell. 141:304–314 10.1016/j.cell.2010.02.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mische S., He Y., Ma L., Li M., Serr M., Hays T.S. 2008. Dynein light intermediate chain: an essential subunit that contributes to spindle checkpoint inactivation. Mol. Biol. Cell. 19:4918–4929 10.1091/mbc.E08-05-0483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro C., Puthalakath H., Adams J.M., Strasser A., Lehmann R. 2004. Egalitarian binds dynein light chain to establish oocyte polarity and maintain oocyte fate. Nat. Cell Biol. 6:427–435 10.1038/ncb1122 [DOI] [PubMed] [Google Scholar]
- Nesvizhskii A.I., Keller A., Kolker E., Aebersold R. 2003. A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 75:4646–4658 10.1021/ac0341261 [DOI] [PubMed] [Google Scholar]
- Pandey J.P., Smith D.S. 2011. A Cdk5-dependent switch regulates Lis1/Ndel1/dynein-driven organelle transport in adult axons. J. Neurosci. 31:17207–17219 10.1523/JNEUROSCI.4108-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey R., Heeger S., Lehner C.F. 2007. Rapid effects of acute anoxia on spindle kinetochore interactions activate the mitotic spindle checkpoint. J. Cell Sci. 120:2807–2818 10.1242/jcs.007690 [DOI] [PubMed] [Google Scholar]
- Parton R.M., Hamilton R.S., Ball G., Yang L., Cullen C.F., Lu W., Ohkura H., Davis I. 2011. A PAR-1-dependent orientation gradient of dynamic microtubules directs posterior cargo transport in the Drosophila oocyte. J. Cell Biol. 194:121–135 10.1083/jcb.201103160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins D.N., Pappin D.J., Creasy D.M., Cottrell J.S. 1999. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 20:3551–3567 [DOI] [PubMed] [Google Scholar]
- Rehberg M., Kleylein-Sohn J., Faix J., Ho T.H., Schulz I., Gräf R. 2005. Dictyostelium LIS1 is a centrosomal protein required for microtubule/cell cortex interactions, nucleus/centrosome linkage, and actin dynamics. Mol. Biol. Cell. 16:2759–2771 10.1091/mbc.E05-01-0069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiner O., Carrozzo R., Shen Y., Wehnert M., Faustinella F., Dobyns W.B., Caskey C.T., Ledbetter D.H. 1993. Isolation of a Miller-Dieker lissencephaly gene containing G protein beta-subunit-like repeats. Nature. 364:717–721 10.1038/364717a0 [DOI] [PubMed] [Google Scholar]
- Roghi C., Allan V.J. 1999. Dynamic association of cytoplasmic dynein heavy chain 1a with the Golgi apparatus and intermediate compartment. J. Cell Sci. 112:4673–4685 [DOI] [PubMed] [Google Scholar]
- Ross J.L., Wallace K., Shuman H., Goldman Y.E., Holzbaur E.L. 2006. Processive bidirectional motion of dynein–dynactin complexes in vitro. Nat. Cell Biol. 8:562–570 10.1038/ncb1421 [DOI] [PubMed] [Google Scholar]
- Sasaki S., Shionoya A., Ishida M., Gambello M.J., Yingling J., Wynshaw-Boris A., Hirotsune S. 2000. A LIS1/NUDEL/cytoplasmic dynein heavy chain complex in the developing and adult nervous system. Neuron. 28:681–696 10.1016/S0896-6273(00)00146-X [DOI] [PubMed] [Google Scholar]
- Schroer T.A., Cheong F.K.Y. 2012. The role of dynactin in dynein-mediated motility. Dyneins. Structure, Function and Disease. King S.M., editor Elsevier; 505–522 [Google Scholar]
- Serano T.L., Cohen R.S. 1995. A small predicted stem-loop structure mediates oocyte localization of Drosophila K10 mRNA. Development. 121:3809–3818 [DOI] [PubMed] [Google Scholar]
- Sharp D.J., Brown H.M., Kwon M., Rogers G.C., Holland G., Scholey J.M. 2000. Functional coordination of three mitotic motors in Drosophila embryos. Mol. Biol. Cell. 11:241–253 10.1091/mbc.11.1.241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheeman B., Carvalho P., Sagot I., Geiser J., Kho D., Hoyt M.A., Pellman D. 2003. Determinants of S. cerevisiae dynein localization and activation: implications for the mechanism of spindle positioning. Curr. Biol. 13:364–372 10.1016/S0960-9822(03)00013-7 [DOI] [PubMed] [Google Scholar]
- Shu T., Ayala R., Nguyen M.D., Xie Z., Gleeson J.G., Tsai L.H. 2004. Ndel1 operates in a common pathway with LIS1 and cytoplasmic dynein to regulate cortical neuronal positioning. Neuron. 44:263–277 10.1016/j.neuron.2004.09.030 [DOI] [PubMed] [Google Scholar]
- Shubeita G.T., Tran S.L., Xu J., Vershinin M., Cermelli S., Cotton S.L., Welte M.A., Gross S.P. 2008. Consequences of motor copy number on the intracellular transport of kinesin-1-driven lipid droplets. Cell. 135:1098–1107 10.1016/j.cell.2008.10.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siller K.H., Doe C.Q. 2008. Lis1/dynactin regulates metaphase spindle orientation in Drosophila neuroblasts. Dev. Biol. 319:1–9 10.1016/j.ydbio.2008.03.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siller K.H., Serr M., Steward R., Hays T.S., Doe C.Q. 2005. Live imaging of Drosophila brain neuroblasts reveals a role for Lis1/dynactin in spindle assembly and mitotic checkpoint control. Mol. Biol. Cell. 16:5127–5140 10.1091/mbc.E05-04-0338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitaram P., Anderson M.A., Jodoin J.N., Lee E., Lee L.A. 2012. Regulation of dynein localization and centrosome positioning by Lis-1 and asunder during Drosophila spermatogenesis. Development. 139:2945–2954 10.1242/dev.077511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D.S., Niethammer M., Ayala R., Zhou Y., Gambello M.J., Wynshaw-Boris A., Tsai L.H. 2000. Regulation of cytoplasmic dynein behaviour and microtubule organization by mammalian Lis1. Nat. Cell Biol. 2:767–775 10.1038/35041000 [DOI] [PubMed] [Google Scholar]
- Splinter D., Razafsky D.S., Schlager M.A., Serra-Marques A., Grigoriev I., Demmers J., Keijzer N., Jiang K., Poser I., Hyman A.A., et al. 2012. BICD2, dynactin, and LIS1 cooperate in regulating dynein recruitment to cellular structures. Mol. Biol. Cell. 23:4226–4241 10.1091/mbc.E12-03-0210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srisawat C., Engelke D.R. 2001. Streptavidin aptamers: affinity tags for the study of RNAs and ribonucleoproteins. RNA. 7:632–641 10.1017/S135583820100245X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffen W., Karki S., Vaughan K.T., Vallee R.B., Holzbaur E.L., Weiss D.G., Kuznetsov S.A. 1997. The involvement of the intermediate chain of cytoplasmic dynein in binding the motor complex to membranous organelles of Xenopus oocytes. Mol. Biol. Cell. 8:2077–2088 10.1091/mbc.8.10.2077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suter B., Steward R. 1991. Requirement for phosphorylation and localization of the Bicaudal-D protein in Drosophila oocyte differentiation. Cell. 67:917–926 10.1016/0092-8674(91)90365-6 [DOI] [PubMed] [Google Scholar]
- Swan A., Nguyen T., Suter B. 1999. Drosophila Lissencephaly-1 functions with Bic-D and dynein in oocyte determination and nuclear positioning. Nat. Cell Biol. 1:444–449 10.1038/15680 [DOI] [PubMed] [Google Scholar]
- Tai C.Y., Dujardin D.L., Faulkner N.E., Vallee R.B. 2002. Role of dynein, dynactin, and CLIP-170 interactions in LIS1 kinetochore function. J. Cell Biol. 156:959–968 10.1083/jcb.200109046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T., Serneo F.F., Higgins C., Gambello M.J., Wynshaw-Boris A., Gleeson J.G. 2004. Lis1 and doublecortin function with dynein to mediate coupling of the nucleus to the centrosome in neuronal migration. J. Cell Biol. 165:709–721 10.1083/jcb.200309025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai J.W., Chen Y., Kriegstein A.R., Vallee R.B. 2005. LIS1 RNA interference blocks neural stem cell division, morphogenesis, and motility at multiple stages. J. Cell Biol. 170:935–945 10.1083/jcb.200505166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallee R.B., McKenney R.J., Ori-McKenney K.M. 2012. Multiple modes of cytoplasmic dynein regulation. Nat. Cell Biol. 14:224–230 10.1038/ncb2420 [DOI] [PubMed] [Google Scholar]
- Van De Bor V., Hartswood E., Jones C., Finnegan D., Davis I. 2005. gurken and the I factor retrotransposon RNAs share common localization signals and machinery. Dev. Cell. 9:51–62 10.1016/j.devcel.2005.04.012 [DOI] [PubMed] [Google Scholar]
- Vendra G., Hamilton R.S., Davis I. 2007. Dynactin suppresses the retrograde movement of apically localized mRNA in Drosophila blastoderm embryos. RNA. 13:1860–1867 10.1261/rna.509007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wainman A., Creque J., Williams B., Williams E.V., Bonaccorsi S., Gatti M., Goldberg M.L. 2009. Roles of the Drosophila NudE protein in kinetochore function and centrosome migration. J. Cell Sci. 122:1747–1758 10.1242/jcs.041798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkie G.S., Davis I. 2001. Drosophila wingless and pair-rule transcripts localize apically by dynein-mediated transport of RNA particles. Cell. 105:209–219 10.1016/S0092-8674(01)00312-9 [DOI] [PubMed] [Google Scholar]
- Wiśniewski J.R., Zougman A., Nagaraj N., Mann M. 2009. Universal sample preparation method for proteome analysis. Nat. Methods. 6:359–362 10.1038/nmeth.1322 [DOI] [PubMed] [Google Scholar]
- Xiang X., Osmani A.H., Osmani S.A., Xin M., Morris N.R. 1995. NudF, a nuclear migration gene in Aspergillus nidulans, is similar to the human LIS-1 gene required for neuronal migration. Mol. Biol. Cell. 6:297–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi J.Y., Ori-McKenney K.M., McKenney R.J., Vershinin M., Gross S.P., Vallee R.B. 2011. High-resolution imaging reveals indirect coordination of opposite motors and a role for LIS1 in high-load axonal transport. J. Cell Biol. 195:193–201 10.1083/jcb.201104076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yingling J., Youn Y.H., Darling D., Toyo-Oka K., Pramparo T., Hirotsune S., Wynshaw-Boris A. 2008. Neuroepithelial stem cell proliferation requires LIS1 for precise spindle orientation and symmetric division. Cell. 132:474–486 10.1016/j.cell.2008.01.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y.V., Li Z., Rizzo N.P., Einstein J., Welte M.A. 2011. Targeting the motor regulator Klar to lipid droplets. BMC Cell Biol. 12:9 10.1186/1471-2121-12-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Zhuang L., Lee Y., Abenza J.F., Peñalva M.A., Xiang X. 2010. The microtubule plus-end localization of Aspergillus dynein is important for dynein-early-endosome interaction but not for dynein ATPase activation. J. Cell Sci. 123:3596–3604 10.1242/jcs.075259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W., Smith J.W., Huang C.M. 2010. Mass spectrometry-based label-free quantitative proteomics. J. Biomed. Biotechnol. 2010:840518. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.