Abstract
To improve signal stability and quantitation, an optically stable, novel class of fluorophore for hybridization analysis of human metaphase chromosomes is demonstrated. Detection of hybridization sites in situ was based on fluorescence from streptavidin-linked inorganic crystals of cadmium selenide [(CdSe)ZnS]. Fluorescence of nanocrystal fluorophores was significantly brighter and more photostable than organic fluorophores Texas Red and fluorescein. Thus, semiconductor nanocrystal fluorophores offer a more stable and quantitative mode of fluorescence in situ hybridization (FISH) for research and clinical applications.
INTRODUCTION
Although semiconductor nanocrystals (quantum dots) have been investigated for several decades (1,2), compatibility of these minute inorganic crystals with interesting electro-optical properties has only recently been demonstrated in biological experiments (3–9). Interest in biomedical applications derives from the fact that quantum dots do not fade upon exposure to light, and multiplexing is facilitated by narrow emission peaks and the property that many probe colors may be excited via a single incident wavelength of light (1,2). Recent applications include labeling of biologically active ligands (3), detection of antibodies and cellular markers (4), non-lethal fluorescent labeling of embryonic cells (6), whole animal imaging of tumors (7) and detection of Y-specific repeats in sperm (9). In addition, protocols for fluorescent detection of oligonucleotides on microbeads have been published (5,8). The recent availability of stable streptavidin conjugates of quantum dots has expedited exploration of additional biomedical protocols in which biotinylated probe species can be tracked by the quantum dot conjugates.
Fluorescence in situ hybridization (FISH) in routine research and clinical use includes applications such as gene mapping and quantitation of gene copy number in tumors with gene amplification, or quantitation of the density of telomere repeats at the ends of human and mammalian chromosomes. Such experiments are compromised by photo-bleaching of organic fluorophores, and the consequent need to image fluorescence immediately. To obviate these drawbacks, we have investigated coated cadmium selenide semiconductor nanocrystals as fluorescence labels for FISH of biotinylated DNA to human metaphase chromosomes under conditions that approximate those commonly found in clinical cytogenetics laboratories. Superior stability of quantum dot fluorophores offers the opportunity to improve quantitation of FISH analysis of human chromosomal changes.
MATERIALS AND METHODS
DNA probes
Total genomic DNA isolated from normal male peripheral blood by standard methods (10), was labeled with biotin-16-dUTP (11). BAC clone CTD-2019C10, encompassing ERBB2/HER2/neu 5′ flanking region, genomic regions spanning exons 1–27 and 3′ flanking regions, was obtained from the Children’s Hospital of Oakland. BAC DNA was isolated by the Qiagen Tip method (Qiagen, Inc.), and nick-translated with biotin-16-dUTP as above. Fixed metaphases from transformed lymphocyte cultures or breast cancer cell line SK-BR-3 (American Type Culture Collection) were prepared on glass slides (12). Slides were heat-denatured [2× SSC, 70% v/v formamide, pH 7.0; 73°C; 5 min (1× SSC is 150 mM NaCl, 15 mM trisodium citrate at pH 7.0)], and quenched in 70%, 90% and 100% v/v ethanol (5 min each) at 4°C.
Hybridization
Biotinylated DNA probe was denatured by boiling for 5 min, then prepared in hybridization mixture at 5 ng/ml in 2× SSC, 50% formamide, pH 7.0, added to denatured cells on slides and sealed with a cover slip and rubber cement. After 12 h at 37°C, cover slips were removed and slides were washed (2× SSC, 50% v/v formamide, 43°C, three washes, 15 min each), and sites of hybridization were detected with Texas Red–streptavidin (Vector A-2006, diluted to 30 µg/ml in 2× SSC, pH 7.0, 3% w/v bovine serum albumin), fluorescein–streptavidin (Vector A-2001 diluted to 30 µg/ml in 2× SSC, pH 7.0, 3% w/v bovine serum albumin) or Qdot605 [Qdot Corporation (CdSe)ZnS nanocrystal–streptavidin conjugate diluted to 40 nM in Qdot buffer]. Post-hybridization and detection, slides were mounted in VectaShield [Vector H-1200 with 1.5 µg/ml DAPI (4,6-diamidino-2-phenylindole)].
BAC FISH experiments were identical to those above with total human DNA except that HER2/neu BAC DNA was detected in lymphocytes and SK-BR-3 cells with Qdot605 at 10 nM in Qdot buffer. FITC detection of HER2/neu BAC DNA followed the same protocol as for total genomic DNA probes above.
Imaging
Slides were imaged on Zeiss and Olympus microscopy systems. Digital photography (Fig. 1) was performed on a Zeiss AxioPlan2 Photomicroscope with a xenon lamp (Osram XBO 150 W CR OFR) fitted with a Hamamatsu C4880–85 CCD camera. Filters were DAPI (DAPI Zeiss Axio: D360/40x, 400 DCLP/D460/50 M), Zeiss SKY Filter Set from ASI (4301-01 BS; 4301-01 X; 4301-01 M). Filters custom-designed for CdSe/ZnS quantum dots were obtained from Chroma Technology (E460SPUV exciter, 475DCXRU dichroic and D605/40 emitter). Digital images were captured through a Zeiss 63× Plan-Apochromat 1.4 oil, ∞/0.17 lens with EasyFISH (version 1.1) software (Applied Spectral Imaging). For quantitative studies, an Olympus BH2-RFCA fluorescence microscope equipped with an Osram HBO 103 w/2 mercury light source and a Quantix CCD camera with a grade 1 KAF 1400 chip/pixel size 6.8 µm2 (Princeton Instruments) was used. Filter sets were Chroma 31000-DAPI (DAPI), 41001 HQ-F (fluorescein), B-M 41004 (Texas Red) and 320007 [Qdot 605 (CdSe)ZnS] custom set as above. Images on the Olympus microscopy system were captured on a NT computer with IP Labs software (Signal Analytics, Version 3.0) set to 4 × 4 pixel resolution. Focusing was accomplished by a remote focusing attachment (Ludl MAC 2Z jystk). Images were captured at exposure times indicated, through an Olympus 100× objective lens S PLAN 100PL 1.25 oil, 160/0.17. Exposure times (Fig. 2A) were 200 ms for FITC, 70 ms for quantum dots and 200 ms for Texas Red. Exposures for Figure 2B were each 120 ms. These exposures avoid CCD pixel saturation. Image files were converted to 8/24 bytes and saved in TIFF format. Densitometry in IP Labs software involved selecting fluorophore image areas corresponding to the DAPI-bright pixels from chromosome band 1q12, and taking area and density measurements via the IP Labs analyze function. Areas representing background fluorescence outside the metaphase were similarly measured. Each relative intensity point was calculated relative to the quantity (sum density/area of 1q12 minus sum density/area adjacent background) and these were graphed as a function of time. Metaphase cells were photo-bleached by full intensity illumination of the mercury lamp beam through the 100× objective lens with dye-matched excitation/emission filters in place.
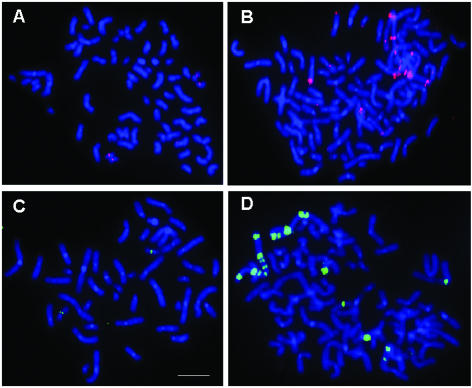
Figure 1.
Specificity of fluorophore–streptavidin detection of biotinylated total human DNA probe in metaphase chromosomes. (A) Control (no fluorophore–streptavidin conjugate); (B) streptavidin–Qdot 605 detection of chromosome 1q12 region (vertical and horizontal arrows); (C) Texas Red–streptavidin detection of biotinylated DNA hybridized to 1q12 (vertical arrows) and (D) FITC–streptavidin detection of 1q12 sites (vertical arrows). Bar in panel C is 10 µm.
Figure 2.
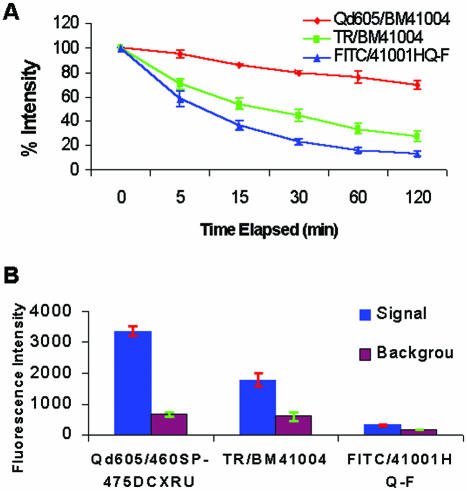
Photostability (panel A) and relative signal intensity (panel B) of FISH hybridization signals detected at human chromosome band 1q12 with streptavidin–fluorophore conjugates. (A) Signal decay upon continuous illumination with fluorescence microscope/mercury illumination in metaphase chromosome band 1q12 during 2 h continuous illumination. Red is Qdot 605, green is Texas Red and blue is FITC. (B) Total intensity of whole interphase nuclei during 120 ms illumination (blue bars) and background (red bars). N = 3 cells in each.
RESULTS AND DISCUSSION
As a model system, FISH experiments were performed with total DNA as probe with human metaphase lymphocytes as hybridization substrate. Probe detection was compared between conventional organic fluorophores (FITC and Texas Red) and inorganic (CdSe)ZnS nanocrystal fluorophores. Because the biotinylated DNA hybridization probes were identical across experiments (Figs. 1A–D and 2), organic fluorophores and inorganic nanocrystal Qdot605 are directly comparable. Figure 1 shows a composite micrograph of the FISH fluorescence signal of hybridized, biotinylated total human DNA probes detected with fluorophore–streptavidin conjugates (red or green) digitally superimposed on chromosome signal (blue) at metaphase of the cell cycle (Fig. 1, panel A–D) in human lymphoblast cell line PB6. The white bar (Fig. 1C) represents 10 µm. Four slides were separately hybridized with biotinylated total human DNA. Detection of biotinylated probe is shown for control prior to fluorophore–streptavidin conjugate (Fig. 1A), biotinylated DNA probe detected with Qdot605–streptavidin conjugate [Fig. 1B (red)], biotinylated DNA probe detected with Texas Red–streptavidin [Fig. 1C (red)], or biotinylated DNA probe detected with streptavidin–FITC [Fig. 1D (green)]. The most intense hybridization signals from total DNA probe correspond to heterochromatic chromosomal regions that contain high-copy DNA repeats, such as human satellite III DNA (13). Because chromosomal region 1q12 is distinctive by size of human chromosome 1 and its DAPI-bright intensity near the centromere, this was selected as target for in situ photo-bleaching measurements of organic and inorganic fluorophores.
Chromosome band 1q12 varies in size in normal human populations (13). Therefore, both chromosome regions 1q12 in each cell were identified in DAPI-stained images. Despite variability between chromosome 1 homologs, their average sum is identical among metaphases of PB6 cell line. This target was therefore valid for quantitative comparison of fluorophores. The number of biotinylated DNA molecules that bound both chromosome bands 1q12 in each cell is identical across experiments.
As seen in Figure 1A, neither green nor red fluorescence was present at the 1q12 site in pre-conjugate controls containing hybridized sites with biotinylated DNA. Only blue DAPI-stained DNA is visible. In Figure 1B, probe hybridization at band 1q12 on both homologous chromosomes 1 was evident by Qdot605–streptavidin detection (vertical and horizontal arrows, red signal). As detected by Texas Red FISH probe (Fig. 1C, vertical arrows, red signal), specific hybridization label was observed at band 1q12. FITC–streptavidin also detected appropriate band 1q12 hybridization signals (Fig. 1D, vertical arrows, green signal). These data demonstrated specificity of hybridization signal and fluorophore detection.
To measure in situ photostability of hybridization signals at region 1q12, a photo-bleaching experiment was performed (Fig. 2A). At to and subsequent times during continuous illumination, an image was digitally captured by the CCD camera. Band 1q12 was identified in the DAPI image, and the integrated density of localized fluorophore at the hybridization site was measured digitally under non-saturating exposure conditions. The sum of pixel density values within band 1q12 divided by area, was defined as average density. For each time point, band 1q12 hybridization signal was measured in both chromosomal homologs in three different cells (six chromosomes total) of the same experimental slide. A randomly chosen area of similar size outside and adjacent to the metaphase, was captured and average background density was subtracted from band 1q12 signal. Time points (Fig. 2A) represent a set of three independently measured cells and error bars.
Decay in fluorescence intensity with time (photostability) of FISH signals at hybridization sites at band 1q12 is shown in Figure 2A. Texas Red values are depicted in green symbols, FITC in blue symbols and Qdot605 (CdSe)ZnS nanocrystal probe values are shown in red. From these data, the intensity of the (CdSe)ZnS probe diminished 5% with 5 min of exposure. This compares favorably with a nearly 42% decrease in FITC signal and a 30% decrease in Texas Red signal at 5 min of illumination. At the 2 h time point, significantly less signal loss was observed for the quantum dot probe (30% loss) than for Texas Red probe (73% loss) or FITC probe (89% loss). This indicates that quantum dot detection is more photostable, and therefore more effective than organic fluorophores for quantitative FISH measurements.
Relative fluorophore intensity was estimated as above in whole nuclei for biotin-labeled total DNA hybridization signal detected by conjugated fluorophores. Figure 2B shows total signal (blue columns) and background signal (red columns) for the three fluorophores on identical short exposures. Texas Red hybridization signal gave an average image density for whole nuclei of 1766 (N = 3 cells). Qdot605 hybridization signal with optimal filters yielded an average image density for whole nuclei of 3362 (N = 3 cells), or about twice the intensity of the Texas Red signal similarly imaged. When compared with fluorescein-labeled nuclei, the quantum dot signal was more than 11-fold that of fluorescein, and about 5-fold greater than background. The quantum dot tag was empirically more photostable, had improved signal-to-noise ratio and showed a brighter intensity than the organic dyes when visualized with fluorophore-optimized optical filters.
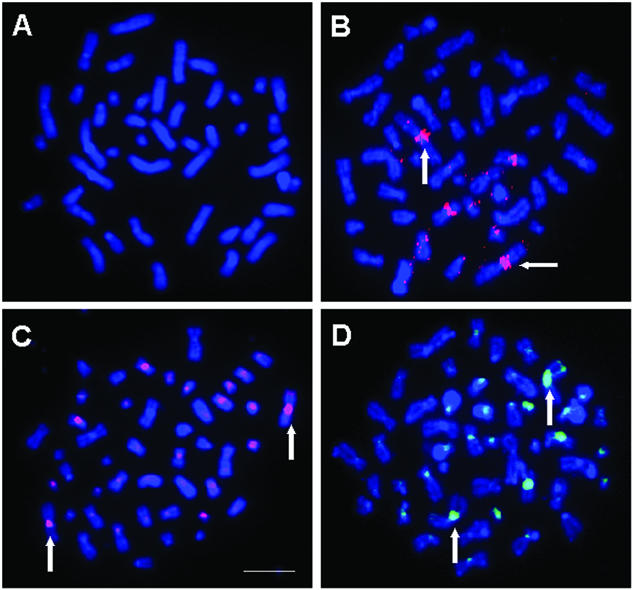
To illustrate that quantum dot probes can be applied to detection of clinically important disease genes, the ERBB2/HER2/neu gene was chosen. BAC CTD-2019C10, a 168 kb genomic probe encompassing human HER2, was detected in low copy human cells (Fig. 3A and C) and in breast cancer cells previously shown (14) with amplified HER2 gene (Fig. 3B and D) with quantum dot–streptavidin (panels A and B) or FITC–streptavidin (panels C and D). These results demonstrate qualitatively the potential for quantum dot detection of a clinically useful locus. Quantitative studies on the HER2 FISH detection with various probes and fluorophores are in progress and will be reported elsewhere.
Figure 3.
Qualitative FISH detection of ERBB2/HER2neu gene with streptavidin conjugates of Qdot605 (Panels A and B) and fluorescein (Panels C and D) in HER2 gene-low copy human lymphocytes (Panels A and C) and HER2 gene-amplified SK-BR-3 breast cancer cells (Panels B and D). Bar in panel C is 10 µm.
Whereas, FISH detection of the Y-specific DNA repeat in human sperm nuclei was previously studied (9), these studies did not quantitatively analyze quantum dot signals, nor did these studies evaluate the comparative merits of quantum dot fluorophores and conventional organic detection labels under comparable experimental conditions.
An unexpected difference in signal distribution was observed between organic labels (Fig. 1C and D) and quantum dot labels (Fig. 1B). Whereas heterochromatic regions of chromosomes 1, 9 and 16 were similarly labeled, centromeric regions of other human chromosomes detected with organic fluorophores, lacked label with quantum dot detected probes. This finding was consistent and reproducible, and is a function of pH and buffer used. Similar effects are noted in FISH experiments with FITC–streptavidin detection with pH 8.3 borate buffer (see Supplementary Material). Unlike whole human DNA FISH probes, the number and distribution of HER2 sites detected with BAC CTD-2019C10 (Fig. 3) appeared to be similar with quantum dots and FITC conjugates in both cells with low copy HER2 genes (PB6) and those with amplified HER2 genes (SK-BR-3). Further work with these and other types of FISH probes is needed to confirm and extend these initial observations with quantum dot-detected FISH probes.
Unlike organic fluorophores, intensity intermittency or flickering of spots within bright regions of hybridization at band 1q12 and in interphase nuclei was observed with Qdot605 probes. This phenomenon is reminiscent of that previously reported in single CdSe nanocrystals in organic glass (15,16), and in quantum dot-labeled transferrin protein studied in aqueous cellular uptake experiments (3). We have observed fluctuation in quantum dot-detected FISH signals of an optically resolved size (∼0.3 µm) with epifluorescence time-lapse imaging. No such flickering was noted in controls detected with Texas Red or FITC.
Previous studies of single CdSe quantum dots revealed 0.02 eV emission fluctuations of ∼100 s and ∼0.5 s at room temperature (16). While apparently random and subject to nonstandard Lèvy statistics in single CdSe nanocrystals (17), this intermittent fluorescence phenomenon is not well understood under conditions described here.
Quantum dot FISH signals observed here appear larger than individual (CdSe)ZnS quantum dots, reported to be 30 Å diameter core with a 6 Å cap in (CdSe)ZnS quantum dots (18,19). With polymer and streptavidin coatings, functionalized (CdSe)ZnS nanoparticles of 15 nm approximate the size of proteins (3,4,18–20). Single nanocrystals should fall below the resolving power of the microscopy system described here. However, a similar imaging system resolved single quantum dots previously (3).
The advantages of quantum dot-labeled FISH probes include photostability and high intensity. Like individual underivatized (CdSe)ZnS quantum dots in laboratory conditions, these properties also hold for (CdSe)ZnS quantum dot–streptavidin conjugates in FISH probes of human metaphase chromosomes studied here under conditions common in clinical genetics and pathology laboratories. These results suggest that semiconductor nanocrystal fluorophores may be a more stable fluorescent tag for FISH applications in pathology and medical genetics diagnostics.
In summary, we have quantitated DNA FISH signals detected with inorganic quantum dot (CdSe)ZnS fluorophores with total human DNA as hybridization probe. We have also demonstrated application of quantum dots to FISH detection of the clinically relevant HER2 locus in breast cancer cells. Quantum dot probes in the total genomic DNA model system are substantially more photostable than Texas Red or fluorescein and show greater intensity. This may open the way for a significantly more quantitative mode of FISH analysis for medical diagnostic applications.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
REFERENCES
- 1.Alivasatos P. (1996) Semiconductor clusters, nanocrystals, and quantum dots. Science, 271, 933–937. [Google Scholar]
- 2.Nirmal M. and Brus,L. (1999) Luminescence photophysics in semiconductor nanocrystals. Acc. Chem. Res., 32, 407–414. [Google Scholar]
- 3.Chan W.C. and Nie,S. (1998) Quantum dot bioconjugates for ultrasensitive nonisotopic detection. Science, 281, 2016–2018. [DOI] [PubMed] [Google Scholar]
- 4.Wu X., Liu,H., Liu,J., Haley,K.N., Treadway,J.A., Larson,J.P., Ge,N., Peale,F. and Bruchez,M.P. (2003) Immunofluorescent labeling of cancer marker HER2 and other cellular targets with semiconductor nanocrystals. Nat. Biotechnol., 21, 41–46. [DOI] [PubMed] [Google Scholar]
- 5.Han M., Gao,X., Su,J.Z. and Nie,S. (2001) Quantum-dot-tagged microbeads for multiplexed optical coding of biomolecules. Nat. Biotechnol., 19, 631–635. [DOI] [PubMed] [Google Scholar]
- 6.Dubertret B., Skourides,P., Norris,D.J., Noireaux,V., Brivanlou,A.H. and Libchaber,A. (2002) In vivo imaging of quantum dots encapsulated in phospholipid micelles. Science, 298, 1759–1762. [DOI] [PubMed] [Google Scholar]
- 7.Akerman M.E., Chan,W.C., Laakkonen,P., Bhatia,S.N. and Ruoslahti,E. (2002) Nanocrystal tagging in vivo. Proc. Natl Acad. Sci. USA, 99, 12617–12621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu H., Sha,M.Y., Wong,E.Y., Uphoff,J., Xu,Y., Treadway,J.A., Truong,A., O’Brien,E., Asquinth,S. and Stubbins,M. et al. (2003) Multiplexed SNP genotyping using the Qbead system: a quantum dot-encoded microsphere-based assay. Nucleic Acids Res., 31, e43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pathak S., Choi,S.-K., Arnheim,N. and Thompson,M.E. (2001) Hydroxylated quantum dots as luminescent probes for in situ hybridization. J. Am. Chem. Soc., 123, 4103–4104. [DOI] [PubMed] [Google Scholar]
- 10.Blin N. and Stafford,D. (1976) A general method for isolation of high molecular weight DNA from eukaryotes. Nucleic Acids Res., 3, 2303–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rigby P.W.J., Dieckmann,M., Rhodes,C. and Berg,P. (1977) Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation. J. Mol. Biol., 113, 237–251. [DOI] [PubMed] [Google Scholar]
- 12.Barch M.J., Knutsen,T. and Spurbeck,J. (1997) In The AGT Cytogenetics Laboratory Manual. 3rd edn. Lippincott-Raven, Philadelphia, p. 43. [Google Scholar]
- 13.Gosden J.R., Lawrie,S.S. and Cooke,H.J. (1981) A cloned repeated sequence in human chromosome heteromorphisms. Cytogenet. Cell Genet., 29, 32–39. [DOI] [PubMed] [Google Scholar]
- 14.Kraus M.H., Popescu,N.C., Amsbaugh,S.C. and King,C.R., (1987) Overexpression of the EGF receptor-related proto-oncogene erbB-2 in human mammary tumor cell lines by different molecular mechanisms. EMBO J., 6, 605–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nirmal M., Dabbousi,B.O., Bawendi,M.G., Macklin,J.J., Trautman,J.K., Harris,T.D. and Brus,L.E. (1996) Fluorescence intermittency in single cadmium selenide nanocrystals. Nature, 383, 802–804. [Google Scholar]
- 16.Blanton S.A., Hines,M.A. and Guyot-Sionnest,P. (1996) Photoluminescence wandering in single CdSe nanocrystals. Appl. Phys. Lett., 69, 3905–3907. [Google Scholar]
- 17.Brokmann X., Hermier,J.P., Messin,G., Desbiolles,P., Bouchard,J.P. and Dahan,M. (2003) Statistical aging and nonergodicity in the fluorescence of single nanocrystals. Phys. Rev. Lett., 90, 120601. [DOI] [PubMed] [Google Scholar]
- 18.Hines M.A. and Guyot-Sionnest,P. (1996) Synthesis and characterization of strongly luminescent ZnS-capped CdSe nanocrystals. J. Phys. Chem., 100, 468–471. [Google Scholar]
- 19.Dabbousi B.O., Rodriguez-Viejo,J., Mikulec,F.V., Heine,J.R., Mattousi,H., Ober,R., Jensen,K.F. and Bawendi,M.G. (1997) (CdSe)ZnS core-shell quantum dots: synthesis and characterization of a size series of highly luminescent nanocrystallites. J. Phys. Chem. B, 101, 9463–9475. [Google Scholar]
- 20.Watson A., Wu,X. and Bruchez,M. (2003) Lighting up cell with quantum dots. Biotechniques, 34, 296–303. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.